Abstract
Eumycetoma is a traumatic fungal infection in tropical and subtropical areas that may lead to severe disability. Madurella mycetomatis is one of the prevalent etiologic agents in arid Northeastern Africa. The source of infection has not been clarified. Subcutaneous inoculation from plant thorns has been hypothesized, but attempts to detect the fungus in relevant material have remained unsuccessful. The present study aims to find clues to reveal the natural habitat of Madurella species using a phylogenetic approach, i.e. by comparison of neighboring taxa with known ecology. Four species of Madurella were included in a large data set of species of Chaetomium, Chaetomidium, Thielavia, and Papulaspora (n = 128) using sequences of the universal fungal barcode gene rDNA ITS and the partial LSU gene sequence. Our study demonstrates that Madurella species are nested within the Chaetomiaceae, a family of fungi that mainly inhabit animal dung, enriched soil, and indoor environments. We hypothesize that cattle dung, ubiquitously present in rural East Africa, plays a significant role in the ecology of Madurella. If cow dung is an essential factor in inoculation by Madurella, preventative measures may involve the use of appropriate footwear in addition to restructuring of villages to reduce the frequency of contact with etiologic agents of mycetoma. On the other hand, the Chaetomiaceae possess a hidden clinical potential which needs to be explored.
Author Summary
Eumycetoma caused by Madurella mycetomatis is a common subcutaneous, mutilating fungal infection endemic in arid climate zones. Still there are many controversies on the route of infection, but traumatic inoculation of the subcutaneous tissue with the thorn or soil causative organism through minor skin trauma is a popular theory. This is due to the fact that, the origin and natural habitat of Madurella species, the prevalent mycetoma agents are still unknown. In order to predict the natural habitat of M. mycetomatis we investigated its phylogenetic relationships to species with known ecology. Two genes phylogeny based on LSU and ITS was performed for the species of the genus Madurella and representative genera from the family of Chaetomiaceae. Our findings confirmed that Madurella species are phylogenetically member of the family Chaetomiaceae. Members of this family are often found in dung and manure-enriched soil. We therefore suggest that animal dung, abundantly present in endemic villages, could be a possible niche for Madurella and plays an essential role in the onset of eumycetoma. This will help in understanding the origin of the disease and could be a base for future in depth study to investigate the presence of Madurella in dung from endemic areas.
Introduction
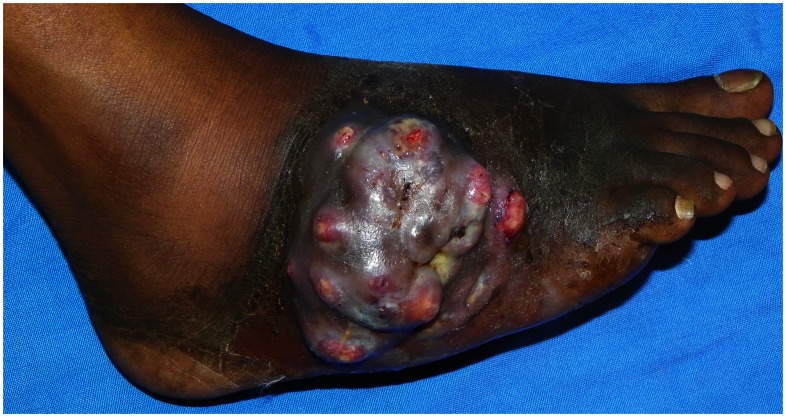
Eumycetoma is a subcutaneous disease with a high morbidity. It is prevalent in tropical and subtropical arid climate zones, with a focus in Northeastern Africa and particularly the Sudan [1]. Patients who develop advanced mycetoma of the extremities frequently become invalids due to the immobilizing nature of the disease (Fig. 1) [2]. Due to lack of social programs and poverty, patients become perpetually dependent on their family. Mycetoma can be caused by a variety of both bacteria (actinomycetoma) and fungi (eumycetoma) and is chronically progressive [1], [2]. eumycetoma is difficult to treat by chemotherapy, surgery frequently leads to mutilation, and relapse is common postoperatively. In the Sudan alone, 25% of the eumycetoma patients underwent amputation of the infected limb because of failure of therapy [3].
Figure 1. Eumycetoma showing granulomatous tumefaction of lateral aspect of right foot with sinus oozing black granules.
In order to reduce the morbidity of this disease, not only is an improvement in chemotherapy required, but also in the preventive measures. These might involve an efficient vaccine, as well as a reduction of contact with the causative agent. Gaining insight in the natural habitat of the prevalent Sudanese agent of mycetoma, Madurella mycetomatis, may lead to strategies to prevent introduction of causative agents into the skin and should reduce the burden of this disease in the endemic communities. However, the natural habitat of the prevalent Sudanese agent of mycetoma, Madurella mycetomatis, is unknown. The classical hypothesis is that aetiologic agents are traumatically introduced via thorn-pricks or with soil particles contaminated by the aetiologic agent, but M. mycetomatis has never been cultured from either thorns or soil. Madurella DNA was demonstrated in 17 out of 74 soil samples, and only in one out of 22 thorns tested [4]. Thus, the thorn-prick hypothesis seems less likely.
Madurella mycetomatis is thus far only known as sterile, melanized mycelium isolated from symptomatic patients. Isolates from subcutaneous infections that consist of dark hyphae are therefore routinely referred to as ‘Madurella’, while those forming compact clumps of cells are traditionally identified as ‘Papulaspora’. Still no form of propagation, either sexual or clonal, is known for these fungi, except for some occasional, undiagnostic phialide-like cells [5]. There are many more causative agents of subcutaneous disorders which lack identifiable sporulation in culture. Today, identification options of such poorly structured fungi have increased with the development of molecular diagnostics. It has become clear that non-sporulating fungi are phylogenetically quite diverse. The melanized species causing black-grain mycetoma worldwide belong to at least two different orders of ascomycetes: the Sordariales and the Pleosporales [6].
In the present study we apply morphology-independent techniques to classify sterile agents of mycetoma in a phylogenetic scaffold of the fungi. This should lead to a better understanding of their ecology and pathology. Non-sporulating clinical isolates, provisionally deposited in two reference laboratories under the generic names Madurella and Papulaspora, were analyzed using the universal fungal barcode gene rDNA partial large subunit (LSU) and the internal transcribed spacer (ITS) regions. Since Madurella mycetomatis is a member of the order Sordariales, Madurella pseudomycetomatis, M. fahalii and M. tropicana most likely belong to the same order [7]. Phylogenies based on the mitochondrial genome confirmed the relationship to the Sordariales. Shared synteny was observed of genes and tRNAs in the mitochondrial genomes of M. mycetomatis and Chaetomium thermophilum [8]. Chaetomium is a large genus of Sordariales with more than 100 described species [9], but only very few species have been sequenced yet. In the present study we sequenced reference and additional clinical isolates of Chaetomium (ITS and LSU). Further members of the family Chaetomiaceae (Sordariales), including representatives of the genera Achaetomium, Aporothielavia, Chaetomidium, and Thielavia were selected to build up a framework of neighboring species to Madurella. Notably nearly all these fungi are ascosporulating only, producing elaborate fruiting bodies which cannot be expressed in human host tissue. Loss of the fruiting body thus immediately leads to sterile, Madurella-like cultures, rather than to a conidial counterpart as is the case in the majority of filamentous fungi. Comparison of ecological habitats of Chaetomiaceae was done in order to predict aspects of possible sources and routes of transmission of Madurella species.
Materials and Methods
Strains analysed
The analysis consists of 128 strains among which 60 strains of Chaetomiaceae contain presently available ex-type strains of described species deposited in the CBS culture collection. A total of 13 sterile filamentous isolates identified as Madurella, and one meristematic isolate, phenotypically identified as Papulaspora sp. were analyzed. The set was complemented with 54 clinical strains identified in this study (Supporting information; table S1). All clinical isolates included in our study were previously isolated from human sources and were taken from the CBS reference collection. Information on strains can be found at (www.cbs.knaw.nl)
DNA extraction
About 10 mm3 fungal mass grown on agar surface were scraped in 2 ml screw cap vial containing 490 µl CTAB-buffer (2% CTAB, 100 mM Tris-HCL, 20 mM EDTA, 1.4 M NaCl) and 6–10 acid washed glass beads. In the subsequent step 10 µl of proteinase K (50 mg/ml) were added and the extraction buffer containing the sample vortexed for 2–5 minutes. The vials were incubated at 60°C for 60 minutes and vortexed again to ensure homogeneity of the sample. 500 µl of SEVAG (Chloroform∶Isoamylalcohol 24∶1) were added and the vials inverted repeatedly for at least two minutes. Vials were centrifuged at 14000 rpm (Eppendorf 5417R, Hamburg, Germany) for 10 minutes and the supernatant collected in new sterile vials with 0.55 volumes of ice cold 2-propanol and inverted several times. The precipitated total nucleic acids were centrifuged at 14000 rpm for 10 minutes. Finally, the pellets were washed with 70% ethanol, air- dried and re-suspended in 100 µl TE buffer.
PCR and sequencing
The internal transcribed spacer (ITS) was amplified using the primers V9G and LS266 [10]. The resulting amplicons were bidirectionally sequenced with primers ITS1 and ITS4 [11]. The partial large ribosomal subunit (28S) was amplified with primer LR0R and LR5 and sequenced with the same primers [12]. A life Technologies Corp. 3730XL Sanger laboratory capillary electrophoresis system was used to retrieve the sequence data.
Alignment and phylogenetic analysis
Trace files retrieved from bidirectional sequencing, were assembled and manually edited using Lasergene Seqman (DNASTAR, USA). A selection of 89 strains from the total data set was used for inferring the phylogenetic tree. Sequences were aligned with MUSCLE using the EMBL-EBI web server (http://www.ebi.ac.uk/Tools/msa/muscle/). A concatenated alignment was assembled for complete ITS (ITS1-5.8S-ITS2) and partial LSU sequences.
Bayesian and maximum likelihood analysis were performed with MrBayes v. 3.1.2 [13], and RAxML 7.2.8 respectively [14], [15]. MrBayes was run for 1 000 000 generations; one tree was saved per 100 of generations and burn-in was set for 25% of the saved trees. The 50% majority consensus tree was calculated and the final tree was edited using MEGA v. 5.05 [16]. Maximum likelihood was conducted using the CIPRES website (www.phylo.org), and GTR (General Time Reversible) model of nucleotide substitution was used; it is the only nucleotide substitution model in the RAxML software.
Results
Phylogenetic analysis
The analyzed data set comprised representative strains of the Chaetomiaceae [Sordariales] of both clinical and environmental origins (Supporting information; table S1).
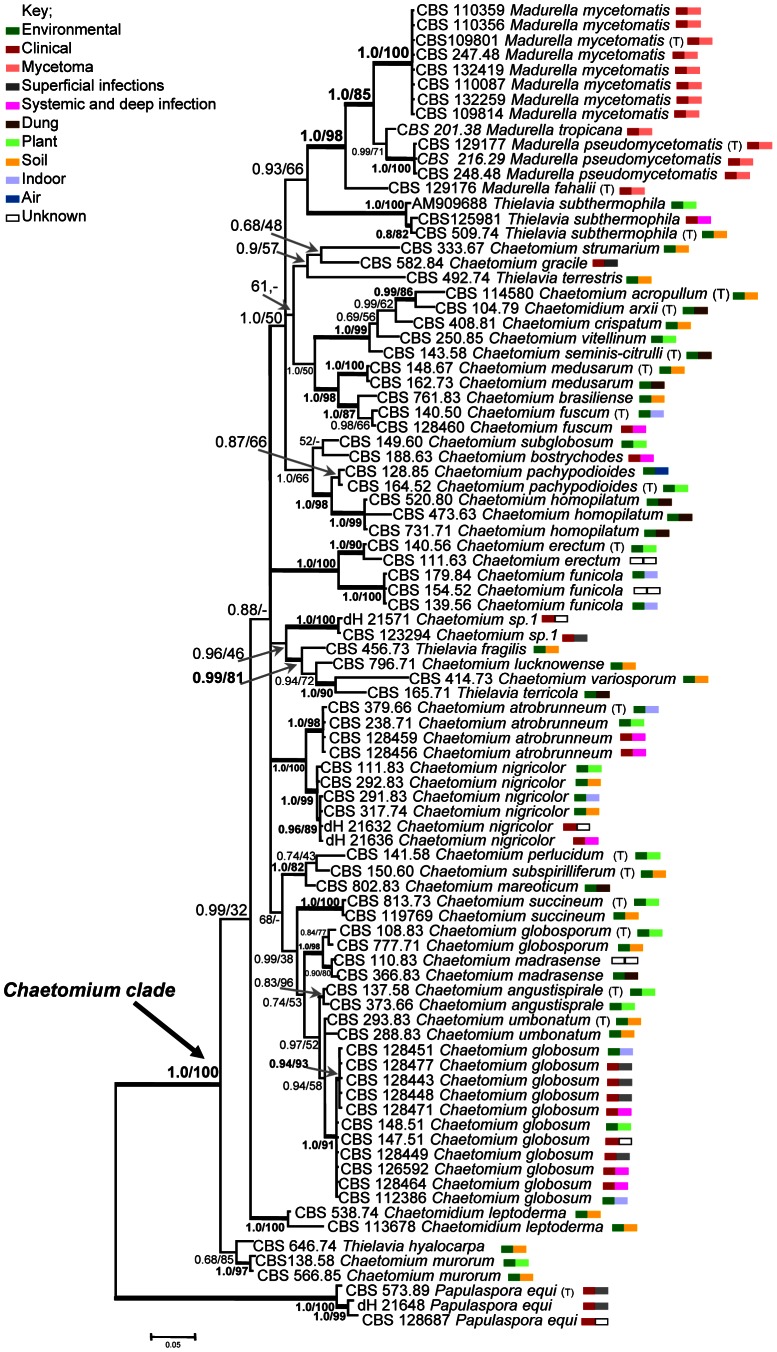
Alignment of the combined genes sequences (ITS, LSU) consisted of 1,356 total characters in which 1029 were constant and 307 were variable. In our two-gene phylogeny most basal and internal branches show high Bayesian inference posterior probability values (BII PP) and maximum-likelihood bootstrap support (ML BS) respectively (Fig. 2). However, some internal branches of the Chaetomiaceae ingroup tree (split 0.88/-) comprising several clusters, e.g. for C. atrobrunneum and C. nigricolor (1.0/100), Chaetomium “sp. 1”, C. lucknowense, C. variosporum, Thielavia terricola and T. fragilis (0.96/46) as well as C. errectum and C. funicola (1.0/100), could not be fully resolved into dichotomies. The ingroup tree comprised a monophyletic cluster with four Madurella species with 1.0, 85% BII PP and ML BS, respectively, basal to Thielavia subthermophila (0.93/66). Madurella clustered within a large clade containing mostly environmental Chaetomium species which were distant from the type species of Chaetomium (C. globosum; Fig. 2). Madurella fahalii was identified as the closest taxon to the Chaetomiaceae at 6.0% ITS divergence from Chaetomium nigricolor. Papulaspora equi, known from three clinical isolates and identified by it is ex-type strain, was resolved basal to the grade comprising the Chaetomium/Chaetomidium/Thielavia/Madurella clades.
Figure 2. Phylogenetic tree resulting from Bayesian and maximum likelihood analysis of the combined ITS/LSU data set.
Branches supported [>0.8 Bayesian probability, >80% Maximum likelihood] are drawn in bold. Papulaspora equi strains [CBS 573.89, CBS 128687, dH 21648] were used to root the tree. Type strain representing correct taxonomy [T].
Sequence identity and classification
The data set contained 38 ex-type and authentic strains. Twenty-two of these were usable to define each as OTU's (Operational taxonomical unit), while 16 were found to be identical to other described species defined by an ex-type isolate. Seven species, as delimited by sequence data, comprised more than one ex-type strain having identical sequences, rendering these species as provisional synonyms. Groups of isolates identified as the classical species Chaetomium globosum, described in the 19th century without deposition of live material, did not contain an ex-type strain. In total, 29 Chaetomium species were judged to be distinct at the LSU/ITS level (Fig. 2), each being separated by several point mutations. Eight strains originating from clinical resources did not show identity to any known Chaetomium species and were therefore reported as ‘unknown Chaetomium sp.’ Three clinical isolates described as ‘Chaetomium sp. 1’, which had provisionally been identified as ‘Papulospora sp.’ on the basis of phenotypic characters, were found within the Chaetomium grade (Fig. 2, Supporting information; table S1).
All Achaetomium species were found to be synonyms of known Chaetomium species including ex-type strains of Achaetomium nepalense, A. thermophilum, and A. strumarium.
Strain origin
The origins of 128 strains analyzed are summarized in table S1 (supporting information). A large quantity (40.6%; n = 52), were of environmental origin; about 7.0% (n = 9) were derived from animal dung, mainly of herbivores such as antelopes, goats, elephants, hares and rodents, but also of carnivores such as foxes. A percentage of 16.4% (n = 21) originated from soil either mixed with dung or decayed plant material, or from rhizosphere; 10.9% (n = 14) were derived from putrid plant material. Several species (C. globosum, C. atrobrunneum) were repeatedly isolated from indoor environments such as mouldy rugs and mattresses.
A total of 54.7% (n = 70) of the overall analyzed strains were from clinical samples. Forty-five out of 112 Chaetomiaceae strains of Chaetomium, Chaetomidium and Thielavia were infection-related, of which 49 strains originated from humans and 5 were veterinary isolates. Five out of eight strains identified as C. atrobrunneum were obtained from deep localizations including sputum, bronchial lavages and brain. Chaetomium globosum was frequently isolated from clinical or veterinary sources (24 strains where information about the origin was available).
In general, the clinical isolates were predominately isolated from the respiratory tract (9.4%, n = 12), possibly as asymptomatic colonizers. A large number of strains (22.7%, n = 29) were isolated from superficial samples including skin, hair, nails and eyes. Five isolates (3.9%) were derived from brain of four humans and one horse, and five (3.9%) strains were recovered from blood and lymph nodes. Infections reported as being subcutaneous were exceptional (0.78%, n = 1 from a wound); none of these were associated with production of grains in tissue.
Within the Chaetomium grade, one unnamed ‘Chaetomium sp. 1’ and four Madurella species were exclusively from clinical origin. Strains of ‘Chaetomium sp. 1’ were mainly associated with eye infections. All 13 strains identified as Madurella were derived from rural patients with subcutaneous eumycetoma with grain production.
Discussion
The genus Madurella, comprising the species M. mycetomatis, M. pseudomycetomatis, M. fahalii and M. tropicana, was found to cluster within the Chaetomiaceae. In contrast to Madurella, most species of this family are able to produce elaborate fruiting bodies with characteristically shaped setae and ascospores. The impressive morphology of the ascomata suggests that species should be easily distinguishable by microscopic morphology, using the available classical, richly illustrated monographs [9], [17]. However, judging from our phylogenetic data (Fig. 2), molecular taxonomy matches poorly with morphology. At the generic level, the distinction between Chaetomium, Achaetomium, Chaetomidium and Thielavia is ambiguous, since several species of these genera clustered amidst Chaetomium species. Sometimes several ex-type strains of described taxa were found to have identical ITS sequences, suggesting that names should be reduced to synonymy. It may be concluded that molecular classification of Chaetomiaceae is significantly different from conventional taxonomy and extensive revision is needed at generic as well as at species levels. The position of Madurella as a derived clade within the family is unambiguous, and unexpected.
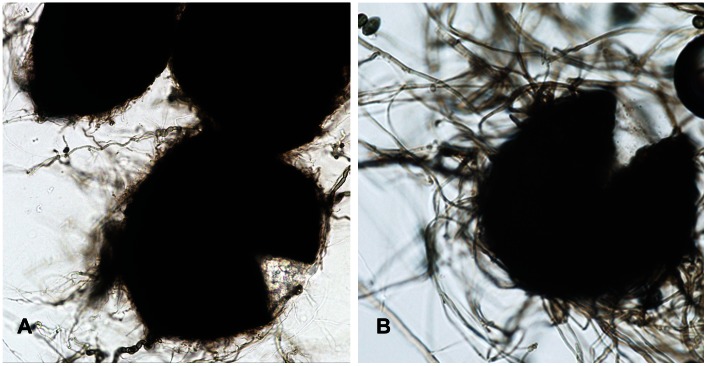
Most members of the Chaetomiaceae lack anamorph sporulation, or some scattered, undiagnostic phialides are present at most [9]. Thus, if strains lose the ability to produce their elaborate ascomata, they cannot be recognized as a Chaetomium species by morphological means, as in Madurella. Most of the clinical Chaetomium strains analyzed in the course of this study produced ascomata in culture, but some had remained sterile. The clinical strains of Chaetomium were responsible for cutaneous or systemic phaeohyphomycoses, but never produced eumycetoma. In contrast, strains of the Madurella subcluster, with four different molecular siblings, were consistently associated with eumycetoma. They were all sterile or produced some undiagnostic, phialide-like cells. Large structures resembling fruiting bodies were occasionally observed in Madurella (Fig. 3), but these did not have the ability to produce ascospores. The Madurella clade is morphologically not so far away from remaining Chaetomiaceae, and the position of Madurella within the Chaetomiaceae thus is explainable. The clade deviates however by producing grains in host subcutaneous tissue.
Figure 3. Structures produced by Madurella mycetomatis and Thielavia subthermophila.
A: black sterile sclerotial bodies of Madurella mycetomatis. B: Thielavia subthermophila fruiting body (ascomata) from which ascospores are produced.
A consistent human pathogen is thus introduced in the family Chaetomiaceae. Traditionally, most species of the family were considered to be insignificant as agents of human disease. Of the ∼100 Chaetomium species described to date only five have repeatedly been associated with infection [5]. The majority of Chaetomium clinical strains analyzed in this study were probably transient colonizers or agents of mild superficial disorders. Twenty seven were involved in onychomycosis or cutaneous and eye infections in otherwise healthy individuals. This matches with literature data [18], [19]. In our data, Chaetomium globosum showed a definite bias towards superficial infection, with 17 out of 29 strains analyzed (supporting information; table S1). The species is able to degrade keratin by production of extracellular keratinases [20]. Fatal, disseminated and cerebral infections by Chaetomiaceae have also been reported. In the literature about 20 deep and disseminated cases were described, nearly all in immunocompromised and severely debilitated patients [21], [22]. Several Chaetomium-like fungi thus show rather pronounced pathology, sometimes with species-specific predilections.
Grain formation in tissue by Chaetomiaceae other than Madurella is not known. A single case of chromoblastomycosis by Chaetomium funicola was reported by Piepenbring et al. [23]. The few subcutaneous cases [24] all showed hyphae in tissue rather than the compact grains of Madurella eumycetoma. In contrast to Madurella, none of the infecting Chaetomiaceae was exclusively clinical; all contained environmental strains as well. If agents of black-grain mycetoma have a relatively limited distribution in the phylogeny of Sordariales, i.e. are clustered within a single family, Chaetomiaceae, one may hypothesize that these fungi are predisposed to human infection and thus are likely to share a set of fundamental virulence factors. Many members of Chaetomiaceae have their natural habitat in soil or on mammal dung. A possible explanation of their recurrent virulence may lie in physiological properties such as growth at the human body temperature of 37°C, and the production of secondary metabolites such as inhibitors of chemokines and TNF-α [25], [26]. Particularly the fatal brain infections, which were repeatedly reported in Achaetomium strumarium (synonym of Chaetomium strumarium) [27], [28], in C. atrobrunneum [19], and in Thielavia subthermophila [21], all belonging to the Chaetomiaceae, are remarkable. The hidden clinical diversity of the Chaetomiaceae urgently needs to be explored.
The role of mammal dung and dung-enriched soil is one of the prime ecological niches in the order Sordariales, and this also holds true for Chaetomium [29] (supporting information; table S1). Some species in the current study exclusively grow in dung, such as Chaetomium homopilatum. Multiple Chaetomium and Thielavia species have been isolated in East Africa from different kinds of dung, ranging from cow and horse to more exotic types of dung such as of elephant and wildebeest [30]. Conversely, the position of Madurella in Chaetomiaceae is informative for the natural habitat of this pathogen. In the highly endemic area in Sudan, M. mycetomatis has as yet not been cultured, whereas the isolation of some other causative agents of mycetoma, Nocardia brasiliensis, Actinomadura madurae, and Streptomyces somaliensis has been successful [31]. The causative agent of eumycetoma Leptosphaeria senegalensis has been recovered from thorns of Acacia species in West and Central Sub-Saharan Africa [32]. Pseudallescheria boydii has been recovered from polluted soil samples all over the world, including the endemic mycetoma regions [33], [34], [35]. For Madurella mycetomatis numerous isolation attempts from environmental sources were without success [4], [36]. Thirumalachar et al. [37] reported M. mycetomatis from soil in India, but the identification was based on scant phenotypic characters only. The difficulty in recovering M. mycetomatis from soil might indicate that pure soil is not the natural habitat for this fungus. Other possible habitats were thorny plant thorns, as plant material was occasionally found in human tissue [36], but this remains exceptional. Based on our study, association with cattle dung now seems to be an alternative option. Madurella mycetomatis apparently needs other culture media for isolation. Enrichment with dung might be a successful strategy. This hypothesis may be extended to Madurella fahalii, M. tropicalis and M. pseudomycetomatis, which are endemic in the arid climate zone of Northeastern Africa and are exclusively known from human mycetoma.
Providing insight into the taxonomic position and possible natural habitat of Madurella species changes our view regarding routes of infection and prevalent risk factors for human mycetoma. The Gezira region in the Sudan is highly endemic for eumycetoma by M. mycetomatis [1]. Most inhabitants live on cattle and camel husbandry and agriculture [38]. Local villages are characterized by an abundance of cattle, goats, sheep, dogs, chickens and donkeys [39]. Cows are raised mainly for their milk and are kept in pens surrounded by walls made of mud or thorny bushes. The floors of the pens are paved with dry feces, thorns and trash [39], and some human settlements are made of dried cow dung. The family house is usually in direct contact with the pen. Inhabitants of the villages mostly are barefoot among the thorny bushes. Traumatic introduction of coprophilic fungi via thorn pricks is thus a plausible scenario. Given the low frequency of Madurella on thorns, contamination of dung and its role as an adjuvant in inoculation seems likely. If cow dung is an essential factor in inoculation by M. mycetomatis, preventative measures may involve the use of appropriate footwear in addition to restructuring of villages by stricter separation of animal husbandry and human settlement to reduce the frequency of contact with mycetoma etiologic agents.
Supporting Information
Name, reported type strains, source, origin, and GenBank accession numbers for the analysed strains. dH: [G.S. de Hoog working collection] UTHSC [University of Texas Health Science Center]. All type strains marked with [T].
(DOCX)
Funding Statement
Wendy W.J. van de Sande was supported by VENI grant 916.11.178 from the Netherlands Society of Scientific Research (NWO). The DNA sequencing work was funded by the Royal Netherlands Academy of Arts and Sciences. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Abbott P (1956) Mycetoma in the Sudan. Trans R Soc Trop Med Hyg 50: 11–24 discussion, 24-30. [DOI] [PubMed] [Google Scholar]
- 2. Fahal AH, Suliman SH (1994) Clinical presentation of mycetoma. Sudan Med J 32: 46–66. [Google Scholar]
- 3. Fahal AH, Hassan MA (1992) Mycetoma. Br J Surg 79: 1138–1141. [DOI] [PubMed] [Google Scholar]
- 4. Ahmed AO, Adelmann D, Fahal A, Verbrugh H, van Belkum A, de Hoog GS (2002) Environmental occurrence of Madurella mycetomatis, the major agent of human eumycetoma in Sudan. J Clin Microbiol 40: 1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Hoog GS, Guarro J, Gene J, Figueras MJ, editors (2000) Atlas of Clinical Fungi. 2nd edition. Washington, D.C.: American Society of Microbiology Press.
- 6. de Hoog GS, Adelmann D, Ahmed AOA, Belkum A (2004) Phylogeny and typification of Madurella mycetomatis, with a comparison of other agents of eumycetoma. Mycoses 47: 121–130. [DOI] [PubMed] [Google Scholar]
- 7. de Hoog GS, van Diepeningen AD, Mahgoub el-S, van de Sande WW (2012) New species of Madurella, causative agents of black-grain mycetoma. J Clin Microbiol 50: 988–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van de Sande WW (2012) Phylogenetic analysis of the complete mitochondrial genome of Madurella mycetomatis confirms its taxonomic position within the order Sordariales. PLoS One 7: e38654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. von Arx JA, Guarro J, Figueras MJ (1986) The Ascomycete genes Chaetomium . Beih Nova Hedw 84: 1–162. [Google Scholar]
- 10. Gerrits van den Ende AHG, de Hoog GS (1999) Variability and molecular diagnostics of the neurotropic species Cladophialophora bantiana . Stud Mycol 43: 152–162. [Google Scholar]
- 11.White T J, Bruns T, Lee S, Taylor J W (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White [ed.], PCR protocols: a guide to methods and applications. New York Academic Press, Inc.: New York. 315–322.
- 12. Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 14. Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- 15. Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57: 758–771. [DOI] [PubMed] [Google Scholar]
- 16. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ames LM (1961) A Monograph of the Chaetomiaceae . US Army Res Develop Ser 2: 1–125. [Google Scholar]
- 18. Hattori N, Adachi M, Kaneko T, Shimozuma M, Ichinohe M, Iozumi K (2000) Case report. Onychomycosis due to Chaetomium globosum successfully treated with itraconazole. Mycoses 43: 89–92. [DOI] [PubMed] [Google Scholar]
- 19. Hubka V, Mencl K, Skorepova M, Lyskova P, Zalabska E (2011) Phaeohyphomycosis and onychomycosis due to Chaetomium spp., including the first report of Chaetomium brasiliense infection. Med Mycol 49: 724–733. [DOI] [PubMed] [Google Scholar]
- 20. Kaul S, Sumbali G (1999) Production of extracellular keratinases by keratinophilic fungal species inhabiting feathers of living poultry birds (Gallus domesticus): A comparison. Mycophatologia 146: 19–24. [Google Scholar]
- 21. Badali H, Chander J, Gupta A, Rani H, Punia RS, De Hoog GS, Meis JF (2011) Fatal cerebral phaeohyphomycosis in an immunocompetent individual due to Thielavia subthermophila . J Clin Microbiol 49: 2336–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guppy KH, Thomas C, Thomas K, Anderson D (1998) Cerebral fungal infections in the immunocompromised host: a literature review and a new pathogen–Chaetomium atrobrunneum: case report. Neurosurgery 43: 1463–1469. [DOI] [PubMed] [Google Scholar]
- 23. Piepenbring M, Caceres MOA, Espino EAA, Kirschner R, Schofer H (2007) Chromoblastomycosis caused by Chaetomium funicola: a case report from Western Panama. Br J Dermatol 157: 1025–1029. [DOI] [PubMed] [Google Scholar]
- 24. Lin Y, Li X, et al. (1995) First Case of Phaeohyphomycosis Caused by Chaetomium Murorum in China. Chin J Dermatol 28: 367–369. [Google Scholar]
- 25. Rether J, Erkel G, Anke T, Sterner O (2004) Inhibition of inducible TNF-alpha expression by oxaspirodion, a novel spiro-compound from the ascomycete Chaetomium subspirale . Biol Chem 385: 829–834. [DOI] [PubMed] [Google Scholar]
- 26. Chan T-M, Chu M (2007) Sch 213766, a novel chemokine receptor CCR-5 inhibitor from Chaetomium globosum . J Antibiot 60: 524–528. [DOI] [PubMed] [Google Scholar]
- 27. Abbott SP, Sigler L, McAleer R, McGough DA, Rinaldi MG, et al. (1995) Fatal cerebral mycoses caused by the ascomycete Chaetomium strumarium . J Clin Microbiol 33: 2692–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aribandi M, Bazan C Iii, Rinaldi MG (2005) Magnetic resonance imaging findings in fatal primary cerebral infection due to Chaetomium strumarium . Australas Radiol 49: 166–169. [DOI] [PubMed] [Google Scholar]
- 29. Zhang N, Castlebury LA, Miller AN, Huhndorf SM, Schoch CL, et al. (2006) An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia 98: 1076–1087. [DOI] [PubMed] [Google Scholar]
- 30. Carter A, Khan RS (1982) New and interesting Chaetomium species from East Africa. Can J Bot 60: 1253–1262. [Google Scholar]
- 31. Aghamirian MR, Ghiasian SA (2009) Isolation and characterization of medically important aerobic actinomycetes in soil of Iran (2006–2007). Open Microbiol J 3: 53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Segretain G, Mariat F (1968) Recherches sur la présence d'agents de mycétomes dans le sol et sur les épineux du Sénégal et de la Mauritanie. Bull Soc Pathol Exot 61: 194–202. [PubMed] [Google Scholar]
- 33. Ajello L (1952) The isolation of Allescheria boydii shear, an etiologic agent of mycetomas, from soil. Am J Trop Med Hyg 1: 227–238. [DOI] [PubMed] [Google Scholar]
- 34. Gugnani HC, Paliwal-Joshi A, Rahman H, Padhye AA, Singh TS, Das TK, Khanal B, Bajaj R, Rao S, Chukhani R (2007) Occurrence of pathogenic fungi in soil of burrows of rats and of other sites in bamboo plantations in India and Nepal. Mycoses 50: 507–511. [DOI] [PubMed] [Google Scholar]
- 35. Harun A, Gilgado F, Chen SC, Meyer W (2010) Abundance of Pseudallescheria/Scedosporium species in the Australian urban environment suggests a possible source for scedosporiosis including the colonization of airways in cystic fibrosis. Med Mycol 48 Suppl 1: S70–76. [DOI] [PubMed] [Google Scholar]
- 36.Fahal AH (2006) Mycetoma — clinicopathological monograph. Khartoum University Press, Khartoum, Sudan.
- 37. Thirumalachar MJ, Padhye AA (1968) Isolation of Madurella mycetomi from soil in India. Hindustan Antibiot Bull 10: 314–318. [PubMed] [Google Scholar]
- 38. Brausch G (1964) Change and Continuity in the Gezira Region of the Sudan. Int Social Science J 16: 340–356. [Google Scholar]
- 39.Kulneff C A (2006) Comparative Study of Urban and Rural Diary Management Systems in Sudan. Uppsala.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Name, reported type strains, source, origin, and GenBank accession numbers for the analysed strains. dH: [G.S. de Hoog working collection] UTHSC [University of Texas Health Science Center]. All type strains marked with [T].
(DOCX)