Abstract
Vibrio parahaemolyticus is a foodborne pathogen that has become a public health concern at the global scale. The epidemiological significance of V. parahaemolyticus infections in Latin America received little attention until the winter of 1997 when cases related to the pandemic clone were detected in the region, changing the epidemic dynamics of this pathogen in Peru. With the aim to assess the impact of the arrival of the pandemic clone on local populations of pathogenic V. parahaemolyticus in Peru, we investigated the population genetics and genomic variation in a complete collection of non-pandemic strains recovered from clinical sources in Peru during the pre- and post-emergence periods of the pandemic clone. A total of 56 clinical strains isolated in Peru during the period 1994 to 2007, 13 strains from Chile and 20 strains from Asia were characterized by Multilocus Sequence Typing (MLST) and checked for the presence of Variable Genomic Regions (VGRs). The emergence of O3:K6 cases in Peru implied a drastic disruption of the seasonal dynamics of infections and a shift in the serotype dominance of pathogenic V. parahaemolyticus. After the arrival of the pandemic clone, a great diversity of serovars not previously reported was detected in the country, which supports the introduction of additional populations cohabitating with the pandemic group. Moreover, the presence of genomic regions characteristic of the pandemic clone in other non-pandemic strains may represent early evidence of genetic transfer from the introduced population to the local communities. Finally, the results of this study stress the importance of population admixture, horizontal genetic transfer and homologous recombination as major events shaping the structure and diversity of pathogenic V. parahaemolyticus.
Author Summary
Infections caused by Vibrio parahaemolyticus have increased significantly over the last two decades, with cases now regularly reported globally. The emergence of cholera at global scale has brought the attention toward other Vibrio diseases in developing countries. This was the situation in Peru, where the investigation of V. cholerae in hospitals and regional public health laboratories after the dramatic emergence of cholera epidemic in 1991 enabled the identification of other pathogenic Vibrio throughout the whole country. The submission of all these bacteria to the Instituto Nacional de Salud (INS, Lima, Peru) for characterization generated an extraordinary repository of records and isolates which have been decisive for sizing the impact of V. parahaemolyticus infections on the population. The present study addresses, for first time, the impact of the arrival of a non-endemic population of V. parahaemolyticus on the genetic structure and virulence attributes of local populations. The detection of the pandemic clone of V. parahaemolyticus to Peru in 1997 changed not only the epidemic dynamics of this pathogen, but also the population structure and genetic variation of native populations through population admixture, horizontal genetic transfer and homologous recombination between native and introduced populations of pathogenic V. parahaemolyticus.
Introduction
Vibrio parahaemolyticus is a Gram-negative halophilic bacterium that naturally inhabits marine and estuarine environments throughout the world. While many strains of V. parahaemolyticus are strictly environmental, some groups are pathogenic and may cause gastroenteritis in human [1]. V. parahaemolyticus is the leading human pathogen of bacterial food-borne diseases associated with the consumption of raw or undercooked seafood [2].
Recently, V. parahaemolyticus has gained notoriety due to global dissemination of infections [3]. The rise of infections was initially linked to the emergence of gastroenteritis throughout Asia associated with a single clone of the O3:K6 serovar in 1996 [4]. The subsequent detection of this clone causing infections in Peru [5], [6], and Chile [7] in 1997 indicated the pandemic expansion of O3:K6 clone infections. Afterwards, gastroenteritis cases associated with the O3:K6 clone were reported in many other countries and different areas of the world, such as the United States, Russia, Mozambique, Mexico, and Spain [3]. In addition to rapid dissemination of the pandemic clone, a parallel rise of human cases associated with other genetic groups has been reported in recent years. Specific genetic groups have been described as being associated with infections in particular geographic regions of the world. Consequently, the presence of local clones and serovars prevail among clinical cases, such as O4:K12 in the Pacific coast area of the United States [8], O4:K8 in the Peruvian coast [5], O4:K13 in Africa [9], and O4:K11 in the northeast of Spain [10].
The majority of clinical cases of V. parahaemolyticus have been associated with strains bearing the thermostable direct hemolysin (tdh) and/or TDH-related hemolysin (trh). Therefore, the presence of one or both hemolysins has been considered to be a conventional marker of V. parahaemolyticus virulence [11], [12]. However, it has been reported recently that not only the presence of hemolysins but also other virulence factors such as the type III secretion system (T3SS) have been involved in the cytotoxicity and enterotoxicity of V. parahaemolyticus [13], [14]. Whole genome sequencing of the clinical V. parahaemolyticus strain RIMD2210633 revealed the presence of two sets of genes encoding two different T3SSs, named T3SS1 and T3SS2, distributed in each chromosome [15]. A functional analysis of these two T3SSs revealed that T3SS1 is involved in cytotoxic activity, while T3SS2 has been related to enterotoxicity [13]. Consequently, the presence of T3SS2 has been consistently associated with pathogenicity of V. parahaemolyticus in humans. The T3SS2 found in the second chromosome of V. parahaemolyticus (T3SS2α) is part of a large pathogenicity island (PAI) of approximately 80 kb that also includes the tdh-genes. In the trh-positive strain TH3996, a novel PAI that is inserted in a distinctive pathogenic island containing a homologous T3SS2 (T3SS2β) has recently been described [16].
Comparative genome analysis of V. parahaemolyticus predicted that RIMD2210633 pathogenesis is associated with the presence of eight pathogenicity islands (VpaI) [17]. However, to date, VpaI that include T3SS2 have only been functionally characterized in infection models [13], [16]. Genomic analyses also revealed that different types of pathogenicity islands and mobile elements are the major structural differences between trh-positive and tdh-positive strains, including the pandemic clone [18].
Vibrio parahaemolyticus has not been routinely investigated in Peru and infections caused by this organism are rarely reported to the surveillance system. The mandatory investigation of V. cholerae in clinical laboratories after the dramatic emergence of cholera epidemic in 1991 in Peru contributed to the identification of other pathogenic Vibrio species. Vibrio strains isolated from hospitals and regional public health laboratories throughout the country were shipped to the Instituto Nacional de Salud (INS, Lima, Peru) for final identification and characterization. This extraordinary repository has been decisive for identification and evaluation of the impact of V. parahaemolyticus infections on the population, especially in remote regions and small villages along the Peruvian coast where seafood consumption constitutes the nutritional base of population.
The arrival of the pandemic clone to Peru in 1997 resulted in a major shift in the epidemic dynamics of V. parahaemolyticus in the region, replacing the seasonal and local self-limited infections attributed to native genetic groups by the generalization of infections exclusively caused by pandemic strains distributed across the country [5], [6]. Due to the environmental nature of V. parahaemolyticus, this overturn in population dominance and the subsequent population admixture may be expected to lead to an unpredictable impact on populations of pathogenic V. parahaemolyticus in Peru. To test this hypothesis, we investigated the population genetics, genomic variation and pathogenic islands distribution in a complete collection of non-pandemic strains recovered from clinical sources in Peru over the pre- and post-emergence of the pandemic clone. Pandemic and non-pandemic clinical strains representative of the entire period of study were subjected to serotyping and genotyping analysis by Multilocus Sequence Typing (MLST) and Variable Genomic Region (VGRs) analysis.
Materials and Methods
Epidemiological Data and Bacterial Strains
Peruvian V. parahaemolyticus strains were recovered from the collection of the National Reference Laboratory for Enteropathogens at the Instituto Nacional de Salud (INS) in Peru. This collection comprised 46 strains from the previously studied period of 1994 to 2005 [6] and 10 additional strains from the period of 2006 to 2007. To extend the comparison, a Chilean group, comprised of 13 strains from Antofagasta and Puerto Montt [19], [7], and 20 Asiatic strains, corresponding to 18 from Japan, one from Bangladesh and one from Thailand [20], [21], were included in the analysis (Table 1). The reference strain to V. alginolyticus ATCC17749 was also added to the panel of strains.
Table 1. Characteristics of V. parahaemolyticus strains included in this study.
| Country | Strain | Isolation Year | Serovar | tdh | trh | GS-PCR | Source | ST | Reference |
| Peru | 324-95, 326-95, 267-95 | 1995 | O4:K8 | + | − | − | Stool | 88 | [5] |
| 288-95 | 1995 | O5:KUT | + | − | − | Stool | 89 | [5] | |
| 090-96, 212-96 | 1996 | O4:K8 | + | − | − | Stool | 265 | [5] | |
| 763-97, 906-97 | 1997 | O3:K6 | + | − | + | Stool | 3 | [5] | |
| 3435-98, 784-98 | 1998 | O3:K6 | + | − | + | Stool | 3 | [5] | |
| 275-99 | 1999 | O3:K58 | + | − | + | Stool | 3 | [5] | |
| 276-99, 278-99, 279-99, 698-99 | 1999 | O3:K6 | + | − | + | Stool | 3 | [5] | |
| 357-99 | 1999 | OUT:KUT | − | − | − | Stool | 19 | [5] | |
| 776-00 | 2000 | O6:KUT | + | − | − | Stool | 93 | [5] | |
| 330-00, 405-00, 429-00, 430-00, 461-00, 462-00, 511-00, 512-00 | 2000 | O3:K6 | + | − | + | Stool | 3 | [5] | |
| 463-01 | 2001 | O1:K33 | − | − | − | Stool | 94 | [5] | |
| 056-01, 498-01, 565-01, 568-01 | 2001 | O3:K6 | + | − | + | Stool | 3 | [5] | |
| vp196-02 | 2002 | O4:K8 | + | − | − | Env. | 265 | [5] | |
| 004-02, 020-02, 169-02, 240-02, 551-02, 552-02, 553-02 | 2002 | O3:K6 | + | − | + | Stool | 3 | [5] | |
| 038-03, 039-03, 131-03, 302-03, | 2003 | O3:K6 | + | − | + | Stool | 3 | [5] | |
| 205-05 | 2005 | O1:KUT | + | − | + | Stool | 3 | [5] | |
| 155-05, 156-05 | 2005 | O1:KUT | − | + | − | Stool | 65 | [5] | |
| 691-05 | 2005 | O4:K8 | + | − | − | Stool | 265 | [5] | |
| 232-06 | 2006 | O4:K8 | + | − | − | Stool | 265 | This study | |
| 301-07, 371-07, | 2007 | O1:KUT | + | − | + | Stool | 3 | This study | |
| 304-07 | 2007 | O3:K30 | + | − | + | Stool | 3 | This study | |
| 369-07, 438-07 | 2007 | O3:KUT | + | − | + | Stool | 3 | This study | |
| 437-07 | 2007 | O3:KUT | + | + | − | Stool | 64 | This study | |
| 1257-07, 1262-07, 245-07 | 2007 | O4:K8 | + | − | − | Stool | 265 | This study | |
| Chile | ATC210, ATC220 | 1998 | O3:K6 | + | − | + | Stool | 3 | [21] |
| PMC48.4 | 2004 | O3:K6 | + | − | + | Stool | 3 | [21] | |
| PMC39.5, PMC42.5 | 2005 | O3:K6 | + | − | + | Stool | 3 | [21] | |
| PMC29.7, PMC41.7, PMC55.7, PMC73.7 | 2007 | O3:K6 | + | − | + | Stool | 3 | [19] | |
| PMC53.7 | 2007 | O3:K59 | − | − | − | Stool | 28 | [19] | |
| PMC38.7 | 2007 | O10:K20 | + | − | − | Stool | 63 | [19] | |
| PMC60.7 | 2007 | O1:KUT | + | + | − | Stool | 64 | [19] | |
| PMC75.7 | 2007 | O1:KUT | − | + | − | Stool | 65 | [19] | |
| Japan | ATCC17802 | 1951 | O1:K1 | − | + | − | Stool | 1 | [21] |
| RIMD2210633 | 1996 | O3:K6 | + | − | + | Stool | 3 | [15] | |
| AQ3857 | 1983 | O1:K1 | + | + | − | Stool | 83 | [20] | |
| AQ3860 | 1983 | O6:K46 | + | + | − | Stool | 83 | [20] | |
| AQ4405 | 1989 | O3:K72 | + | + | − | Stool | 83 | [20] | |
| AQ4433 | 1989 | O1:KUT | + | + | − | Stool | 83 | [20] | |
| AQ4704 | 1992 | O1:KUT | + | + | − | Stool | 83 | [20] | |
| AQ4969 | 1994 | O1:KUT | + | + | − | Stool | 83 | [20] | |
| AQ4966 | 1994 | O1:KUT | + | + | − | Stool | 83 | [20] | |
| AQ4781 | 1992 | O1:KUT | + | + | − | Stool | 84 | [20] | |
| AQ4815 | 1993 | O1:K59 | + | + | − | Stool | 84 | [20] | |
| AT4 | 1987 | O4:K37 | − | + | − | Env. | 85 | [20] | |
| AQ4889 | 1993 | O4:K12 | + | + | − | Stool | 86 | [20] | |
| AQ3810 | 1983 | O3:K6 | + | − | − | Stool | 87 | [20] | |
| AQ3855 | 1983 | O6:K18 | + | + | − | Stool | 90 | [20] | |
| AQ4901 | 1993 | O3:K6 | − | + | − | Stool | 91 | [20] | |
| KX-V132 | 1995 | O3:KUT | + | + | − | Stool | 92 | [20] | |
| AQ4037 | 1985 | O3:K6 | − | + | − | Stool | 96 | [20] | |
| Thailand | 275 | 1990 | O1:K1 | + | + | − | Stool | 82 | [20] |
| Bangladesh | U5474 | 1980 | O3:K6 | + | − | − | Stool | 14 | [21] |
Detection of Genetic Markers and Serotyping
DNA extraction of V. parahaemolyticus strains was performed using an overnight culture in trypticase soy broth at 37°C using the Chelex-100 method [22]. Strains were confirmed by the presence of the V. parahaemolyticus species-specific genes toxR as described previously [23]. Additionally, the presence of the genes tdh and trh was determined according to procedures described by Tada et al. [24]. Group-specific PCR for the detection of the toxRS sequence of strains belonging to the pandemic clone of V. parahaemolyticus was performed as previously described [6], [25]. Serotyping Lipopolysaccharide (O) and capsular (K) serotypes were determined by a commercially available antisera scheme (Denka Seiken Corp., Tokyo, Japan).
Genetic Relatedness and Population Structure
MLST analysis was performed as previously described [21], based on internal fragments of seven housekeeping genes: recA, gyrB, dnaE, dtdS, pntA, pyrC, and tnaA. Sequences of both strands were determined by custom sequencing (Macrogen Inc., Seoul, South Korea). All chromatograms were assembled, manually edited and trimmed in Bionumerics 5.1 (Applied-Maths, Kortrijk, Belgium). Allele numbers were assigned to each strain by comparing the nucleotide sequence at each locus to all known corresponding alleles available at the V. parahaemolyticus MLST Database (http://pubmlst.org/vparahaemolyticus/). Novel sequence variants and sequence types (ST) were deposited in this database, and ST assignment was performed using the MLST website tools (http://pubmlst.org). Nucleotide sequences of MLST locus corresponding to STs generated from the study were also deposited in GenBank under accession numbers KC542949–KC543109.
Population genetic relationships among V. parahaemolyticus strains included in this study were performed based on the MLST allelic profiles by a minimum spanning tree analysis implemented in BioNumerics 5.1 software (Applied-Maths, Sint Maartens-Latem, Belgium). Strains were grouped according to priority rules adopted from the BURST algorithm [26], with the highest priority given to profiles with the largest numbers of single locus variants or double-locus variants in the case of a match. Clonal complexes were defined as groups with a maximum neighbor distance of one change and a minimum size of two strains.
Characterization of V. parahaemolyticus Variable Genomic Regions
Variable genomic regions (VGRs) were identified by comparative genome analysis based on a complete genome sequence (RIMD2210633) and two draft genomes of V. parahaemolyticus (AQ4037 and AQ3810) retrieved from the NCBI database (http://www.ncbi.nlm.nih.gov/). Genomic sequence data were analyzed by MUMmer 3.0 [27] to identify non-redundant genomic sequences. The identified sequences were listed as VGRs and used in further analyses. To determine the presence and distribution of VGRs among the strains included in this study, we carried out 31 different PCR assays using specific primers targeting the 23 regions identified by comparative analysis (Table 2). The PCR assays were performed in a 50-µl reaction volume containing 10 ng of genomic DNA, 1 U of Taq DNA polymerase (Roche, Mannheim, Germany), 0.2 µM of each primer (Sigma-Aldrich, Sigma-Aldrich, St. Louis, MO), and 200 µM of each deoxynucleoside triphosphate (Roche, Mannheim, Germany). PCR cycling conditions consisted of an initial denaturation step at 94°C for 3 min followed by 30 cycles of denaturation at 94°C for 50 s, 50 s at the annealing temperature, which was variable for each region (see Table 2), and extension step at 72°C for 1 min. A final extension step consisted of 10 min at 72°C.
Table 2. Primers used and V. parahaemolyticus genomic regions investigated.
| ID | Chromose | Genome reference | Gene target | Primer Sequence (5′ - 3′) | Product size (bp) | Annealing T° (Extension time) | Associated genetic region |
| HGT1 | chr1 | RIMD 2210633 | VP0390 | TTTTCATCCATATCCATCGTCC | 579 | 62 (1 min) | (VPaI-1) |
| VP0392 | GCTTCCTCTTACCCTTTTCA | ||||||
| HGT1A | VP0389 | AAGCGGAGTCGTGTTTTTGTGT | 1723 | 64 (2 min) | |||
| VP0392 | TCCTGCCTTCGCGCTATCTTTG | ||||||
| HGT2 | chr1 | RIMD 2210633 | VP1072 | AAAGAGTTGACCGCTGTAAGG | 1677 | 62 (1.3 min) | (VPaI-3) |
| VP1073 | CGCTGTAACGTCATTGGTCTA | ||||||
| HGT3 | chr1 | RIMD 2210633 | VP1394 | ACGGCACCATCAGATTCATACCA | 1116 | 62 (1.3 min) | T6SS |
| VP1396 | ATAGCACCGCCCAATAGACCTGT | ||||||
| HGT3A | VP1408 | GAGATAACCGCACAACCAAG | 1065 | 62 (1.3 min) | |||
| VP1409 | CTGATCAAAAAACCGAGGCT | ||||||
| HGT4 | chr1 | RIMD 2210633 | VP1561 | GAGCGCTTCAAAGTGGGGATTAC | 1142 | 62 (1.3 min) | Phage f237 |
| VP1563 | CTGCCATTGGATTGATAGAAACC | ||||||
| HGT5 | chr1 | RIMD 2210633 | VP1796 | TGGGCTGAAAACAATAACCTCT | 1026 | 62 (1.3 min) | Super Integron |
| VP1798 | TGCTGCAGCCAAACTAATACC | ||||||
| HGT5A | VP1845 | TCGATCATAGGCCGCATAACAG | 1293 | 62 (1.3 min) | |||
| VP1848 | TTCGCATCCAACTTCATCAACC | ||||||
| HGT6 | chr1 | RIMD 2210633 | VP1886 | TGGGCTACGTCTTTGCTACTGA | 1577 | 62 (1 min) | cold-shock proteins, |
| VP1888 | ATACCATGATGCCAAAGAGACG | Ribonuclease R | |||||
| HGT6A | VP1889 | TGGTGGTGAAGACTTGTTTG | 594 | 62 (1 min) | |||
| VP1890 | TCTTTCTCTGGCTCGTCATT | ||||||
| HGT7 | chr1 | RIMD 2210633 | VP2139 | GAAGCGCGTTGGCAGGAATAC | 1066 | 64 (1.3 min) | |
| VP2142 | TGGAAGGGGGACGGATACGAT | (VPaI-4) | |||||
| HGT8 | chr1 | RIMD 2210633 | VP2903 | AAAGAAAGGTGGGAGGGTGAAG | 753 | 62 (1 min) | (VPaI-5) |
| VP2905 | TTTGACAAGGAGAACGGAGGTG | ||||||
| HGT9 | chr2 | RIMD 2210633 | VPA0434 | ATCTCAAACCCCCGCTCAAAAG | 770 | 62 (1 min) | putative site-specific |
| VPA0435 | TAAAAGACACCGCCTACGACTGC | recombinase | |||||
| HGT10 | chr2 | RIMD 2210633 | VPA1261 | GTTTTGCCACAGCCATTATTTC | 963 | 62 (1 min) | (VPaI-6) |
| VPA1262 | TTCCCTTTCGATCTCCAACACC | ||||||
| HGT12 | chr1 | RIMD 2210633 | VP0639 | GGGCATGTTAAAGCGTTAGT | 948 | 62 (1 min) | (VPaI-2) |
| VP0641 | TAGGTAAGTGTATCGAGCGT | ||||||
| HGT13 | chr1 | RIMD 2210633 | VP0657 | ACAGCGGCACAGAATATTAC | 792 | 62 (1 min) | type IV pilin |
| VP0658 | GTTGGCTCGTTTAATTTGGT | ||||||
| HGT14 | chr1 | RIMD 2210633 | VP1365 | AGGTTGTGAAAGGGGATTAG | 1010 | 62 (1.3 min) | hypothetical proteins |
| VP137 | GACAACAAACAACGAGACCA | ||||||
| HGT15 | chr1 | RIMD 2210633 | VP2693 | AACAATACGACGATCAGGAC | 856 | 62 (1 min) | MSHA pilin |
| VP2695 | TACACCACCGACAGGAATCA | ||||||
| HGT16 | chr2 | RIMD 2210633 | VPA0891 | AGCCAGAATACCCAAAGGGA | 1065 | 64 (1.3 min) | Phage related genes |
| VPA0893 | GCCACCAAACGAAAACACA | ||||||
| HGT16A | VPA0904 | GGAAACGGTGGCTTTTTAATG | 1051 | 62 (1.3 min) | |||
| VPA0908 | GAGCTTTGAGGGAGTGAAAT | ||||||
| HGT17 | chr2 | RIMD 2210633 | VPA0953 | CTCTACTAATTCAACGACCACT | 766 | 62 (1 min) | biofilm-associated surface |
| VPA0954 | CATATCTTTGCGGGTCATCT | proteins | |||||
| HGT18 | chr2 | RIMD 2210633 | VPA1564 | TAACCAAAAGAGCGCAAAAG | 859 | 62 (1.3 min) | hypothetical protein |
| VPA1565 | AACATCAAAACCAACACCAG | ||||||
| HGT19 | chr2 | RIMD 2210633 | VPA1702 | TCAAAGTGGTCGCAGCATAA | 1026 | 62 (1.3 min) | hypothetical protein |
| VPA1704 | AAGACCAATGGCGAAGAAA | ||||||
| HGT20 | chr1 | AQ3810 | A79_3286 | GGAAGCGAACAACATGAGAT | 922 | 62 (1.3 min) | Type I restriction system |
| A79_3288 | GAATGAAGAAATAGGTGAGGG | ||||||
| HGT20A | A79_3276 | CCACTTCTGCACATCTTCAA | 627 | 62 (1 min) | |||
| A79_3277 | GCATTTTTCTCTCGCCCATT | ||||||
| HGT21 | chr1 | AQ3810 | A79_2542 | CACCCTAAGCATATCAAACA | 701 | 62 (1 min) | Integron cassette |
| A79_2543 | GACAGCAGAAGAAATATCCA | ||||||
| HGT22 | chr1 | AQ3810 | A79_5185 | ATAGTCAGGTTTTCATCGGG | 941 | 62 (1 min) | hypothetical protein |
| A79_5186 | TTAAGGAAGAGGGGAAAGCA | ||||||
| T3SS2α | chr2 | RIMD 2210633 | VPA1338 | CGTCGTGGCGTGATCTCTACTT | 1083 | 64 (1 min) | T3SS2α (VPaI-7) |
| VPA1339 | ATTGCAGCAGCCATTACGAACA | ||||||
| VPA1355 | TTTAACCCCAACGATTCAGTGC | 709 | 62 (1 min) | ||||
| VPA1356 | CATGCGTAGTCAAGCCCGTTAT | ||||||
| T3SS2β | chr2 | AQ4037, | vscN | TTGGGGTGATTTCGACTTTT | 767 | 60 (1 min) | T3SS2β |
| TH3996 | vscC | GTGGCGTTTATGATCTTGTT | |||||
| vopD2 | GACGCCAATGAAATACCAAA | 1068 | 60 (1 min) | ||||
| vopB | CAGAAGAAGGCAGAACAAAA |
Evolutionary Analysis
Phylogenetic relationships were inferred using ClonalFrame v1.1 software [28]. MLST loci sequences of unique STs were input into ClonalFrame using the default options. Two independent ClonalFrame runs were performed consisting of 500,000 iterations. The first 100,000 iterations in each run were discarded, and the phylogeny and additional model parameters were sampled every 100 generations in the last 400,000 iterations. The phylograms sampled from the two different runs were concatenated and summarized in a 50% majority rule consensus tree constructed by ClonalFrame GUI [28]. The convergence of the Markov Chain Monte Carlo (MCMC) in both runs was proven based on the Gelman-Rubin test as implemented in ClonalFrame [29].
To visualize potential associations between the phylogeny of housekeeping genes and the distribution of VGRs, we performed a transversal clustering analysis. For this purpose, the concatenated sequences of 67 strains were grouped by the mean of their phylogenetic relationship using ClonalFrame (50% majority consensus), while the VGR data were grouped using a UPGMA algorithm mean of their value (0 = absent and 1 = present), resulting in a data matrix ordered according to both phylogenetic relationship and VGR clustering.
Results
Epidemiology Trends and Laboratory Investigation of V. parahaemolyticus
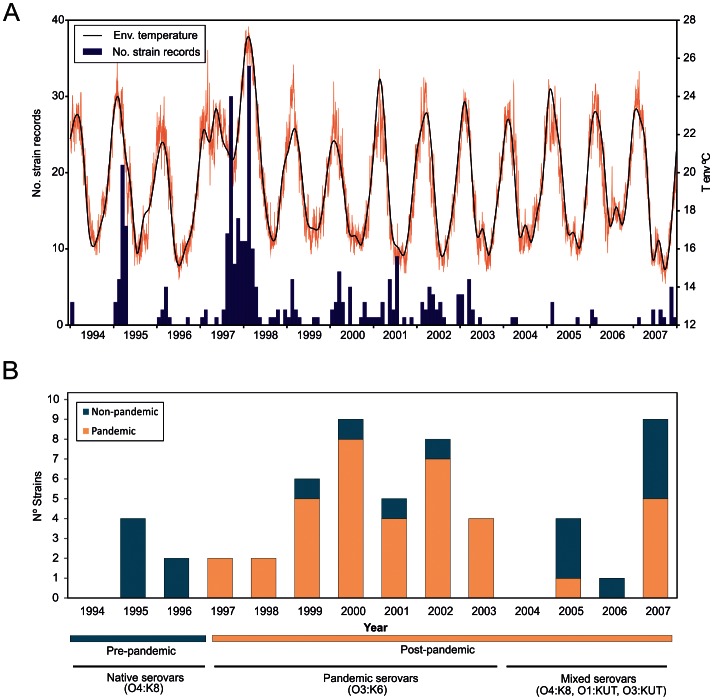
A total of 324 V. parahaemolyticus strain records were retrieved from the INS database from 1994 to 2007. These strains were obtained from gastroenteritis cases that occurred in different regions of Peru and were subsequently submitted to the INS in Lima for identification and storage. The overall distribution of V. parahaemolyticus cases over the 14 years of study (Figure 1) showed a characteristic seasonal pattern with annual peaks of incidence concurring with the warmest months. This epidemic pattern was uniquely disturbed in the course of the austral winter of 1997 when V. parahaemolyticus cases dramatically increased coincidently with an anomalous rise of temperature. The highest incidence of cases was observed from July 1997 to May 1998 with two peaks during September 1997 and February 1998. The epidemic dynamics were restored from 1999 onwards.
Figure 1. Distribution of V. parahaemolyticus over the study period.
(A) Number of V. parahaemolyticus strains received at the National Institute of Health in Peru between 1994 and 2007 and environmental temperature over the same period. (B) Rate of pandemic versus non-pandemic strains over the study period; information on the dominant serotypes of each period is provided below the X-axis.
From the 324 cases recorded over the period 1994–2007, only 56 V. parahaemolyticus strains could be recovered from the INS collection. Distinct serovar dominances were detected over different periods of the study. From 1994 to 1996, infections were associated with serovars O4:K8 and O5:KUT. This serovar dominance abruptly changed during the winter of 1997, with the emergence of infections caused by strains belonging to the pandemic clone. Finally, an undefined pattern of serovar dominance was detected after the emergence of the pandemic clone in 1997–98. In this post-pandemic period, pandemic strains were identified together with O4:K8 strains as well as multiple serovars not previously detected (O1:K33, O1:KUT, O3:K30, O3:K58, O3:KUT, O5:KUT, O6:KUT and OUT:KUT). Pandemic strains represented the largest group of strains (n = 38, 67.9%) detected from 1997 onwards, and O3:K6 was the most frequent serovar among the pandemic strains (n = 31, 55.4%), although serovars O3:K58 and O3:KUT were also identified in the pandemic group. Serovar O4:K8 was the second most frequent group of strains over the whole period (n = 11, 19.6%), whereas the remaining serovars (n = 14) represented 25% of the total strains.
Genetic Structure of V. parahaemolyticus
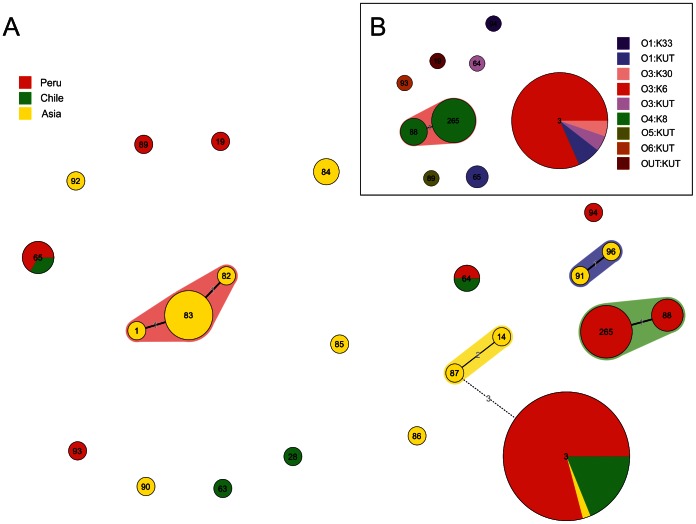
The population genetic structure of V. parahaemolyticus strains representative of Peru, Chile, and Asian countries was analyzed by MLST (Fig. 2). MLST profiles of V. parahaemolyticus (n = 89) were categorized into 23 sequence types (STs). The recA gene of strains included within the ST265 (n = 8, O4:K8 serotype) showed an unexpected large PCR product of approximately 1500 bp (773 bp in the original) with a nucleotide identity diverging 18–19% from the characteristic sequence of V. parahaemolyticus.
Figure 2. Minimum spanning tree (MST) of V. parahaemolyticus.
A) MST of 89 MLST allelic profiles of the whole dataset included in this study. The colors are based on the origin of isolation. B) MST of the Peruvian subset that includes 56 strains. The colors are based on the serotyping group. Each circle represents an MLST genotype, and its size is proportional to the number of strains. The numbers along the branch links of MLST genotypes denote the amount of allelic differences.
The Peruvian group was split into 9 different STs (Fig. 2B). Minimum spanning tree analysis identified a single clonal complex consisting of ST88 (n = 3) and ST265 (n = 8) both of which included O4:K8 strains differing in a single locus. All of the strains belonging to the pandemic complex were grouped in ST3 (n = 24), whereas the remaining strains were included in 6 unrelated STs. Minimum spanning tree of the 23 STs resulting from the MLST analysis of the 89 strains showed a similar topography and group discrimination. Peruvian, Chilean and Asian pandemic strains shared the sequence of the 7 loci and were assigned to a single ST (ST3). Strains from Peru and Chile shared two additional STs: ST65 composed by O1:KUT strains and ST64 including strains belonging to O3:KUT and O1:KUT serovars. Three other clonal complexes (CC) were identified among trh+ Asian strains: one CC included STs 1, 83 and 82; a second group consisted of ST96 and ST91; and a third CC was shared by STs 87 and 14. The remaining strains were clustered in different and unrelated STs with differences in more than 5 loci.
Evolutionary History
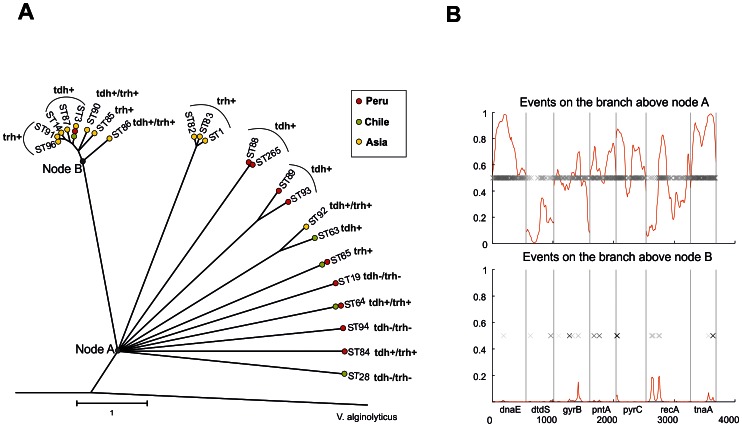
The nucleotide sequences of 23 unique STs identified among the V. parahaemolyticus, as well as homologous sequences of V. alginolyticus (outgroup), were concatenated, and their phylogenetic relationships were inferred by ClonalFrame. The clonal genealogy inferred from the data revealed a star-like topology of V. parahaemolyticus delineated into two evident lineages, with the rest of the STs remaining unresolved (Figure 3). The pandemic lineage consisted of all of the pandemic strains from Asia, Chile and Peru (ST3), as well as other STs that included Asian pre-pandemic strains of diverse serotypes and the three variants of hemolysin-related genotypes. A second lineage consisted of trh+ strains of STs 1, 82 and 83, all of them from Asia.
Figure 3. Phylogenetic tree and recombination events.
A). The phylogenetic relationship of 22 V. parahaemolyticus STs was generated using ClonalFrame by 50% majority-rule consensus visualized using MEGA4. V. alginolyticus was used as an outgroup. Nodes A and B correspond to the recombination events presented in Figure 4B. Red, green and yellow circles denote the place of isolation from Peru, Chile and Asia, respectively. Coalescent units are indicated by the scale bar. B) ClonalFrame representation of the recombination event probability occurring in MLST concatenated is represented on the x-axis with the red line indicating the probability at each locus for an import event on a scale from 0 to 1 (y-axis). Each inferred substitution in the graph is represented by a cross, the intensity of which indicates the posterior probability for that substitution.
The genealogy showed the influence of recombination on the genetic diversification of V. parahaemolyticus with a central node (node A) from which all of the branches diverged. This specific topology indicated an unresolved phylogeny due to the non-identification of a common ancestor. ST88 and ST265, comprising all the O4:K8 strains were grouped in a single lineage. ST89 and ST93, which included Peruvian strains, also shared a common linage, as well as ST92 and ST63, sequence types composed of Asian and Chilean strains, respectively. For each branch of the reconstructed genealogy, ClonalFrame identified fragments that were likely imported. The influence of mutation and recombination events in the generation of polymorphisms was further investigated in the two cluster nodes (A and B) indicated in Figure 3A. Node A showed a high probability of importing events consistent with recombination substitutions, while divergence in node B, which included the Pandemic lineage, most likely originated as a result of point mutations (Fig. 3B). ClonalFrame analysis shows that the relative impact of recombination versus point mutation expressed as a ratio (r/m) was approximately 1.45, and that the relative frequency of recombination in comparison to point mutation (ρ/θ) was approximately 0.20.
VGR Distribution
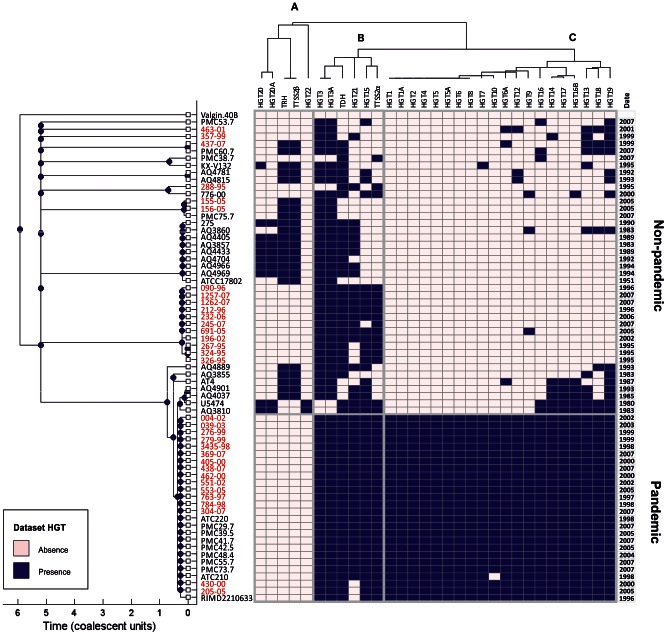
To extend our analysis of the phylogenetic relationships and to understand the impact of genomic variation in the evolutionary history of V. parahaemolyticus, the distribution of VGRs was assessed by PCR assays in the 67 strains. A transversal clustering analysis was performed to visualize the evolutionary significance of the presence of genomic regions within the strain collection (Fig. 4).
Figure 4. Transversal clustering of phylogenetic relationships and genomic variation.
Clustering was built using a ClonalFrame tree of MLST allele sequences of 67 V. parahaemolyticus strains, while the VGR data were grouped using a UPGMA algorithm, resulting in a data matrix ordered according to both phylogenetic relationship and the prevalence of VGRs. Solid boxes and open boxes in the appropriate column indicate VGR presence or absence, respectively. Peruvian subset names are shown in red.
ClonalFrame majority-rule consensus tree building with the whole set of strains revealed the same group structure as resolved by the clonal genealogy. However, analysis of VGR data showed a specific clustering of VGRs with three different groups (A, B, and C) (Fig. 4). Cluster A grouped five different genomic regions almost exclusive for trh+ strains with the exception of two tdh+ strains (U5474 and AQ3810). The presence of TTSS2β was only detected in strains bearing the trh gene. This cluster also included regions HGT20 and HGT22 encoding for the Type I restriction system, which was only identified in trh+ Asian strains. Cluster B included 6 genomic regions; two of them (HGT3 and HGT3A) correspond to ORFs coding the type 6 secretion system (T6SS1), which were present in all of the clinical strains with the exception of 4 tdh+/trh− non-pandemic strains isolated in Peru, Chile, and Asia. T3SS2α was singularly detected in those strains with genotype tdh+/trh− but was not present in those strains positive for the tdh and trh gene. Cluster C consisted of the majority of VGRs (20 genomic regions), which were uniformly distributed among almost all the pandemic strains. Some of these regions were partially present in all Asian strains phylogenetically related to pandemic strains, while those were less frequent in the remaining non-pandemic strains and distributed with an undefined pattern. Non-pandemic Peruvian strains showed a shift in the distribution of the VGRs included within this cluster from the pre-pandemic period to 1999 and afterwards. Genomic regions such as HGT6A (ribonuclease R), HGT12 (VPaI2), HGT9 (site-specific recombinase), HGT14 (hypothetical protein), HGT16B (phage genes), HGT13 (type IV pilin), HGT18 (hypothetical protein) and HGT19 (hypothetical protein), which are characteristic of pandemic strains, were exclusively identified in those non-pandemic strains from Peru isolated after the arrival of the pandemic clone in 1997. Finally, none of the VGRs investigated were found in V. alginolyticus ATCC17749.
Discussion
Vibrio parahaemolyticus represents an intriguing food-borne pathogen and poses a significant threat to public health in Peru. However, despite this concern for public health, the knowledge of the molecular epidemiology and genetic structure of this pathogen remains incomplete.
Epidemiology of V. parahaemolyticus in Peru have been historically associated with sporadic outbreaks linked with seafood consumption. Since 1983, clinical strains were dominated by the presence of local serovars such as O4:K8 that was isolated from both clinical and environmental samples [30]. These infections were characteristically related to auto-limiting outbreaks detected along the coastal regions over summer months. This epidemic pattern shifted in 1997 with the unexpected arrival of pandemic V. parahaemolyticus to Peru [5]. The presence of the O4:K8 serovar in pre- and post-pandemic periods allowed for the identification of this group as the dominant population among clinical strains in Peru and the cause of recurrent outbreaks during the warmest months [31], [5]. The totality of strains recovered from infections in 1997 and 1998 belonged exclusively to the pandemic clone. After the period of the infections associated with the pandemic clone in 1997–98, the dominance of this clone in clinical infections began to decline and a mix of different serovars began to emerge. This specific epidemiological trend of arrival and rapid decline of pandemic V. parahaemolyticus infection in Peru clearly contrasts with the infection dynamics found in Chile, the neighboring country where infections associated with the pandemic clone started first in 1997 and subsequently in 2004, and where the pandemic clone has dominated the clinical isolations since the second epidemic radiation in 2004 [32]. The arrival of the pandemic clone to Peru provided a unique opportunity for testing the potential impact of an introduced genetic group on the structure and genetic variability of local pathogenic populations. One particular feature of the epidemiology of V. parahaemolyticus in Peru after the appearance of the pandemic clone was the sudden apparition of diverse serovars not detected previously. Serovars O1:K33, O1:KUT, O3:K30, O3:K58, O3:KUT, O5:KUT, O6:KUT and OUT:KUT had not been isolated prior to 1998. Similar results were found in a previous retrospective study carried out in Peru covering the 1993–2002 period [6], which reported the presence of serovars O3:K68, O3:K58, OUT:K6, O6:K18, O11:KUT, O11:K15 and OUT:KUT after 1998. The flourishing of serovars and genotypes in Peru may be related to the particular vehicle for the introduction and propagation of pandemic strains. The epidemic dissemination of this pandemic clone along the coast of Peru corresponded with the expansion and dynamics of the poleward propagation and the receding of tropical waters linked to the 1997 El Niño event [5], [33].
Another important aspect to be considered to understand the possible genetic impact of the pandemic clone on Peruvian local populations is the comparative analysis of phylogeny inferred from MLST sequences and the distribution of variable regions in the panel of strains. The pattern of VGR distribution forming cluster C showed a conserved presence for all regions among strains belonging to the pandemic clone, which provides an additional evidence for the highly clonal nature of this phylotype [34], [18]. On the contrary, a sparse presence of VGRs in cluster C was observed, showing an undefined pattern among non-pandemic strains. A detailed analysis of variations and phylotypes showed that VGRs in cluster C are only present in strains isolated after the arrival of the pandemic clone in 1997. This specific pattern of distribution and the connection between these strains and the arrival of the pandemic clone may suggest a common origin for all of these groups. However, the presence of a region coding hypothetical genes (VPA0434 and VPA0435) in one O4:K8 strain also raises the possibility of a local horizontal transfer from pandemic strains to local Vibrio communities in Peru.
Horizontal gene transfer appears to be the major force shaping both the genomic variation and virulence of V. parahaemolyticus, as evidenced in previous studies linking the acquisition of pathogenicity islands with the emergence of the pandemic clone [17], [18]. The results of transversal clustering analysis revealed that the presence of T3SS, T6SS and mannose-sensitive hemagglutinin (MSHA) pilus were clustered together in most clinical isolates showing a clear association with the pathogenicity of these strains, suggesting that these regions are conserved in pathogenic strains and could be a good marker of pathogenicity.
The evolutionary history of pathogenic lineages of V. parahaemolyticus has been analyzed previously by different approaches [21], [17], [35]. However, the previous analysis of population structure was not sufficiently integrated within the epidemic dynamics prevailing in a specific region so that there could be an adequate evaluation of the shift in genotype and population dominance over different periods. ClonalFrame genealogy inferred a star-shaped tree with long terminal branches showing a primary diversification affecting all of the STs, likely as a result of recombination events [36]. A defined lineage grouped all of the pandemic strains as well as most of the Asian genotypes showing a hierarchical pattern recently evolved from a common ancestor, likely due to the course of successive point mutations. The overall results evidenced the influence of recombination events in the diversification of most pathogenic V. parahaemolyticus genotypes.
Homologous recombination in housekeeping genes has been found naturally in Vibrio, and it is an important driver of diversification in this genus [37]. ClonalFrame analyses of the whole concatenated dataset showed rates of recombination of 1.45 and 0.20 for r/m and ρ/θ, respectively. These data suggest an intermediate rate of recombination among the strains characterized [38]. This estimate of recombination frequency suggests that recombination is relatively rare compared to other species, such as Streptococcus uberis (ρ/θ, 9.0) [39] and Clostridium perfringens (ρ/θ, 3.2) [40], but it is approximately similar to that observed for other groups, such as lineage I of Listeria monocytogenes (ρ/θ, 0.13) [41], This particular feature may be related to the exclusive use of pathogenic strains in the study, which are characterized by a clonal diversification [38].
To conclude, the results of this study describe the epidemiological impact caused by the introduction of the pandemic clone in Peru on the epidemiology and structure of the local population of V. parahaemolyticus. The presence of genomic regions characteristic of the pandemic clone in other non-pandemic strains provides early evidence of genetic transfer from the introduced population to the local communities. Additionally, the genetic relationships based on MLST and VGR analyses support the epidemiological connection between pandemic and non-pandemic strains isolated in both Peru and Chile. The phylogenetic and genomic analysis performed allowed us to determine the recent origin of the pandemic clone lineage, probably caused by successive acquisition of genomic regions, as well as the influence of recombination events in the diversification of non-pandemic pathogenic V. parahaemolyticus. Ultimately, these results provide a preliminary framework about evolutionary history of V. parahaemolyticus. Recent advances in high throughput sequencing are revolutionizing the field of population genetics of human pathogens. Application of fine-scale analysis based on whole genome sequences in future studies of pathogenic bacteria will contribute to improve our knowledge of the epidemic dynamics and routes of dispersion of Vibrio diseases [42], [43].
Acknowledgments
It is a pleasure to acknowledge the excellent technical assistance of Silvia Carles and laboratory professionals of Instituto Nacional de Salud, Peru. We also thank Mitsuaki Nishibuchi and Romilio Espejo for providing some of the strains included in this study.
Funding Statement
This work was partially supported by project 2007/CP381 from the Department for International Cooperation of the Regional Government of Galicia, Spain. This project finished in 2008. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Broberg CA, Calder TJ, Orth K (2011) Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect 13: 992–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Su YC, Liu C (2007) Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol 24: 549–558. [DOI] [PubMed] [Google Scholar]
- 3. Nair GB, Ramamurthy T, Bhattacharya SK, Dutta B, Takeda Y, et al. (2007) Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin Microbiol Rev 20: 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Okuda J, Ishibashi M, Hayakawa E, Nishino T, Takeda Y, et al. (1997) Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J Clin Microbiol 35: 3150–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martinez-Urtaza J, Huapaya B, Gavilan RG, Blanco-Abad V, Ansede-Bermejo J, et al. (2008) Emergence of Asiatic Vibrio diseases in South America in phase with El Niño. Epidemiology 19: 829–37. [DOI] [PubMed] [Google Scholar]
- 6. Gil AI, Miranda H, Lanata CF, Prada A, Hall ER, et al. (2007) O3:K6 serotype of Vibrio parahaemolyticus identical to the global pandemic clone associated with diarrhea in Peru. Int J Infect Dis 11: 324–8. [DOI] [PubMed] [Google Scholar]
- 7. González-Escalona N, Cachicas V, Acevedo C, Rioseco ML, Vergara JA, et al. (2005) Vibrio parahaemolyticus diarrhea, Chile, 1998 and 2004. Emerg Infect Dis 11: 129–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. DePaola A, Ulaszek J, Kaysner CA, Tenge BJ, Nordstrom JL, et al. (2003) Molecular, serological, and virulence characteristics of Vibrio parahaemolyticus isolated from environmental, food, and clinical sources in North America and Asia. Appl Environ Microbiol 69: 3999–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ansaruzzaman M, Chowdhury A, Bhuiyan NA, Sultana M, Safa A, et al. (2008) Characteristics of a pandemic clone of O3:K6 and O4:K68 Vibrio parahaemolyticus isolated in Beira, Mozambique. J Med Microbiol 57: 1502–7. [DOI] [PubMed] [Google Scholar]
- 10. Martinez-Urtaza J, Lozano-Leon A, DePaola A, Ishibashi M, Shimada K, et al. (2004) Characterization of pathogenic Vibrio parahaemolyticus isolates from clinical sources in Spain and comparison with Asian and North American pandemic isolates. J Clin Microbiol 42: 4672–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bej AK, Patterson DP, Brasher CW, Vickery MC, Jones DD, et al. (1999) Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J Microbiol Methods 36: 215–25. [DOI] [PubMed] [Google Scholar]
- 12. Rosec JP, Simon M, Causse V, Boudjemaa M (2009) Detection of total and pathogenic Vibrio parahaemolyticus in shellfish: comparison of PCR protocols using pR72H or toxR targets with a culture method. Int J Food Microbiol 129: 136–45. [DOI] [PubMed] [Google Scholar]
- 13. Park KS, Ono T, Rokuda M, Jang MH, Okada K, et al. (2004) Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect Immun 72: 6659–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kodama T, Rokuda M, Park KS, Cantarelli VV, Matsuda S, et al. (2007) Identification and characterization of VopT, a novel ADP-ribosyltransferase effector protein secreted via the Vibrio parahaemolyticus type III secretion system 2. Cell Microbiol 9: 2598–609. [DOI] [PubMed] [Google Scholar]
- 15. Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, et al. (2003) Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361: 743–9. [DOI] [PubMed] [Google Scholar]
- 16. Okada N, Iida T, Park KS, Goto N, Yasunaga T, et al. (2009) Identification and characterization of a novel type III secretion system in trh-positive Vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect Immun 77: 904–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boyd EF, Cohen AL, Naughton LM, Ussery DW, Binnewies TT, et al. (2008) Molecular analysis of the emergence of pandemic Vibrio parahaemolyticus. BMC Microbiol 8: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen Y, Stine OC, Badger JH, Gil AI, Nair GB, et al. (2011) Comparative genomic analysis of Vibrio parahaemolyticus: serotype conversion and virulence. BMC Genomics 6: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harth E, Matsuda L, Hernández C, Rioseco ML, Romero J, et al. (2009) Epidemiology of Vibrio parahaemolyticus outbreaks, southern Chile. Emerg Infect Dis 2009 15: 163–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nakaguchi Y, Ishizuka T, Ohnaka S, Hayashi T, Yasukawa K, et al. (2004) Rapid and specific detection of tdh, trh1, and trh2 mRNA of Vibrio parahaemolyticus by transcription-reverse transcription concerted reaction with an automated system. J Clin Microbiol 42: 4284–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gonzalez-Escalona N, Martinez-Urtaza J, Romero J, Espejo RT, Jaykus LA, et al. (2008) Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J Bacteriol 190: 2831–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blanco-Abad V, Ansede-Bermejo J, Rodriguez-Castro A, Martinez-Urtaza J (2008) Evaluation of different procedures for the optimized detection of Vibrio parahaemolyticus in mussels and environmental samples. Int J Food Microbiol 129: 229–36. [DOI] [PubMed] [Google Scholar]
- 23. Kim YB, Okuda J, Matsumoto C, Takahashi N, Hashimoto S, et al. (1999) Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol 37: 1173–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tada J, Ohashi T, Nishimura N, Shirasaki Y, Ozaki H, et al. (1992) Detection of the thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol Cell Probes 6: 477–87. [DOI] [PubMed] [Google Scholar]
- 25. Matsumoto C, Okuda J, Ishibashi M, Iwanaga M, Garg P, et al. (2000) Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J Clin Microbiol 38: 578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG (2004) eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186: 1518–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, et al. (2004) Versatile and open software for comparing large genomes. Genome Biol 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Didelot X, Falush D (2007) Inference of bacterial microevolution using multilocus sequence data. Genetics 175: 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gelman A, Rubin DB (1992) Inference from iterative simulation using multiple sequences. Statistical Science 7: 457–511. [Google Scholar]
- 30.Guerra Soto HE. (1988) Contribucion al hallazgo de especies de Vibrio de origen humano y ambiental y su significado epidemiologico en Peru [PhD thesis]. Universidad Nacional Mayor de San Marcos.
- 31. Begue RE, Meza R, Castellares G, Cabezas C, Vasquez B, et al. (1995) Outbreak of diarrhea due to Vibrio parahaemolyticus among military personnel in Lima, Peru. Clin Infect Dis 21: 1513–4. [DOI] [PubMed] [Google Scholar]
- 32. García K, Torres R, Uribe P, Hernández C, Rioseco ML, et al. (2009) Dynamics of clinical and environmental Vibrio parahaemolyticus strains during seafood-related summer diarrhea outbreaks in southern Chile. Appl Environ Microbiol 75: 7482–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ansede-Bermejo J, Gavilan RG, Triñanes J, Espejo RT, Martinez-Urtaza J (2010) Origins and colonization history of pandemic Vibrio parahaemolyticus in South America. Mol Ecol 19: 3924–37. [DOI] [PubMed] [Google Scholar]
- 34. Hurley CC, Quirke A, Reen FJ, Boyd EF (2006) Four genomic islands that mark post-1995 pandemic Vibrio parahaemolyticus isolates. BMC Genomics 7: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Han H, Wong HC, Kan B, Guo Z, Zeng X, et al. (2008) Genome plasticity of Vibrio parahaemolyticus: microevolution of the ‘pandemic group’. BMC Genomics 9: 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martin DP, Lemey P, Posada D (2011) Analysing recombination in nucleotide sequences. Mol Ecol Resour 11: 943–55. [DOI] [PubMed] [Google Scholar]
- 37. Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK (2005) Chitin induces natural competence in Vibrio cholerae. Science 310: 1824–7. [DOI] [PubMed] [Google Scholar]
- 38. Vos M, Didelot X (2009) A comparison of homologous recombination rates in bacteria and archaea. ISME J 3: 199–208. [DOI] [PubMed] [Google Scholar]
- 39. Lang P, Lefébure T, Wang W, Zadoks RN, Schukken Y, et al. (2009) Gene content differences across strains of Streptococcus uberis identified using oligonucleotide microarray comparative genomic hybridization. Infect Genet Evol 9: 179–188. [DOI] [PubMed] [Google Scholar]
- 40. Rooney AP, Swezey JL, Friedman R, Hecht DW, Maddox CW (2006) Analysis of core housekeeping and virulence genes reveals cryptic lineages of Clostridium perfringens that are associated with distinct disease presentations. Genetics 172: 2081–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. den Bakker HC, Didelot X, Fortes ED, Nightingale KK, Wiedmann M (2008) Lineage specific recombination rates and microevolution in Listeria monocytogenes. BMC Evol Biol 8: 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, et al. (2009) Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477: 462–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Didelot X, Bowden R, Wilson DJ, Peto TE, Crook DW (2012) Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet 13: 601–12. [DOI] [PMC free article] [PubMed] [Google Scholar]