Abstract
Safflower (Carthamus tinctorious L.) is valued as a source of high quality vegetable oil. 20 ISSR primers were used to assess the genetic diversity of 18 accessions of safflower collected from different geographical regions of Iran. The ISSR primers combinations revealed 57.6 % polymorphism, among 338 genetic loci amplified from the accessions. The sum of effective number of alleles and observed number of alleles were 29.76 and 36.77, respectively. To understand genetic relationships among these cultivars, Jacquards’ similarity coefficient and UPGMA clustering algorithm were applied to the ISSR marker data set. ISSR markers grouped accessions into two main clusters and four sub clusters. Also, the principal coordinate analysis (PCoA) supported the cluster analysis results. The results showed these genotypes have high genetic diversity, and can be used for alternative safflower breeding program.
Keywords: Carthamus tinctorius L., Genetic diversity, PCoA and ISSR
Introduction
Safflower (Carthamus tinctorius L. 2n = 2× = 24), is a member of the family Asteraceae. It is an annual and predominantly self-pollinated species and it is one of the world’s oldest oil seed crops (Sehgal et al. 2009). It is believed to have been domesticated somewhere in the Fertile Crescent region over 4,000 years ago (Knowles and Ashri 1995). On the basis of morphological variability existing in C. tinctorius, Knowles (1969) proposed seven ‘centers of similarity’ (the Far East, India-Pakistan, the Middle East, Egypt, Sudan, Ethiopia and Europe). Safflower lines native to each ‘center’ are remarkably similar in height, branching, spines, flower color and head size; however, consistent morphological differences are maintained between the centers (Chapman and Burke 2007).
Most of the genetic diversity that local and traditional varieties posses is being lost currently, so evaluation on the genetic diversity of safflower accessions available in different geographical regions will help to provide valuable information on the conservation and ultization of safflower germplasm (Yang et al. 2007). In Iran, research efforts have been undertaken to diversify the farming systems and the government is encouraging the cultivation of various oilseed crops, including safflower (Mohammadi and Pourdad 2009). In the past few years, the area under safflower cultivation has increased to 15,000 ha, mostly under rain-fed conditions (Mohammadi and Pourdad 2009). An analysis of the variability would be of great value in planning a successful breeding program (Mary and Gopalan 2006).
There is a range of molecular methods available to study genetic diversity. Amplified fragment length polymorphism (AFLP), isozymes, simple sequence repeats (SSR), random amplified microsatellite polymorphisms (RAMP), random amplified polymorphic DNA (RAPD) and inter simple sequence repeats (ISSR) have all been used to determine genetic diversity in plant populations (Wang et al. 1994; Godwin et al. 1997; Hollngsworth et al. 1998; Blair et al. 1999; Amsellem et al. 2000; Mengoni et al. 2000). Microsatellites or simple sequence repeats are tandemly repeated mono-, di-, tri-, tetra- or pentanucleotide units (Sehgal et al. 2009).
Inter-simple sequence repeats (ISSR) PCR using primers based on di-, tetr- or penta-nucleotide repeats have now become a routine among the researchers (Zietkiewicz et al. 1994). For its advantages of simple procedure, low-cost, good stability and high reproducibility, ISSR markers has been successfully used in genetic mapping (Casaoli et al. 2001; Cekic et al. 2001; Tanyolac 2003), germplasm identification (Nagaoka and Ogihara 1997; Potter et al. 2002) and genetic diversity analysis (Ash et al. 2003; Wu et al. 2005). Until now, few studies have been carried on the genetic variations of C. tinctorius L. using ISSR markers (Amiri et al. 2001; Guo et al. 2003). The objectives of the presented research here are to asses of genetic variability of Iranian landrace of safflower and to study their genetic relationships using ISSR markers.
Materials and methods
The collection of promising genotypes for different purposes was planted in 2011 at the Experimental Station of the Faculty of Agriculture of Ferdowsi University of Mashhad. Experiment was laid out in a randomized complete block design. In every block, there were three rows and in each row 25 seeds were sown. Each row was 3.5 m long, and the path between rows was 50 cm. The names of the 18 cultivars investigated are given in Table 1.
Table 1.
Accessions of safflower used in this study, their origin and collection areas
| No. | Abbreviation | Collection area | Origin |
|---|---|---|---|
| 1 | IR 1 | Gholestan province-gorgan (local 1) | Iran |
| 2 | IR 2 | Gholestan province-gorgan (local 3) | Iran |
| 3 | IR 3 | Gholestan province-gorgan (local 2) | Iran |
| 4 | IR 4 | Khorasan razavi province-mashhad (local 2) | Iran |
| 5 | IR 5 | Khorasan razavi province-mashhad (local 1) | Iran |
| 6 | IR 6 | Khorasan razavi province-mashhad (local 3) | Iran |
| 7 | IR 7 | Isfahan province-isfahan (local 4) | Iran |
| 8 | IR 8 | Isfahan province-isfahan (local 1) | Iran |
| 9 | IR 9 | Isfahan province-isfahan (local 2) | Iran |
| 10 | IR 10 | Isfahan province-isfahan (local 3) | Iran |
| 11 | IR 11 | Ghuilan province-rasht(118) | Iran |
| 12 | IR 12 | Ghuilan province-rasht (127) | Iran |
| 13 | IR 13 | East azarbaijan-marand | Iran |
| 14 | IR 14 | IL111 | Iran |
| 15 | IR 15 | Markazi province-arak | Iran |
| 16 | IR 16 | Fars province-darab | Iran |
| 17 | IR 17 | Fars province-darab (DAR4) | Iran |
| 18 | IR18 | PI-537598-tehran province | Iran |
DNA extraction
Genomic DNA was extracted from young leaves following the cetyl tri methyl ammonium (CTAB) procedure described by Saghai-Maroof et al. (1984). Extracted DNA concentration was quantified by using the NanoDrop spectrophotometer and qualified using agarose gel electrophoresis.
ISSR analysis
Twenty ISSR primers, (4 of them were 5′ anchored and 16 primers were 3′ anchored), were used for the PCR. Each 25 μl reaction volume contained 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 2.5 mM MgCl2, 0.24 mM dNTPs, 0.1 % gelatin, 2 % formamide, 5 μM primer, 0.5 U Taq polymerase (Fermentas), and 20 ng of genomic DNA.
The amplified products were separated on 1.4 % agarose gels and stained with ethidium bromide. Images were photographed, captured by Gel Doc 2000TM (Bio-Rad, USA). Amplified products were scored for the presence (1) or absence (0) of bands and binary matrices were assembled for the ISSR markers. The binary matrices were subjected to statistical analyses using NTSYS-pc software version 2.1 (Rohlf 2000).
Data analysis
Jacquard’s similarity coefficient was employed to compute pairwise genetic similarities. Similarity matrix was used for the cluster analysis and construction of dendrogram using unweighted pair-group method (UPGMA) (Sneath and Sokal 1973). Genotypic data were analyzed using POPGENE (Yeh et al. 1999) and GenAlex (Peakall and Smouse 2007) to calculate the observed number of alleles, effective number of alleles, observed heterozygosity, expected heterozygosity.
For individual primer/primer combination, observed number of alleles (Na), number of polymorphic bands (P) and percentage polymorphism (%P) were calculated using POPGENE software ver 32. Effective number of all four individual primer combination (P), determines the ability of a marker system on per assay basis to distinguish number of individuals primer combination (Belaj et al. 2003), was calculated as Ae = 1/(1 − h) = 1/Σpi 2 Where, pi is frequency of the i allele in a locus and h = 1 − Σpi 2th is heterozygosity in a locus. The Shannon’s diversity index for each primer combination was calculated as H p = −∑pi log p, where p is the frequency of a given band in a regional accession.
Result and discussion
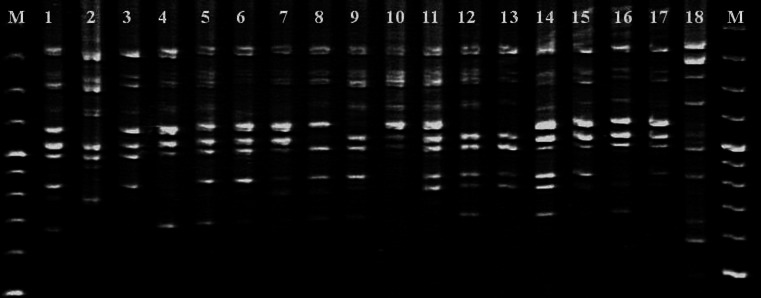
Twenty ISSR primers (Table 2) produced 338 bands (Fig. 1) across the 18 accessions, of which 284 were polymorphic (Table 2). The number of amplified fragments varied from 8 (primer number 5) to 19 (primer number 16) across the genotypes. The average polymorphic bands per primer were 14.2. The percentage of polymorphism for primers ranged from 66 to 100, with an average polymorphism percent of 83.8 (Table 2).
Table 2.
The 20 ISSR primers used for genetic diversity analysis of safflower accessions. Observed number of alleles (Na), Effective number of alleles (Ne), Nei’s (1973) gene diversity (h), Shannon’s Information index (I). P, number of polymorphic loci
| Primer No. | Primer sequence | Na | Ne | H | I | P | % P |
|---|---|---|---|---|---|---|---|
| 1 | 5′-BDB(TCC)5-3′ | 1.7500 | 1.3791 | 0.2402 | 0.3715 | 12 | 75.00 |
| 2 | 5′-HVH(TCC) 5-3′ | 1.7500 | 1.5280 | 0.3012 | 0.4404 | 12 | 75.00 |
| 3 | 5′-DBD(AC) 7-3′ | 1.8750 | 1.5057 | 0.3065 | 0.4628 | 14 | 87.50 |
| 4 | 5′- (TCC) 5RY-3′ | 1.9333 | 1.4527 | 0.2699 | 0.4143 | 14 | 93.33 |
| 5 | 5′-VHVG(TG) 7-3′ | 1.6667 | 1.2948 | 0.1908 | 0.3013 | 8 | 66.67 |
| 6 | 5′- (AG) 8YC-3′ | 2.0000 | 1.7226 | 0.4015 | 0.5842 | 18 | 100.00 |
| 7 | 5′- (AC) 8YG-3′ | 2.0000 | 1.7226 | 0.4015 | 0.5842 | 18 | 100.00 |
| 8 | 5′- (AG) 8T-3′ | 1.8571 | 1.5055 | 0.2993 | 0.4505 | 12 | 85.71 |
| 9 | 5′- (AG) 8C-3′ | 1.7647 | 1.3367 | 0.2119 | 0.3325 | 13 | 76.47 |
| 10 | 5′- (GA) 8T-3′ | 1.7059 | 1.4457 | 0.2590 | 0.3857 | 12 | 70.59 |
| 11 | 5′- (GA) 8C-3′ | 1.8095 | 1.4586 | 0.2793 | 0.4226 | 17 | 80.95 |
| 12 | 5′- (CT) 8G-3′ | 1.8667 | 1.5280 | 0.3039 | 0.4540 | 13 | 86.67 |
| 13 | 5′- (CA) 8G-3′ | 1.7647 | 1.5749 | 0.3183 | 0.4597 | 13 | 76.47 |
| 14 | 5′- (TC) 8C-3′ | 2.0000 | 1.5520 | 0.3291 | 0.5000 | 18 | 100.00 |
| 15 | 5′- (TC) 8G-3′ | 1.7143 | 1.3294 | 0.2088 | 0.3252 | 10 | 71.43 |
| 16 | 5′- (AC) 8C-3′ | 1.9048 | 1.6003 | 0.3396 | 0.4997 | 19 | 90.48 |
| 17 | 5′- (TG) 8G-3′ | 1.9375 | 1.6153 | 0.3554 | 0.5251 | 15 | 93.75 |
| 18 | 5′- (AG) 8YT-3′ | 1.8889 | 1.4057 | 0.2523 | 0.3941 | 16 | 88.89 |
| 19 | 5′- (GA) 8YC-3′ | 1.7500 | 1.4760 | 0.2793 | 0.4158 | 15 | 75.00 |
| 20 | 5′- (CT) 8RG -3′ | 1.8333 | 1.3338 | 0.2136 | 0.3390 | 15 | 83.33 |
R=A/T, Y=G/C, B=T/G/C, D=A/T/G, H=A/T/C, V=3A/G/C
Fig. 1.
ISSR amplification profile for primer 2 on Carthamus tinctorius accessions. Numbers represent the accessions according to Table 1. M: 1 kb DNA ladder
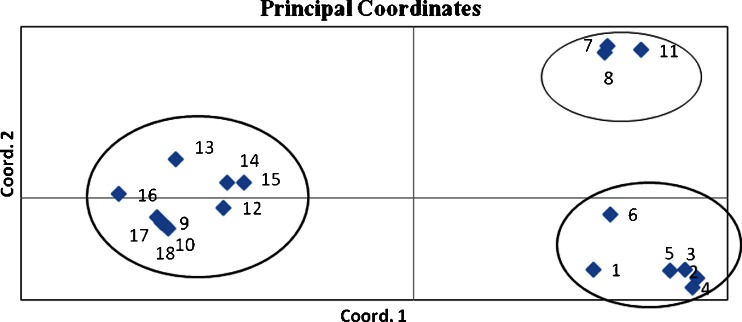
The genetic similarity values varied from 0.427 for IR11 versus IR 14 to 0.71 for IR 3 and IR 4 (Fig. 2). A dendrogram based on UPGMA analysis with ISSR data is presented in Fig. 2. According to dendrogram, genotypes were separated into two main clusters and further within four subclusters. Sub clusters one (I) included khorasan razavi and golestan province accessions (IR1, IR2, IR3, IR4, IR5, IR6), while sub cluster two (II) included guilan and Isfahan provine accessions (IR7, IR8 and IR 9) (Fig. 2). Other accessions form two others sub clusters. Genetic relationship among 18 accessions was also visualized by performing principle coordinate analysis (PCoA) based on ISSR data (Fig. 3). The first two Eigen values accounted for 61.98 % of variation observed in the genotypes. Two-dimensional plot generated from PCoA also supported the clustering pattern of UPGMA dendrogram (Fig. 3).
Fig. 2.
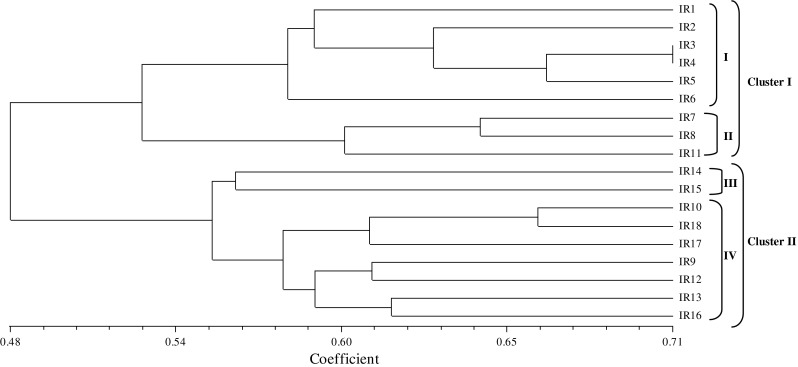
Dendrogram of ISSR analysis on 18 accessions of safflower based on Jacquard similarity coefficient, using the Unweight Pair-Group Method
Fig. 3.
Principle coordinate analysis for 20 ISSR primers applied on 18 safflower accessions. Numbers represent the accessions according to Table 1
Our data indicated that ISSR technology can detect considerable polymorphisms (76.4 %) in our genotypes, suggesting that it will be useful in safflower germplasm characterization and fingerprinting purposes. This study provides fundamental evidence that ISSR marker is a simple, informative, reproducible and suitable approach to evaluation of molecular diversity and phylogenetic relationships in safflower. Various measures of genetic variation are presented in the Table 2. The number of alleles observed across ISSR primers studied varied from 1.6 (primer 5) to 2 (primers 6, 7, 14) (Table 2). This measure provides complementary information to that of polymorphism. Nei’s gene diversity (H) values showed overall 19.08 % to 40.15 % heterozygosity. Similarly, the Shannon’s information indices (I) were 0.30 to 0.58, show a gene diversity measurement with an average of 0.4 ± 0.08 (Table 2) which indicate the high polymorphism across the loci in Iranian safflowers.
The cluster analysis based on ISSR data showed that there was a considerable agreement between geographic origin and their genomic similarities. Similar results were obtained in the study by Arzani and Rezaei (2011). Similarities in genotypes grouped in the same cluster could also appear because of participating a common lineage, convergent evolution and selection of superior genotypes by farmers. All of C. tinctorius genotypes were placed in cluster II with most of Isfahan, markazi and fars province. This could show the close relationship of these accessions. This could be due to the exchange of plant materials across the regions during the safflower cultivation. Some aspects of interrelation among materials studied that were not recognizable by cluster, revealed by the principal coordinate analysis (PCOA). Sum of the first three PCOs could be represented most of (66.1 %) the total variation in the original dimensions. Therefore, this result demonstrates proper distribution of ISSR markers through entire genome and confirms the results of cluster analysis. Also the result showed these genotypes have high genetic diversity, thus, for success in safflower breeding programs use to recommended Iranian safflower local.
References
- Amiri RM, Azdi SB, Ghanadha MR, Abd MC. Detection of DNA polymorphism in landrace populations of safflower in Iran using RAPD-PCR technique. Iran J Agri Sci. 2001;32:737–745. [Google Scholar]
- Amsellem L, Noyer JL, Lebourgeois T, Hossaertmckey M (2000) Comparison of genetic diversity of the invasive weed Rubus alceifolius Poir. (Rosaceae) in its native range and in areas of introduction, using amplified fragment length polymorphism (AFLP) markers. Mol Ecol 9:443–455 [DOI] [PubMed]
- Arzani A, Rezaei AM. Genetic variation in safflower (Carthamus tinctorious L.) for seed quality-related traits and Inter-Simple Sequence Repeat (ISSR) markers. Int J Mol Sci. 2011;12:2664–2677. doi: 10.3390/ijms12042664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash GJ, Raman R, Crump NS. An investigation of genetic variation in Carthamus lanatus in New South Wales, Australia, using intersimple sequence repeats(ISSR) analysis. Weed Res (Oxford) 2003;43:208–213. doi: 10.1046/j.1365-3180.2003.00335.x. [DOI] [Google Scholar]
- Belaj A, Satovic Z, Cipriani G, Baldoni L, Testolin R, Rallo L, Trujillo I. Comparative study of the discriminating capacity of RAPD, AFLP and SSR markers and of their effectiveness in establishing genetic relationships in olive. Theor Appl Genet. 2003;107:736–744. doi: 10.1007/s00122-003-1301-5. [DOI] [PubMed] [Google Scholar]
- Blair MW, Panaud O, Mccouch SR. Inter-simple sequence repeat (ISSR) amplification for analysis of microsatellite motif frequency and fingerprinting in rice (Oryza sativa L.) Theor Appl Genet. 1999;98:780–792. doi: 10.1007/s001220051135. [DOI] [Google Scholar]
- Casaoli M, Mattion C, Cherubini M, Villani F. A genetic linkage map of European chestnut (Castanea sativa Mill.) based on RAPD, ISSR and isozyme markers. Theor Appl Genet. 2001;102:1190–1199. doi: 10.1007/s00122-001-0553-1. [DOI] [Google Scholar]
- Cekic C, Battey NH, Wilkinson MJ. The potential of ISSR-PCR primer-pair combinations for genetic. Theor Appl Genet. 2001;103:540–546. doi: 10.1007/PL00002907. [DOI] [Google Scholar]
- Chapman MA, Burke JM (2007) DNA sequence diversity and the origin of cultivated safflower (Carthamus tinctorius L.; Asteraceae). BMC Plant Biol. doi:10.1186/1471-2229-7-60 [DOI] [PMC free article] [PubMed]
- Godwin ID, Aitken EAB, Smith LW. Application of inters simple sequence repeat (ISSR) markers to plant genetics. Electrophoresis. 1997;18:1524–1528. doi: 10.1002/elps.1150180906. [DOI] [PubMed] [Google Scholar]
- Guo ML, Jiang W, Zhang ZZ, Zhang G, Mao JF, Yin M, Su ZW. Randomly amplified polymorphic DNA technique in molecular identification of germplasms of Carthamus tinctorius L. Acad J Sec Mil Med Univ. 2003;24(10):1116–1119. [Google Scholar]
- Hollngsworth PM, Tebbitt M, Watson KS, Gornall RJ. Conservation genetics of an artic species, Saxifgra rivularis L. Bot J Linn Soc. 1998;128:1–14. [Google Scholar]
- Knowles PF. Centers of plant diversity and conservation of crop germplasm: safflower. Econ Bot. 1969;23:324–329. doi: 10.1007/BF02860678. [DOI] [Google Scholar]
- Knowles PF, Ashri A (1995) Evolution of crop plants, 2nd edn. Longman, Harlow 31:47–50
- Mary SS, Gopalan A. Dissection of genetic attributes yield traits of fodder cowpea in F3 and F4. J Appl Sci Res. 2006;2(6):805–808. [Google Scholar]
- Mengoni A, Gor A, Bazzicalupo M. Use of RAPD and microsatellite (SSR) variation to assess genetic relationships among populations of tetraploid alfalfa, Medicago sativa. Plant Breed. 2000;119:311–317. doi: 10.1046/j.1439-0523.2000.00501.x. [DOI] [Google Scholar]
- Mohammadi R, Pourdad SS. Estimation, interrelationships and repeatability of genetic variability parameters in spring safflower using multi-environment trial data. Euphytica. 2009;165(3):313–324. doi: 10.1007/s10681-008-9789-z. [DOI] [Google Scholar]
- Nagaoka T, Ogihara Y. Applicability of inter-simple sequence repeat markers in wheat for use as DNA markers in comparison to RFLP and RAPD markers. Theor Appl Genet. 1997;94(5):597–602. doi: 10.1007/s001220050456. [DOI] [Google Scholar]
- Peakall R, Smouse PE. GenAlEx V6.1: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes. 2007;6:288–295. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter D, Gao FY, Aliello G, Leslie C, McGranahan G. Intersimple sequence repeat markers for fingerprinting and determining genetic relationships of walnut (Juglans regia) cultivars. Am Soc Horticult Sci. 2002;127(1):75–81. [Google Scholar]
- Rohlf FG. NTsys-pc numerical taxonomy and multivariate system version 2.0. New York: Appl Biostat. Inc; 2000. [Google Scholar]
- Saghai-Maroof MA, Biyashev RM, Yang GP, Zhang Q, Allard RW. Extraordarily polymorphic microsatellite DNA in barly: species diversity, chromosomal locations, and population dynamics. Proc Natl Acad Sci. 1984;91:5466–5470. doi: 10.1073/pnas.91.12.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal D, Rajpal VR, Raina SN, Sasanuma T, Sasakuma T. Assaying polymorphism at DNA level for genetic diversity diagnostics of the safflower (Carthamus tinctorius L.) world germplasm resources. Genetics. 2009;135:457–470. doi: 10.1007/s10709-008-9292-4. [DOI] [PubMed] [Google Scholar]
- Sneath PHA, Sokal RR (1973) Numerical Taxonomy: The principles and practice of. Numerical classification. San Francisco: Freeman 573:566–576
- Tanyolac B. Inter-simple sequence repeat (ISSR) and RAPD variation among wild barely (Hordeum vulgare subsp. spontaneum) populations from west Turkey. Genet Resour Crop Evol. 2003;50(6):611–614. doi: 10.1023/A:1024412814757. [DOI] [Google Scholar]
- Wang Z, Weber JL, Zhang G, Tanksley SD. Survey of plant short tandem DNA repeats. Theor Appl Genet. 1994;88:1–6. doi: 10.1007/BF00222386. [DOI] [PubMed] [Google Scholar]
- Wu W, Zheng YL, Chen L, Wei YM, Yang RW, Yan ZH (2005) Evaluation of genetic relationships in the genus Houttuynia Thunb. in China based on RAPD and ISSR markers. Biochem Syst Ecol 33:1141–1157
- Yang YX, Wu W, Zheng YL, Chen L, Liu RJ, Huang CY (2007) Genetic diversity and relationships among safflower (Carthamus tinctorius L.) analyzed by inter-simple sequence repeats (ISSRs). Genet Resour Crop Evol 54:1043–1051
- Yeh FC, Yang RC, Boyle T (1999) POPGENE, the user-friendly shareware for population genetic analysis. Molecular biology and biotechnology center, University of Alberta, Canada
- Zietkiewicz E, Rafalski A, Labuda D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics. 1994;20:176–183. doi: 10.1006/geno.1994.1151. [DOI] [PubMed] [Google Scholar]