Abstract
Aims
DDX3 is an RNA helicase that has antiapoptotic properties, and promotes proliferation and transformation. In addition, DDX3 was shown to be a direct downstream target of HIF-1α (the master regulatory of the hypoxia response) in breast cancer cell lines. However, the relation between DDX3 and hypoxia has not been addressed in human tumors. In this paper, we studied the relation between DDX3 and the hypoxic responsive proteins in human breast cancer.
Methods and Results
DDX3 expression was investigated by immunohistochemistry in breast cancer in comparison with hypoxia related proteins HIF-1α, GLUT1, CAIX, EGFR, HER2, Akt1, FOXO4, p53, ERα, COMMD1, FER kinase, PIN1, E-cadherin, p21, p27, Transferrin receptor, FOXO3A, c-Met and Notch1. DDX3 was overexpressed in 127 of 366 breast cancer patients, and was correlated with overexpression of HIF-1α and its downstream genes CAIX and GLUT1. Moreover, DDX3 expression correlated with hypoxia-related proteins EGFR, HER2, FOXO4, ERα and c-Met in a HIF-1α dependent fashion, and with COMMD1, FER kinase, Akt1, E-cadherin, TfR and FOXO3A independent of HIF-1α.
Conclusions
In invasive breast cancer, expression of DDX3 was correlated with overexpression of HIF-1α and many other hypoxia related proteins, pointing to a distinct role for DDX3 under hypoxic conditions and supporting the oncogenic role of DDX3 which could have clinical implication for current development of DDX3 inhibitors.
Introduction
In the Western world, one in eight women will develop breast cancer during their life and breast cancer is causing about 458.000 deaths worldwide per year [1], [2]. Aggressive forms of breast cancer are frequently refractory to treatment [3], even to established targeted therapy, and thus have a high risk of relapse and formation of distant metastases [4]. Identification of molecular pathways involved in aggressive forms of breast cancer is therefore important to design novel targeted therapeutic agents to counteract tumor progression and metastasis.
DDX3, also known as DDX3X because of its location on the X chromosome, is a member of the DEAD-box RNA helicase family which is involved in transcription, RNA splicing, nuclear export of mRNA and translation initiation [5], [6]. Initially, DDX3 was studied because of its manipulation by viruses like hepatitis C (HCV) and human immunodeficiency virus (HIV) [7], [8]. Recently DDX3 has been associated with cancer [9]. Conflicting evidence exists with regard to its tumor enhancing or repressing properties. Nevertheless, DDX3 was proven to have antiapoptotic properties [10], [11], promotes proliferation and cellular transformation[9], [12]–[14]. Recently, novel compounds were developed which could potentially inhibit DDX3 activity[15]–[20].
A recent in vitro study [21] showed that DDX3 is a direct downstream target of HIF-1α, the predominant factor in the mammalian hypoxia response [22]. Hypoxia is an important event in breast carcinogenesis[23]–[26], causing a more aggressive phenotype with increased invasiveness and proliferation, formation of metastases, resistance to therapy [27] and poorer survival [28], [29].
However, no data are yet available on the relation between DDX3 and hypoxia in human breast cancer, or any other human tumors specimens. Therefore, we set out to correlate expression of DDX3 and HIF-1α in a large set of human invasive breast cancers. Furthermore, we correlated DDX3 expression to expression of various other proteins upstream of HIF-1α like EGFR [30], HER2 [31], Akt1[32]–[34], p53[35]–[39], COMMD1 [40], [41], FER kinase [42], PIN1 [43] and FOXO4 [44]. Also we assessed proteins downstream of HIF-1α such as ERα [45], [46] Transferrin receptor (TfR) [47], FOXO3A [48] and Notch1 [49], [50]. Finally, we included proteins that have been associated with HIF-1α without clear functional relationship like E-cadherin [51], p21 [52], c-Met [53], [54] and p27 [55].
Materials and Methods
Patients
Representative paraffin embedded tissue blocks of 422 breast cancer patients collected between 2004 and 2007 were taken from the archive of the Department of Pathology of the University Medical Centre in Utrecht and routinely processed to four tissue microarrays (TMA) as described before [56], [57].
Clinicopathological data including tumor stage, histological data (type, grade, mitotic index (MAI), estrogen receptor alpha (ERα) and human epidermal growth factor receptor 2 (HER2)) status was collected from patient files (Table 1). Protein expression data by immunohistochemistry of HIF-1α, FOXO3A, FOXO4, PIN1, Akt1, COMMD1, p53, p21, p27, EGFR, E-cadherin, GLUT1 and CAIX was derived from previous studies[34], [40], [58]–[62].
Table 1. Patient characteristics.
| N (422) | missing | ||||||||||
| Mean age (range) | 61.0 | (28–88) | 0 | ||||||||
| Tumor size | |||||||||||
| ≤20 mm | 212 | 50% | 3 | ||||||||
| ≤50 mm | 181 | 43% | |||||||||
| >50 mm | 26 | 6% | |||||||||
| Lymph node status | |||||||||||
| Positive* | 193 | 48% | 18 | ||||||||
| Negative** | 211 | 52% | |||||||||
| Histological type | |||||||||||
| ductal | 343 | 82% | 1 | ||||||||
| lobular | 42 | 10% | |||||||||
| other | 36 | 9% | |||||||||
| Grade | |||||||||||
| I | 80 | 20% | 26 | ||||||||
| II | 145 | 37% | |||||||||
| III | 171 | 43% | |||||||||
| Mitotic index (range) | 17.2 | (0–196) | 0 | ||||||||
| Estrogen receptor# | |||||||||||
| Positive | 335 | 79% | 0 | ||||||||
| Negative | 87 | 21% | |||||||||
| Progesterone receptor# | |||||||||||
| Positive | 247 | 59% | 1 | ||||||||
| Negative | 174 | 41% | |||||||||
| HER2 receptor | |||||||||||
| Positive | 44 | 10% | 0 | ||||||||
| Negative | 378 | 90% | |||||||||
Positive = ≥N1mi.
Negative = N0 or N0(i+) (according to TNM 7th edition, 2010).
10% cut-off.
Use of anonymous or coded left over material for scientific purposes is part of the standard treatment contract with patients in the UMCU [63].
Immunohistochemistry
Sections of 4 µm were cut, mounted on SuperFrost slides (Menzel&Glaeser, Brunswick, Germany), deparaffinized and rehydrated. Endogenous peroxidase was then blocked for 15 min with a buffer solution containing 0.3% hydrogenperoxide. Antigens were retrieved by boiling for 20 min in 10 mM citrate buffer (pH 6.0) (for DDX3, c-Met, TfR, FER kinase and Notch1), cooled and washed in PBS. Nonspecific binding sites were blocked with a 2% normal goat serum, 1% BSA in PBS (pH 7.4) (Notch1). TMAs were subsequently incubated in a humidified chamber for 1 hour with polyclonal rabbit anti-DDX3 R648 [64] diluted 1∶1000, TfR 1∶300 (13–6800, Invitrogen, Breda, The Netherlands) and FER kinase 1∶300 (clone 5D2, Cell Signaling Technologies, USA). Primary antibodies against c-Met 1∶100 (18-2257, Zymed, Invitrogen) and Notch1 1:100 (Cell Signaling Technologies, USA) were incubated overnight at 4°C. Subsequently, sections were washed in PBS and incubated for 30 min with secondary antibodies (Brightvision, Immunologic, Duiven, The Netherlands) washed with PBS and developed with diaminobenzidine. Slides were counterstained with hematoxylin, dehydrated and cover-slipped. Appropriate positive and negative controls were used throughout.
Scoring of Immunohistochemistry
Scoring was done by a single experienced pathologist (PJvD). Intensity of cytoplasmic DDX3, FER kinase and membranous E-cadherin, TfR and c-Met was scored semi-quantitatively from 0–3 and percentages of cells with nuclear DDX3 and Notch1 expression were estimated. Out of three cores from the same patient, the maximum cytoplasmic DDX3 score was used for further analysis.
DDX3 scores 1 and 2 were grouped as low DDX3 expression and evaluated against high DDX3 expression (scores 3). For E-cadherin, TfR, c-Met and FER kinase scores 0 and 1 were defined as low expression versus score 2 and 3 as high expression. For HIF-1α, the 1% threshold was used as before [59].
Statistics
Expression levels of DDX3 and the other proteins were compared by chi-square test or t-test whenever applicable. Logistic regression or ANCOVA was used for multivariate analysis to determine dependence of these relations on HIF-1α.
Since EGFR and HER2 are upstream regulators of HIF-1α via PI-3K/AKT, we also assessed the relation of EGFR and HER2 with DDX3 independent of Akt1 and HIF-1α. In lobular breast cancer there is very little or no expression of E-cadherin, so the lobular cancers were excluded in analysis with respect to E-cadherin.
Pearson correlation coefficient was determined for correlation analysis.
All statistical analyses were carried out with SPSS 17.0 for Windows. (SPSS Inc., Chicago, IL, USA), regarding two-sided p-values below 0.05 as significant.
Results
DDX3 staining could be evaluated in 366 of the 422 breast cancer cases. The drop outs were caused by damaged or detached cores during cutting, mounting, or staining, or did not contain tumor. All breast cancer cases showed some expression of DDX3 of which 127 (35%) showed strong cytoplasmic DDX3 expression.(table 2).
Table 2. Expression of DDX3 in relation to oxygen sensing proteins.
| Cytoplasmic DDX3 | Multivariate | |||||||
| N (%) | Low (%) | High (%) | OR | p valuea | OR | p valueb | ||
| 366 | 239 | 127 | ||||||
| HIF-1α | ≤1% | 214 (66) | 155 (75) | 59 (51) | 2.83 | <0.001 | 2.52 | 0.001 |
| >1% | 108 (34) | 52 (25) | 56 (49) | |||||
| GLUT1 | negative | 123 (39) | 94 (46) | 29 (27) | 2.36 | 0.001 | 1.94 | 0.021 |
| positive | 190 (61) | 110 (54) | 80 (73) | |||||
| CAIX | negative | 62 (19) | 49 (24) | 13 (11) | 2.39 | 0.012 | 2.23 | 0.042 |
| positive | 260 (81) | 159 (76) | 101 (89) | |||||
chi-square test.
logistic regression.
HIF-1α overexpression correlated with expression of CAIX, GLUT1, EGFR, HER2, Akt1, FER kinase, ERα, FOXO4, TfR, c-Met as expected (data not shown). Strong cytoplasmic DDX3 expression was associated with overexpression of the master regulator of the hypoxia response HIF-1α (OR = 2.83; p<0.001) (figure 1) and its downstream proteins GLUT1 (OR = 2.36; p = 0.001) and CAIX (2.39; p = 0.012). In logistic regression, HIF-1α (OR = 2.52; p = 0.001), GLUT1 (OR = 1.94; p = 0.021), CAIX (OR = 2.23; p = 0.042) predicted cytoplasmic DDX3 levels independently.(table 2).
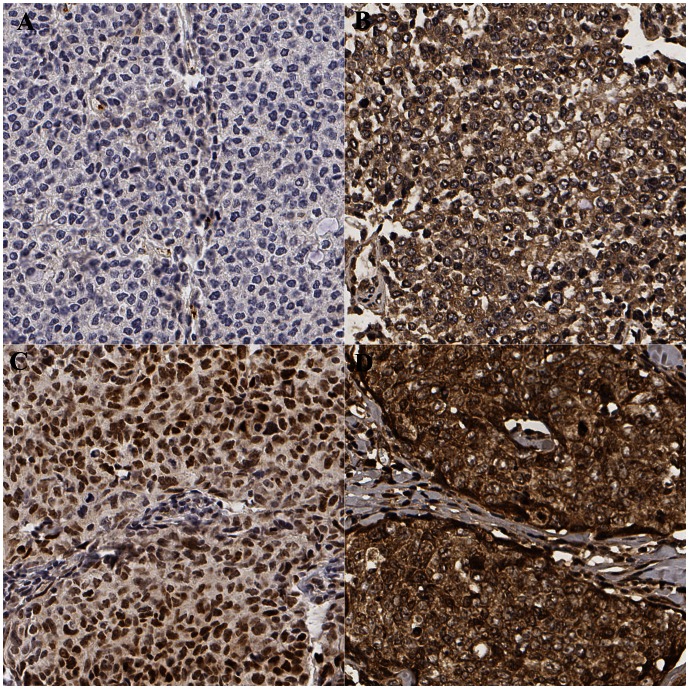
Figure 1. Examples of DDX3 and HIF-1α staining.
Breast cancer photomicrographs are taken at 20X. A. low HIF-1α expression (0%); B. low DDX3 expression (1), same patient as in A; C. high HIF-1α expression (90%); D. high DDX3 expression (3), same patient as in C.
The HIF-1α transcription regulators HER2 (OR = 3.04; p = 0.001), EGFR (OR = 2.01; p = 0.013) and Akt1 (83% vs. 95%; p = 0.001) were correlated with DDX3 expression. Association of EGFR and HER2 with DDX3 was dependent on HIF-1α.(table 3).
Table 3. Expression of DDX3 in relation to regulators of HIF-1α.
| Cytoplasmic DDX3 | Correction for HIF-1α | ||||||||||
| N (%) | Low (%) | High (%) | OR | p valuea | OR | p valueb | |||||
| 366 | 239 | 127 | |||||||||
| EGFR | negative | 299 (83) | 204 (86) | 95 (76) | 2.01 | 0.013 | 1.61 | 0.134 | |||
| positive | 62 (17) | 32 (14) | 30 (24) | ||||||||
| HER2 | negative | 325 (89) | 222 (93) | 103 (81) | 3.04 | 0.001 | 1.88 | 0.092 | |||
| positive | 41 (11) | 17 (7) | 24 (19) | ||||||||
| p53 | negative | 80 (81) | 45 (82) | 35 (80) | 1.16 | 0.802 | 1.42 | 0.571 | |||
| positive | 19 (19) | 10 (18) | 9 (20) | ||||||||
| COMMD1 | low | 23 (29) | 17 (41) | 6 (15) | 3.90 | 0.013 | 5.45 | 0.006 | |||
| high | 57 (71) | 24 (59) | 33 (85) | ||||||||
| FER kinase | low | 203 (57) | 161 (70) | 42 (34) | 4.49 | <0.001 | 4.10 | <0.001 | |||
| high | 152 (43) | 70 (30) | 82 (66) | ||||||||
| PIN1 | low | 58 (71) | 32 (78) | 26 (63) | 2.05 | 0.225 | 1.73 | 0.305 | |||
| high | 24 (29) | 9 (22) | 15 (37) | ||||||||
| Cytoplasmic DDX3 | Cytoplasmic DDX3 | ||||||||||
| N | Low | High | p value c | N | Low | High | p value d | ||||
| Akt1 | 80 | 83% | 95% | 0.001 | 75 | 86% | 95% | 0.026 | |||
| FOXO4 | 75 | 30% | 16% | 0.035 | 61 | 21% | 17% | 0.600 | |||
chi-square test.
logistic regression.
student’s t-test.
ANCOVA.
Proteins known to regulate HIF-1α in a PI-3K/AKT independent fashion were associated with DDX3 as well; COMMD1 (OR = 3.90; p = 0.013), FER kinase (OR = 4.49; p<0.001), FOXO4 (30% vs. 16%; p = 0.035), but not p53 (OR = 1.16; p = 0.802) and PIN1 (OR = 2.05; p = 0.225).(table 3). Logistic regression indicated a HIF-1α independent relation between cytoplasmic DDX3 on the one hand and COMMD1 (OR = 5.45; p = 0.006) and FER kinase (OR = 4.10; p<0.001) on the other.(table 3).
DDX3 was further associated with ERα (OR = 0.48; p = 0.005), E-cadherin (OR = 2.84; p = 0.005), TfR (OR = 2.77; p<0.001), c-Met (OR = 1.72; p = 0.042) and FOXO3A (83% vs. 94%; p = 0.021). (table 4) After correction for HIF-1α expression, E-cadherin (OR = 2.91; p = 0.009), TfR (OR = 2.01; p = 0.007) and FOXO3A (78% vs. 95%; p = 0.007) were still associated with DDX3.(table 4). Table 5 shows the Pearson correlation analysis results.
Table 4. Expression of DDX3 in relation to various other hypoxia induced proteins.
| N (%) | Low (%) | High (%) | OR | p valuea | OR | p valueb | ||||
| 366 | 239 | 127 | ||||||||
| ERα | negative | 81 (22) | 42 (18) | 39 (31) | 0.48 | 0.005 | 0.67 | 0.166 | ||
| positive | 285 (78) | 197 (82) | 88 (69) | |||||||
| E-cadherin* | low | 48 (17) | 38 (23) | 10 (9) | 2.84 | 0.005 | 2.91 | 0.009 | ||
| high | 227 (83) | 130 (77) | 97 (91) | |||||||
| p21 | low | 46 (46) | 28 (51) | 18 (41) | 1.50 | 0.418 | 1.26 | 0.625 | ||
| high | 53 (54) | 27 (49) | 26 (59) | |||||||
| TfR | low | 221 (63) | 161 (72) | 60 (48) | 2.77 | <0.001 | 2.01 | 0.007 | ||
| high | 128 (37) | 63 (28) | 65 (52) | |||||||
| c-Met | negative | 264 (77) | 181 (81) | 83 (71) | 1.72 | 0.042 | 1.62 | 0.096 | ||
| positive | 77 (23) | 43 (19) | 34 (29) | |||||||
| Cytoplasmic DDX3 | Cytoplasmic DDX3 | |||||||||
| N | Low | High | p value c | N | Low | High | p value d | |||
| p27 | 99 | 42% | 44% | 0.683 | 77 | 40% | 45% | 0.507 | ||
| FOXO3A | 86 | 83% | 94% | 0.021 | 71 | 78% | 95% | 0.007 | ||
| Notch1 | 305 | 63% | 55% | 0.064 | 282 | 61% | 56% | 0.299 | ||
chi-square test.
logistic regression.
student’s t-test.
ANCOVA.
in ductal breast cancer.
TfR = Transferrin receptor.
Table 5. DDX3 correlations with the most important hypoxia related proteins.
| N | ra | p value | |
| HIF-1α | 322 | 0.276 | <0.001 |
| GLUT1 | 313 | 0.186 | 0.001 |
| CAIX | 322 | 0.136 | 0.015 |
| HER2 | 366 | 0.185 | <0.001 |
| ERα | 366 | -0.132 | 0.011 |
Pearson correlation coefficient.
Discussion
The aim of this study was to investigate the relation between DDX3 and the hypoxic response in human breast cancer in the light of in vitro results pointing to regulation of DDX3 by HIF-1α. We indeed show a positive correlation between HIF-1α and DDX3 overexpression in a large series of human breast cancer cases, as well as an association between DDX3 overexpression and various other hypoxia related proteins.
However, we have established a correlation between DDX3 overexpression and nuclear HIF-1α overexpression which supports the direct regulation of DDX3 by HIF-1α found in vitro [21], but this is obviously no more than an association at this point no proof for a causal relationship. Immunohistochemistry has some limitations like being inherently a more qualitative than quantitative method, and semiquantitative scoring and dichotomization with non-optimal reproducibility. To compensate for these issues we standardized the IHC procedure, used control tissue throughout, scored three samples per patient, studied a large cohort of breast cancer patients and results obtained from dichotomized parameters were confirmed by correlation analysis for the most important parameters with the DAKO score of DDX3 (table 5). Patient features in this study corresponded with known clinicopathological characteristics in breast cancer (table 1) [65].
Furthermore, DDX3 correlated with EGFR, HER2, FOXO4, ERα and c-Met in a HIF-1α dependent way. Also, we found a positive correlation with COMMD1, FER kinase, Akt1, E-cadherin, TfR and FOXO3A independent of HIF-1α. COMMD1 down regulates HIF-1α by competition with HSP90β [41], or down regulates the transcriptional activity of HIF-1α [40]. However, we could not detect an association with COMMD1 and HIF-1α expression or its downstream targets: E-cadherin, TfR, p21, p27 or c-Met. Nonetheless, COMMD1 correlates with DDX3 independent of HIF-1α.
FER kinases help cells to withstand stress, including hypoxia, via up regulation of HIF-1α [42]. We found a strong relation with FER kinase with both HIF-1α and DDX3. After correction for the effect FER kinase has on HIF-1α, a strong relation between FER kinase and DDX3 remained, implying a HIF-1α dependent and independent relation.
DDX3 was shown to down regulate E-cadherin [12], but in the present study we show a positive correlation, for which we have no obvious explanation. TfR is also under transcriptional control of HIF-1α [47]. TfR is overexpressed in many cancers, which could be attributed to the increased need for iron as a cofactor of the ribonucleotide reductase enzyme involved in DNA synthesis of rapidly dividing cells. Thus, the HIF-1α independent relation between DDX3 and TfR corroborates previous reports on the oncogenic properties of DDX3. Nuclear expression of FOXO3A in breast cancer is associated with anti-apoptotic signaling via Akt1, an aggressive phenotype and poor survival [66]. In response to hypoxia, FOXO3A accumulates in a HIF-1α dependent way to inhibit HIF-1α induced apoptosis [48]. Although we did not find a relation between HIF-1α and FOXO3A we did find a relation between FOXO3A and DDX3, independent of HIF-1α and Akt1. Perhaps DDX3 and FOXO3A function in a concerted survival response after stress stimuli.
EGFR, HER2 and Akt1 regulate HIF-1α transcription in a PI-3K/AKT dependent fashion[30]–[33]. As expected, the positive correlation between DDX3, on the one hand and EGFR and HER2 on the other was HIF-1α dependent. Moreover, HER2 and EGFR regulation of HIF-1α was Akt1 dependent. Furthermore, in 93% of patient samples with high expression of DDX3 and HIF-1α, Akt1 was highly expressed of which 71% of these patients also had EGFR or HER2 overexpression. This fits with a concerted HER2/EGFR-Akt1-HIF-1α-DDX3 pathway, which is consistent with previous reports [21], [34].
In conclusion, ten of eighteen proteins analyzed by IHC showed a similar HIF-1α related effect as described in the literature. All these ten HIF-1α related proteins were associated with expression of DDX3 as well, indicating an important role for DDX3 in the hypoxia response via HIF-1α, and underlying the oncogenic role of DDX3. Since hypoxic tumor regions are typically resistant to current therapy [27], this emphasizes the potential of DDX3 inhibitors, perhaps in combination with HER2 and/or EGFR inhibitors.
Funding Statement
This work was supported by the Dr. Saal van Swanenberg foundation [GMB], the Dutch Cancer Society (UU 2010-4856)[GMB] and FAMRI [VR]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, et al. (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127: 2893–2917. [DOI] [PubMed] [Google Scholar]
- 2.Altekruse S, Kosary C, Krapcho M, Neyman N, Aminou R, et al. (2010) SEER Cancer Statistics Review, 1975–2007, National Cancer Institute. Bethesda, MD, based on November 2009 SEER data submission, posted to the SEER web site. http://seercancergov/csr/1975_2007/Accessed 2012 Nov.
- 3. Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, et al. (2008) Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol 26: 1275–1281. [DOI] [PubMed] [Google Scholar]
- 4. Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, et al. (2007) Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 13: 4429–4434. [DOI] [PubMed] [Google Scholar]
- 5. Rocak S, Linder P (2004) DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 5: 232–241. [DOI] [PubMed] [Google Scholar]
- 6. Lorsch JR (2002) RNA chaperones exist and DEAD box proteins get a life. Cell 109: 797–800. [DOI] [PubMed] [Google Scholar]
- 7. Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT (2004) Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119: 381–392. [DOI] [PubMed] [Google Scholar]
- 8. Owsianka AM, Patel AH (1999) Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology 257: 330–340. [DOI] [PubMed] [Google Scholar]
- 9. Huang JS, Chao CC, Su TL, Yeh SH, Chen DS, et al. (2004) Diverse cellular transformation capability of overexpressed genes in human hepatocellular carcinoma. Biochem Biophys Res Commun 315: 950–958. [DOI] [PubMed] [Google Scholar]
- 10. Sun M, Song L, Li Y, Zhou T, Jope RS (2008) Identification of an antiapoptotic protein complex at death receptors. Cell Death Differ 15: 1887–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Y, Wang H, Wang Z, Makhija S, Buchsbaum D, et al. (2006) Inducible resistance of tumor cells to tumor necrosis factor-related apoptosis-inducing ligand receptor 2-mediated apoptosis by generation of a blockade at the death domain function. Cancer Res 66: 8520–8528. [DOI] [PubMed] [Google Scholar]
- 12. Botlagunta M, Vesuna F, Mironchik Y, Raman A, Lisok A, et al. (2008) Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene 27: 3912–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shih JW, Tsai TY, Chao CH, Wu Lee YH (2007) Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene 27: 700–714. [DOI] [PubMed] [Google Scholar]
- 14. Lee CS, Dias AP, Jedrychowski M, Patel AH, Hsu JL, et al. (2008) Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res 36: 4708–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kondaskar A, Kondaskar S, Kumar R, Fishbein JC, Muvarak N, et al. (2010) Novel, Broad Spectrum Anti-Cancer Agents Containing the Tricyclic 5:7:5-Fused Diimidazodiazepine Ring System. ACS Med Chem Lett 2: 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maga G, Falchi F, Garbelli A, Belfiore A, Witvrouw M, et al. (2008) Pharmacophore modeling and molecular docking led to the discovery of inhibitors of human immunodeficiency virus-1 replication targeting the human cellular aspartic acid-glutamic acid-alanine-aspartic acid box polypeptide 3. J Med Chem 51: 6635–6638. [DOI] [PubMed] [Google Scholar]
- 17. Kumar R, Ujjinamatada RK, Hosmane RS (2008) The first synthesis of a novel 5:7:5-fused diimidazodiazepine ring system and some of its chemical properties. Org Lett 10: 4681–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Radi M, Falchi F, Garbelli A, Samuele A, Bernardo V, et al. (2012) Discovery of the first small molecule inhibitor of human DDX3 specifically designed to target the RNA binding site: towards the next generation HIV-1 inhibitors. Bioorg Med Chem Lett 22: 2094–2098. [DOI] [PubMed] [Google Scholar]
- 19. Belon CA, High YD, Lin TI, Pauwels F, Frick DN (2010) Mechanism and specificity of a symmetrical benzimidazolephenylcarboxamide helicase inhibitor. Biochemistry 49: 1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yedavalli VS, Zhang N, Cai H, Zhang P, Starost MF, et al. (2008) Ring expanded nucleoside analogues inhibit RNA helicase and intracellular human immunodeficiency virus type 1 replication. J Med Chem 51: 5043–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Botlagunta M, Krishnamachary B, Vesuna F, Winnard PT Jr, Bol GM, et al. (2011) Expression of DDX3 is directly modulated by hypoxia inducible factor-1 alpha in breast epithelial cells. PLoS ONE 6: e17563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Majmundar AJ, Wong WJ, Simon MC (2010) Hypoxia-inducible factors and the response to hypoxic stress. Molecular cell 40: 294–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bos R, Zhong H, Hanrahan CF, Mommers EC, Semenza GL, et al. (2001) Levels of hypoxia-inducible factor-1 alpha during breast carcinogenesis. J Natl Cancer Inst 93: 309–314. [DOI] [PubMed] [Google Scholar]
- 24. Bos R, Diest PJv, Groep Pvd, Greijer AE, Hermsen MAJA, et al. (2003) Protein expression of B-cell lymphoma gene 6 (BCL-6) in invasive breast cancer is associated with cyclin D1 and hypoxia-inducible factor-1[alpha] (HIF-1[alpha]). Oncogene 22: 8948–8951. [DOI] [PubMed] [Google Scholar]
- 25. Bos R, van Diest PJ, van der Groep P, Shvarts A, Greijer AE, et al. (2004) Expression of hypoxia-inducible factor-1alpha and cell cycle proteins in invasive breast cancer are estrogen receptor related. Breast Cancer Res 6: R450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bos R, van Diest PJ, de Jong JS, van der Groep P, van der Valk P, et al. (2005) Hypoxia-inducible factor-1alpha is associated with angiogenesis, and expression of bFGF, PDGF-BB, and EGFR in invasive breast cancer. Histopathology 46: 31–36. [DOI] [PubMed] [Google Scholar]
- 27. Greijer AE, de Jong JS, Scheffer GL, Shvarts A, van Diest PJ, et al. (2005) Hypoxia-induced acidification causes mitoxantrone resistance not mediated by drug transporters in human breast cancer cells. Cellular Oncology 27: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Semenza GL (2000) Hypoxia, clonal selection, and the role of HIF-1 in tumor progression. Crit Rev Biochem Mol Biol 35: 71–103. [DOI] [PubMed] [Google Scholar]
- 29. Vaupel P (2004) The role of hypoxia-induced factors in tumor progression. Oncologist 9 Suppl 510–17. [DOI] [PubMed] [Google Scholar]
- 30. Peng XH, Karna P, Cao Z, Jiang BH, Zhou M, et al. (2006) Cross-talk between epidermal growth factor receptor and hypoxia-inducible factor-1alpha signal pathways increases resistance to apoptosis by up-regulating survivin gene expression. J Biol Chem 281: 25903–25914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laughner E, Taghavi P, Chiles K, Mahon PC, Semenza GL (2001) HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1alpha (HIF-1alpha) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol Cell Biol 21: 3995–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mottet D, Dumont V, Deccache Y, Demazy C, Ninane N, et al. (2003) Regulation of hypoxia-inducible factor-1alpha protein level during hypoxic conditions by the phosphatidylinositol 3-kinase/Akt/glycogen synthase kinase 3beta pathway in HepG2 cells. J Biol Chem 278: 31277–31285. [DOI] [PubMed] [Google Scholar]
- 33. Brugarolas J, Kaelin WG (2004) Dysregulation of HIF and VEGF is a unifying feature of the familial hamartoma syndromes. Cancer Cell 6: 7–10. [DOI] [PubMed] [Google Scholar]
- 34. Gort EH, Groot AJ, van de Ven TLPD, van der Groep P, Verlaan I, et al. (2006) Hypoxia-inducible factor-1 alpha expression requires PI 3-kinase activity and correlates with Akt1 phosphorylation in invasive breast carcinomas. Oncogene 25: 6123–6127. [DOI] [PubMed] [Google Scholar]
- 35. Blagosklonny MV, An WG, Romanova LY, Trepel J, Fojo T, et al. (1998) p53 inhibits hypoxia-inducible factor-stimulated transcription. J Biol Chem 273: 11995–11998. [DOI] [PubMed] [Google Scholar]
- 36. Yamakuchi M, Lotterman CD, Bao C, Hruban RH, Karim B, et al. (2010) P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci U S A 107: 6334–6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, et al. (2000) Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev 14: 34–44. [PMC free article] [PubMed] [Google Scholar]
- 38. Schmid T, Zhou J, Kohl R, Brune B (2004) p300 relieves p53-evoked transcriptional repression of hypoxia-inducible factor-1 (HIF-1). Biochem J 380: 289–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sano M, Minamino T, Toko H, Miyauchi H, Orimo M, et al. (2007) p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature 446: 444–448. [DOI] [PubMed] [Google Scholar]
- 40. van de Sluis B, Mao X, Zhai Y, Groot AJ, Vermeulen JF, et al. (2010) COMMD1 disrupts HIF-1alpha/beta dimerization and inhibits human tumor cell invasion. J Clin Invest 120: 2119–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van de Sluis B, Groot AJ, Vermeulen J, van der Wall E, van Diest PJ, et al. (2009) COMMD1 Promotes pVHL and O2-Independent Proteolysis of HIF-1alpha via HSP90/70. PLoS ONE 4: e7332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Salem Y, Shpungin S, Pasder O, Pomp O, Taler M, et al. (2005) Fer kinase sustains the activation level of ERK1/2 and increases the production of VEGF in hypoxic cells. Cell Signal 17: 341–353. [DOI] [PubMed] [Google Scholar]
- 43. Yuan WC, Lee YR, Huang SF, Lin YM, Chen TY, et al. (2011) A Cullin3-KLHL20 Ubiquitin ligase-dependent pathway targets PML to potentiate HIF-1 signaling and prostate cancer progression. Cancer Cell 20: 214–228. [DOI] [PubMed] [Google Scholar]
- 44. Tang TT, Lasky LA (2003) The forkhead transcription factor FOXO4 induces the down-regulation of hypoxia-inducible factor 1 alpha by a von Hippel-Lindau protein-independent mechanism. J Biol Chem 278: 30125–30135. [DOI] [PubMed] [Google Scholar]
- 45. Stoner M, Saville B, Wormke M, Dean D, Burghardt R, et al. (2002) Hypoxia induces proteasome-dependent degradation of estrogen receptor alpha in ZR-75 breast cancer cells. Mol Endocrinol 16: 2231–2242. [DOI] [PubMed] [Google Scholar]
- 46. Cho J, Kim D, Lee S, Lee Y (2005) Cobalt chloride-induced estrogen receptor alpha down-regulation involves hypoxia-inducible factor-1alpha in MCF-7 human breast cancer cells. Mol Endocrinol 19: 1191–1199. [DOI] [PubMed] [Google Scholar]
- 47. Tacchini L, Bianchi L, Bernelli-Zazzera A, Cairo G (1999) Transferrin receptor induction by hypoxia. HIF-1-mediated transcriptional activation and cell-specific post-transcriptional regulation. J Biol Chem 274: 24142–24146. [DOI] [PubMed] [Google Scholar]
- 48. Bakker WJ, Harris IS, Mak TW (2007) FOXO3a is activated in response to hypoxic stress and inhibits HIF1-induced apoptosis via regulation of CITED2. Molecular cell 28: 941–953. [DOI] [PubMed] [Google Scholar]
- 49. Qiang L, Wu T, Zhang HW, Lu N, Hu R, et al. (2012) HIF-1 alpha is critical for hypoxia-mediated maintenance of glioblastoma stem cells by activating Notch signaling pathway. Cell Death and Differentiation 19: 284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gustafsson MV, Zheng X, Pereira T, Gradin K, Jin S, et al. (2005) Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell 9: 617–628. [DOI] [PubMed] [Google Scholar]
- 51. Krishnamachary B, Zagzag D, Nagasawa H, Rainey K, Okuyama H, et al. (2006) Hypoxia-inducible factor-1-dependent repression of E-cadherin in von Hippel-Lindau tumor suppressor-null renal cell carcinoma mediated by TCF3, ZFHX1A, and ZFHX1B. Cancer Res 66: 2725–2731. [DOI] [PubMed] [Google Scholar]
- 52. Koshiji M, Kageyama Y, Pete EA, Horikawa I, Barrett JC, et al. (2004) HIF-1 alpha induces cell cycle arrest by functionally counteracting Myc. Embo Journal 23: 1949–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pennacchietti S, Michieli P, Galluzzo M, Mazzone M, Giordano S, et al. (2003) Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell 3: 347–361. [DOI] [PubMed] [Google Scholar]
- 54. Hayashi M, Sakata M, Takeda T, Tahara M, Yamamoto T, et al. (2005) Up-regulation of c-met protooncogene product expression through hypoxia-inducible factor-1alpha is involved in trophoblast invasion under low-oxygen tension. Endocrinology 146: 4682–4689. [DOI] [PubMed] [Google Scholar]
- 55. Horree N, Gort EH, van der Groep P, Heintz AP, Vooijs M, et al. (2008) Hypoxia-inducible factor 1 alpha is essential for hypoxic p27 induction in endometrioid endometrial carcinoma. J Pathol 214: 38–45. [DOI] [PubMed] [Google Scholar]
- 56. Moelans CB, de Weger RA, van Blokland MT, Ezendam C, Elshof S, et al. (2009) HER-2/neu amplification testing in breast cancer by multiplex ligation-dependent probe amplification in comparison with immunohistochemistry and in situ hybridization. Cellular oncology : the official journal of the International Society for Cellular Oncology 31: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van der Groep P, Bouter A, van der Zanden R, Menko FH, Buerger H, et al.. (2004) Re: Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst 96: 712–713; author reply 714. [DOI] [PubMed]
- 58. Vleugel MM, Greijer AE, Shvarts A, van der Groep P, van Berkel M, et al. (2005) Differential prognostic impact of hypoxia induced and diffuse HIF-1alpha expression in invasive breast cancer. J Clin Pathol 58: 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. van der Groep P, Bouter A, Menko FH, van der Wall E, van Diest PJ (2008) High frequency of HIF-1alpha overexpression in BRCA1 related breast cancer. Breast Cancer Res Treat 111: 475–480. [DOI] [PubMed] [Google Scholar]
- 60. Brenkman AB, de Keizer PL, van den Broek NJ, van der Groep P, van Diest PJ, et al. (2008) The peptidyl-isomerase Pin1 regulates p27kip1 expression through inhibition of Forkhead box O tumor suppressors. Cancer Res 68: 7597–7605. [DOI] [PubMed] [Google Scholar]
- 61. Gort EH, Suijkerbuijk KP, Roothaan SM, Raman V, Vooijs M, et al. (2008) Methylation of the TWIST1 promoter, TWIST1 mRNA levels, and immunohistochemical expression of TWIST1 in breast cancer. Cancer Epidemiol Biomarkers Prev 17: 3325–3330. [DOI] [PubMed] [Google Scholar]
- 62. Vermeulen JF, van de Ven RAH, Ercan C, van der Groep P, van der Wall E, et al. (2012) Nuclear Kaiso Expression Is Associated with High Grade and Triple-Negative Invasive Breast Cancer. PLoS ONE 7: e37864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. van Diest PJ, Savulescu J (2002) For and against: No consent should be needed for using leftover body material for scientific purposes. BMJ 325: 648–651. [PubMed] [Google Scholar]
- 64. Angus AG, Dalrymple D, Boulant S, McGivern DR, Clayton RF, et al. (2010) Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein. J Gen Virol 91: 122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li CI, Uribe DJ, Daling JR (2005) Clinical characteristics of different histologic types of breast cancer. Br J Cancer 93: 1046–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chen J, Gomes AR, Monteiro LJ, Wong SY, Wu LH, et al. (2010) Constitutively nuclear FOXO3a localization predicts poor survival and promotes Akt phosphorylation in breast cancer. PLoS ONE 5: e12293. [DOI] [PMC free article] [PubMed] [Google Scholar]