Background: S. cerevisiae ribonucleotide reductase (RNR) comprises α and ββ′ subunits in an (α2)m(ββ′)n active holoenzyme.
Results: C-terminal tail mutants of ββ′ prevent radical transfer across the α-β interface and consequently deoxynucleotide production in vivo.
Conclusion: The ββ′ C-terminal tails interact only with the proximal α within each α/β(α/β′) pair.
Significance: The predisposition of the ββ′ C-terminal tails in active RNR in vivo is established.
Keywords: Iron, Nucleoside Nucleotide Biosynthesis, Radicals, Ribonucleotide Reductase, Tyrosine, Diferric Tyrosyl Cofactor, Long Range Radical Transfer
Abstract
The small subunit (β2) of class Ia ribonucleotide reductase (RNR) houses a diferric tyrosyl cofactor (Fe2III-Y•) that initiates nucleotide reduction in the large subunit (α2) via a long range radical transfer (RT) pathway in the holo-(α2)m(β2)n complex. The C-terminal tails of β2 are predominantly responsible for interaction with α2, with a conserved tyrosine residue in the tail (Tyr356 in Escherichia coli NrdB) proposed to participate in cofactor assembly/maintenance and in RT. In the absence of structure of any holo-RNR, the role of the β tail in cluster assembly/maintenance and its predisposition within the holo-complex have remained unknown. In this study, we have taken advantage of the unusual heterodimeric nature of the Saccharomyces cerevisiae RNR small subunit (ββ′), of which only β contains a cofactor, to address both of these issues. We demonstrate that neither β-Tyr376 nor β′-Tyr323 (Tyr356 equivalent in NrdB) is required for cofactor assembly in vivo, in contrast to the previously proposed mechanism for E. coli cofactor maintenance and assembly in vitro. Furthermore, studies with reconstituted-ββ′ and an in vivo viability assay show that β-Tyr376 is essential for RT, whereas Tyr323 in β′ is not. Although the C-terminal tail of β′ is dispensable for cofactor formation and RT, it is essential for interactions with β and α to form the active holo-RNR. Together the results provide the first evidence of a directed orientation of the β and β′ C-terminal tails relative to α within the holoenzyme consistent with a docking model of the two subunits and argue against RT across the β β′ interface.
Introduction
Ribonucleotide reductase (RNR)2 catalyzes the reduction of nucleoside 5′-diphosphates to the corresponding deoxynucleotides in all organisms (1, 2). The class Ia RNRs are composed of α and β subunits. In Escherichia coli, the active quaternary structure is α2β2 (3, 4), whereas in eukaryotic RNRs including Saccharomyces cerevisiae, the active structure is likely α2β2 and/or (α2)3β2 and (α2)3(β2)3 (5–9). α2 contains the active site where nucleoside 5′-diphosphates are reduced and the allosteric effector binding sites that control which substrate is reduced and the rate of the overall reduction (1, 10). β2 houses a diferric tyrosyl radical cofactor (Fe2III-Y•) assembled from FeII, O2, and a reducing equivalent into the active Fe2III-Y• cofactor (∼1 Y•/β2) that is essential to nucleotide reduction. During each turnover, the Y• in β2 oxidizes a cysteine in the active site of α2 that is 35 Å removed from the β metal center β2 (see Fig. 1), which initiates nucleotide reduction (11).
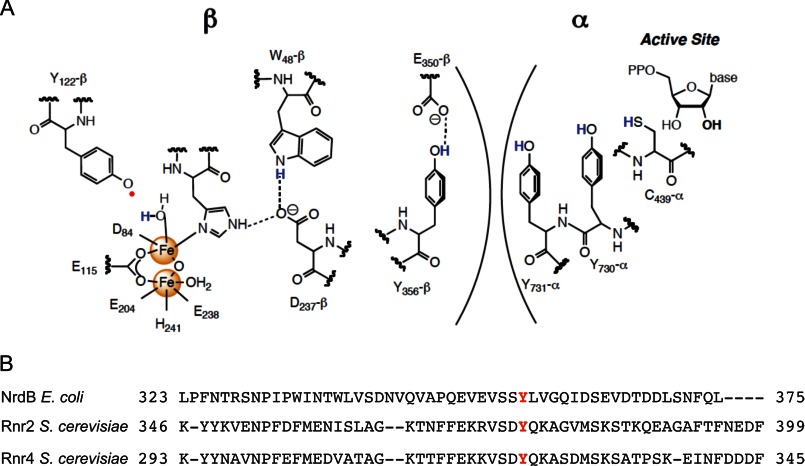
FIGURE 1.
The proposed long range RT pathway involves a conserved tyrosine residue in the C-terminal tail of class Ia RNR small subunit. A, the proposed long range RT pathway between the α2 and β2 subunits (only one α and one β are shown) of the E. coli class Ia RNR. An essential tyrosine in the RT pathway, Tyr356 in E. coli, corresponds to Tyr376 and Tyr323 in β and β′, respectively. B, sequence alignment of the C-terminal tails of the RNR small subunits from E. coli (NrdB) and S. cerevisiae (Rnr2(β) and RNR4(β′)).
Atomic resolution structures of E. coli α2 and of β2 from the Eklund group (12, 13) led them to propose a docking model for the active α2β2 complex, which in conjunction with biochemical studies (14–18) demonstrated the importance of the C-terminal tail of each β (15 and 8 amino acids in the prokaryotic and eukaryotic RNRs, respectively) for the interaction with α. Although their model had the tail from each β associated with the corresponding α, an equally probable model could have the tail of β associated with the adjacent α (see Fig. 2, right panel). Unfortunately, the C termini (30–35 amino acids) of all β2 structures are disordered (19), and thus, the molecular details of the tail with respect to both itself and α remain unknown.
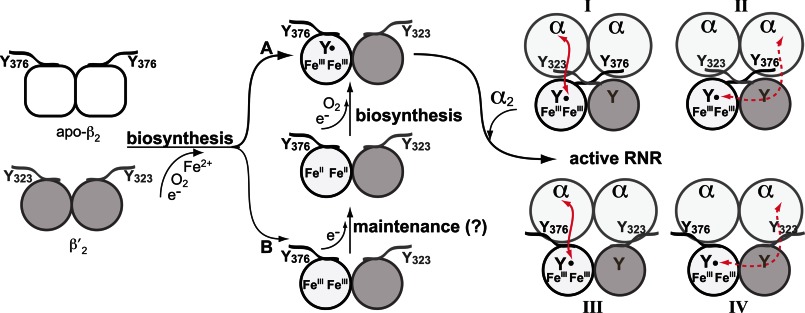
FIGURE 2.
Models depicting the roles of β and β′ C-terminal tails containing the conserved tyrosine residue (β-Tyr376 and β′-Tyr323) in cluster biosynthesis/maintenance and association between ββ′ and α2 to perform RT within the RNR holoenzyme. Biosynthesis of the Fe2III-Y• cofactor in ββ′ involves delivery of two FeII ions into β that in the presence of O2 and a reducing equivalent provide an active FeIIIFeIV species, which oxidizes β-Tyr183 in ββ′ to the Tyr183• (left panel, pathway A). Cofactor can self-assemble in vitro when apo-β2 and apo-β′2 are mixed, which spontaneously form a heterodimer that then binds FeII and self-assembles with O2 and reductant. Substoichiometric Y•/ββ′ levels with stoichiometric Fe2III clusters are obtained both from in vitro self-assembly and from ββ′ isolated from yeast cells, indicating that some ββ′ is in an inactive Fe2III (met) state (left panel, pathway B). Conversion of the Fe2III to active cofactor can occur by a maintenance pathway that requires a reductant to generate the Fe2II state, which can then assemble to form Fe2III-Y• (middle panel). The conserved tyrosine residues at the C-terminal tails, β-Tyr376 and β′-Tyr323, have been proposed to be involved in FeII delivery, in electron transfer required for cofactor biosynthesis and maintenance, and in RT between β and α to generate a thiyl radical in α. The C-terminal tails of β and β′ could cross over to interact with the distant α of the neighboring αβ pair (right panel, I and II) or interact exclusively with the proximal α of the same αβ pair (right panel, III and IV). Red lines indicate the putative RT pathway between α and β, which can occur between proximal (I and III) or distant (II and IV) α β subunits. The latter case requires a RT pathway across the ββ′ interface. Only model III is supported by experimental results in this study.
Within this C-terminal tail resides a conserved tyrosine residue, Tyr356 in E. coli β2, that has been proposed to play an important role in electron transfer in Fe2III-Y• cluster assembly from both the Fe2II and the Fe2III (met) state in β2 (1) and in the long range RT to initiate nucleotide reduction in α2 (see Figs. 1A and Fig. 2, left and center panels). In the former case, efforts to regenerate Y• from the met state of the cofactor using a Y356A mutant gave only low levels of Y• relative to the wild-type (WT) control. Thus, electron transfer required for reducing the Fe2III state was prohibited by this mutation. Deletion of the C-terminal tail also gave poor recovery of Y• in efforts to assemble the cofactor from the Fe2II state (20). In the latter case, recent studies using unnatural tyrosine analogs site-specifically in place of Tyr356 revealed that Tyr356• plays an essential role in Cys439 oxidation in α2 (21) (see Fig. 1A).
The RNR of the budding yeast S. cerevisiae possesses several unique properties that have made it feasible to investigate the functions proposed for the C-terminal tail of β2 and more specifically the conserved tyrosine residue (Tyr356, see Fig. 1B) within the tail. In contrast with the small β2 subunit of most organisms, the yeast small subunit is a heterodimer in vivo, ββ′, encoded by the RNR2 and RNR4 genes, respectively (22–25). β′ is structurally homologous to β but lacks three iron ligands, and as a consequence, contains no metallo-cofactor (18, 22, 25–27). Although β′ is catalytically inactive, it is required for converting β into a conformation that is competent for iron loading and Y• formation in vitro and in vivo (23, 24, 28). Importantly, recombinant apo-β2 and β′2 rapidly form apo-ββ′ in vitro (24), and although cluster assembly from FeII, O2, and reductant is inefficient (0.25 Y•/ββ′), these properties allow us to study reconstitution in vitro with mutant small subunits.
Although β is essential for cell viability, cells lacking β′ (Δrnr4) are viable, albeit with extremely low Y• content and RNR activity in some yeast strain backgrounds (e.g. S288C) (29). These strains are also hypersensitive to the Y• quenching reagent hydroxyurea (28). We have been able to take advantage of this viability and our ability to permeabilize WT and Δrnr4 cells to take up proteins (e.g. α,β′) to provide a robust assay for in vivo Fe2III-Y• cofactor assembly by the addition of β′2 and assay for RNR activity by the addition of β′2 followed by α2 (28) herein.
In this study, we have also used EPR spectroscopy on whole cells and permeabilized cells to show in vivo that the C-terminal 8 amino acids of β′, rich in carboxylates, are not required for binding and delivery of iron to β and that consequently this tail does not function as a molecular chaperone as we originally proposed from in vitro studies (18). We also demonstrate that β-Tyr376 is not on an essential electron transfer pathway to deliver the reducing equivalent to generate the Fe2III-Y• cofactor from the Fe2II or met (Fe2III tyrosyl radical reduced) state either in vitro or in vivo. Moreover, studies using plasmid shuffle strains reveal that β-Tyr376 is nevertheless essential for the long range RT and cell survival, whereas β′-Tyr323 is not essential. Finally, although the β′ mutant lacking the C-terminal 8 amino acids (β′-Δ8aa) is capable of heterodimer formation with β and supporting Fe2III-Y• cofactor assembly in β, the resulting ββ′ is catalytically inactive (< 0.5% WT activity), suggesting an essential role of the β′ tail for interactions with α2 to form an active holo-enzyme. Our results for the first time provide in vivo evidence for the predisposition of the C-terminal tails of β and β′ relative to α in the active RNR; these tails interact only with the α in the α/β (α/β′) pair in the holo-RNR and do not cross over to interact with the adjacent α (see Fig. 2, right panel).
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Media
Yeast strains and plasmids used in this study are listed in Tables 1 and 2, respectively. Rich YPD medium contains 1% yeast extract, 2% peptone, and 2% glucose. Synthetic, defined 6.7 g/liter yeast nitrogen base without amino acids (Difco), 2% glucose, complete supplemental mixtures (CSM), or CSM with dropout of amino acid(s) (MP Biomedicals). Solid media contain 1.5% agar (Difco) as a solidifying agent. YP- and CSM-raffinose media contain 2% raffinose as the sole carbon source, which maintains the GAL1 promoter at residual activity level (uninduced state). YP- and CSM-Raff/Gal media contain 2% raffinose and 0.5% galactose that activate the GAL1 promoter (induced state).
TABLE 1.
Yeast strains used in this study
| Strains | Genotype | Parental Strain |
|---|---|---|
| Y300 | MATa can1–100 ade2–1 his3–11,15 leu2–3,112 trp1-, ura3–1 | Y300 |
| MHY593 | MATa rnr2::KanMX6, pMH881 (URA3CENRNR2) | Y300 |
| MHY20 | MATa rnr4::LEU2, pMH140 (URA3CENRNR4) | Y300 |
| RNR2-WT shuffle | MATa rnr2::KanMX6, pMH881 (URA3CENRNR2) pMH811 (pRS314-PRNR2-3xMyc-RNR2) | Y300 |
| AXY853 | MATa rnr2::KanMX6, pMH881 (URA3CENRNR2) pMH1669 (pRS314-PRNR2-3xMyc-rnr2 (Y376F)) | Y300 |
| RNR4-WT shuffle | MATa rnr4::LEU2, pMH140 (URA3CENRNR4)pMH569 (pRS413-PRNR4-HA-RNR4) | Y300 |
| AXY854 | MATa rnr4::LEU2, pMH140 (URA3CENRNR4) pMH1668 (pRS413-PRNR4-HA-rnr4 (Y323F)) | Y300 |
| BY4741 | MATa his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 | BY4741 |
| GalRNR2 | MATa rnr2::HisMX6-PGAL1-RNR2 | BY4741 |
| AXY1619 | MATa rnr2::HisMX6-PGAL1-RNR2 pRS415 | BY4741 |
| AXY1620 | MATa rnr2::HisMX6-PGAL1-RNR2 pRS415-PRNR2-3xMyc-RNR2 | BY4741 |
| AXY1621 | MATa rnr2::HisMX6-PGAL1-RNR2 pRS415-PRNR2-3xMyc-rnr2 (Y376F) | BY4741 |
TABLE 2.
Plasmids used in this study
| Plasmid | Features | Reference |
|---|---|---|
| pY1A | T7 promoter-driven yeast RNR1 ORF | 31 |
| p(His)6-Y2 | T7 promoter-driven yeast RNR2 ORF, N-terminal His6 tag pET-14b vector | 31 |
| pHis-Y4 | T7 promoter-driven yeast RNR4 ORF, N-terminal His6 tag pET-14b vector | 31 |
| pHis-Y4Δ | T7 promoter-driven yeast RNR4 ORF, C-terminal 8 a.a. truncated, N-terminal His6 tag, pET-14b vector | 18 |
| p(His)6-Y2-Y376F | T7 promoter-driven yeast RNR2 ORF, N-terminal His6 tag pET-14b vector; site-directed mutagenesis Y376F introduced to p(His)6-Y2 | This study |
| pHis-Y4-Y323F | T7 promoter-driven yeast RNR4 ORF, N-terminal His6 tag pET-14b vector; site-directed mutagenesis Y323F introduced to pHis-Y4 | This study |
| pMH813 | pRS415-PRNR2-3xMyc-RNR2, CEN LEU2 | 25 |
| pMH1669 | pRS415-PRNR2-3xMyc-rnr2 (Y376F), CEN LEU2 | This study |
| pMH569 | pRS413-PRNR4-HA-RNR4, CEN HIS3 | 25 |
| pMH1668 | pRS413-PRNR4-HA-rnr4 (Y323F), CEN HIS3 | This study |
Plasmids pMH813 (pRS415-PRNR2-3×Myc-RNR2) and pMH569 (pRS413-PRNR4-HA-RNR4) contain an N-terminal in-frame triple-Myc and HA epitope between the promoter and coding sequences of RNR2 and RNR4, respectively (25). The rnr2-Y376F and rnr4-Y323F harboring plasmids pMH1669 and pMH1668 were constructed by using site-directed mutagenesis on pMH813 and pMH569 and were introduced into the plasmid shuffle strains MHY593 (MATa, rnr2::KanMX6, pMH881 (URA3CENRNR2)) and MHY20 (rnr4::LEU2, pMH140 (URA3CENRNR4)), respectively, as described (25).
The GalRNR2 strain was constructed by replacing the endogenous promoter of RNR2 with the GAL1 promoter as described (28, 30) and maintained in YP-Raff/Gal medium. Plasmids pMH813 (RNR2) and pMH1669 (rnr2-Y376F), as well as the vector control pRS415, were introduced into the GalRNR2 strain, and the transformants were selected and maintained on CSM-Leu Raff/Gal plates, resulting in AXY1619 (GalRNR2 pRS415), AXY1620 (GalRNR2 pRS415-PRNR2-3×Myc-RNR2), and AXY1621(GalRNR2 pRS415-PRNR2-3×Myc-rnr2(Y376F)) strains.
E. coli BL21 (DE3) cells transformed with previously constructed pET-14b vectors p(His)6-RNR2 (β), pHis-RNR4 (β′), and pHis-RNR4Δ8aa (β′Δ8aa) (18) were used to express His6-β, His6-β′, and His6-β′Δ8aa, respectively. Site-directed mutagenesis studies were performed using the QuikChange kit (Stratagene) to introduce Y376F and Y323F mutations into p(His)6-β and p(His)6-β′, respectively, and the changes were confirmed by sequencing. E. coli BL21 CodonPlus (DE3) RIL cells transformed with pY1A (31) were used to express α.
Purification of Recombinant α, His6-β, His6-β′, His6-β′Δ8aa, His6-β-Y376F, and His6-β′-Y323F
E. coli transformants harboring the desired expression vectors were grown in LB medium with antibiotics (ampicillin or kanamycin) at 37 °C to an A600 of 0.4–0.8 before cooling the cultures to 16 °C by adjusting the temperature setting of the incubator. Induction of subunit expression was carried out by the addition of 0.5 mm (for β and β′) or 1 mm (for α) isopropyl β-d-1-thiogalactopyranoside to the medium, and growth was continued for 16 h. The proteins were purified as described previously (18, 24, 31). α was purified using a DEAE Sephadex column and a dATP affinity column (31) and had a specific activity of 102 nmol min−1 mg−1 when assayed in the presence of a 10× molar excess of FLAG-tagged ββ′ (specific activity 3000 nmol min−1 mg−1) that was isolated from yeast Δcrt1 MHY619 cells (29). Wild-type (WT) and mutant β and β′ were both N-terminally His6-tagged with the following N terminus: MGSSHHHHHHSSGLVPRGSH-native protein, and were purified by using an nickel-nitrilotriacetic acid column as described (18). All purified proteins were homogeneous as judged by SDS-PAGE.
Reconstitution of Fe2III-Y• Cofactor in ββ′
Reconstitution of Fe2III-Y• with β2 and β′2 was carried out using previously reported protocols (32). Briefly, purified β2 and β′2 were degassed on a Schlenk line and brought into an anaerobic box (MBraun) in a cold room. The β2 and β′2 were mixed for 2 min before the addition of 3 eq of ferrous iron. The resulting reaction mixture (137 μl) contained 44 μm each of β2 and β′2, 50 mm HEPES at pH 7.6, 5% glycerol and was incubated in the glove box for 10 min. The sample was then removed from the box, and 165 μl of O2-saturated buffer was added and mixed immediately at room temperature, resulting in 20 μm ββ′. The sample (260 μl) was transferred to EPR tubes, and the rest of the sample was frozen immediately and used for subsequent activity assays.
Permeabilized Δrnr4 Yeast Cells Reconstituted with RNR4 and Analyzed by EPR Spectroscopy
A suspension of Δrnr4 cell (60 μl, 32 A600/ml) permeabilized as described previously (28) was added to 200 μl of 0.1 m potassium Pi buffer (pH 7.5) containing 0.6 m sorbitol, 50 mm DTT, and 6.5 μm His6-β′2 or His6-(β′Δ8aa)2. The reaction mixture was incubated at 30 °C for 3 min before being transferred into an EPR tube and rapidly frozen in liquid nitrogen. Our previous study has found that rapid and efficient Fe2III-Y• cofactor assembly (0.5–0.6 Y•/ββ′ in contrast with 0.25–0.3 Y•/ββ′ in vitro) in permeabilized Δrnr4 cells requires high levels (50 mm) of DTT, although its specific role in the process is unclear (28).
EPR spectra were acquired on a Bruker EMX X-band spectrometer at 30 K using an Oxford Instruments liquid helium cryostat. Acquisition parameters for reconstitution in permeabilized cells and whole cell EPR with intact cells at 9.4 GHz were 0.2-milliwatt power, 1.8 × 105 gain, 2-G modulation amplitude, and 100-kHz modulation frequency. Spin quantitation was performed by double integration of the signal using E. coli NrdB as a standard in which the Y• concentration was determined by the dropline correction method (33). Analysis was carried out using WinEPR software (Bruker).
RNR Activity Assays
Activity of ββ′ in Δrnr4 permeabilized cells was measured as described previously (28). Typically, a solution (180 μl) containing 0.1 m potassium Pi, pH 7.5, 0.6 m sorbitol, 3 mm ATP, 10 mm NaF, 50 mm DTT, 4.4 μm α (specific activity 102 nmol min−1 mg−1), and 7.7 μm His6-β′2 or His6-(β′Δ8aa)2 was mixed with 1.5 A600 Δrnr4 permeabilized cells (180 μl) at 4 °C and warmed at 30 °C for 1 min. The reaction was started by the addition of 1 mm [5-3H]CDP (ViTrax, 17 Ci/mmol, 5790 cpm/nmol). Aliquots (58 μl) were removed at 0, 5, and 10 min. The reaction was stopped by placing the sample in a boiling water bath for 2 min. Each sample was then adjusted to pH 8.5 by the addition of 0.5 m of Tris-HCl (final concentration 50 mm) and incubated with 10 units of alkaline phosphatase (Roche Applied Science, from calf intestine) for 2 h, and the amount of dC was analyzed by the method of Steeper and Steuart (34) as revised in Ref. 35.
For activity determination of the reconstituted recombinant ββ′, a typical reaction mixture of 160 μl contained 50 mm HEPES, pH 7.5, 15 mm MgSO4, 1 mm EDTA, 3 mm ATP, 50 mm DTT, 5 μm α2, 1 μm ββ′, and 1 mm [5-3H]CDP (4000–8000 cpm/nmol). Aliquots (50 μl) were removed at 0, 5, and 10 min and analyzed as described above.
Western Blotting for β Protein Levels in Yeast Cell Extracts
Yeast cells (2 × 107) from log-phase cultures were treated with 200 μl of 0.1 m NaOH for 5 min at room temperature (36). The cells were pelleted by a brief centrifugation, resuspended in 200 μl of SDS loading buffer, and lysed by boiling in a sand bath for 5 min. Lysate from 1 × 106 cells (10 μl) was resolved by 10% SDS-PAGE, and the proteins were transferred to nitrocellulose membranes and probed with primary and secondary antibodies. The dilutions for antibodies were: polyclonal anti-β (31) at 1:200,000, monoclonal anti-Myc 9E10 (Covance) at 1:1,000, and goat-anti-mouse and goat-anti-rabbit (Jackson ImmunoResearch Laboratories) at 1:10,000. Blots were developed by using SuperSignal West Femto maximum sensitivity substrate (Thermo Scientific). A CCD camera (ChemiDoc XRS Bio-Rad) was used to record the blotting signals. The proteins were quantified by analyses of the signal intensities with Quantity One (Bio-Rad).
RESULTS
The C-terminal Tail of β′ Is Dispensable for Fe2III-Y• Formation in β
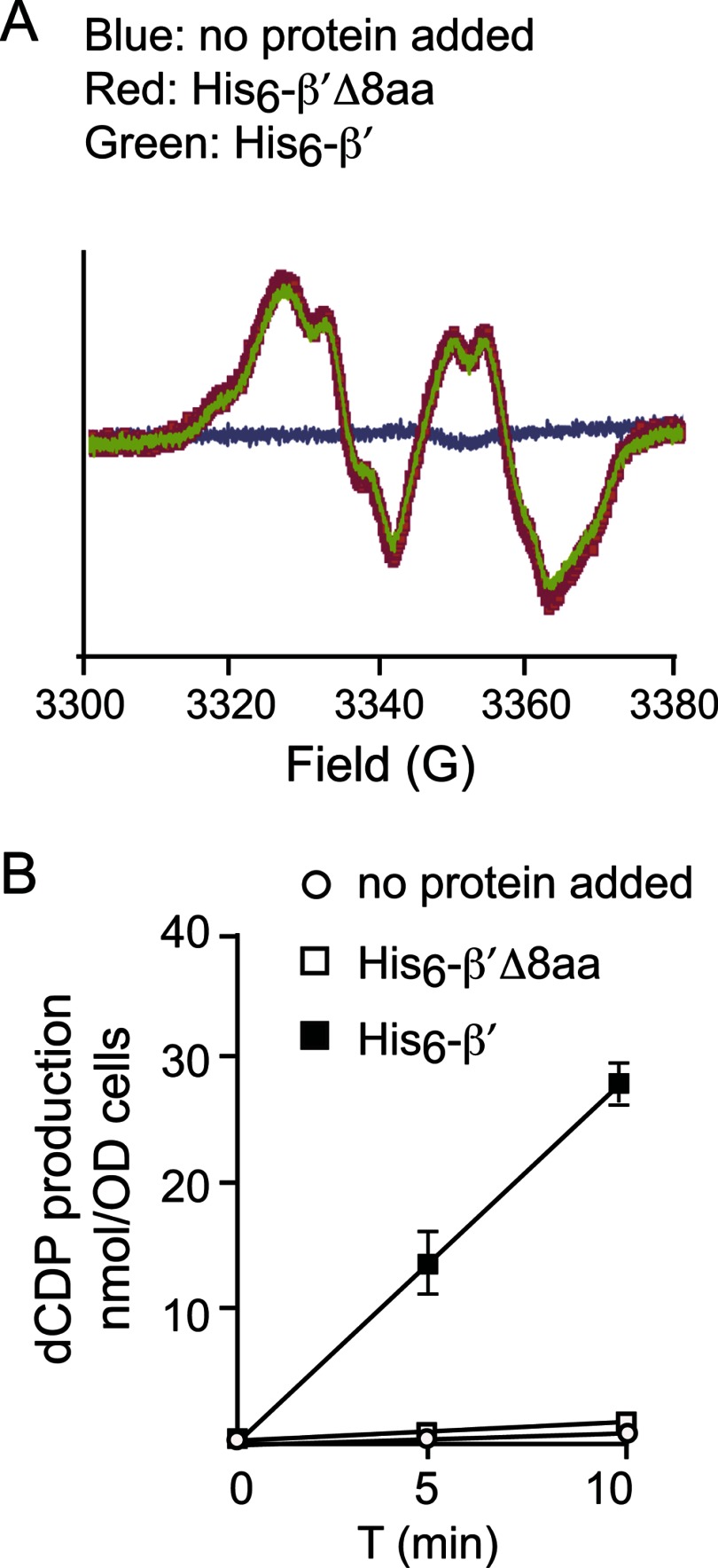
We have previously proposed that β′ might use the aspartate and glutamate residues within its C-terminal tail (Figs. 1B and Fig. 2) to bind and deliver FeII to β in the ββ′ heterodimer in a manner similar to that of copper loading of Sod1 by the chaperone protein Ccs1 (38, 39). To test this hypothesis, we previously constructed the β′-Δ8aa mutant that lacks the last 8 amino acid residues of β′ (18). In vitro cluster assembly experiments using apo ββ′ and ββ′-Δ8aa, Fe2+, and O2 gave the same levels of Y•/ββ′, suggesting that the tail did not play an essential role in this process. However, in contrast to the self-assembly studies in E. coli and mouse β2, the yield of active yeast ββ′ is poor (18). Thus, we have used the same constructs (18) to introduce β′2 into permeabilized Δrnr4 cells that contain very low activity (<1% of WT), high levels of β2, and undetectable Y• determined by whole cell EPR analysis (28). The addition of β′2 to permeabilized Δrnr4 cells resulted in rapid heterodimer formation and Fe2III-Y• formation within 3 min (Fig. 3A, reproduced from supplemental Fig. 2B in Ref. 28). The (β′Δ8aa)2 mutant gave the same amount of Fe2III-Y• as β′2, suggesting that inside the cell, the β′ C-terminal tail is not involved in iron delivery and cofactor formation.
FIGURE 3.
Reconstitution of the Fe2III-Y• cluster and RNR activity in permeabilized Δrnr4 cells by the addition of recombinant β′ and β′Δ8aa. A, EPR analyses of permeabilized Δrnr4 cells in the presence of exogenous His6-β′ (green), His6-β′Δ8aa (red), or no protein addition (blue). B, specific activities of ββ′ were assayed using permeabilized Δrnr4 cells in the presence of exogenous His6-β′ (filled squares) and α, His6-β′Δ8aa (open squares) and α, or without any added protein (open circles).
The C-terminal Tail of β′ Is Essential for RNR Enzyme Activity, Indicating a Role in Interaction with α2
In the experiments described above, the permeabilized Δrnr4 cells, subsequent to treatment with β′2 for 10–20 s, were then incubated with 4.4 μm α2 and 1 mm [3H] CDP to assay for ββ′ activity. Deoxy-CDP was produced at 3 nmol min−1 per A600 cells (Fig. 3B, filled squares). In contrast, with β′2-Δ8aa under otherwise identical conditions, the activity was <0.5% of that of the WT β′2 (Fig. 3B, open squares). Thus, although the C-terminal tail of β′2 is dispensable for iron delivery and subsequent Fe2III-Y• assembly in β, it plays a crucial role in nucleotide reduction within the active holo-enzyme. The results indicate that an active complex is not formed at the concentrations of (β′Δ8aa)2 (∼6.5 μm) examined. Our previous studies have established that endogenous concentrations of β and β′ in WT cells are 0.5–1.0 μm and that concentration of β in Δrnr4 cells is 5–10 μm (24). Thus, the C-terminal 8 amino acids of β′ are critical for association with α2 and formation of holo-enzyme.
Distinct Effects of Mutations of the E. coli NrdB Tyr356 Counterparts in β and β′ on Yeast Viability
Tyr356 (Tyr376 and Tyr323 in β and β′, respectively, in S. cerevisiae, Figs. 1B and Fig. 2) within the C-terminal tail of the small subunit is conserved in all class I RNRs. One proposed function for this residue is its involvement in the delivery of the required reducing equivalent for assembly and maintenance of the Fe2III-Y• cofactor (20, 40). A second proposed function, established in the E. coli Ia RNR, is its involvement in the pathway of RT that is essential for nucleotide reduction (14, 15, 41, 42) (Fig. 1A).
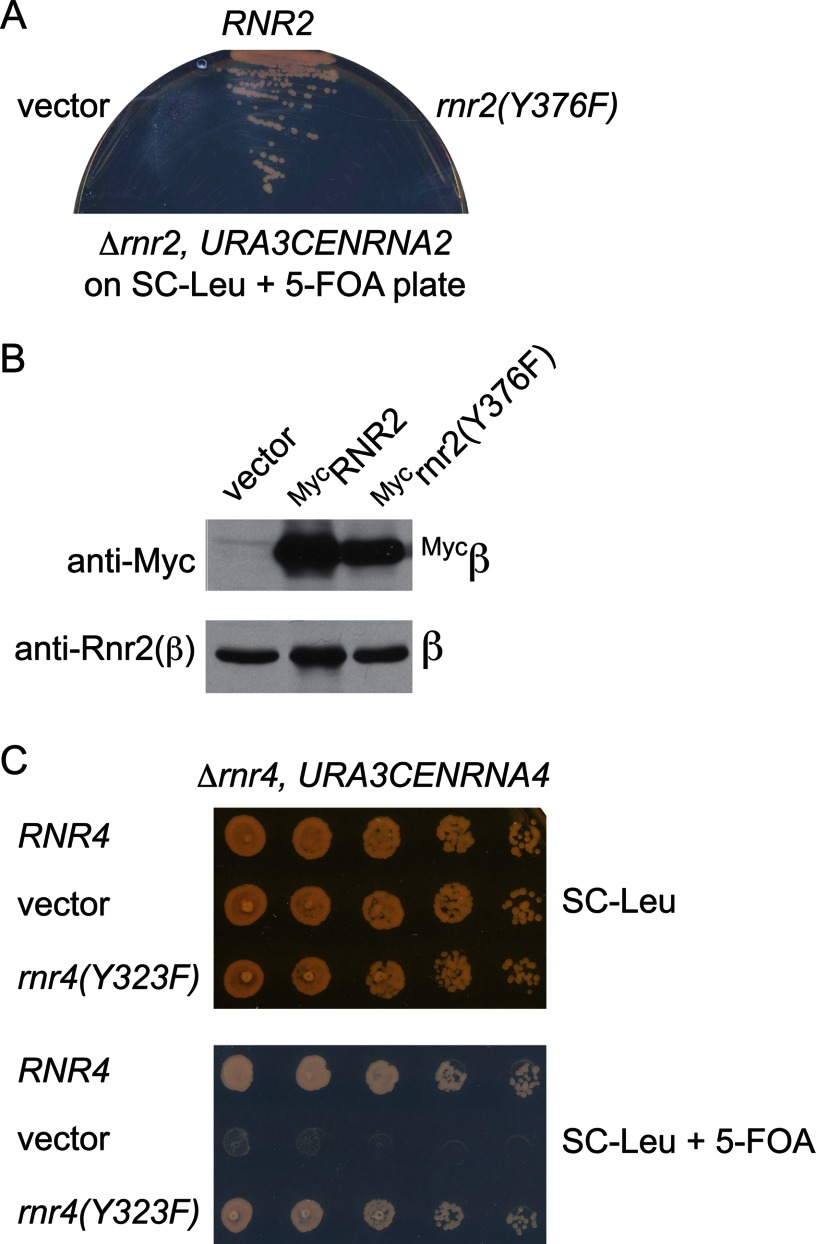
To determine the functional importance of this tyrosine residue in β or β′, we constructed rnr2(Y376F) and rnr4(Y323F) mutant-containing plasmids and transformed them into the RNR2 and RNR4 shuffle strains, respectively. Each shuffle strain contains a chromosomal deletion of the gene being tested and is kept alive by a WT copy of the cognate gene on an URA3-marked plasmid. Survival of the transformants on media containing 5-fluoroorotic acid (5-FOA), a reagent that is converted by the URA3 gene product to the cytotoxic chemical 5-fluorouracil that kills the Ura+ cells, indicates that the mutant-bearing plasmids can provide the essential function in the absence of the WT copy on the URA3 plasmid. The rnr2(Y376F) mutant failed to grow on a 5-FOA-containing plate (Fig. 4A). Western blotting confirmed the expression of the rnr2(Y376F) protein in the shuffle strain transformant prior to 5-FOA selection (Fig. 4B), indicating that the mutant protein was expressed but failed to replace the WT protein function. In contrast, the rnr4(Y323F) mutant was viable and exhibited no obvious difference in growth from the WT control strain on a 5-FOA plate (Fig. 4C). These results are consistent with the proposed essential role of β-Tyr376 either in Fe2III-Y• assembly/maintenance or in the RT pathway, or both, and also indicate that β′-Tyr323 is not required for either event.
FIGURE 4.
Comparison of growth phenotypes of β(Y376F) and β′(Y323F) mutants lacking the conserved tyrosine residue in their C-terminal tails. A, the β-Tyr376 residue is required for mitotic growth. RNR2 shuffle strains (Δrnr2, URA3CENRNR2) harboring a CENLEU2 vector expressing WT-RNR2 or the rnr2(Y376F) mutant were streaked onto a SC-Leu plate containing 5-FOA to eject the URA3 plasmid. Growth on the 5-FOA plate indicates that the copy of RNR2 on the CENLEU2 plasmid can provide the essential RNR2 function for mitotic survival. B, Western blots show comparable levels of the wild-type MycRnr2 and MycRnr2(Y376F) mutant proteins expressed in the RNR2 shuffle strain before 5-FOA selection. C, the β′-Tyr323 is not required for mitotic growth. Serial dilutions (1:10, starting from 1 × 106 cells) of the RNR4 shuffle strain (Δrnr4, URA3CENRNR4) harboring a CENHIS3 vector expressing the wild-type RNR4 or the rnr4(Y323F) mutant were plated on a SC-His plate containing 5-FOA to eject the URA3 plasmid. The plate was photographed after 3 days of incubation at 30 °C.
Measurement of in Vitro Fe2III-Y• Cofactor Formation and Enzyme Activity of the C-terminal Tail Tyrosine Mutants of ββ′
Vegetative growth of yeast cells bearing the β(Y376F) and β′(Y323F) mutants offers a qualitative rather than quantitative measurement of their in vivo activity as the threshold of RNR activity to maintain viability or optimal growth in different yeast strains may vary. For instance, the Δrnr4 mutation is lethal in the W303 background (26) but viable in the S288C background despite very low Y• content (not detectable by whole cell EPR) (28, 29). To gain mechanistic insight into the lethality resulting from the β(Y376F) mutant and to further rule out a role of β′-Tyr323 in RT, we have directly monitored in vitro the Fe2III-Y• cluster assembly and activity of the ββ′ complexes formed from WT β, β′, and the β(Y376F) and β′(Y323F) mutants.
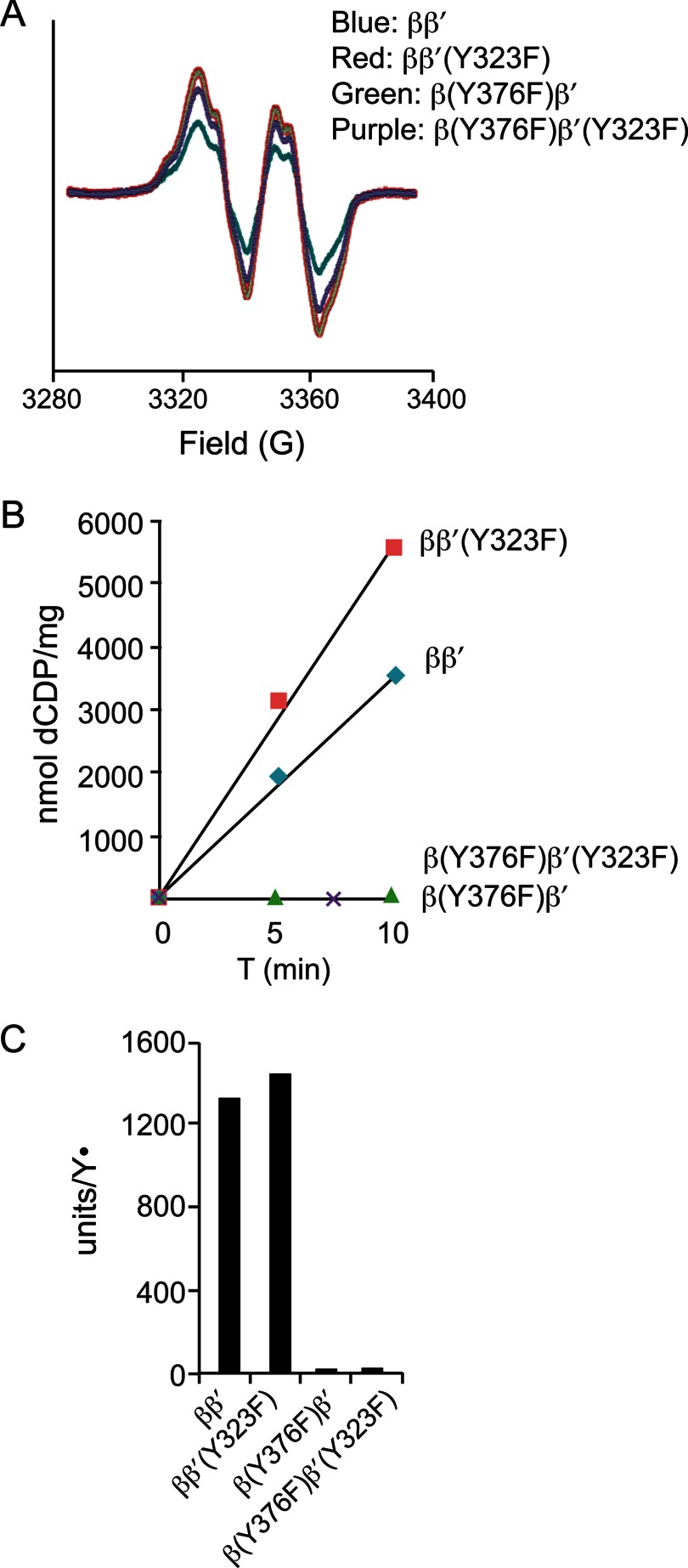
N-terminally His6-tagged β2, β′2, β2(Y376F), and β′2(Y323F) were expressed in E. coli, purified to >95% homogeneity, and subjected to complex formation and cofactor reconstitution in four different combinations: ββ′, ββ′(Y323F), β(Y376F)β′, and β(Y376F)β′(Y323F) as described under “Experimental Procedures.” EPR analyses of the resulting products show Y• formation in all four experiments (Fig. 5A). Spin quantitation gave 0.25 Y• in WT ββ′ and ranged from 0.35 to 0.43 Y• with the mutant combinations. Thus, neither β-Tyr376 nor β′-Tyr323 is required for cofactor assembly in vitro. These reconstituted small subunits were then used to evaluate whether the C-terminal tyrosine residue is directly involved in the RT pathway by enzyme activity assays in the presence of an excess of α2. Both ββ′ and ββ′(Y323F) were capable of converting CDP to dCDP, with specific activities proportional to the Y• contents (Fig. 4, B and C). By contrast, neither β(Y376F)β′ nor β(Y376F)β′(Y323F) exhibited any detectable catalytic activity despite comparable Y• levels in ββ′ and ββ′(Y323F) (Fig. 5, B and 5C). These results demonstrate that β-Tyr376 plays an essential role in the RT pathway, whereas β′-Tyr323 is dispensable in this process.
FIGURE 5.
In vitro reconstitution of the Fe2III-Y• cluster with purified recombinant wild-type and mutant β and β′ proteins. A, EPR analyses of 20 μm reconstituted ββ′ complex from WT β with WT β′ (blue), WT β with β′(Y323F) (red), β(Y376F) with WT β′ (green), and β-Y376F with β′(Y323F) (purple). B, enzyme activity assays of reconstituted WT and mutant ββ′ complexes using radioactive CDP as a substrate in the presence of an excess of α2. C, specific activities of WT and mutant ββ′ complexes normalized by Y• content.
The β-Tyr376 Residue Is Not Required for Fe2III-Y• Cluster Formation in Vivo, Ruling out a Role in Electron Delivery in Cofactor
The β(Y376F) mutant is catalytically inactive and cannot support mitotic growth of yeast cells, although it is capable of forming the Fe2III-Y• cofactor in vitro. The self-assembly of the cofactor in vitro requires Fe2+ and O2 with the reducing equivalent supplied by the Fe2+. In vivo, we have proposed that this reducing equivalent is supplied by YfaE, a 2Fe2S cluster ferredoxin in E. coli (40, 43), and Dre2, an FeS requiring protein in S. cerevisiae (28). We wanted to test the proposal that β-Tyr376 is part of the electron transfer pathway to deliver the reducing equivalent for cluster assembly inside the cell. Because cells harboring β(Y376F) as the only source of β are unviable, we chose to examine a GalRNR2 strain in which β expression from the chromosomal RNR2 locus is controlled by the GAL1 promoter. The GalRNR2 cells propagate normally in galactose-containing media (GAL induced state) but are unable to grow in glucose-containing media (GAL repressed). When the trisaccharide raffinose is used as the sole carbon source for yeast cells, the GAL promoter is neither induced nor repressed, allowing basal levels of transcription (i.e. GAL uninduced or derepressed state). Consistent with low GAL promoter activity, GalRNR2 cells cultured in raffinose media grow slowly with very low levels of endogenous β protein (Fig. 6A, lanes 2–4) and Y• radical content (Fig. 6B, blue).
FIGURE 6.
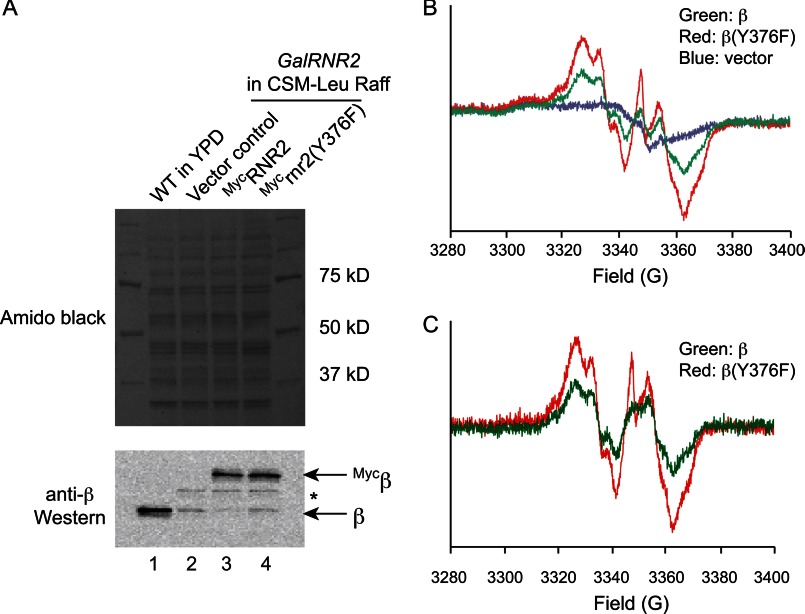
Residue β-Tyr376 is not required for Fe2III-Y• cluster formation in vivo. A, Western blot showing comparable levels of ectopically expressed wild-type Mycβ and Mycβ(Y376F) mutant proteins in the GalRNR2 strain. Cells were inoculated from a CSM-Leu-Raff/Gal plate to 100 ml of CSM-Leu-Raff liquid medium and grown to a density of A600 ∼1 over 16–20 h before being harvested, which depleted the endogenous, untagged β expressed from chromosomal RNR2 locus. 1 × 106 cells were used for Western blotting, and the rest of cells (2 × 109) were used for EPR. Amido Black staining of the nitrocellulose membrane prior to Western blot shows equal loading of cell lysate (upper panel); Western blot using anti-β antibody detects both the untagged and 3×Myc-tagged β proteins (lower panel). Lane 2 contains protein extracts from an untagged wild-type strain showing β expression level from the native RNR2 promoter. The band between Mycβ and β (marked by *) is a cross-reacting protein. B, whole cell EPR spectra of the GalRNR2 cells harboring the empty vector (blue), Mycβ (green), and Mycβ(Y376F) (red) in the GalRNR2 cells grown in a raffinose-containing medium. C, EPR spectra after subtraction of the spectrum of vector control (blue in B) from Mycβ (green) and Mycβ(Y376F) (red).
To study cluster assembly in β-mutants, we then transformed CENLEU2 (low copy number, 1–3 copies/cell) (37) plasmids with the ability to express N-terminally Myc-tagged β WT and β(Y376F) from the native RNR2 promoter into the GalRNR2 cells. GalRNR2 transformants harboring the empty CENLEU2 vector were used as a negative control. All sets of transformants were inoculated from CSM-Leu-Raff/Gal plate (GAL induced) to 100 of ml CSM-Leu-Raff liquid medium (GAL uninduced) and grown to a density of A600 ∼1 over 16–20 h. At this time, the endogenous, untagged β expressed from GalRNR2 was depleted to residual levels (Fig. 6A, lanes 2–4), and the ectopically expressed Myc-β and Myc-β(Y376F) are present at levels that are 7–14-fold higher (Fig. 6A, lanes 3 and 4). These levels are comparable with that of the endogenous β in a WT control strain (Fig. 6A, compare lane 1 with lanes 3 and 4). The moderately (∼60%) higher level of Myc-β(Y376F) is likely due to mild checkpoint activation resulting from the absence of a functional β.
The elevated levels of ectopically expressed Myc-β and Myc-β(Y376F) over the endogenous β under the experimental conditions allowed us to determine and compare Fe2III-Y• cluster content between the two strains using whole cell EPR spectroscopy (Fig. 6B). The Y• was detectable in both the Myc-β-expressing cells (green) and the Myc-β(Y376F)-expressing cells (red) but was not detectable in cells carrying the control vector (blue).
The EPR spectrum from the vector control was used as a background signal and was subtracted from both the Myc-β and the Myc-β(Y376F) samples. The analysis shown in Fig. 5C revealed that Myc-β(Y376F)-expressing cells have a Y• content that is 60% higher than that from the Myc-β-expressing cells. When the Y• signals were normalized for levels of the Myc-β and Myc-β(Y376F) proteins, respectively (Fig. 6A), there was no difference in steady-state Fe2III-Y• cluster content between wild-type β and the β(Y376F) mutant. The results together demonstrate that β-Tyr376 is not required for Fe2III-Y• cluster formation in vivo. It should be noted, however, that the in vivo rates of cluster assembly cannot be analyzed under the experimental conditions.
DISCUSSION
In this study, we provide evidence that the C-terminal tail of β′ does not cross over to the neighboring αβ pair thus providing, insights into the geometry of ββ′ C-terminal tails relative to α2 within the holoenzyme. The unusual heterodimeric structure of yeast small subunit ββ′ with only a single Fe2III-Y• cluster per ββ′ have provided a unique system to investigate the RT pathway and the quaternary structure of the RNR holoenzyme in an active complex inside the cell. Although β′ on its own does not form a Fe2III-Y• cluster (18, 24, 26, 31), it is conceivable that Tyr323 of β′ could participate in the RT process as a stepping stone between the α2 and ββ′ during the turnover cycle (18). In this proposal, the C terminus of β′ would not interact with the α of its own α/β′ pair, but instead would interact with the α of the neighboring α/β pair, which contains the Fe2III-Y• cofactor. Thus, this proposal posits that the C-terminal tail of β′ would cross over at the subunit interface (Fig. 2, right panel, I and II). However, our observation that the β(Y376F) mutant is catalytically inactive and causes cell lethality, whereas the β′(Y323F) mutant has no effect on either yeast vegetative growth or enzyme activity, strongly suggests that the tails of β and β′ do not cross over to interact with the adjacent α within the holo-complex. Instead, each β and β′ tail interacts with the proximal α in the same α/β (α/β′) pair (Fig. 2, right panel, III and IV).
In the studies of Ge et al. (35) in 2003, single turnover experiments using the E. coli α2β2 with one Y•/β2 observed formation of two dCDPs with the same rate constant (35). At the time, two hypotheses were proposed for this unusual observation that a single Y• could service both active sites in α2. One hypothesis was that in addition to the established RT pathway between the proximal α and β (Fig. 2, right panel, I and III), there was a second RT pathway involving both Trp48 and Tyr356 in β with the electron transfer occurring across the β-β monomer interface via the two Trp48 residues to generate a thiyl radical in the second α (Fig. 2, right panel, II and IV). A second hypothesis involved rapid dissociation of β2 from α2 and their subsequent rapid reassociation, both faster than turnover, to allow the second dCDP to be generated quickly in the second α of the α2. Our results with the β(Y376F) and β′(Y323F) mutants suggest that the former hypothesis is unlikely for the yeast RNR. If RT could cross the β-β′ interface and generate thiyl radical in the active site of the adjacent α according to the first model, then the β-Y376F mutant would not have completely inactivated the enzyme as observed. Thus, our studies support the model in which RT occurs only within one α/β pair (Fig. 2, right panel, III).
In vitro reconstitution of the Fe2III-Y• cofactor uses excess ferrous iron to provide reducing equivalent required for its formation (11). However, in vivo, a ferredoxin-like protein YfaE in E. coli (43) and an FeS cluster-containing protein Dre2 (28) in S. cerevisiae have been proposed to provide electrons for the Fe2III-Y• formation and maintenance pathways. Before the identification of YfaE, Coves et al. (20) showed in vitro that a reduced met-β could regenerate the active Fe2III-Y•-containing β and that the C-terminal tail Tyr356 was important for this process (20). The C-terminal Tyr356 was proposed to mediate electron transfer for the maintenance of the cofactor and potentially for cofactor assembly. Our whole cell EPR results with the β(Y376F) mutant in the GalRNR2 cells at uninduced state clearly demonstrate for the first time that β-Tyr376 is not required for Fe2III-Y• formation in vivo, and thus, its essential function is likely due to its role in the RT pathway.
This work was supported, in whole or in part, by National Institutes of Health Grants GM29595 (to J. S.), CA125574 (to M. H.), and GM81393 (to J. S. and M. H.).
This article was selected as a Paper of the Week.
- RNR
- ribonucleotide reductase
- α2
- ribonucleotide reductase large subunit
- β2
- ββ′ ribonucleotide reductase small subunit homodimer and heterodimer
- Fe2III-Y•
- diferric tyrosyl radical
- RT
- radical transfer
- CSM
- complete supplemental mixture(s)
- 5-FOA
- 5-fluoroorotic acid
- aa
- amino acids.
REFERENCES
- 1. Nordlund P., Reichard P. (2006) Ribonucleotide reductases. Annu. Rev. Biochem. 75, 681–706 [DOI] [PubMed] [Google Scholar]
- 2. van der Donk W. A., Yu G., Pérez L., Sanchez R. J., Stubbe J., Samano V., Robins M. J. (1998) Detection of a new substrate-derived radical during inactivation of ribonucleotide reductase from Escherichia coli by gemcitabine 5′-diphosphate. Biochemistry 37, 6419–6426 [DOI] [PubMed] [Google Scholar]
- 3. Ando N., Brignole E. J., Zimanyi C. M., Funk M. A., Yokoyama K., Asturias F. J., Stubbe J., Drennan C. L. (2011) Structural interconversions modulate activity of Escherichia coli ribonucleotide reductase. Proc. Natl. Acad. Sci. U.S.A. 108, 21046–21051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Minnihan E. C., Ando N., Brignole E. J., Olshansky L., Chittuluru J., Asturias F. J., Drennan C. L., Nocera D. G., Stubbe J. (2013) Generation of a stable, aminotyrosyl radical-induced α2β2 complex of Escherichia coli class Ia ribonucleotide reductase. Proc. Natl. Acad. Sci. U.S.A. 110, 3835–3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rofougaran R., Vodnala M., Hofer A. (2006) Enzymatically active mammalian ribonucleotide reductase exists primarily as an α6β2 octamer. J. Biol. Chem. 281, 27705–27711 [DOI] [PubMed] [Google Scholar]
- 6. Wang J., Lohman G. J., Stubbe J. (2009) Mechanism of inactivation of human ribonucleotide reductase with p53R2 by gemcitabine 5′-diphosphate. Biochemistry 48, 11612–11621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rofougaran R., Crona M., Vodnala M., Sjöberg B. M., Hofer A. (2008) Oligomerization status directs overall activity regulation of the Escherichia coli class Ia ribonucleotide reductase. J. Biol. Chem. 283, 35310–35318 [DOI] [PubMed] [Google Scholar]
- 8. Fairman J. W., Wijerathna S. R., Ahmad M. F., Xu H., Nakano R., Jha S., Prendergast J., Welin R. M., Flodin S., Roos A., Nordlund P., Li Z., Walz T., Dealwis C. G. (2011) Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat. Struct. Mol. Biol. 18, 316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kashlan O. B., Cooperman B. S. (2003) Comprehensive model for allosteric regulation of mammalian ribonucleotide reductase: refinements and consequences. Biochemistry 42, 1696–1706 [DOI] [PubMed] [Google Scholar]
- 10. Hofer A., Crona M., Logan D. T., Sjöberg B. M. (2012) DNA building blocks: keeping control of manufacture. Crit. Rev. Biochem. Mol. Biol. 47, 50–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cotruvo J. A., Stubbe J. (2011) Class I ribonucleotide reductases: metallocofactor assembly and repair in vitro and in vivo. Annu. Rev. Biochem. 80, 733–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Uhlin U., Eklund H. (1994) Structure of ribonucleotide reductase protein R1. Nature 370, 533–539 [DOI] [PubMed] [Google Scholar]
- 13. Nordlund P., Sjöberg B. M., Eklund H. (1990) Three-dimensional structure of the free radical protein of ribonucleotide reductase. Nature 345, 593–598 [DOI] [PubMed] [Google Scholar]
- 14. Climent I., Sjöberg B. M., Huang C. Y. (1991) Carboxyl-terminal peptides as probes for Escherichia coli ribonucleotide reductase subunit interaction: kinetic analysis of inhibition studies. Biochemistry 30, 5164–5171 [DOI] [PubMed] [Google Scholar]
- 15. Climent I., Sjöberg B. M., Huang C. Y. (1992) Site-directed mutagenesis and deletion of the carboxyl terminus of Escherichia coli ribonucleotide reductase protein R2. Effects on catalytic activity and subunit interaction. Biochemistry 31, 4801–4807 [DOI] [PubMed] [Google Scholar]
- 16. Cohen E. A., Gaudreau P., Brazeau P., Langelier Y. (1986) Specific inhibition of herpesvirus ribonucleotide reductase by a nonapeptide derived from the carboxy terminus of subunit 2. Nature 321, 441–443 [DOI] [PubMed] [Google Scholar]
- 17. Dutia B. M., Frame M. C., Subak-Sharpe J. H., Clark W. N., Marsden H. S. (1986) Specific inhibition of herpesvirus ribonucleotide reductase by synthetic peptides. Nature 321, 439–441 [DOI] [PubMed] [Google Scholar]
- 18. Ge J., Perlstein D. L., Nguyen H. H., Bar G., Griffin R. G., Stubbe J. (2001) Why multiple small subunits (Y2 and Y4) for yeast ribonucleotide reductase? Toward understanding the role of Y4. Proc. Natl. Acad. Sci. U.S.A. 98, 10067–10072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eklund H., Uhlin U., Färnegårdh M., Logan D. T., Nordlund P. (2001) Structure and function of the radical enzyme ribonucleotide reductase. Prog. Biophys. Mol. Biol. 77, 177–268 [DOI] [PubMed] [Google Scholar]
- 20. Covès J., Delon B., Climent I., Sjöberg B. M., Fontecave M. (1995) Enzymic and chemical reduction of the iron center of the Escherichia coli ribonucleotide reductase protein R2. The role of the C-terminus. Eur. J. Biochem. 233, 357–363 [DOI] [PubMed] [Google Scholar]
- 21. Minnihan C. C., Nocera D. G., Stubbe J. (2013) Reversible long-range radical transfer in class Ia ribonucleotide reductase. Acc. Chem. Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chabes A., Domkin V., Larsson G., Liu A., Gräslund A., Wijmenga S., Thelander L. (2000) Yeast ribonucleotide reductase has a heterodimeric iron-radical-containing subunit. Proc. Natl. Acad. Sci. U.S.A. 97, 2474–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sommerhalter M., Voegtli W. C., Perlstein D. L., Ge J., Stubbe J., Rosenzweig A. C. (2004) Structures of the yeast ribonucleotide reductase Rnr2 and Rnr4 homodimers. Biochemistry 43, 7736–7742 [DOI] [PubMed] [Google Scholar]
- 24. Perlstein D. L., Ge J., Ortigosa A. D., Robblee J. H., Zhang Z., Huang M., Stubbe J. (2005) The active form of the Saccharomyces cerevisiae ribonucleotide reductase small subunit is a heterodimer in vitro and in vivo. Biochemistry 44, 15366–15377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. An X., Zhang Z., Yang K., Huang M. (2006) Cotransport of the heterodimeric small subunit of the Saccharomyces cerevisiae ribonucleotide reductase between the nucleus and the cytoplasm. Genetics 173, 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang M., Elledge S. J. (1997) Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol. Cell. Biol. 17, 6105–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang P. J., Chabes A., Casagrande R., Tian X. C., Thelander L., Huffaker T. C. (1997) Rnr4p, a novel ribonucleotide reductase small-subunit protein. Mol. Cell. Biol. 17, 6114–6121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y., Liu L., Wu X., An X., Stubbe J., Huang M. (2011) Investigation of in vivo diferric tyrosyl radical formation in Saccharomyces cerevisiae Rnr2 protein: requirement of Rnr4 and contribution of Grx3/4 AND Dre2 proteins. J. Biol. Chem. 286, 41499–41509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ortigosa A. D., Hristova D., Perlstein D. L., Zhang Z., Huang M., Stubbe J. (2006) Determination of the in vivo stoichiometry of tyrosyl radical per ββ′ in Saccharomyces cerevisiae ribonucleotide reductase. Biochemistry 45, 12282–12294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 31. Nguyen H. H., Ge J., Perlstein D. L., Stubbe J. (1999) Purification of ribonucleotide reductase subunits Y1, Y2, Y3, and Y4 from yeast: Y4 plays a key role in diiron cluster assembly. Proc. Natl. Acad. Sci. U.S.A. 96, 12339–12344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Atkin C. L., Thelander L., Reichard P., Lang G. (1973) Iron and free radical in ribonucleotide reductase. Exchange of iron and Mossbauer spectroscopy of the protein B2 subunit of the Escherichia coli enzyme. J. Biol. Chem. 248, 7464–7472 [PubMed] [Google Scholar]
- 33. Bollinger J. M., Jr., Tong W. H., Ravi N., Huynh B. H., Edmondson D. E., Stubbe J. (1995) Use of rapid kinetics methods to study the assembly of the diferric-tyrosyl radical cofactor of Escherichia coli ribonucleotide reductase. Methods Enzymol. 258, 278–303 [DOI] [PubMed] [Google Scholar]
- 34. Steeper J. R., Steuart C. D. (1970) A rapid assay for CDP reductase activity in mammalian cell extracts. Anal. Biochem. 34, 123–130 [DOI] [PubMed] [Google Scholar]
- 35. Ge J., Yu G., Ator M. A., Stubbe J. (2003) Pre-steady-state and steady-state kinetic analysis of E. coli class I ribonucleotide reductase. Biochemistry 42, 10071–10083 [DOI] [PubMed] [Google Scholar]
- 36. Kushnirov V. V. (2000) Rapid and reliable protein extraction from yeast. Yeast 16, 857–860 [DOI] [PubMed] [Google Scholar]
- 37. Koshland D., Kent J. C., Hartwell L. H. (1985) Genetic analysis of the mitotic transmission of minichromosomes. Cell. 40, 393–403 [DOI] [PubMed] [Google Scholar]
- 38. Culotta V. C., Klomp L. W., Strain J., Casareno R. L., Krems B., Gitlin J. D. (1997) The copper chaperone for superoxide dismutase. J. Biol. Chem. 272, 23469–23472 [DOI] [PubMed] [Google Scholar]
- 39. Lamb A. L., Torres A. S., O'Halloran T. V., Rosenzweig A. C. (2001) Heterodimeric structure of superoxide dismutase in complex with its metallochaperone. Nat. Struct. Biol. 8, 751–755 [DOI] [PubMed] [Google Scholar]
- 40. Hristova D., Wu C. H., Jiang W., Krebs C., Stubbe J. (2008) Importance of the maintenance pathway in the regulation of the activity of Escherichia coli ribonucleotide reductase. Biochemistry 47, 3989–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yee C. S., Chang M. C., Ge J., Nocera D. G., Stubbe J. (2003) 2,3-difluorotyrosine at position 356 of ribonucleotide reductase R2: a probe of long-range proton-coupled electron transfer. J. Am. Chem. Soc. 125, 10506–10507 [DOI] [PubMed] [Google Scholar]
- 42. Yee C. S., Seyedsayamdost M. R., Chang M. C., Nocera D. G., Stubbe J. (2003) Generation of the R2 subunit of ribonucleotide reductase by intein chemistry: insertion of 3-nitrotyrosine at residue 356 as a probe of the radical initiation process. Biochemistry 42, 14541–14552 [DOI] [PubMed] [Google Scholar]
- 43. Wu C.-H., Jiang W., Krebs C., Stubbe J. (2007) YfaE, a ferredoxin involved in diferric-tyrosyl radical maintenance in Escherichia coli ribonucleotide reductase. Biochemistry 46, 11577–11588 [DOI] [PubMed] [Google Scholar]