Abstract
Contrary to the traditional view that bacterial populations are clonal, single-cell analysis reveals that phenotypic heterogeneity is common in bacteria. Formation of distinct bacterial lineages appears to be frequent during adaptation to harsh environments, including the colonization of animals by bacterial pathogens. Formation of bacterial subpopulations is often controlled by epigenetic mechanisms that generate inheritable phenotypic diversity without altering the DNA sequence. Such mechanisms are diverse, ranging from relatively simple feedback loops to complex self-perpetuating DNA methylation patterns.
Keywords: Bacterial Genetics, DNA Methylation, DNA Methyltransferase, Epigenetics, Escherichia coli, Gene Regulation, DNA Methylation Pattern, Genetic Switch, Phase Variation, Reversible Bistability
Introduction
The term “epigenesis” was introduced into contemporary biology by Conrad Waddington, a British visionary embryologist, to describe how cell lineages are formed during the development of multicellular eukaryotes (1, 2). During differentiation of eukaryotic tissues, genetically identical cells diversify into distinct lineages by inheritable changes in gene expression without loss or alteration of the DNA sequence. Many decades after Waddington, a universally accepted definition of epigenetics remains to be agreed upon. However, a tentative definition may be that epigenetics addresses the study of cell lineage formation by non-mutational mechanisms.
Most textbooks and reviews on epigenetic gene regulation concern only eukaryotes. One reason may be the enormous success of eukaryotic epigenetics and its implications for human disease. In addition, bacteria have been traditionally viewed as clonal populations of genetically identical cells with phenotypes merely reflecting their genetic constitution. This view is, however, naïve. Certain bacterial genera undergo complex developmental programs that involve cell differentiation. Spore formation by Bacillus subtilis (3), differentiation of Rhizobium into nitrogen-fixing bacteroids (4), asymmetric cell division in Caulobacter (5), formation of fruiting bodies by Myxococcus (6), heterocyst formation in cyanobacteria (7), and biofilm formation in many bacterial species (8, 9) are well known examples of bacterial development. In all of these phenomena, bacterial cells with distinct morphological and physiological properties are formed while the genome DNA sequence remains intact.
Formation of phenotypically distinct cells in populations made of genetically identical bacteria is not restricted to developmental programs. In the last few decades, the introduction of single-cell analysis in bacteriology has revealed many examples of subpopulation formation. For instance, clonal populations of bacteria can sometimes bifurcate into two distinct states, a phenomenon known as bistability (10, 11). Reversible bistability, traditionally known as phase variation, is also common (12). Transition at high frequency between two or more phenotypic states (13) can occur through mutations at genomic repeat sequences (14, 15) or via site-specific recombination (16–19). In other cases, bistability and phase variation are controlled by epigenetic mechanisms with strikingly different levels of complexity, from the propagation of simple feedback loops to the formation of DNA methylation patterns reminiscent of chromatin modification in eukaryotic cells (20–22).
Subpopulation formation can often be observed in the laboratory. However, it may be especially relevant in natural environments, either as an adaptive strategy (e.g. to evade the immune system and other host defenses during bacterial infection) or as a bet-hedging strategy that may facilitate survival if environmental changes occur (23). Relevant examples of phenotypic heterogeneity in natural environments are the formation of “persisters” (dormant bacterial cells resistant to antibiotics) (24, 25), the formation of lineages during Salmonella colonization of animals (26–28), and the bistable expression of extracellular matrix genes during biofilm formation by B. subtilis (9).
Even though subpopulation formation can be seen as the execution of intrinsic bacterial programs, it often involves stochastic events. For instance, random fluctuations in gene expression, a phenomenon known as “noise,” can establish cell-to-cell differences in an isogenic population of bacteria (29). These quantitative differences can become qualitative (30) in the sense that expression above a critical threshold will provide a distinct signal, and expression below the threshold will provide a different signal (21, 31). Propagation of these signals by feedback loops enables the formation of epigenetic lineages (Fig. 1).
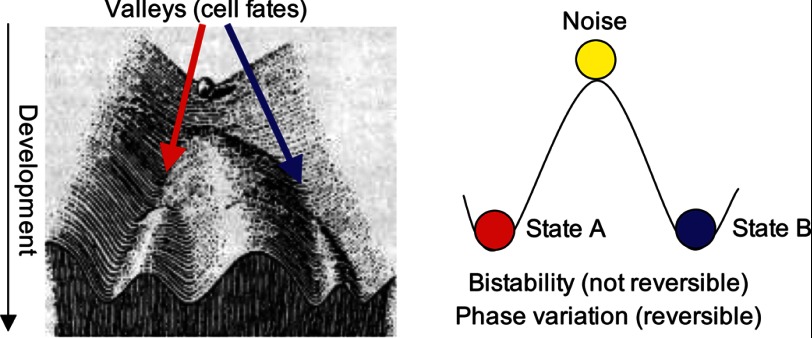
FIGURE 1.
Left panel, Waddington's artistic drawing of an “epigenetic landscape” as a ball that falls to stable valleys from unstable ridges (adapted from Ref. 1). Right panel, bistability viewed as the fall of a ball from an unstable state on a ridge to a stable state in a valley. In phase variation, the valley state is metastable, and the ball periodically returns to the ridge.
Formation of Epigenetic Lineages by a Positive Feedback Loop
Bistable gene expression occurs when a unimodal pattern of gene expression becomes bimodal, bifurcating into two distinct patterns (10, 32). Bistability can be generated either by a positive feedback loop or by a double-negative feedback loop (22, 33). A classical example of bistability generated by a positive feedback loop was described in the Escherichia coli lac operon (34). When added at high concentrations, the gratuitous inducer isopropyl β-d-thiogalactopyranoside (IPTG)3 fully derepresses the lac operon. At low concentrations, however, IPTG is unable to induce a naïve (uninduced) culture. However, if a fully induced culture is transferred to medium containing low concentrations of IPTG, a subpopulation of cells is able to maintain the fully induced state (34). Maintenance occurs because fully induced cells have a high level of β-galactoside permease in their membrane. The permease transports IPTG, providing a high internal concentration of inducer, which maintains full induction (32, 34). The positive feedback loop in this system is that a high level of permease is required to concentrate IPTG in the cell, and high internal IPTG levels are required for high levels of permease synthesis (34). In other cells, however, a decrease in the internal concentration of inducer will reduce permease synthesis, which in turn will cause further reduction in the internal concentration of IPTG, driving the cell toward the repressed state via binding of the LacI repressor. The overall consequence is that a fully induced population bifurcates into two bistable states: fully induced and uninduced (repressed) (32–34).
Errors made during transcription can also provide signals for epigenetic switching in the E. coli lac operon (35). An increased error rate during transcription, caused either by mutations that reduce transcription fidelity of RNA polymerase or by the absence of transcription fidelity factors GreA and GreB, increases switching of the lac operon from the off state (uninduced) to the on state (induced) (35). The interpretation is that errors in lacI mRNA synthesis cause a transient decrease in the Lac repressor level, which permits switching to the on state (35, 36). Note that an uninduced E. coli cell contains ∼10 molecules of the Lac repressor, an amount small enough to make the system noisy and therefore metastable. Perturbation of this delicate equilibrium by transcription inaccuracy can switch the system to the on state. Even though the decrease in the Lac repressor concentration is transient, synthesis of permease will generate a positive feedback loop that will maintain in the on state in certain cells (34). Lac bistability is not observed in cells containing a 10-fold higher Lac repressor level, consistent with the hypothesis that switching occurs only under conditions in which repressor levels are subsaturating.
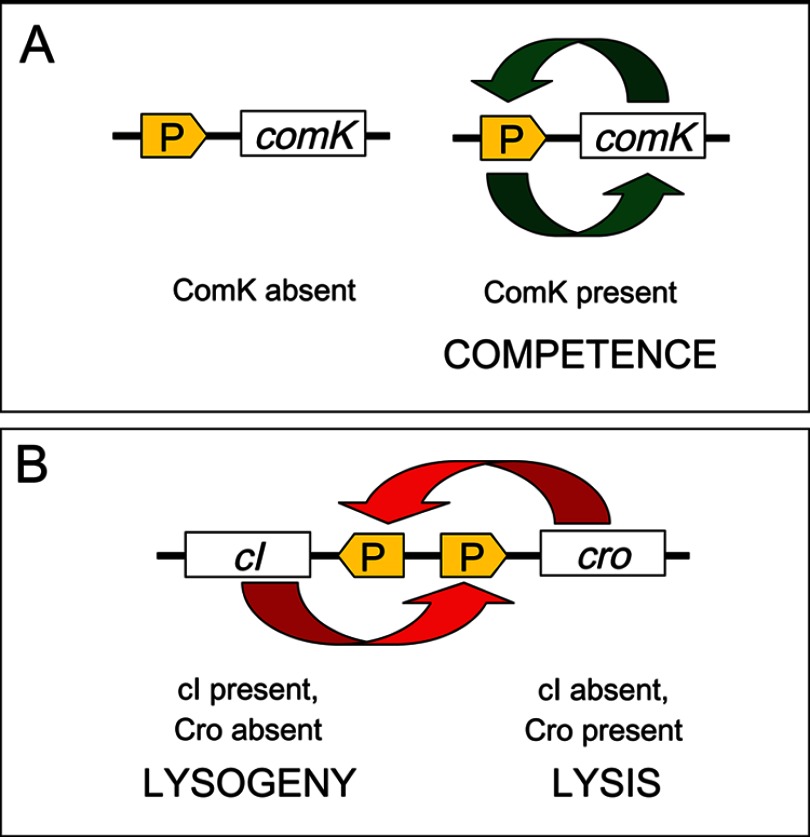
Another classical example of bistability occurs in B. subtilis. Upon entry into stationary phase, a fraction of B. subtilis cells acquire the capacity to take up DNA, a phenomenon known as competence (10). A crucial factor for competence development is accumulation of ComK, which activates genes required for DNA uptake as well as the comK gene itself (37). During exponential growth, ComK is synthesized but degraded. When the culture approaches stationary phase, a quorum sensing-related factor stabilizes ComK (38, 39). At that moment, a competition is initiated between several repressors and ComK for binding to regulatory regions of the comK promoter (40, 41). Binding of ComK initiates a positive feedback loop, leading to increased synthesis of ComK and subsequent transcription of competence genes. Binding of the repressors inhibits comK expression and prevents competence. A crucial property for bifurcation of the population into two subpopulations is that the level of ComK in individual cells fluctuates, generating stochastic noise. When the ComK level reaches a threshold in a B. subtilis cell, a quantitative difference becomes qualitative: the ComK positive feedback loop will be activated, and competence will develop (42–44). Development of competence thus occurs in cells that undergo a small but critical increase in ComK concentration (Fig. 2). In turn, comK will be repressed in cells in which the ComK level remains below the threshold, and they will not develop competence (Fig. 2) (43).
FIGURE 2.
A, competence development in B. subtilis, an example of bistability created by a positive feedback loop. B, the lysis/lysogeny decision in bacteriophage λ, an example of bistability created by a double-negative feedback loop.
Formation of Epigenetic Lineages by a Double-negative Feedback Loop
Infection of E. coli by bacteriophage λ can follow two developmental programs. One is lysis of the bacterial cell; the other is lysogeny, a symbiosis-like association in which the phage enters a dormant state. Although the lysis/lysogeny decision is influenced by the physiological state of the cell and by environmental factors, the fate of individual infections is unpredictable and may be considered stochastic (33, 45, 46). Phage λ has two repressors, cI and Cro, each of which represses expression of the other. At the onset of infection, both repressors are produced, and the lysis/lysogeny decision may be viewed as a repressor race: the repressor that first occupies specific regulatory DNA sites in λ DNA will repress synthesis of its antagonist (45). If the winner is Cro, synthesis of cI will be repressed, and λ will lyse the host cell (Fig. 2). If the winner is cI, synthesis of Cro will be repressed, and λ will lysogenize the cell (Fig. 2) (45). Note that the outcomes of a positive feedback loop and a double-negative feedback loop are analogous (22, 33). In the case of λ, preventing the synthesis of Cro by cI is equivalent to positive autoregulation of cI and vice versa.
Phase Variation via DNA Methylation Patterns
A common epigenetic mechanism to regulate switches involves the formation of DNA methylation patterns (47, 48). This occurs when a methylation sequence on DNA overlaps the binding site for a protein, and methylation of that sequence is blocked (49, 50). For example, most GATC sites in the E. coli chromosome are fully methylated except for a short time following DNA replication, in which they are hemimethylated. However, a few sites are stably unmethylated due to binding of proteins at sites that overlap or are adjacent to a GATC site, competing with Dam for binding and blocking methylation (47, 48, 51). Two such GATC sites in the pap (pyelonephritis-associated pili) operon of uropathogenic E. coli orchestrate Pap pilus phase variation (52, 53). The core of the Pap switch consists of two sets of binding sites, 1–3 and 4–6, within the pap promoter region for the global regulator known as the leucine-responsive regulatory protein, Lrp (54). Lrp appears to be predominantly a tetramer of dimers (octamer), with three Lrp dimers binding to three pap sites, leaving one dimer unbound (Fig. 3) (55–57).
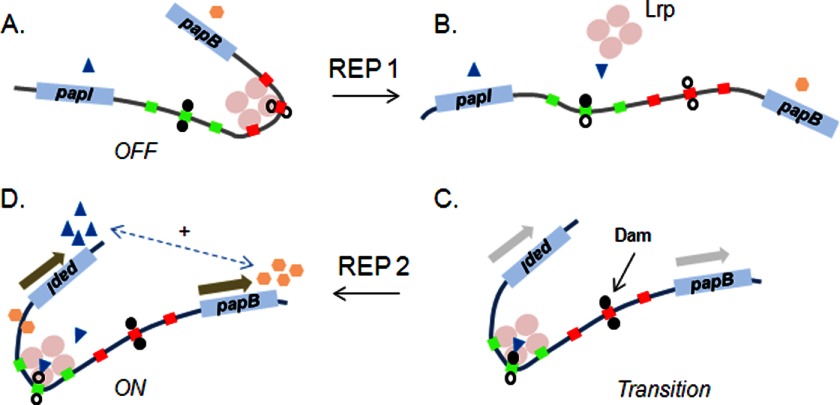
FIGURE 3.
Model for the pap off-to-on state transition, an example of a bistable switch controlled by DNA methylation patterns. A, in the phase off state, an octamer of Lrp (a tetramer of dimers; only one tetramer is depicted) binds cooperatively to promoter-proximal sites 1–3 (red boxes). Lrp binding to sites 1–3 inhibits further binding of Lrp to sites 4–6 (green boxes) by mutual exclusion. B, immediately following passage of the replication fork (REP 1), the two daughter chromosomes become hemimethylated. Only the daughter chromosome methylated on the top strand is shown (filled circle above Lrp-binding site 5). C, two stochastic events occur in which PapI facilitates Lrp binding to sites 4–6, and Dam (DNA adenine methylase) methylates both strands of the proximal GATC site. Binding of Lrp at sites 4–6 reduces the affinity of Lrp for sites 1–3 by mutual exclusion and facilitates activation of pap transcription via cAMP-catabolite gene activator protein/RNA polymerase binding (not shown). Methylation of GATCprox reduces the affinity of PapI/Lrp for sites 1–3 and is required for transition to the on phase (98). D, one additional round of DNA replication (REP 2) completes transition to the phase on state, in which GATCdist is fully unmethylated. The on phase is self-perpetuating due to a bidirectional feedback loop between PapB and PapI (dashed arrow). The PapB level rises due to activation of transcription of the first gene of the pap operon, papB. PapB binds near the papI promoter, increases the PapI level via activation of papI transcription, and helps maintain the on state via binding of PapI/Lrp to sites 4–6.
A GATC site is present within site 2 (GATCprox) and site 5 (GATCdist); methylation of these sites affects Lrp binding, as discussed below. Lrp binds cooperatively to a set of three pap sites, but occupancy of all six Lrp sites occurs infrequently due to a mutual exclusion mechanism that requires negative DNA writhe (supercoils) (58). Lrp binding to sites 1–3 (Fig. 3, red boxes) blocks methylation of GATCprox and also blocks pap transcription because the RNA polymerase σ70-binding site is in this region (Fig. 3A) (59). In contrast, binding of Lrp to sites 4–6 (Fig. 3, green boxes) blocks methylation of GATCdist and helps to activate pap transcription (60). The role of Lrp in activating transcription may be to bend DNA, facilitating binding of catabolite gene activator protein to the RNA polymerase α-subunit (61).
Transition from the phase off state to the phase on state requires two pap-encoded regulators, PapI and PapB. PapI increases the affinity of Lrp for pap sites 2 and 5 via an ACGATC sequence present in each site (52, 56, 62). PapB, the product of the first gene of the pap operon, binds near the papI promoter and activates papI transcription, forming a positive feedback loop (Fig. 3D, dashed arrow) (63). Methylation of GATCprox is required for the off-to-on transition because it lowers the affinity of PapI/Lrp for site 2, increasing the probability that PapI/Lrp will bind to sites 4–6 and initiate transition to the on phase (58). For this to occur, Lrp bound at sites 1–3 in off phase cells must dissociate to allow methylation of GATCprox by Dam. This likely occurs as the replication fork passes through the pap regulatory region, and a hemimethylated GATCdist site is generated (Fig. 3B). The affinity of PapI/Lrp for hemimethylated pap sites 4–6 is significantly higher than for the fully methylated DNA (52, 56). If PapI/Lrp binds to site 5 before Dam methylates the daughter strand, cooperative binding of Lrp/PapI to sites 4–6 will occur to initiate transition to the phase on state. Evidence indicates that a dimer of Lrp and a monomer of PapI bind to pap site 5 (56). This transition is also dependent on dissociation of Lrp from sites 1–3 and methylation of GATCprox: increasing the off rate (kdis) of Lrp at sites 1–3 increases the off-to-on rate (64).
Dam methylase is highly processive, such that ∼130 Dam molecules can efficiently methylate ∼20,000 genomic GATC sites (65). Thus, when Dam methylates GATCprox, it should have a high propensity to methylate the adjacent GATCdist site before dissociating from DNA. This would block PapI/Lrp binding to site 5 and block transition to the on phase (60). Recent work has shown that the presence of a poly(A) tract 5′ to the two pap GATC sites decreases the processivity of methylation by reducing the rate of methyl transfer (kchem) (66). This may be necessary to allow PapI/Lrp to compete with Dam for access to hemimethylated GATCdist sites following DNA replication (67).
The phase on-to-off transition, which occurs at an ∼100-fold higher rate than the off-to-on transition (47), has not been analyzed in detail. Following DNA replication, cells in the phase on state contain a hemimethylated GATCdist site and a fully unmethylated GATCprox site. If Dam methylates GATCdist, binding of PapI/Lrp will be inhibited, providing an opportunity for Lrp binding at sites 1–3 due to release of mutual exclusion. Notably, binding of Lrp to site 2 is unaffected by methylation of GATCprox (52); therefore, the key step must be competition of Dam and PapI/Lrp for binding at site 5. Formation of the phase off DNA methylation pattern requires two rounds of DNA replication to convert a fully methylated GATCprox site to a fully unmethylated site.
The on and off pap transcription states are each self-perpetuating and heritable. In the off state, GATCdist is fully methylated, preventing PapI/Lrp binding to sites 4–6 (Fig. 3A). Conversely, in the on state, PapI expression is high due to the PapB positive regulatory feedback, and GATCprox is fully methylated, preventing PapI/Lrp binding to sites 1–3 (Fig. 3D). In addition, it is likely that both the off and on states are stabilized by mutual exclusion (58).
The pap switch is modulated by additional transcription factors that are environmentally responsive, including H-NS, RimJ, and CpxR. Transcription of pap is blocked at 23 °C by H-NS, which binds to the pap regulatory region and blocks GATC methylation (68). H-NS also modulates Pap switching at 37 °C in response to high osmolarity and other environmental conditions (69, 70). This may occur by altering PapI/Lrp binding to pap regulatory sites, but the mechanistic details are unknown. RimJ, which acetylates ribosomal protein S5, inhibits transition to the on state in response to temperature and other environmental conditions by an unknown mechanism (71). The CpxAR two-component regulatory system responds to cell envelope stress by phosphorylation of CpxR. Phosphorylated CpxR binds specifically to the pap regulatory region, competes with Lrp, and blocks pap transcription, which may protect cells from further cell envelope damage (72–74).
Other Switches Regulated by DNA Methylation Patterns
Many methylation-dependent phase variation systems have been identified since the initial discovery of the Pap system. Some of these systems, such as foo, clp, and pef, which all encode pili, are designed similarly to the pap switch (75–77). Remarkably, the latter two systems have a reversed architecture in which the PapI homologs ClpI and PefI act as negative regulators. Other methylation-controlled switches use DNA-binding proteins other than Lrp, including OxyR and Fur. The best characterized system is agn43, which controls the expression of antigen 43 (78, 79), an outer membrane protein that plays a role in biofilm formation and pathogenesis (80, 81). OxyR binds three GATC sites in the agn43 regulatory region. Binding of OxyR blocks methylation of the three GATC sites and inhibits agn43 transcription, forming the off phase. Transition to the on phase occurs following DNA replication if Dam can methylate both strands of the three GATC sites before OxyR rebinds to the sites (50, 82, 83). Notably, the poly(A) tracts adjacent to the GATC sequences in pap and its relatives are not present in agn43, and thus, Dam should processively methylate the three agn43 GATC sites if they are not bound by OxyR (84). The on-to-off switch can occur after DNA replication, when the three GATC sites are hemimethylated. OxyR has a higher affinity for agn43 DNA containing hemimethylated GATC sites versus fully methylated GATC sites (84, 85). Thus, if OxyR binds to the GATC region before Dam fully methylates the GATC sites, a phase off intermediate state will ensue, and after one more round of replication to convert the hemimethylated GATC sites to fully unmethylated sites, the phase off transition will be complete. On-to-off transition is affected by the local concentration of OxyR; the addition of three or more OxyR-binding sites upstream of agn43 biases cells toward the off phase (84).
A number of phase variation switches appear to be regulated by mechanisms reminiscent of agn43. These include the gtr switch on the P22 bacteriophage (86) and the chromosomal switch locus STM2209-STM2208 (opvAB) (87), each controlling modification of cell surface lipopolysaccharide of Salmonella, both of which are controlled by OxyR and Dam. In enteroaggregative E. coli, the sci1 type VI secretion system is controlled by a phase switch in which the iron regulatory protein Fur blocks Dam methylation of sci1 GATC sites, forming phase off and on methylation patterns (88).
Phasevarions: Formation of Epigenetic Lineages by Phase Variation of DNA Methylase Synthesis
Certain restriction-modification systems show phase variation, and a common mechanism for switching between off and on states is expansion and contraction of nucleotide repeats (89). Phase variation of restriction-modification systems may generate subpopulations of bacterial cells differing in their susceptibility to phage infection and in their ability to acquire foreign DNA. In addition, DNA adenine methylation by certain phase-variable restriction-modification systems regulates expression of specific genes (90). These systems, known as “phasevarions,” conserve their restriction-modification activity but have additionally acquired epigenetic regulatory capacity (91, 92). In some phasevarions, the gene encoding the restriction enzyme is inactivated by mutation, whereas the modification gene (mod) remains active. Hence, in these mutant type III restriction-modification systems, the Mod enzyme is a functional analog of solitary methyltransferases (e.g. Dam).
In the human pathogens Haemophilus influenzae, Neisseria meningitidis, and Neisseria gonorrhoeae, DNA adenine methylation by Mod enzymes has been shown to regulate gene expression, and the loci under Mod control include genes with roles in envelope structure, virulence, and stress responses (92). Because synthesis of Mod DNA methylase is phase-variable, isogenic subpopulations contain two types of bacterial cells. One population contains N6-methyladenine in the genome, whereas the other subpopulation does not. As a consequence, each lineage shows a distinct pattern of gene expression that affects all DNA methylation-sensitive loci.
Whereas individual phase variation systems, such as pap and agn43, generate heterogeneity of a single phenotypic trait, cell lineages under phasevarion control differ in multiple phenotypic traits. The capacity of phasevarions to generate bacterial lineages may be further extended in bacterial species that contain multiple mod alleles, each with slightly different DNA-binding domains (92). Independent switching in the synthesis of several Mod proteins can be expected to generate multiple gene expression patterns, thus increasing the phenotypic heterogeneity of the population.
Hierarchical Epigenetic Networks
Phase variation of certain genetic loci causes bistable expression of other genes, extending phenotypic heterogeneity to cell functions encoded outside the phase variation locus. An example of this kind occurs in the Salmonella enterica std operon, which encodes fimbriae for attachment to the intestinal mucosa (93). Transcription of std is controlled by a LysR-like regulator known as HdfR and by two products of the std operon, StdE and StdF (94). Production of Std fimbriae in isogenic populations of Salmonella is subject to phase variation; the switching mechanism remains to be deciphered. However, it is well established that the StdE and StdF gene products regulate expression of genes outside the std operon, including the cluster of virulence genes known as Salmonella pathogenicity island 1, SPI-1 (95). Because SPI-1 expression is prevented by StdE/StdF, cells that produce Std fimbriae do not synthesize the SPI-1-encoded apparatus and vice versa (95). One may thus predict that phase variation of the std operon in the animal intestine will split Salmonella populations into two lineages, one able to invade the intestinal mucosa (causing acute disease) and one able to attach to the intestinal epithelium (causing latent infection). Depending on the host physiological conditions and the host response, one of the two subpopulations will be able to colonize the animal, whereas the other will be eliminated. Whatever the outcome, bet-hedging will increase the chances that a fraction of the Salmonella population survives. This model fits well with the view that colonization of animals by Salmonella involves subpopulation formation at several stages (26–28), and the same may be true for other human pathogens (25, 96, 97). Subpopulations may differ in their susceptibility to antibacterial drugs, thus explaining why certain bacterial infections are difficult or impossible to eradicate.
Footnotes
- IPTG
- isopropyl β-d-thiogalactopyranoside.
REFERENCES
- 1. Waddington C. H. (1957) The Strategy of the Genes, George Allen and Unwin, London [Google Scholar]
- 2. Goldberg A. D., Allis C. D., Bernstein E. (2007) Epigenetics: a landscape takes shape. Cell 128, 635–638 [DOI] [PubMed] [Google Scholar]
- 3. Errington J. (2003) Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1, 117–126 [DOI] [PubMed] [Google Scholar]
- 4. Gibson K. E., Kobayashi H., Walker G. C. (2008) Molecular determinants of a symbiotic chronic infection. Annu. Rev. Genet. 42, 413–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kirkpatrick C. L., Viollier P. H. (2012) Decoding Caulobacter development. FEMS Microbiol. Rev. 36, 193–205 [DOI] [PubMed] [Google Scholar]
- 6. Kaiser D. (2008) Myxococcus–from single-cell polarity to complex multicellular patterns. Annu. Rev. Genet. 42, 109–130 [DOI] [PubMed] [Google Scholar]
- 7. Flores E., Herrero A. (2010) Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat. Rev. Microbiol. 8, 39–50 [DOI] [PubMed] [Google Scholar]
- 8. Stewart P. S., Franklin M. J. (2008) Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6, 199–210 [DOI] [PubMed] [Google Scholar]
- 9. Chai Y., Chu F., Kolter R., Losick R. (2008) Bistability and biofilm formation in Bacillus subtilis. Mol. Microbiol. 67, 254–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dubnau D., Losick R. (2006) Bistability in bacteria. Mol. Microbiol. 61, 564–572 [DOI] [PubMed] [Google Scholar]
- 11. Veening J. W., Smits W. K., Kuipers O. P. (2008) Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62, 193–210 [DOI] [PubMed] [Google Scholar]
- 12. van der Woude M. W. (2011) Phase variation: how to create and coordinate population diversity. Curr. Opin. Microbiol. 14, 205–211 [DOI] [PubMed] [Google Scholar]
- 13. Levinson G., Gutman G. A. (1987) High frequencies of short frameshifts in poly-CA/TG tandem repeats borne by bacteriophage M13 in Escherichia coli K-12. Nucleic Acids Res. 15, 5323–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stern A., Brown M., Nickel P., Meyer T. F. (1986) Opacity genes in Neisseria gonorrhoeae: control of phase and antigenic variation. Cell 47, 61–71 [DOI] [PubMed] [Google Scholar]
- 15. Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. (1966) Frameshift mutations and the genetic code. Cold Spring Harb. Symp. Quant. Biol. 31, 77–84 [DOI] [PubMed] [Google Scholar]
- 16. Eisenstein B. I., Sweet D. S., Vaughn V., Friedman D. I. (1987) Integration host factor is required for the DNA inversion that controls phase variation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 84, 6506–6510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pearce U. B., Stocker B. A. (1967) Phase variation of flagellar antigens in Salmonella: abortive transduction studies. J. Gen. Microbiol. 49, 335–349 [DOI] [PubMed] [Google Scholar]
- 18. Sohanpal B. K., Kulasekara H. D., Bonnen A., Blomfield I. C. (2001) Orientational control of fimE expression in Escherichia coli. Mol. Microbiol. 42, 483–494 [DOI] [PubMed] [Google Scholar]
- 19. Zieg J., Hilmen M., Simon M. (1978) Regulation of gene expression by site-specific inversion. Cell 15, 237–244 [DOI] [PubMed] [Google Scholar]
- 20. Casadesús J., Low D. (2006) Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 70, 830–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davidson C. J., Surette M. G. (2008) Individuality in bacteria. Annu. Rev. Genet. 42, 253–268 [DOI] [PubMed] [Google Scholar]
- 22. Ferrell J. E., Jr. (2002) Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 14, 140–148 [DOI] [PubMed] [Google Scholar]
- 23. Beaumont H. J., Gallie J., Kost C., Ferguson G. C., Rainey P. B. (2009) Experimental evolution of bet hedging. Nature 462, 90–93 [DOI] [PubMed] [Google Scholar]
- 24. Balaban N. Q., Merrin J., Chait R., Kowalik L., Leibler S. (2004) Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 [DOI] [PubMed] [Google Scholar]
- 25. Dhar N., McKinney J. D. (2007) Microbial phenotypic heterogeneity and antibiotic tolerance. Curr. Opin. Microbiol. 10, 30–38 [DOI] [PubMed] [Google Scholar]
- 26. Helaine S., Thompson J. A., Watson K. G., Liu M., Boyle C., Holden D. W. (2010) Dynamics of intracellular bacterial replication at the single cell level. Proc. Natl. Acad. Sci. U.S.A. 107, 3746–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cummings L. A., Wilkerson W. D., Bergsbaken T., Cookson B. T. (2006) In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol. Microbiol. 61, 795–809 [DOI] [PubMed] [Google Scholar]
- 28. Bailly-Bechet M., Benecke A., Hardt W. D., Lanza V., Sturm A., Zecchina R. (2011) An externally modulated, noise-driven switch for the regulation of SPI1 in Salmonella enterica serovar Typhimurium. J. Math. Biol. 63, 637–662 [DOI] [PubMed] [Google Scholar]
- 29. Silva-Rocha R., de Lorenzo V. (2010) Noise and robustness in prokaryotic regulatory networks. Annu. Rev. Microbiol. 64, 257–275 [DOI] [PubMed] [Google Scholar]
- 30. Anderson P. W. (1972) More is different. Science 177, 393–396 [DOI] [PubMed] [Google Scholar]
- 31. Veening J. W., Stewart E. J., Berngruber T. W., Taddei F., Kuipers O. P., Hamoen L. W. (2008) Bet-hedging and epigenetic inheritance in bacterial cell development. Proc. Natl. Acad. Sci. U.S.A. 105, 4393–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laurent M., Charvin G., Guespin-Michel J. (2005) Bistability and hysteresis in epigenetic regulation of the lactose operon. Since Delbrück, a long series of ignored models. Cell. Mol. Biol. 51, 583–594 [PubMed] [Google Scholar]
- 33. Casadesús J., D'Ari R. (2002) Memory in bacteria and phage. Bioessays 24, 512–518 [DOI] [PubMed] [Google Scholar]
- 34. Novick A., Weiner M. (1957) Enzyme induction as an all-or-none phenomenon. Proc. Natl. Acad. Sci. U.S.A. 43, 553–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gordon A. J., Halliday J. A., Blankschien M. D., Burns P. A., Yatagai F., Herman C. (2009) Transcriptional infidelity promotes heritable phenotypic change in a bistable gene network. PLoS Biol. 7, e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Satory D., Gordon A. J., Halliday J. A., Herman C. (2011) Epigenetic switches: can infidelity govern fate in microbes? Curr. Opin. Microbiol. 14, 212–217 [DOI] [PubMed] [Google Scholar]
- 37. van Sinderen D., Luttinger A., Kong L., Dubnau D., Venema G., Hamoen L. (1995) comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol. Microbiol. 15, 455–462 [DOI] [PubMed] [Google Scholar]
- 38. Magnuson R., Solomon J., Grossman A. D. (1994) Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell 77, 207–216 [DOI] [PubMed] [Google Scholar]
- 39. Turgay K., Hahn J., Burghoorn J., Dubnau D. (1998) Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17, 6730–6738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hoa T. T., Tortosa P., Albano M., Dubnau D. (2002) Rok (YkuW) regulates genetic competence in Bacillus subtilis by directly repressing comK. Mol. Microbiol. 43, 15–26 [DOI] [PubMed] [Google Scholar]
- 41. Hamoen L. W., Kausche D., Marahiel M. A., van Sinderen D., Venema G., Serror P. (2003) The Bacillus subtilis transition state regulator AbrB binds to the −35 promoter region of comK. FEMS Microbiol. Lett. 218, 299–304 [DOI] [PubMed] [Google Scholar]
- 42. Smits W. K., Eschevins C. C., Susanna K. A., Bron S., Kuipers O. P., Hamoen L. W. (2005) Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol. Microbiol. 56, 604–614 [DOI] [PubMed] [Google Scholar]
- 43. Smits W. K., Kuipers O. P., Veening J. W. (2006) Phenotypic variation in bacteria: the role of feedback regulation. Nat. Rev. Microbiol. 4, 259–271 [DOI] [PubMed] [Google Scholar]
- 44. Dandach S. H., Khammash M. (2010) Analysis of stochastic strategies in bacterial competence: a master equation approach. PLoS Comp. Biol. 6, e1000985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Johnson A. D., Poteete A. R., Lauer G., Sauer R. T., Ackers G. K., Ptashne M. (1981) λ Repressor and cro–components of an efficient molecular switch. Nature 294, 217–223 [DOI] [PubMed] [Google Scholar]
- 46. Munsky B., Khammash M. (2010) Identification from stochastic cell-to-cell variation: a genetic switch case study. IET Systems Biol. 4, 356–366 [DOI] [PubMed] [Google Scholar]
- 47. Blyn L. B., Braaten B. A., Low D. A. (1990) Regulation of pap pilin phase variation by a mechanism involving differential dam methylation states. EMBO J. 9, 4045–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang M. X., Church G. M. (1992) A whole genome approach to in vivo DNA-protein interactions in E. coli. Nature 360, 606–610 [DOI] [PubMed] [Google Scholar]
- 49. van der Woude M., Hale W. B., Low D. A. (1998) Formation of DNA methylation patterns: nonmethylated GATC sequences in gut and pap operons. J. Bacteriol. 180, 5913–5920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Waldron D. E., Owen P., Dorman C. J. (2002) Competitive interaction of the OxyR DNA-binding protein and the Dam methylase at the antigen 43 gene regulatory region in Escherichia coli. Mol. Microbiol. 44, 509–520 [DOI] [PubMed] [Google Scholar]
- 51. Ringquist S., Smith C. L. (1992) The Escherichia coli chromosome contains specific, unmethylated dam and dcm sites. Proc. Natl. Acad. Sci. U.S.A. 89, 4539–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hernday A. D., Braaten B. A., Low D. A. (2003) The mechanism by which DNA adenine methylase and PapI activate the Pap epigenetic switch. Mol. Cell 12, 947–957 [DOI] [PubMed] [Google Scholar]
- 53. van der Woude M. W., Braaten B. A., Low D. A. (1992) Evidence for global regulatory control of pilus expression in Escherichia coli by Lrp and DNA methylation: model building based on analysis of pap. Mol. Microbiol. 6, 2429–2435 [DOI] [PubMed] [Google Scholar]
- 54. Nou X., Braaten B., Kaltenbach L., Low D. A. (1995) Differential binding of Lrp to two sets of pap DNA binding sites mediated by Pap I regulates Pap phase variation in Escherichia coli. EMBO J. 14, 5785–5797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. de los Rios S., Perona J. J. (2007) Structure of the Escherichia coli leucine-responsive regulatory protein Lrp reveals a novel octameric assembly. J. Mol. Biol. 366, 1589–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kawamura T., Vartanian A. S., Zhou H., Dahlquist F. W. (2011) The design involved in PapI and Lrp regulation of the pap operon. J. Mol. Biol. 409, 311–332 [DOI] [PubMed] [Google Scholar]
- 57. Leonard P. M., Smits S. H., Sedelnikova S. E., Brinkman A. B., de Vos W. M., van der Oost J., Rice D. W., Rafferty J. B. (2001) Crystal structure of the Lrp-like transcriptional regulator from the archaeon Pyrococcus furiosus. EMBO J. 20, 990–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hernday A., Krabbe M., Braaten B., Low D. (2002) Self-perpetuating epigenetic pili switches in bacteria. Proc. Natl. Acad. Sci. U.S.A. 99, 16470–16476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Weyand N. J., Low D. A. (2000) Regulation of Pap phase variation. Lrp is sufficient for the establishment of the phase off pap DNA methylation pattern and repression of pap transcription in vitro. J. Biol. Chem. 275, 3192–3200 [DOI] [PubMed] [Google Scholar]
- 60. Hernday A., Braaten B., Low D. (2004) The intricate workings of a bacterial epigenetic switch. Adv. Exp. Med. Biol. 547, 83–89 [DOI] [PubMed] [Google Scholar]
- 61. Weyand N. J., Braaten B. A., van der Woude M., Tucker J., Low D. A. (2001) The essential role of the promoter-proximal subunit of CAP in pap phase variation: Lrp- and helical phase-dependent activation of papBA transcription by CAP from −215. Mol. Microbiol. 39, 1504–1522 [DOI] [PubMed] [Google Scholar]
- 62. Kaltenbach L. S., Braaten B. A., Low D. A. (1995) Specific binding of PapI to Lrp-pap DNA complexes. J. Bacteriol. 177, 6449–6455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Forsman K., Göransson M., Uhlin B. E. (1989) Autoregulation and multiple DNA interactions by a transcriptional regulatory protein in E. coli pili biogenesis. EMBO J. 8, 1271–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Graveline R., Mourez M., Hancock M. A., Martin C., Boisclair S., Harel J. (2011) Lrp-DNA complex stability determines the level of ON cells in type P fimbriae phase variation. Mol. Microbiol. 81, 1286–1299 [DOI] [PubMed] [Google Scholar]
- 65. Boye E., Marinus M. G., Løbner-Olesen A. (1992) Quantitation of Dam methyltransferase in Escherichia coli. J. Bacteriol. 174, 1682–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Peterson S. N., Reich N. O. (2006) GATC flanking sequences regulate Dam activity: evidence for how Dam specificity may influence pap expression. J. Mol. Biol. 355, 459–472 [DOI] [PubMed] [Google Scholar]
- 67. Peterson S. N., Reich N. O. (2008) Competitive Lrp and Dam assembly at the pap regulatory region: implications for mechanisms of epigenetic regulation. J. Mol. Biol. 383, 92–105 [DOI] [PubMed] [Google Scholar]
- 68. White-Ziegler C. A., Angus Hill M. L., Braaten B. A., van der Woude M. W., Low D. A. (1998) Thermoregulation of Escherichia coli pap transcription: H-NS is a temperature-dependent DNA methylation blocking factor. Mol. Microbiol. 28, 1121–1137 [DOI] [PubMed] [Google Scholar]
- 69. White-Ziegler C. A., Davis T. R. (2009) Genome-wide identification of H-NS-controlled, temperature-regulated genes in Escherichia coli K-12. J. Bacteriol. 191, 1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. White-Ziegler C. A., Villapakkam A., Ronaszeki K., Young S. (2000) H-NS controls pap and daa fimbrial transcription in Escherichia coli in response to multiple environmental cues. J. Bacteriol. 182, 6391–6400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. White-Ziegler C. A., Black A. M., Eliades S. H., Young S., Porter K. (2002) The N-acetyltransferase RimJ responds to environmental stimuli to repress pap fimbrial transcription in Escherichia coli. J. Bacteriol. 184, 4334–4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hernday A. D., Braaten B. A., Broitman-Maduro G., Engelberts P., Low D. A. (2004) Regulation of the Pap epigenetic switch by CpxAR: phosphorylated CpxR inhibits transition to the phase ON state by competition with Lrp. Mol. Cell 16, 537–547 [DOI] [PubMed] [Google Scholar]
- 73. Hung D. L., Raivio T. L., Jones C. H., Silhavy T. J., Hultgren S. J. (2001) Cpx signaling pathway monitors biogenesis and affects assembly and expression of P pili. EMBO J. 20, 1508–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Raivio T. L. (2005) Envelope stress responses and Gram-negative bacterial pathogenesis. Mol. Microbiol. 56, 1119–1128 [DOI] [PubMed] [Google Scholar]
- 75. Harel J., Daigle F., Forget C., Tessier M. C., Crost C., Martin C. (2000) Phase variation of F165(1) (Prs-like) fimbriae from Escherichia coli causing septicaemia in animals. Can. J. Microbiol. 46, 1101–1107 [DOI] [PubMed] [Google Scholar]
- 76. Martin C. (1996) The clp (CS31A) operon is negatively controlled by Lrp, ClpB, and l-alanine at the transcriptional level. Mol. Microbiol. 21, 281–292 [DOI] [PubMed] [Google Scholar]
- 77. Nicholson B., Low D. (2000) DNA methylation-dependent regulation of Pef expression in Salmonella typhimurium. Mol. Microbiol. 35, 728–742 [DOI] [PubMed] [Google Scholar]
- 78. Owen P., Meehan M., de Loughry-Doherty H., Henderson I. (1996) Phase-variable outer membrane proteins in Escherichia coli. FEMS Immunol. Med. Microbiol. 16, 63–76 [DOI] [PubMed] [Google Scholar]
- 79. van der Woude M. W., Henderson I. R. (2008) Regulation and function of Ag43 (flu). Annu. Rev. Microbiol. 62, 153–169 [DOI] [PubMed] [Google Scholar]
- 80. Danese P. N., Pratt L. A., Dove S. L., Kolter R. (2000) The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37, 424–432 [DOI] [PubMed] [Google Scholar]
- 81. Lüthje P., Brauner A. (2010) Ag43 promotes persistence of uropathogenic Escherichia coli isolates in the urinary tract. J. Clin. Microbiol. 48, 2316–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Henderson I. R., Owen P. (1999) The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and OxyR. J. Bacteriol. 181, 2132–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wallecha A., Munster V., Correnti J., Chan T., van der Woude M. (2002) Dam- and OxyR-dependent phase variation of agn43: essential elements and evidence for a new role of DNA methylation. J. Bacteriol. 184, 3338–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kaminska R., van der Woude M. W. (2010) Establishing and maintaining sequestration of Dam target sites for phase variation of agn43 in Escherichia coli. J. Bacteriol. 192, 1937–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Correnti J., Munster V., Chan T., Woude M. (2002) Dam-dependent phase variation of Ag43 in Escherichia coli is altered in a seqA mutant. Mol. Microbiol. 44, 521–532 [DOI] [PubMed] [Google Scholar]
- 86. Broadbent S. E., Davies M. R., van der Woude M. W. (2010) Phase variation controls expression of Salmonella lipopolysaccharide modification genes by a DNA methylation-dependent mechanism. Mol. Microbiol. 77, 337–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Cota I., Blanc-Potard A. B., Casadesús J. (2012) STM2209-STM2208 (opvAB): a phase variation locus of Salmonella enterica involved in control of O-antigen chain length. PLoS ONE 7, e36863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Brunet Y. R., Bernard C. S., Gavioli M., Lloubès R., Cascales E. (2011) An epigenetic switch involving overlapping Fur and DNA methylation optimizes expression of a type VI secretion gene cluster. PLoS Genet. 7, e1002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Fox K. L., Srikhanta Y. N., Jennings M. P. (2007) Phase variable type III restriction-modification systems of host-adapted bacterial pathogens. Mol. Microbiol. 65, 1375–1379 [DOI] [PubMed] [Google Scholar]
- 90. Fox K. L., Dowideit S. J., Erwin A. L., Srikhanta Y. N., Smith A. L., Jennings M. P. (2007) Haemophilus influenzae phasevarions have evolved from type III DNA restriction systems into epigenetic regulators of gene expression. Nucleic Acids Res. 35, 5242–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Srikhanta Y. N., Dowideit S. J., Edwards J. L., Falsetta M. L., Wu H. J., Harrison O. B., Fox K. L., Seib K. L., Maguire T. L., Wang A. H., Maiden M. C., Grimmond S. M., Apicella M. A., Jennings M. P. (2009) Phasevarions mediate random switching of gene expression in pathogenic Neisseria. PLoS Pathog. 5, e1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Srikhanta Y. N., Fox K. L., Jennings M. P. (2010) The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nat. Rev. Microbiol. 8, 196–206 [DOI] [PubMed] [Google Scholar]
- 93. Chessa D., Winter M. G., Jakomin M., Bäumler A. J. (2009) Salmonella enterica serotype Typhimurium Std fimbriae bind terminal α(1,2)fucose residues in the cecal mucosa. Mol. Microbiol. 71, 864–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jakomin M., Chessa D., Bäumler A. J., Casadesús J. (2008) Regulation of the Salmonella enterica std fimbrial operon by DNA adenine methylation, SeqA, and HdfR. J. Bacteriol. 190, 7406–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. López-Garrido J., Casadesús J. (2012) Crosstalk between virulence loci: regulation of Salmonella enterica pathogenicity island 1 (SPI-1) by products of the std fimbrial operon. PLoS ONE 7, e30499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. García-Del Portillo F. (2008) Heterogeneity in tissue culture infection models: a source of novel host-pathogen interactions? Microbes Infect. 10, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 97. Rotem E., Loinger A., Ronin I., Levin-Reisman I., Gabay C., Shoresh N., Biham O., Balaban N. Q. (2010) Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc. Natl. Acad. Sci. U.S.A. 107, 12541–12546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Braaten B. A., Nou X., Kaltenbach L. S., Low D. A. (1994) Methylation patterns in pap regulatory DNA control pyelonephritis-associated pili phase variation in E. coli. Cell 76, 577–588 [DOI] [PubMed] [Google Scholar]