Abstract
Proteases regulate a large number of biological processes in plants, such as metabolism, physiology, growth, and defense. In this study, we carried out virus-induced gene silencing assays with pepper cDNA clones to elucidate the biological roles of protease superfamilies. A total of 153 representative protease genes from pepper cDNA were selected and cloned into a Tobacco rattle virus-ligation independent cloning vector in a loss-of-function study. Silencing of 61 proteases resulted in altered phenotypes, such as the inhibition of shoot growth, abnormal leaf shape, leaf color change, and lethality. Furthermore, the silencing experiments revealed that multiple proteases play a role in cell death and immune response against avirulent and virulent pathogens. Among these 153 proteases, 34 modulated the hypersensitive cell death response caused by infection with an avirulent pathogen, and 16 proteases affected disease symptom development caused by a virulent pathogen. Specifically, we provide experimental evidence for the roles of multiple protease genes in plant development and immune defense following pathogen infection. With these results, we created a broad sketch of each protease function. This information will provide basic information for further understanding the roles of the protease superfamily in plant growth, development, and defense.
Introduction
Proteases catalyze the hydrolytic cleavage of peptide bonds, which are present in all living organisms and play crucial roles in many biological processes [1]. Higher plants are autotrophic organisms that can synthesize all of their organic molecular components from inorganic nutrients without digestion of heterotrophic protein. However, hundreds of proteases are encoded by the plant genome, suggesting that these enzymes have essential roles in various plant processes including responses to developmental and environmental cues, metabolism, and immunity [2]–[4].
Proteases are classified into five families according to their catalytic activity, namely the cysteine, serine, threonine, metallo- and aspartic proteases based on their nucleophile and oxyanion stabilizer [5]. Depending on the catalytic mechanism, serine, cysteine, and threonine proteases use a portion of their amino acids as the catalytic site for the nucleophile, whereas metallo- and aspartic proteases use an activated water molecule as the nucleophile [6].
Several members of the serine, cysteine, and threonine protease families have reported regulatory roles in the development and morphogenesis of different stages of the plant life cycle. For instance, defective kernel 1 (DEK1), a cysteine protease, is involved in an epidermal cell fate stage, and a dek1 mutant in maize (Zea mays) causes a lethal defect in which kernels lack an aleurone layer [7], [8]. Two other cysteine proteases, ubiquitin C-terminal hydrolase1 (UCH1) and 2 (UCH2), regulate branching in Arabidopsis thaliana. The uch1/uch2 mutant strain exhibits reduced branched primary inflorescence under short day conditions [9]. In addition, the serine protease abnormal leaf shape 1 (ALE1) function in embryo cuticle deposition as evidenced by the lack of cuticle in embryos of ale1 mutants [10].
Along with these roles in development, many aspartic and metalloproteases have been associated with immune defense in plants. Constitutive disease resistance 1 (CDR1) from Arabidopsis, an aspartic protease localized in the apoplast of plant cells, induces local and systemic defense responses. Overexpression of CDR1 caused dwarfing and resistance to the virulent pathogen Pseudomonas syringae and CDR1 deficiency resulted in increased susceptibility to infection by this pathogen [2], [11]. Aspartic proteases (APs) from Solanum tuberosum exhibit antimicrobial activity induced by abiotic and biotic stress by interacting directly with microbial cell surface proteins followed by membrane permeabilization [12]. Furthermore, APs from barley, which are specifically expressed in nuclear cells during degeneration, is reported to be involved in programmed cell death [13]. Moreover, matrix metalloproteases 2 isolated from Glycine max is induced by biotic stresses such as fungal or bacterial pathogens [14]. Altogether these reports suggest that proteases may play roles in the immune response against pathogenic infection in plants.
Functional characterization of plant protease gene family members has been conducted in a fragmentary scale to uncover a specific function for each protease. To understand the broad role of proteases in plants, we isolated the protease superfamily from pepper (Capsicum annuum) expressed sequenced tags (ESTs, http://genepool.kribb.re.kr/pepper/) on the basis of MEROPS (http://merops.sanger.ac.uk/) classification [15], [16]. We analyzed multiple pepper ESTs to gain a better understanding of proteases involved in plant development and immune defense. In an effort to identify protease function, a Tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) technique was adapted as an easy and powerful method for silencing uncharacterized genes on a large scale [17]. To date, VIGS has been used successfully in studies of Nicotiana benthamiana, Solanum lycopersicum, Solanum bulbocastanum, C. annuum, Arabidopsis, and Papaver somniferum among other plants [18]–[23].
In this study, we present the phenotypic profile of N. benthamiana plants following silencing of multiple proteases. We further characterized some selected proteases with roles in pathogen-induced plant cell death. This study of 153 different protease genes from pepper plant could provide an overall sketch of the biological roles of plant proteases in growth, development, and defense against pathogens.
Materials and Methods
Plant Growth and Agroinfiltration
N. benthamiana plants were grown in a 4.09 inch soil pot. The pots were grown in a growth chamber at 22°C under a cycle of 16 h light and 8 h dark. TRV-based VIGS on N. benthamiana was performed as described by Dong et al. [24]. The phenotypic changes of protease-silenced plants were observed at 4 or 5 weeks after infiltration.
Dataset
The database searched for annotated proteases was pepper EST database (http://genepool.kribb.re.kr/pepper/) [15]. Functional annotation for protease domain prediction was performed using Hmmpfam and Hmmsmart with E-value set at less than e-5. The annotated proteases were classified based on MEROPS database classification (http://merops.sanger.ac.uk/) [16]. In addition, Arabidopsis, Populus trichocarpa proteases gene family database and N. benthamiana, S. lycopersicum, Solanum phureja genome database were used for ortholog search [25], [26], [3], [27], [28].
Cloning of Pepper EST for VIGS
Pepper ESTs were amplified with: 5′-GACGACAAGACCCT (adaptor sequence) - GTAATACGACTCACTATAGGGC (pBluescript SK- specific sequence: ESTs were packaged into pBluescript SK-; Stratagene) - 3′ and 5′-GAGGAGAAGAGCCCT (adaptor sequence) - CGCTCTAGAACTAGTGGATCC (pBluescript SK- specific sequence)-3′. The PCR products were purified with DNA Clean and Concentrator™ (Zymo Research) to remove primers and nonspecific PCR products. A total 15 fmol of purified PCR product was treated with T4 DNA polymerase (Novagen) in 10×reaction buffer containing 5 mM dATP at 22°C for 30 min and 70°C for 20 min for inactivation of T4 DNA polymerase. The TRV-LIC vector was digested with PstI and treated with T4 DNA polymerase similarly with dTTP instead of dATP [24]. A total 15 fmol of T4 DNA polymerase treated PCR products and TRV-LIC vector were mixed and incubated at 65°C for 1 min and slowly cooled down to room temperature. Then, the mixture was transformed into E. coli DH5α competent cells. Transformants were tested by PCR amplification using primers 5′-TGTTACTCAAGGAAGCACGATGAGCT- 3′ and 5′- CAGGCACGGATCTACTTAAAGAACGTAG- 3′.PCR products with EST insertions were confirmed by DNA sequencing (NICEM, http://nicem.snu.ac.kr/).
RNA Extraction and qRT-PCR Analysis
Total RNA was extracted from leaves of N. benthamiana plant using TRI Reagent (Molecular Research Center) according to the manufacturer’s instruction. First-strand cDNA was synthesized using 5 µg of total RNA in a mixture with anchor primer (oligo-dT) and SUPERSCRIPT® II Reverse-Transcriptase (Invitrogen) according to the manufacturer’s protocol. RT-PCR products were used for quantitative RT-PCR to monitor gene expression levels in protease-silenced plants. Triplicate samples from 3 independently silenced plants of cDNA were analyzed using Rotor-Gene 6000 apparatus (QIAGEN) with SYBR Green (Invitrogen) according to manufacturer’s instructions. All calculations and statistical analyses were demonstrated as described by the manufacturer. Primers for qRT-PCR of specific proteases were presented in Table S3. To normalize the expression levels, transcript level of NbActin gene was used as a control.
Pathogen Preparation and Inoculation
The bacterial strains used for this study were P. syringae pv. tomato T1 which causes hypersensitive response (HR) cell death and P. syringae pv. tabaci which causes wild fire disease on N. benthamiana [29], [30]. Both bacterial pathogens were grown in yeast extract peptone (YEP) medium and resuspended in 10 mM sterile MgCl2 solution. N. benthamiana leaves were pressure infiltrated using a needless syringe with bacterial concentration of OD600 = 0.2 (P. syringae pv. tomato T1) and OD600 = 0.005 (P. syringae pv. tabaci). Changes of the HR cell death and symptom development in bacteria-infiltrated plants were observed for 3 and 7 days, respectively [31].
P. syringae pv. tabaci cultures (OD600 = 0.005, approx. 1×108 CFU ml−1) were resuspended in 10 mM MgCl2 and infiltrated into the protease-silenced leaves using a 1 ml needless syringe. The growth of bacterial in the leaf was measured on four replicate plants. Two leaf discs (1 cm in diameter) per plant were collected from the P. syringae pv. tabaci infiltrated region of each protease-silenced and control plants. Numbers of the bacteria were measured by grinding the leaf discs in 10 mM MgCl2 and plating serial dilutions LB agar plants containing 100 ug ml−1 rifampicin on a daily basis for 2 dpi.
Staining with Trypan Blue
To detect the cell death, leaves were stained with lactophenol/trypan blue (10 ml glycerol, 10 ml lactic acid, 10 g phenol and 10 mg trypan blue dissolved in 10 ml distilled water). Pathogen-infected leaves were vacuum-infiltrated two-times for 5 min in staining solution and incubated for overnight. Stained leaves were de-stained in 2.5 g ml−1 chloral hydrate (TOKYO KASEI). The de-stained leaves were mounted in 50% glycerol and photographed under a digital microscope (DIMIS M, Siwon Optical Technology, http://www.dimis.co.kr/) [32].
Measurement of Ion Leakage
One and two days after infiltration with P. syringae pv. tomato T1, two leaf discs (1 cm in diameter) were floated in 5 ml of distilled water for 4 h at room temperature. Electrical conductivity was measured using a conductivity meter (EC215, HANNA instruments).
Results
Isolation of Putative Protease Genes in Pepper ESTs
The pepper EST database, which is a useful collection for functional genomics, consists of 122,582 ESTs including 22,011 unigenes, 11,225 consensus sequences and 5,585 full-length cDNAs have been published [15]. From the pepper EST database, putative protease genes (totaling 939 entries, data not shown) were selected computationally using Hmmpfam and Hmmsmart for protease domain prediction based on classifications from MEROPS. This protease database, displays hierarchical classifications and homologous proteases grouped into species, families, and clans based on evolutionary relationships. MEROPS can also be used to obtain information about the substrate and inhibitors of particular protease [16].
To select representative protease genes, sequence alignments using TBLASTX, BLASTX, and BLASTP were performed to avoid silencing multiple genes with homology. Ultimately, 153 pepper cDNAs were selected as representatives of protease genes for VIGS. The 153 pepper cDNA is presented with its EST ID, MEROPS ID, e-value, and the accession numbers of corresponding from N. benthamiana, S. lycopersicum, S. phureja, Arabidopsis genome database and other organisms are summarized in Table S1 [25]–[28]. The selected protease genes consisted of 15 aspartic, 27 cysteine, 28 metalloproteases, 72 serine, and 11 threonine proteases as classified by MEROPS. All of the selected cDNAs were cloned into the TRV-LIC vector for the VIGS assay [24].
Phenotypic Analysis of Protease-silenced Plants
VIGS experiments were performed in N. benthamiana plants with the selected 153 cDNAs. Since pepper and N. benthamiana belong to the same solanaceae family, coding sequences from both plants exhibit a high degree of similarity. Previous studies have also demonstrated successful heterologous silencing of tomato genes in N. benthamiana [24]. Therefore, it is not surprising that several genes from pepper have been characterized in N. benthamiana using the VIGS assay [33], [30], [32]. The pepper ESTs investigated in this study are listed with their corresponding N. benthamiana proteases in Table 1.
Table 1. List of pepper EST referred in this study with its corresponding proteases in N. benthamiana.
| N. benthamiana genome | |||
| Pepper EST IDa | Classificationb | Accession numberc | Description |
| Ncn10583 | M18 | NbS00031969g0005.1 | Aspartyl aminopeptidase protein |
| Ncn1321 | T01 | NbS00028089g0021.1 | Proteasome subunit alpha |
| Ncn1998 | S33 | NbS00039951g0005.1 | Proline iminopeptidase |
| Ncn2132 | M24 | NbS00024034g0008.1 | Xaa Pro dipeptidase |
| Ncn2134 | T01 | NbS00053635g0009.1 | Proteasome subunit beta type |
| Ncn2155 | C15 | NbS00016770g0011.1 | Pyrrolidone carboxylate peptidase |
| Ncn2258 | A01 | NbS00031482g0006.1 | Aspartic proteinase |
| Ncn2901 | A01 | NbS00004524g0008.1 | Aspartic proteinase nepenthesin 1 |
| Ncn3606 | S33 | NbS00008723g0009.1 | Hydrolase alpha/beta fold |
| Ncn4597 | S10 | NbS00017915g0109.1 | Niben044Scf00017915∶116939.122486 |
| Ncn4707 | M16 | NbS00026030g0004.1 | Peptidase M16 family |
| Ncn5036 | S01 | NbS00055986g0001.1 | Serine protease |
| Ncn5964 | A01 | NbS00059099g0001.1 | Aspartic proteinase nepenthesin 1 |
| Ncn6721 | M24 | NbS00011972g0104.1 | Niben044Scf00011972∶154949.175101 |
| Ncn7004 | C26 | NbS00007507g0016.1 | Glutamine amidotransferase |
| Ncn7288 | T01 | NbS00043198g0009.1 | Proteasome subunit alpha type |
| Ncn8326 | M18 | NbS00026037g0013.1 | Aspartyl aminopeptidase |
| Ncn881 | M24 | NbS00006055g0014.1 | Proliferation associated protein 2G4 |
| Ncn8849 | T01 | NbS00021500g0008.1 | Proteasome subunit alpha type |
| Ncn9390 | A01 | NbS00031201g0007.1 | Nucellin aspartic protease fragment |
| Ncn964 | S01 | NbS00023513g0009.1 | Serine protease Do |
| Ncn9826 | M24 | NbS00009510g0004.1 | Xaa pro aminopeptidase |
| N n7292 | C19 | NbS00037871g0005.1 | Ubiquitin carboxyl terminal hydrolase |
Pepper (C. annuum) EST database (http://genepool.kribb.re.kr/pepper/).
Based on MEROPS database (http://merops.sanger.ac.uk/) classification.
N. benthamiana genome database (http://solgenomics.net/, Niben.genome.v0.4.4).
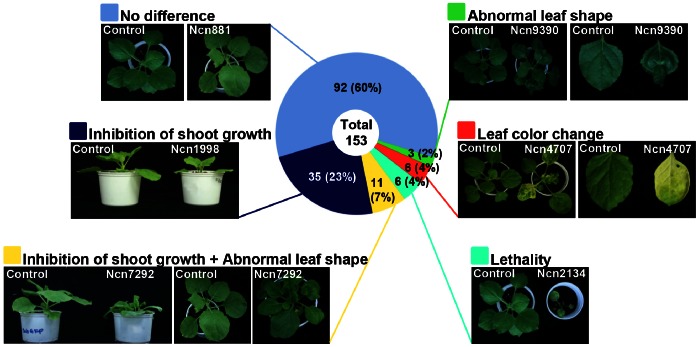
The developmental phenotypes of protease-silenced plants were observed 3 or 4 weeks after VIGS and representative examples were shown (Fig. 1 and Table S2). These phenotypes were grouped into six classes, including no difference, inhibition of shoot growth, inhibition of shoot growth with abnormal leaf shape, lethality, leaf color change, and abnormal leaf shape (Fig. 1). The largest class, of which proliferation-association protein 1 silencing is an example (Ncn881, Fig. 1), showed no phenotypic differences compared to the control plant and was composed of 92 proteases. The second largest class contained 35 proteases and gene silencing resulted in severe growth retardation relative to control plants (Fig. S1A), shown in the silencing phenotype of prolyl aminopeptidase (Ncn1998, Fig. 1). The third largest class consisted of 11 proteases and the silencing phenotype was characterized by severe stunting with crinkled leaves (Fig. S1B). This is evidenced by an ortholog of At5g22030 (Arabidopsis ubiquitin-specific protease 8) (Ncn7292, Fig. 1). The fourth largest class, composed with 6 proteases (Fig. S1C), was characterized by lethality as shown by the silencing phenotype of proteasome subunit beta 3 (Ncn2134, Fig. 1). The fifth class contained 6 proteases (Fig. S1D) and silencing resulted in photobleaching and yellowing of leaves as shown by chloroplast (stromal) processing peptidase silencing (Ncn4707, Fig. 1). This altered phenotype showed similar developmental phenotypes to those in previous studies of down-regulation of the gene product [34], [35]. The smallest class was made up of 3 proteases (Fig. S1E) that, when silenced, lead to disruption of leaf development resulting crinkled leaves as shown by silencing an ortholog of At1g49050 (aspartyl protease family) protein-type peptidase (Ncn9390, Fig. 1).
Figure 1. Phenotypic classification of protease-silenced plants.
Protease-silenced plants are categorized into 6 classes. Each class was given with a specific color square box on left side of the class description. Circular diagram indicates the percentage each class. Representative protease described in each class is shown. These phenotype changes had been observed for 3 or 4 weeks and the picture are taken at 3 or 4 weeks after silencing. For every protease gene, 4 plants were silenced at each experiment. Similar results were obtained from at least three independent experiments. One representative experiment is shown.
Nearly half of the members exhibited altered phenotypes in aspartic, cysteine and metalloproteases family (aspartic: 9 out of 15, cysteine: 14 out of 27, metallo-: 13 out of 28) (Table S2). However, the serine protease family was the least affected group with only 16 out of 72 members displaying a phenotypic change compared to that of control plants.
Interestingly, 9 out of 11 threonine protease family members produced altered developmental phenotypes, such as severe stunting, crinkled leaves, and lethality. Consistant with these results, previous studies have demonstrated that threonine proteases play a crucial role in the 26S proteasome and regulation of all aspects of plant development [36], [37]. These data suggest that threonine proteases have an essential role in plant growth and development.
Temporal Change in Hypersensitive Response (HR) Cell Death in Protease-silenced Plants
To investigate the role of proteases in immune defense, an avirulent pathogen P. syringae pv.tomato T1 capable of inducing HR cell death in N. bethamiana was introduced into protease-silenced plants [38]. Three weeks after silencing, plants were inoculated with a suspension of P. syringae pv. tomato T1 at a density of OD600 = 0.2. HR cell death was observed at days 1 and 2 post-inoculation (1 dpi and 2 dpi, respectively) in TRV-ΔGFP control plants. After 1 dpi, 26 protease-silenced plants showed temporally advanced HR cell death compared to that of control plants while the control plant showed HR cell death at 2 dpi. This group consisted of 11 cysteine, 7 metallo-, 5 serine, 2 threonine, and 1 aspartic protease family members (Fig. 2A and Fig. S2A). Cysteine protease deficiency produced the greatest effect on HR cell death in response to avirulent pathogen. This result is consistent with previous reports that demonstrated a crucial role for cysteine proteases in the plant-pathogen/pest interaction [39], [40]. After 2 dpi, delayed HR cell death was observed in 5 serine, 2 aspartic, and 1 cysteine protease-silenced plants. Delay of the HR cell death was until 3 dpi. Interestingly, none of the metallo- and threonine protease family members exhibited delayed HR cell death in gene-silenced N. benthamiana plants (Fig. 2B and Fig. S2B).
Figure 2. Change of HR cell death in protease-silenced plant following avirulent bacterial pathogen inoculation.
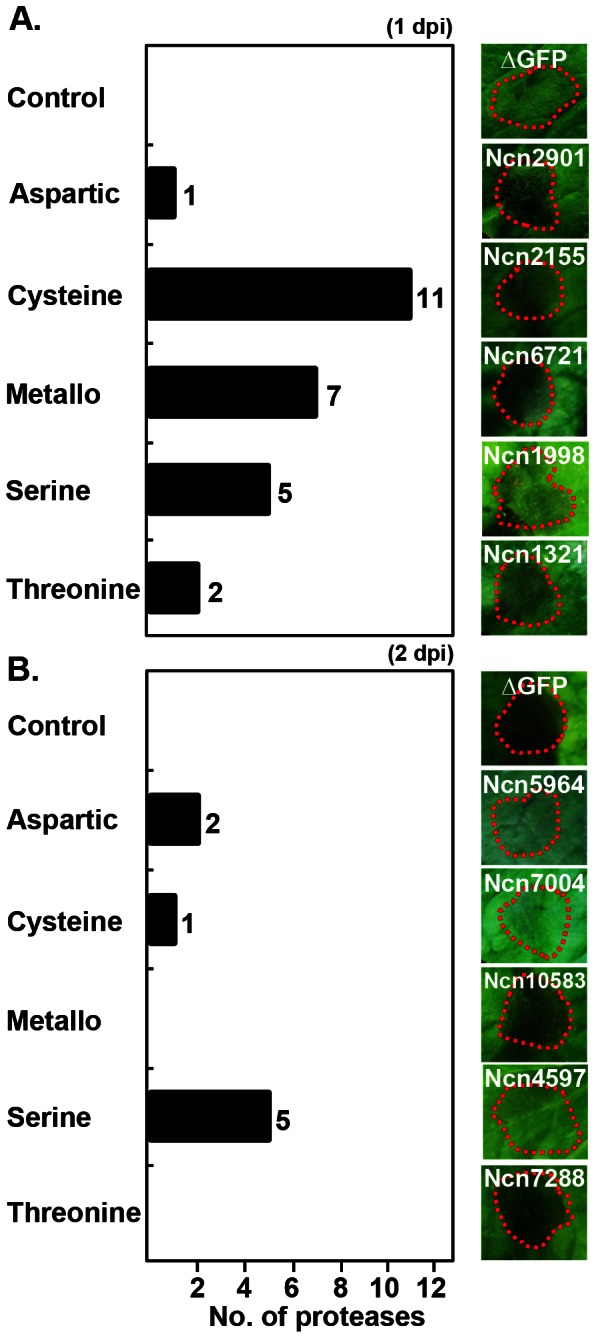
Protease-silenced plants were infiltrated with avirulent bacterial pathogen P. syringae pv. tomato T1 (OD600 = 0.2). The HR cell death symptoms were taken at 1 dpi (A) and 2 dpi (B). The bar graph indicates the numbers of showing enhanced (A) or delayed (B) HR cell death from each protease family (left panel). The right panel shows the representative member from each family representing enhanced (A) and delayed (B) HR cell death. For every protease gene, 6 sections per 1 leaf were infected with the pathogen and total 2 leaves were used for 1 plant. Total 4 plants were infiltrated at each experiment. Similar results were obtained from at least three independent experiments. Red dotted line indicates the site of P. syringae pv. tomato T1 infection. One representative experiment is shown.
Change of Disease Symptom Development in Protease-silenced Plants
To better understand the role of proteases in the immune response to pathogen invasion, protease-silenced N. benthamiana plants were also infected with a virulent bacterial strain. P. syringae pv. tabaci, a fire blight pathogen isolated from tobacco plant, produces a monocyclic ß-lactem named tabtoxin and causes loss of chlorophyll, accumulation of ammonia and finally inactivation of glutamine synthetase in plant [41], [42], [29]. Appearance of the susceptible host plant infected with P. syringae tabaci starts with water-soaking in the apoplast of the infected tissues, localized tissue necrosis and various tissue discolorations [29]. Protease-silenced and control plants were inoculated with P. syringae pv. tabaci at an OD600 = 0.005. Disease symptoms were observed at day 3 post-inoculation (3 dpi) in both groups. Sixteen protease-silenced plants constituting 7 cysteine, 5 serine, 3 metallo- and 1 threonine proteases showed delayed development of disease symptoms (Fig. 3 and Fig. S3). Disease symptom was delayed until 4 to 5 dpi. Aspartic proteases were not found in this experimental group.
Figure 3. Change of disease symptom development in protease-silenced plant following virulent bacterial pathogen inoculation.
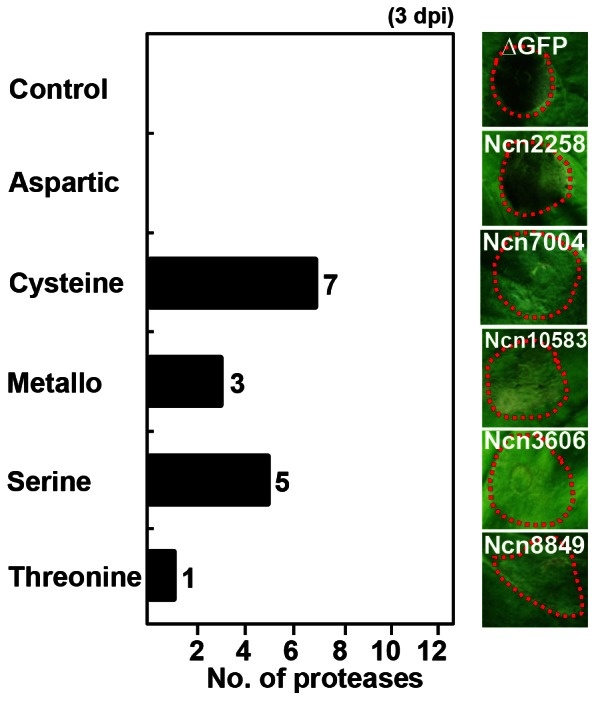
Protease-silenced plants were infiltrated with virulent bacterial pathogen P. syringae pv. tabaci (OD600 = 0.005). The disease symptoms induced by the pathogen were taken at 3 dpi. The bar graph indicates the numbers of showing delayed disease development from each protease family (left panel). The right panel shows the representative member from each family which showed delayed symptom. For every protease gene, 6 sections per 1 leaf were infected with the pathogen and total 2 leaves were used for 1 plant. Total 4 plants were infiltrated at each experiment. Similar results were obtained from at least three independent experiments. Red dotted line indicates the site of P. syringae pv. tabaci infection. One representative experiment is shown.
Characterization of Selected Protease-silenced Plants following Pathogens Infection
To confirm the level of gene silencing in each protease subfamily, quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was conducted using primers that anneal outside the sequence used for silencing. Our data indicate that the level of mRNA transcript for all protease members was decreased at least 70% by gene silencing compared to control plants (Fig. S4).
Three protease subfamilies, which showed modulated pathogen responses when silenced, were selected for further characterization, including M18 (metallo- type, aminopeptidase family), M24 (metallo- type, methionyl aminopeptidase family) and S01 (serine type, chymotrypsin family). M18 family is consisted with a member showing delayed symptom development whereas M24 and S01 families are each constituted with a member resulted in enhanced and delayed HR cell death response compared to the control. Silencing of each representative member showed enhanced and delayed HR cell death and delayed disease symptom onset compared to the control, respectively.
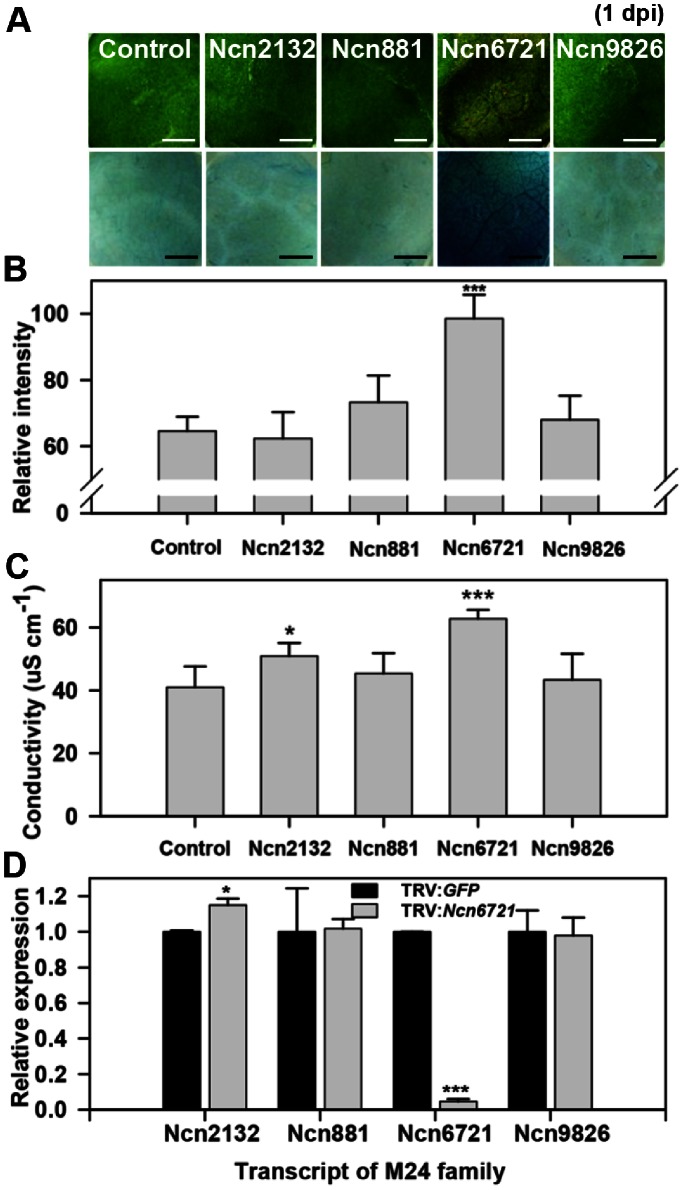
The M24 and S01 protease subfamilies possessed with a member showing enhanced and delayed HR cell death after infection with an avirulent pathogen, P. syringae pv. tomato T1, compared to control plants at 1 dpi and 2 dpi (upper panel of Fig. 4A and Fig. 5A). The control plants represented the HR cell death at 2 dpi, while the enhanced HR cell death of Ncn6721 was carried out at 1 dpi while the HR cell death of Ncn5036 was delayed until 3 dpi. Trypan blue staining was performed to assess cell death in the M24 and S01 protease subfamily member (lower panel of Fig. 4A and Fig. 5A). Our data reveal that cell death following Ncn6721-silencing was 20% higher than the control plants and other silenced member from the M24 protease subfamily at 1 dpi (Fig. 4B), suggesting a critical role for this gene product in plants. In contrast, cell death in Ncn5036-silenced plants was 20% lower than the control and Ncn964-silenced plants at 2 dpi (Fig. 5B). The ion leakage of the pathogen-infiltration area was significantly high in Ncn6721- and low in Ncn5036-silenced plant than that of the control plant (Fig. 4C and 5C).
Figure 4. Enhanced HR cell death identified in M24 protease subfamily-silenced plant.
A. HR cell death from M24 protease subfamily-silenced N. benthamiana plant following non-host pathogen infection (P. syringae pv. tomato T1, OD600 = 0.2). HR symptoms were taken at 1 dpi (upper panel). Trypan blue staining at 1 dpi on the same area as the photographs was taken (lower panel). Scale bars = 0.3 mm. B. Relative intensity of trypan blue stained N. benthamiana plants. Values are calculated as means and standard deviation (SD) for three plants (n = 3) of one infiltration experiment. Relative intensity was calculated to the control of Fig. 5A. C. Ion leakage from leaf discs of M24 protease subfamily-silenced N. benthamiana plant following non-host pathogen infection. Values are means±SD (n = 3). D. Transcript levels of M24 protease subfamily members were examined in Ncn6721-silenced plant. Values are means±SD (n = 3). The values were normalized to NbActin and were calculated to the control. Similar results were obtained from at least two experiments. One representative experiment is shown. Asterisks indicate significant differences relative to the control as determined by Student’s t test (*P<0.05, **P<0.01, ***P<0.001).
Figure 5. Delayed HR cell death obtained in S01 protease subfamily-silenced plant.
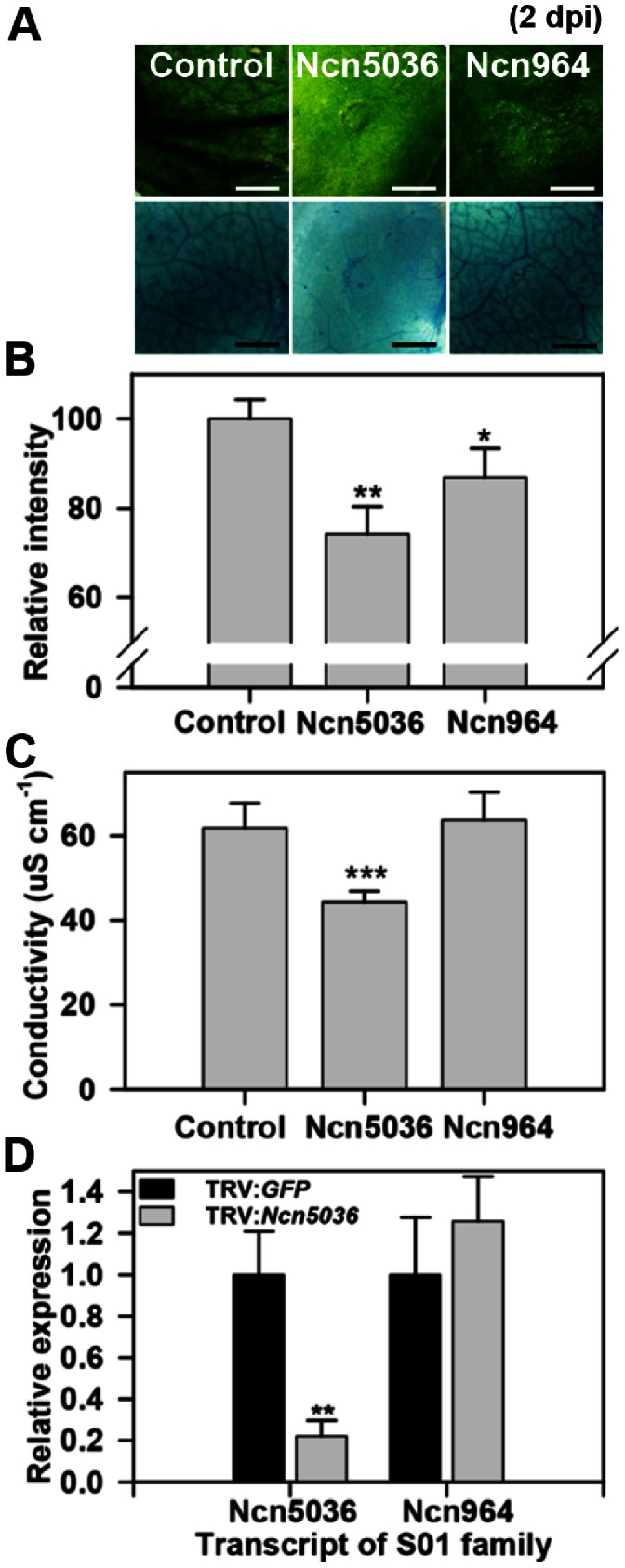
A. HR cell death of S01 protease subfamily-silenced N. benthamiana plant following non-host pathogen infection (P. syringae pv. tomato T1, OD600 = 0.2). HR symptoms were taken at 2 dpi (upper panel). Trypan blue staining at 2 dpi on the same area as the photographs was taken (lower panel). Scale bars = 0.3 mm. B. Relative intensity of trypan blue stained N. benthamiana plants. Values are means±SD (n = 3). Relative intensity was calculated to the control. C. Ion leakage from leaf discs of S01 protease subfamily-silenced N. benthamiana plant following non-host pathogen infection. Values are means±SD (n = 3). D. Transcript level of S01 protease subfamily member was examined in Ncn5036- silenced plant. Values are means±SD (n = 3). The values were normalized to NbActin and were calculated to the control. Similar results were obtained from at least two experiments. One representative experiment is shown. Asterisks indicate significant differences relative to the control as determined by Student’s t test (*P<0.05, **P<0.01, ***P<0.001).
Gene-specific silencing was confirmed with gene-specific primers in the Ncn6721-silenced plants. Among the closest paralogs, the transcript levels of Ncn2132, Ncn881 and Ncn9826 were not reduced in Ncn6721-silenced plants (Fig. 4D). Additionally, the level of Ncn964 mRNA was not decreased in Ncn5036-silenced plants (Fig. 5D). The transcript levels of other M24 and S01 protease members in each protease-silenced plant are shown in Fig. S5. Interestingly, Ncn6721 is predicted to encode methionyl aminopeptidase 2 (MAP2), a cytosolic enzyme responsible for protein N-terminal methionine excision (NME). This cytoplasmic NME machinery displays the same specificity in the plant and animal kingdom and has been established to be required for normal development [43]. Functional studies of MAP2 in plants during development were performed in Arabidopsis [44]. Our finding indicates that MAP2 may play a role in pathogen-induced HR cell death in plants. Moreover, Ncn5036 is described as an At5g40200-type peptidase and predicted to encode degradation of periplasmic proteins 9 (DegP9), a serine type ATP-dependent protease. In Arabidopsis, DegP9 protease is involved in the degradation of D1 protein from photosystem II following photoinhibition and is up-regulated during senescence [45]–[47]. Several recent studies have confirmed that plant chloroplasts are associated with plant pathogen-induced cell death [48]. This result indicates that DegP9 is associated with HR cell death in plants.
The M18 protease subfamily contains a member that is involved in delayed disease symptom development onset following infection with the virulent bacterial pathogen P. syringae pv. tabaci (Fig. 6). Disease symptom of Ncn10583-silenced plant was carried out at 4 to 5 dpi, whereas the control and Ncn8326-silenced plant showed disease symptom at 3 dpi (upper panel of Fig. 6A). Trypan blue analysis revealed that Ncn10583-silenced plants showed 30% less death than the control and Ncn8326-silenced plants at 3 dpi (lower panel of Fig. 6A and Fig. 6B). The bacterial cell growth of Ncn10583-silenced plants was one and a half-fold lower than in control-silenced plants (Fig. 6C). Gene-specific silencing of Ncn10583 was also confirmed by qRT-PCR. The transcript level of the most closely related protease Ncn8326 was not reduced in Ncn10583-silenced plants (Fig. 6D). The level of Ncn10583 mRNA in Ncn8326-silenced plants is shown in Fig. S5. Ncn10583 is an At5g60160-like peptidase and predicted to encode a Zn-dependent exopeptidase superfamily protein also known as aspartyl aminopeptidase (AAP), which is distributed widely in animals and plants and involved in a variety of physiologically important processes [49]. Most studies of AAP have been limited to mammals and focused on its functions and enzymatic activity [50], [51]. Nevertheless, our data imply that AAP plays a role as a susceptibility factor of plants against virulent bacterial pathogens.
Figure 6. Delayed pathogen-induced disease symptom represented in M18 protease subfamily-silenced plant.
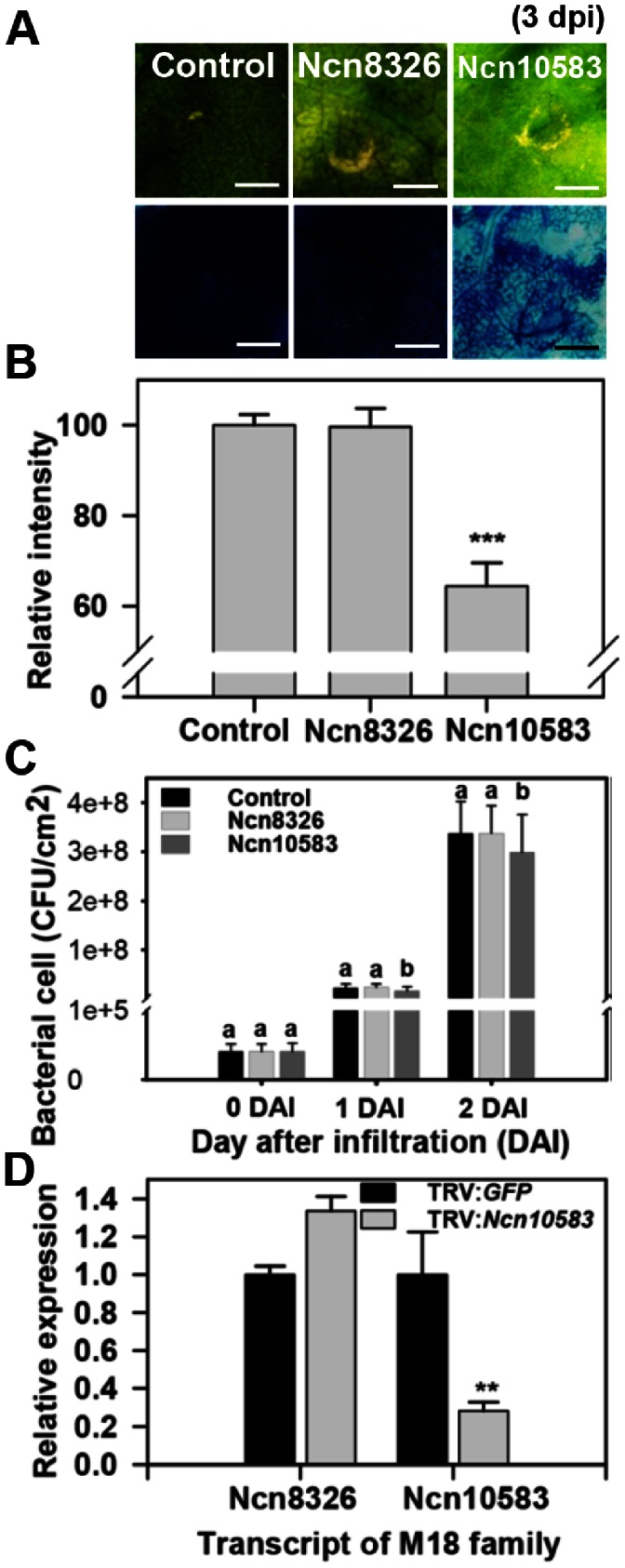
A. Disease symptom of M18 protease subfamily-silenced N. benthamiana plant following bacterial host pathogen infection (P. syringae pv. tabaci, OD600 = 0.005). Disease symptoms were taken at 3 dpi (upper panel). Trypan blue staining is in the same area as the photographs were taken at 3 dpi (lower panel). Scale bars = 0.3 mm. B. Relative intensity of trypan blue stained N. benthamiana plants. Relative intensity was calculated to the control. Values are means±SD (n = 3). C. Bacterial cell growth in control and M19 protease subfamily-silenced N. benthamiana plants infiltrated with P. syringae pv. tabaci (1×108 CFU ml−1 in MgCl2) was determined at 0, 1 and 2 dpi. Values are means±SD (n = 3). Different letters indicate significant differences at the 95% level by Duncan’s multiple range tests. D. Transcript level of M18 protease subfamily member was examined in Ncn10583- silenced plant. Values are means±SD (n = 3). The values were normalized to NbActin and were calculated to the control. Similar results were obtained from at least two experiments. One representative experiment is shown. Asterisks indicate significant differences relative to the control as determined by Student’s t test (*P<0.05, **P<0.01, ***P<0.001).
Discussion
This study presents a larger view of the role of protease in plants. Some of our results pertaining to the function of these enzymes were consistent with previous reports. Among the 61 proteases identified to be involved in the plant developmental process by loss-of function studies, 39 belong to the serine, cysteine, or threonine protease families. Several members from these families have been demonstrated to play roles in plant growth and development [7], [8], [10], [9].
Based on the EST reference dataset, our analysis of representative proteases provided further insight into the role of these enzymes in plant development. Our data demonstrate that silencing of Ncn1998 (serine type) resulted in inhibition of shoot growth (Fig. 1, Fig. S1A, and Table S2). Ncn1998 is predicted to encode a prolyl aminopeptidase (PAP) with a significant degree of sequence identity to other solanaceous PAP proteins in other species, namely S. lycopersicum PAP (88%, Solyc11g044310.1.1), N. benthamiana PAP (74%, NbS00039951g0005.1), Medicago truncatula PAP (92%, XP003593347.1), and Vitis vinifera PAP (84%, XP002271289.2). Despite the ubiquity of PAP in nature, its role remains poorly understood in plants. To date, studies have only shown that the mRNA level of PAP is increased in the shoots of triticale plants in response to suboptimal conditions such as drought and high saline [52]. Our results are the first to indicate that PAP may function in plant growth and development, including the control of shoot apical meristem formation. More specifically, proline, a product of PAP, has various physiological roles in plants, animals, and micro-organisms [53], [54]. Our results suggest that decreased expression of PAP may affect proline metabolism and cause an altered phenotype in Ncn1998-silenced plants. Our experiment was limited to observing the pathology of the silenced plant for only 3 or 4 weeks; therefore, either a longer observation period or examination of knock out mutants may needed to determine the role of PAP in reproductive growth processes such as floral development.
Multiple proteases have roles in the pathogenesis and immune defense of plants. Our study identified 34 proteases that function in avirulent pathogen-induced HR cell death; 7 metallo- and 12 cysteine family members represented these proteases. Moreover, we identified 16 proteases, of which 7 were members of the cysteine family, involved in the development of pathogen-induced disease symptoms. Many members of the aspartic, cysteine, and metalloprotease families have been reported as associated with plant defense responses [2], [14], [39], [40], [11]. Our results are in close agreement with previous reports. Interestingly, silencing of only 3 aspartic proteases affected HR cell death in response to infection by an avirulent pathogen, while aspartic proteases did not appear to play a role in disease symptom development in plants.
To expand our knowledge of the involvement of proteases in pathogen infection, one protease that showed delayed HR cell death and disease symptom development after silencing was selected for further study (Fig. 2B and Fig. 3). Ncn7004, an ortholog of At5g38200 in Arabidopsis, is predicted to encode a Class 1 glutamine amidotransferase (GAT1)-like protein and has significant similarity in other species, including S. lycopersicum (92%, Solyc02g086300.2.1), V. vinifera (93%, XP002279823.1), N. benthamiana (90%, NbS00007507g0016.1), and Arabidopsis (89%, NP564885.1).
GAT generates ammonia from glutamine amide nitrogen and transfers it to acceptors [55]. One bacterial pathogen we used to infect GAT-1 like protein (Ncn7004)-silenced plants was P. syringae pv. tabaci, which produces tabtoxin to inhibit the plant enzyme glutamine synthetase (GS) [56]. GS is a key enzyme in the photorespiratory nitrogen cycle. Inhibiting this enzyme in plants results in chlorosis, which arises due to necrotic lesions caused by bacteria on leaves [57]. Moreover, tabtoxin-induced symptoms are associated with the accumulation of ammonia under photorespiration [58]. Challenging Ncn7004-silenced plants with virulent bacterial pathogen resulted in delayed disease onset, possibly due to blocking glutamine amide nitrogen from supplying ammonia. This decreased level of ammonia may merely slow the development of disease since ammonia accumulation was not enough to cause the symptom. To date, studies on GAT have mainly focused on its biochemical and structural properties [55], [59]. Thus, our finding on the function of GAT may provide additional insight into the molecular basis of disease symptom development in tobacco plants.
In addition, the Ncn7004-silenced plant showed other phenotypes besides cell death and defense responses. The Ncn7004-silenced plant displayed inhibited shoot growth and abnormal leaf shape (Fig. S1B). Ncn7004 consists of a GAT type 1 domain and could function similar to what has been reported previously, such as the effect of lower nitrogen metabolism on normal plant growth [60]. Moreover, GAT links nitrogen metabolism to the biosynthetic pathways of several important compounds [55]. Nitrogen supply is also known to affect plant disease development [60]. In previous studies, GAT was reported clustered with At1g15040, which belongs to the same subfamily as GAT1 (GATase 1_2) in Arabidopsis. In the Arabidopsis genome, there are 30 putative genes encoding GAT1 and a previous study suggested that At1g15040 has specific functions in plant branching control [61]. Our results also reveal that Ncn7004 might have roles in plant developmental processes. Furthermore, HR cell death is a kind of programmed cell death (PCD), which is important throughout plant development stages such as senescence, vacuole formation for the degradation of cellular contents, normal seed development, leaf sculpting, cell death in the root cap, and shaping the sexual and nonsexual organs of the flower [62]. GAT1-like protein may play both processes, roles in plant developmental and pathogen-induced cell death processes, as PCD. Together with previous studies, our results suggest that GAT1-like protein (Ncn7004) is closely associated with plant development and the regulation of cell death in response to pathogen invasion in plant immunity.
A number of plant proteases have been implicated in HR development. Some of them were reported as have roles in regulating HR and some of them were firstly described in this study. Orchestration of the multiple proteases in HR cell death is not quite clear yet but each protease may has distinct and diverse roles. From previous studies, some proteases can act at the level of signal perception, transduction and execution in defense response [63]. There are some evidences that type I metacaspase is a initiator or activator of type II metacaspase for progression of PCD [64]. Moreover, AtMC1 (Arabidopsis metacaspase 1) works as a positive regulator, whereas AtMC2 function as a negative regulator of cell death [65]. Even with these studies, there are still more to be found in the events of the chain leading to cell death. Our data only have given the evidence for one-to-one interaction with a protease and a pathogen. Comprehensive studies on the functions of these proteases may be required to uncover the multiple roles of proteases in the complicated machinery of HR cell death in plants. Therefore, our data might have given evidence for the starting point of the cell death event as the diverse roles of protease in HR cell death.
In this study, we provide a phenotypic sketch of protease function in plant developmental processes, pathogen-induced disease symptom development, and HR cell death. Our studies, together with previously published data, provide additional insights into the unknown role of proteases and clues for further investigation into the molecular functions of the protease super family in plants.
Supporting Information
Altered phenotypes of protease-silenced plants. Protease-silenced plants showing altered phenotypes are categorized into 5 classes. The phenotypes are A. Inhibition of shoot growth. B. Inhibition of shoot growth with abnormal leaf shape. C. Lethality. D. Leaf color change. E. Crinkled leaves. These phenotype changes had been observed for 3 or 4 weeks and the picture are taken at 3 or 4 weeks after silencing. For every protease gene, 4 plants were silenced at each experiment. Similar results were obtained from at least three independent experiments. One representative experiment is shown.
(TIF)
Enhanced and delayed HR responses in protease silenced-plant following incompatible pathogen infection. Protease-silenced plants were infiltrated with non-host bacterial pathogen P. syringae pv. tomato T1 (OD600 = 0.2). The HR cell death symptoms were taken at 1 dpi (A) and 2 dpi (B). The phenotypes indicate enhanced (A) or delayed HR (B) responses. For every protease gene, 6 sections per 1 leaf were infected with the pathogen and total 2 leaves were used for 1 plant. Total 4 plants were infiltrated at each experiment and done it at least three repeated tests. Red dotted line indicates the site of P. syringae pv. tomato T1 infection. One representative experiment is shown.
(TIF)
Delayed symptom development in protease-silenced plant following compatible pathogen infection. Protease-silenced plants were infiltrated with host bacterial pathogen P. syringae pv. tabaci (OD600 = 0.005). The disease symptoms induced by the pathogen were taken at 3 dpi. For every protease gene, 6 sections per 1 leaf were infected with the pathogen and total 2 leaves were used for 1 plant. Total 4 plants were infiltrated at each experiment and done it at least three repeated tests. Red dotted line indicates the site of P. syringae pv. tabaci infection. One representative experiment is shown.
(TIF)
Confirmation of gene-specific silencing in M24, S01 and M18 proteases subfamily. Silencing confirmation of M24 (A), S01 (B) and M18 (C) protease subfamily members with gene specific primers were confirmed by quantitative RT-PCR. The values were normalized to NbActin and were calculated to the control. Values are means±SD (n = 3). Similar results were obtained from at least two experiments. One representative experiment is shown. Asterisks indicate significant differences relative to the control as determined by Student’s t test (*P<0.05, **P<0.01, ***P<0.001).
(TIF)
Transcript levels of M24, S01 and M18 protease subfamily members in protease-silenced plants. A. Transcript levels of M24 protease subfamily members were examined in Ncn2132-silenced plants. B. Transcript levels of M24 protease subfamily members were examined in Ncn881-silenced plants. C. Transcript levels of M24 protease subfamily members were examined in Ncn9826- silenced plants. D. Transcript level of S01 protease subfamily members was examined in Ncn964-silenced plants. E. Transcript level of M18 protease subfamily members was examined in Ncn8326-silenced plants. Values are means±SD (n = 3). The values were normalized to NbActin and were calculated to the control. Similar results were obtained from at least two experiments. One representative experiment is shown.
(TIF)
List of selected 153 proteases with its corresponding proteases from different organisms. aClassification abbreviations : (A) = Aspartic, (C) = Cysteine, (M) = Metallo-, (S) = Serine, (T) = Threonine proteases family based on MEROPS classification system (http://merops.sanger.ac.uk/). The corresponding proteases in the b N. benthamiana genome (http://solgenomics.net/, Niben.genome.v0.4.4), dtomato genome (http://solgenomics.net/, ITAG2.40), epotato genome (http://solgenomics.net/PGSC DM v3.4), f Arabidopsis genome (http://www.arabidopsis.org/, TAIR10) and gother organims from the Genbank database (http://www.ncbi.nlm.nih.gov/genbank/). cNumbers in () indicates the additional accession number which corresponds to the pepper EST ID.
(DOCX)
List of 153 protease-silenced plants with developmental phenotypes and responses to incompatible and compatible pathogen. a Developmental phenotypes abbreviations : N.D. = No difference, ISG = Inhibition of shoot growth, ISG+ALS = Inhibition of shoot growth with abnormal leaf shape, L = Lethality, LCC = Leaf color changed, ALS = Abnormal leaf shape. bResponse to incompatible pathogens abbreviations :+ = Delayed HR, − = Enhanced HR, N.D. = No difference, L = Lethality. cResponse to compatible pathogens abbreviations :+ = Delayed symptom, N.D. = No difference, L = Lethality.
(DOCX)
Primer information for quantitative RT-PCR analysis.
(DOCX)
Acknowledgments
We thank professor, S.P. Dinesh-Kumar (UC Davis) for providing the TRV-LIC vector, JungEun Kim and Cheol-Goo Hur (Omics Research Center, KRIBB) for the bioinformatics work, Maribel García-Lorenzo (Umeå University, Sweden) for providing accession numbers of Populus and Arabidopsis proteases and Sophien Kamoun (The Sainsbury Laboratory) for the critical advice on the manuscript.
Funding Statement
This work was supported by grants from the National Research Foundation of the Korea Ministry of Education, Science and Technology (project number 2010-0015105), and the Agricultural Genome Center of Biogreen21 for the Next Generation Program, RDA of the Korean Government (project number PJ008199012012). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Turk B, Turk du SA, Turk V (2012) Protease signalling: the cutting edge. EMBO J 31: 1630–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baek KH, Choi D (2008) Roles of plant proteases in pathogen defense. Plant Pathology Journal 24: 367–374. [Google Scholar]
- 3. Garcia-Lorenzo M, Sjodin A, Jansson S, Funk C (2006) Protease gene families in Populus and Arabidopsis. BMC Plant Biol 6: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van der Hoorn RA (2008) Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol 59: 191–223. [DOI] [PubMed] [Google Scholar]
- 5.Barrett AJ, Rawlings ND, Woessner JF (2004) Handbook of proteolytic enzymes. Amsterdam; Boston: Elsvier Academic Press.
- 6. Turk B (2006) Targeting proteases: successes, failures and future prospects. Nat Rev Drug Discov 5: 785–799. [DOI] [PubMed] [Google Scholar]
- 7. Becraft PW, Asuncion-Crabb Y (2000) Positional cues specify and maintain aleurone cell fate in maize endosperm development. Development 127: 4039–4048. [DOI] [PubMed] [Google Scholar]
- 8. Johnson KL, Degnan KA, Ross Walker J, Ingram GC (2005) AtDEK1 is essential for specification of embryonic epidermal cell fate. Plant J 44: 114–127. [DOI] [PubMed] [Google Scholar]
- 9. Yang P, Smalle J, Lee S, Yan N, Emborg TJ, et al. (2007) Ubiquitin C-terminal hydrolases 1 and 2 affect shoot architecture in Arabidopsis. Plant J 51: 441–457. [DOI] [PubMed] [Google Scholar]
- 10. Tanaka H, Watanabe M, Sasabe M, Hiroe T, Tanaka T, et al. (2007) Novel receptor-like kinase ALE2 controls shoot development by specifying epidermis in Arabidopsis. Development 134: 1643–1652. [DOI] [PubMed] [Google Scholar]
- 11. Xia Y, Suzuki H, Borevitz J, Blount J, Guo Z, et al. (2004) An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J 23: 980–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mendieta JR, Pagano MR, Munoz FF, Daleo GR, Guevara MG (2006) Antimicrobial activity of potato aspartic proteases (StAPs) involves membrane permeabilization. Microbiology 152: 2039–2047. [DOI] [PubMed] [Google Scholar]
- 13. Chen FQ, Foolad MR (1997) Molecular organization of a gene in barley which encodes a protein similar to aspartic protease and its specific expression in nucellar cells during degeneration. Plant Molecular Biology 35: 821–831. [DOI] [PubMed] [Google Scholar]
- 14. Liu YQ, Dammann C, Bhattacharyya MK (2001) The matrix metalloproteinase gene GmMMP2 is activated in response to pathogenic infections in soybean. Plant Physiology 127: 1788–1797. [PMC free article] [PubMed] [Google Scholar]
- 15. Kim HJ, Baek KH, Lee SW, Kim J, Lee BW, et al. (2008) Pepper EST database: comprehensive in silico tool for analyzing the chili pepper (Capsicum annuum) transcriptome. BMC Plant Biol 8: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rawlings ND, Barrett AJ, Bateman A (2012) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 40: D343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robertson D (2004) VIGS vectors for gene silencing: many targets, many tools. Annu Rev Plant Biol 55: 495–519. [DOI] [PubMed] [Google Scholar]
- 18. Brigneti G, Martin-Hernandez AM, Jin H, Chen J, Baulcombe DC, et al. (2004) Virus-induced gene silencing in Solanum species. Plant J 39: 264–272. [DOI] [PubMed] [Google Scholar]
- 19. Chung E, Seong E, Kim YC, Chung EJ, Oh SK, et al. (2004) A method of high frequency virus-induced gene silencing in chili pepper (Capsicum annuum L. cv. Bukang). Mol Cells 17: 377–380. [PubMed] [Google Scholar]
- 20. Hileman LC, Drea S, Martino G, Litt A, Irish VF (2005) Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy). Plant J 44: 334–341. [DOI] [PubMed] [Google Scholar]
- 21. Liu Y, Schiff M, Dinesh-Kumar SP (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786. [DOI] [PubMed] [Google Scholar]
- 22. Ratcliff F, Martin-Hernandez AM, Baulcombe DC (2001) Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant Journal 25: 237–245. [DOI] [PubMed] [Google Scholar]
- 23. Wang CC, Cai XZ, Wang XM, Zheng Z (2006) Optimisation of tobacco rattle virus-induced gene silencing in Arabidopsis. Functional Plant Biology 33: 347–355. [DOI] [PubMed] [Google Scholar]
- 24. Dong Y, Burch-Smith TM, Liu Y, Mamillapalli P, Dinesh-Kumar SP (2007) A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4–1 and -2 in floral development. Plant Physiology 145: 1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana (2000) Nature. 408: 796–815. [DOI] [PubMed] [Google Scholar]
- 26. Bombarely A, Rosli HG, Vrebalov J, Moffett P, Mueller LA, et al. (2012) A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol Plant Microbe Interact 25: 1523–1530. [DOI] [PubMed] [Google Scholar]
- 27. Sato S, Tabata S, Hirakawa H, Asamizu E, Shirasawa K, et al. (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu X, Pan S, Cheng S, Zhang B, Mu D, et al. (2011) Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195. [DOI] [PubMed] [Google Scholar]
- 29. Melotto M, Underwood W, He SY (2008) Role of stomata in plant innate immunity and foliar bacterial diseases. Annu Rev Phytopathol 46: 101–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yeom SI, Seo E, Oh SK, Kim KW, Choi D (2012) A common plant cell-wall protein HyPRP1 has dual roles as a positive regulator of cell death and a negative regulator of basal defense against pathogens. Plant J 69: 755–768. [DOI] [PubMed] [Google Scholar]
- 31. Kang S, Oh SK, Kim JJ, Choi D, Baek KH (2010) NMMP1, a matrix metalloprotease in Nicotiana benthamiana has a role in protection against bacterial infection. Plant Pathology Journal 26: 402–408. [Google Scholar]
- 32. Yi SY, Lee DJ, Yeom SI, Yoon J, Kim YH, et al. (2010) A novel pepper (Capsicum annuum) receptor-like kinase functions as a negative regulator of plant cell death via accumulation of superoxide anions. New Phytol 185: 701–715. [DOI] [PubMed] [Google Scholar]
- 33. Goodin MM, Zaitlin D, Naidu RA, Lommel SA (2008) Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol Plant Microbe Interact 21: 1015–1026. [DOI] [PubMed] [Google Scholar]
- 34. Wan JX, Bringloe D, Lamppa GK (1998) Disruption of chloroplast biogenesis and plant development upon down-regulation of a chloroplast processing enzyme involved in the import pathway. Plant Journal 15: 459–468. [Google Scholar]
- 35. Zhong R, Wan JX, Jin RG, Lamppa G (2003) A pea antisense gene for the chloroplast stromal processing peptidase yields seedling lethals in Arabidopsis: survivors show defective GFP import in vivo. Plant Journal 34: 802–812. [DOI] [PubMed] [Google Scholar]
- 36. Schaller A (2004) A cut above the rest: the regulatory function of plant proteases. Planta 220: 183–197. [DOI] [PubMed] [Google Scholar]
- 37. Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590. [DOI] [PubMed] [Google Scholar]
- 38. Yoon J, Chung WI, Choi D (2009) NbHB1, Nicotiana benthamiana homeobox 1, is a jasmonic acid-dependent positive regulator of pathogen-induced plant cell death. New Phytol 184: 71–84. [DOI] [PubMed] [Google Scholar]
- 39. Shindo T, Van der Hoorn RA (2008) Papain-like cysteine proteases: key players at molecular battlefields employed by both plants and their invaders. Mol Plant Pathol 9: 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Solomon M, Belenghi B, Delledonne M, Menachem E, Levine A (1999) The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell 11: 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chakravarthy S, Velasquez AC, Ekengren SK, Collmer A, Martin GB (2010) Identification of Nicotiana benthamiana genes involved in pathogen-associated molecular pattern-triggered immunity. Mol Plant Microbe Interact 23: 715–726. [DOI] [PubMed] [Google Scholar]
- 42. Hann DR, Rathjen JP (2007) Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana. Plant J 49: 607–618. [DOI] [PubMed] [Google Scholar]
- 43. Giglione C, Boularot A, Meinnel T (2004) Protein N-terminal methionine excision. Cell Mol Life Sci 61: 1455–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ross S, Giglione C, Pierre M, Espagne C, Meinnel T (2005) Functional and developmental impact of cytosolic protein N-terminal methionine excision in Arabidopsis. Plant Physiology 137: 623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Adam Z, Adamska I, Nakabayashi K, Ostersetzer O, Haussuhl K, et al. (2001) Chloroplast and mitochondrial proteases in Arabidopsis. A proposed nomenclature. Plant Physiology 125: 1912–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Andersson A, Keskitalo J, Sjodin A, Bhalerao R, Sterky F, et al. (2004) A transcriptional timetable of autumn senescence. Genome Biol 5: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kieselbach T, Funk C (2003) The family of Deg/HtrA proteases: from Escherichia coli to Arabidopsis. Physiologia Plantarum 119: 337–346. [Google Scholar]
- 48. Liu Y, Ren D, Pike S, Pallardy S, Gassmann W, et al. (2007) Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant J 51: 941–954. [DOI] [PubMed] [Google Scholar]
- 49. Schomburg L, Kollmus H, Friedrichsen S, Bauer K (2000) Molecular characterization of a puromycin-insensitive leucyl-specific aminopeptidase, PILS-AP. Eur J Biochem 267: 3198–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wilk S, Wilk E, Magnusson RP (1998) Purification, characterization, and cloning of a cytosolic aspartyl aminopeptidase. J Biol Chem 273: 15961–15970. [DOI] [PubMed] [Google Scholar]
- 51. Yokoyama R, Kawasaki H, Hirano H (2006) Identification of yeast aspartyl aminopeptidase gene by purifying and characterizing its product from yeast cells. FEBS J 273: 192–198. [DOI] [PubMed] [Google Scholar]
- 52. Szawlowska U, Grabowska A, Zdunek-Zastocka E, Bielawski W (2012) TsPAP1 encodes a novel plant prolyl aminopeptidase whose expression is induced in response to suboptimal growth conditions. Biochem Biophys Res Commun 419: 104–109. [DOI] [PubMed] [Google Scholar]
- 53. Mahon CS, O’Donoghue AJ, Goetz DH, Murray PG, Craik CS, et al. (2009) Characterization of a multimeric, eukaryotic prolyl aminopeptidase: an inducible and highly specific intracellular peptidase from the non-pathogenic fungus Talaromyces emersonii. Microbiology 155: 3673–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Takagi H (2008) Proline as a stress protectant in yeast: physiological functions, metabolic regulations, and biotechnological applications. Appl Microbiol Biotechnol 81: 211–223. [DOI] [PubMed] [Google Scholar]
- 55. Massiere F, Badet-Denisot MA (1998) The mechanism of glutamine-dependent amidotransferases. Cell Mol Life Sci 54: 205–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Turner JG, Taha RR (1984) Contribution of tabtoxin to the pathogenicity of Pseudomonas syringae pv tabaci. Physiological Plant Pathology 25: 55–69. [Google Scholar]
- 57. Barta TM, Kinscherf TG, Willis DK (1992) Regulation of tabtoxin production by the lemA gene in Pseudomonas syringae. Journal of Bacteriology 174: 3021–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Turner JG, Debbage JM (1982) Tabtoxin-induced symptoms are associated with the accumulation of ammonia formed during photo-respiration. Physiological Plant Pathology 20: 223–233. [Google Scholar]
- 59. Willemoes M, Molgaard A, Johansson E, Martinussen J (2005) Lid L11 of the glutamine amidotransferase domain of CTP synthase mediates allosteric GTP activation of glutaminase activity. FEBS J 272: 856–864. [DOI] [PubMed] [Google Scholar]
- 60. Snoeijers SS, Perez-Garcia A, Joosten MHAJ, De Wit PJGM (2000) The effect of nitrogen on disease development and gene expression in bacterial and fungal plant pathogens. European Journal of Plant Pathology 106: 493–506. [Google Scholar]
- 61.Zhu H, Kranz RG (2012) A nitrogen regulated glutamine amidotransferase (GAT1_2.1) represses shoot branching in Arabidopsis. Plant Physiology. [DOI] [PMC free article] [PubMed]
- 62. Rogers HJ (2005) Cell death and organ development in plants. Curr Top Dev Biol 71: 225–261. [DOI] [PubMed] [Google Scholar]
- 63. van der Hoorn RA, Jones JD (2004) The plant proteolytic machinery and its role in defence. Curr Opin Plant Biol 7: 400–407. [DOI] [PubMed] [Google Scholar]
- 64. Piszczek E, Gutman W (2007) Caspase-like proteases and their role in programmed cell death in plants. Acta Physiologiae Plantarum 29: 391–398. [Google Scholar]
- 65. Coll NS, Epple P, Dangl JL (2011) Programmed cell death in the plant immune system. Cell Death Differ 18: 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Altered phenotypes of protease-silenced plants. Protease-silenced plants showing altered phenotypes are categorized into 5 classes. The phenotypes are A. Inhibition of shoot growth. B. Inhibition of shoot growth with abnormal leaf shape. C. Lethality. D. Leaf color change. E. Crinkled leaves. These phenotype changes had been observed for 3 or 4 weeks and the picture are taken at 3 or 4 weeks after silencing. For every protease gene, 4 plants were silenced at each experiment. Similar results were obtained from at least three independent experiments. One representative experiment is shown.
(TIF)
Enhanced and delayed HR responses in protease silenced-plant following incompatible pathogen infection. Protease-silenced plants were infiltrated with non-host bacterial pathogen P. syringae pv. tomato T1 (OD600 = 0.2). The HR cell death symptoms were taken at 1 dpi (A) and 2 dpi (B). The phenotypes indicate enhanced (A) or delayed HR (B) responses. For every protease gene, 6 sections per 1 leaf were infected with the pathogen and total 2 leaves were used for 1 plant. Total 4 plants were infiltrated at each experiment and done it at least three repeated tests. Red dotted line indicates the site of P. syringae pv. tomato T1 infection. One representative experiment is shown.
(TIF)
Delayed symptom development in protease-silenced plant following compatible pathogen infection. Protease-silenced plants were infiltrated with host bacterial pathogen P. syringae pv. tabaci (OD600 = 0.005). The disease symptoms induced by the pathogen were taken at 3 dpi. For every protease gene, 6 sections per 1 leaf were infected with the pathogen and total 2 leaves were used for 1 plant. Total 4 plants were infiltrated at each experiment and done it at least three repeated tests. Red dotted line indicates the site of P. syringae pv. tabaci infection. One representative experiment is shown.
(TIF)
Confirmation of gene-specific silencing in M24, S01 and M18 proteases subfamily. Silencing confirmation of M24 (A), S01 (B) and M18 (C) protease subfamily members with gene specific primers were confirmed by quantitative RT-PCR. The values were normalized to NbActin and were calculated to the control. Values are means±SD (n = 3). Similar results were obtained from at least two experiments. One representative experiment is shown. Asterisks indicate significant differences relative to the control as determined by Student’s t test (*P<0.05, **P<0.01, ***P<0.001).
(TIF)
Transcript levels of M24, S01 and M18 protease subfamily members in protease-silenced plants. A. Transcript levels of M24 protease subfamily members were examined in Ncn2132-silenced plants. B. Transcript levels of M24 protease subfamily members were examined in Ncn881-silenced plants. C. Transcript levels of M24 protease subfamily members were examined in Ncn9826- silenced plants. D. Transcript level of S01 protease subfamily members was examined in Ncn964-silenced plants. E. Transcript level of M18 protease subfamily members was examined in Ncn8326-silenced plants. Values are means±SD (n = 3). The values were normalized to NbActin and were calculated to the control. Similar results were obtained from at least two experiments. One representative experiment is shown.
(TIF)
List of selected 153 proteases with its corresponding proteases from different organisms. aClassification abbreviations : (A) = Aspartic, (C) = Cysteine, (M) = Metallo-, (S) = Serine, (T) = Threonine proteases family based on MEROPS classification system (http://merops.sanger.ac.uk/). The corresponding proteases in the b N. benthamiana genome (http://solgenomics.net/, Niben.genome.v0.4.4), dtomato genome (http://solgenomics.net/, ITAG2.40), epotato genome (http://solgenomics.net/PGSC DM v3.4), f Arabidopsis genome (http://www.arabidopsis.org/, TAIR10) and gother organims from the Genbank database (http://www.ncbi.nlm.nih.gov/genbank/). cNumbers in () indicates the additional accession number which corresponds to the pepper EST ID.
(DOCX)
List of 153 protease-silenced plants with developmental phenotypes and responses to incompatible and compatible pathogen. a Developmental phenotypes abbreviations : N.D. = No difference, ISG = Inhibition of shoot growth, ISG+ALS = Inhibition of shoot growth with abnormal leaf shape, L = Lethality, LCC = Leaf color changed, ALS = Abnormal leaf shape. bResponse to incompatible pathogens abbreviations :+ = Delayed HR, − = Enhanced HR, N.D. = No difference, L = Lethality. cResponse to compatible pathogens abbreviations :+ = Delayed symptom, N.D. = No difference, L = Lethality.
(DOCX)
Primer information for quantitative RT-PCR analysis.
(DOCX)