Background: Intrinsically disordered neuropeptide hormones are synthesized as larger precursors with an α-helical prodomain.
Results: The prodomain promotes productive import of the hormone domain into the endoplasmic reticulum (ER) by its propensity to adopt an α-helical structure.
Conclusion: Impaired ER import of intrinsically disordered proteins can be restored by α-helical prodomains.
Significance: Secondary structure of the nascent chain can regulate translocation into the ER.
Keywords: Endoplasmic Reticulum (ER), Intrinsically Disordered Proteins, Neuropeptide, Neuroscience, Protein Folding
Abstract
Different neuropeptide hormones, which are either too small to adopt a stable conformation or are predicted to be intrinsically disordered, are synthesized as larger precursors containing a prodomain in addition to an N-terminal signal peptide. We analyzed the biogenesis of three unstructured neuropeptide hormones and observed that translocation of these precursors into the lumen of the endoplasmic reticulum (ER) is critically dependent on the presence of the prodomain. The hormone domains could be deleted from the precursors without interfering with ER import and secretion, whereas constructs lacking the prodomain remained in the cytosol. Domain-swapping experiments revealed that the activity of the prodomains to promote productive ER import resides in their ability to adopt an α-helical structure. Removal of the prodomain from the precursor did not interfere with co-translational targeting of the nascent chain to the Sec61 translocon but with its subsequent productive translocation into the ER lumen. Our study reveals a novel function of prodomains to enable import of small or intrinsically disordered secretory proteins into the ER based on their ability to adopt an α-helical conformation.
Introduction
A major fraction of secretory proteins is targeted to the ER4 via N-terminal signal peptides, which mediate transport of the nascent chain to the Sec61 translocon and initiate translocation into the ER lumen (for reviews, see Refs. 1–8). Non-secretory proteins can be targeted to the ER by fusing ER signal peptides to their N termini, revealing that the sorting information is encoded in the signal peptide, whereas the remaining part of the protein seems not to significantly regulate this pathway. Interestingly, an exceptionally diverse set of signal peptide sequences can target proteins for ER import. The physiological relevance of this diversity is not fully understood; however, it has been demonstrated that the translocation efficiency can be modulated in a signal peptide sequence-specific manner (for reviews, see Refs. 9–12). Context-specific regulation of protein translocation might enable the cell to prevent an overload of protein folding and quality control pathways in the ER lumen (13–15). In addition, it might be implicated in dual targeting of secretory proteins to other cellular compartments to expand their physiological function (16, 17).
In previous work using pathogenic prion protein mutants and model substrates, we identified an additional element to regulate translocation efficiency: the secondary structure of the nascent chain (18, 19). Specifically, we found that intrinsically disordered proteins (IDPs) equipped with an N-terminal ER signal peptide are not efficiently translocated into the ER. ER import of these proteins could be restored by fusing α-helical domains to the IDPs (19). In this context, it is important to note that the nascent chain can adopt a stable secondary structure already in the ribosomal exit tunnel (20–25). Furthermore, it has been shown previously that polypeptide structure within the ribosomal exit tunnel can modulate translocation of distal parts of the nascent chain (26).
The absence of a stable secondary structure does not necessarily imply a non-functional state of a protein. Instead, it became evident that IDPs and intrinsically disordered domains (IDDs) exert defined physiological activities and play a major role in several protein classes, including transcription factors, scaffold proteins, and signaling molecules (for reviews, see Refs. 27–32). In this study, we analyzed the biogenesis of three neuropeptide hormones that are predicted to be intrinsically disordered. Interestingly, these neuropeptide hormones are synthesized as larger precursors containing a prodomain in addition to an N-terminal ER signal peptide. Our analysis revealed that the prodomains have a vital function in hormone biogenesis by promoting import of the precursor into the ER lumen. This activity is linked to the propensity of the prodomain to adopt an α-helical structure.
EXPERIMENTAL PROCEDURES
Plasmid Constructions
The full-length cDNA clones of human somatostatin (Som) (GenBankTM accession number BC032625), human thyrotropin-releasing hormone (TRH) (GenBank accession number BC074889), and human gonadotropin-releasing hormone (GnRH) (GenBank accession number BC126463) were ordered from Imagenes. All mutants used in this study were generated by standard PCR cloning techniques. GnRHΔpro and GnRHΔhor were created by deletion of amino acids (aa) 37–92 or aa 24–33, respectively. Somatostatin lacking the prodomain was generated by deletion of aa 25–88. In the case of somatostatin lacking the hormone domain, aa 89–116 were deleted. SomΔpro→helix-syn was created by an exchange of aa 25–88 for a synthetic peptide consisting of 10 EAAAK repeats that form a standard α-helix (25). In the case of SomΔpro-helix-PrP and SomΔpro-IDD-Sho, aa 130–230 of the mouse prion protein (PrP; NCBI Reference Sequence NP_035300.1) or aa 25–124 of human shadoo (Sho; NCBI Reference Sequence NP_001012526.2) were fused to SomΔpro. TRHΔpro and TRHΔhor were generated by deletion of aa 26–62 or deletion of aa 63–99, respectively. In the case of TRHΔpro→proSom and TRHΔpro→helix-PrP, aa 26–62 were replaced by the prodomain (aa 25–88) of somatostatin or aa 173–219 of mouse PrP, respectively. For TRHΔpro→helix-PrP, amino acid Asn at position 196 of the PrP domain was replaced by Gln to delete the second glycosylation site. TRHΔpro→IDD-PrP was created by substituting aa 26–62 of TRH with the defined unstructured domain of mouse PrP (aa 23–115). In this construct, the amino acids Trp and Asn at positions 31/32 were replaced by Asn and Phe to generate a glycosylation acceptor site (NFT). GnRHΔpro→helix-PrP aa 37–92 of GnRH were replaced by aa 173–219 of mouse PrP. GnRHΔpro→IDD-Sho was generated by an exchange of aa 37–92 of GnRH for aa 69–124 of human Sho. ΔSP mutants were generated by deleting the ER signal peptide except for the first methionine of the respective protein. For detection, all constructs were equipped with a V5 tag (5′-ggt aaa ccg ata ccg aac cct ctg ctc ggt ctg gat agc acg-3′) at the C-terminal end. All constructs were inserted into the pcDNA3.1/Zeo(+) vector for mammalian expression.
Cell Culture, Transfection, and Secretion Analysis
Mouse N2a (ATCC number Ccl 131) cells were cultivated in modified Eagle's medium (Invitrogen) containing 10% fetal calf serum, 100 units/ml penicillin, 100 μg/ml streptomycin (PAA Laboratories) and 2 mm l-glutamine (PAA Laboratories). Cells were transfected 24 h after plating by a liposome-mediated method using Lipofectamine with PLUS Reagent (Invitrogen) according to the manufacturer's instructions. To examine secretion of proteins into the cell culture supernatant, cells were cultivated in cell culture medium without supplements for 3 h at 37 °C. The medium was collected, and proteins were precipitated with TCA and analyzed by Western blotting.
Cell Lysis and Western Blot Analysis
Cells were rinsed twice with ice-cold PBS, scraped off the plate, pelleted by centrifugation, and lysed in cold detergent buffer (0.5% Triton X-100, 0.5% sodium deoxycholate in PBS) supplemented with proteinase inhibitor (1:500; Roche Applied Science). Lysates were incubated on ice for 20 min before addition of Laemmli sample buffer with β-mercaptoethanol (140 mm). Protein concentrations in the cell lysates were determined with a Bio-Rad protein assay. Following SDS-PAGE, proteins were transferred to nitrocellulose, and membranes were blocked by incubation in PBS-T (PBS with 0.1% Tween 20) containing 5% dry milk for 1 h at room temperature and incubated with mouse monoclonal anti-V5 antibody (Invitrogen) in PBS-T overnight at 4 °C. After washing with PBS-T, blots were incubated with horseradish peroxidase-conjugated anti-IgG antibody in PBS-T for 1 h at room temperature and finally washed with PBS-T. Protein signals were visualized using Amersham Biosciences ECL Western blotting detection reagent (GE Healthcare) and Fuji film (Super RX). Signals were quantified using Image Quant TL software.
Metabolic Labeling and Immunoprecipitation
Transfected cells were starved for 30 min in methionine-free modified Eagle's medium (Invitrogen) and subsequently pulse-labeled for 15 min with 300 μCi/ml l-[35S]methionine (Hartmann Analytics; >37 TBq/mmol) in methionine-free modified Eagle's medium. After labeling, the cells were washed twice and either harvested immediately (pulse) or after a further incubation in complete medium for 1 h (chase). Cells were harvested and lysed as described under “Cell Lysis and Western Blot Analysis,” and transfected constructs in the lysate or medium fraction were immunoprecipitated using the mouse monoclonal anti-V5 antibody (Invitrogen). Immunopellets were analyzed by SDS-PAGE and autoradiography.
Cell-free Translation and Proteinase K Protection Assay
In vitro transcription and translation was performed using the TnT T7 Quick Coupled Transcription/Translation System (Promega), 0.2 μg of plasmid DNA as a template, and l-[35S]methionine (Hartmann Analytics; >37 TBq/mmol) according to the manufacturer's instructions in the presence or absence of canine pancreatic microsomal membranes (Promega). Following translation, samples were centrifuged for 10 min at 18,000 × g. Pellet fractions were resuspended in microsomal resuspension buffer (0.1 m KOAc, 50 mm Hepes, 5 mm MgOAc, and 0.25 m sucrose) and analyzed by SDS-PAGE and autoradiography. If indicated, proteinase K (0.5 mg/ml; Roche Applied Science) was added to the microsomal resuspension buffer, and samples were incubated for 30 min on ice. Proteinase K digestion was stopped by the addition of 3 mm PMSF (Sigma). All samples were analyzed by SDS-PAGE and autoradiography. Signals were quantified using a phosphorimaging system (Typhoon) and the quantification software Image Quant TL.
In Vitro Targeting Assay
Truncated constructs lacking a stop codon were generated by PCR with full-length or mutant cDNA as template and extended primers. The 5′-primer added a T7 site followed by a high initiation efficiency 5′-untranslated region and an optimized AUG context (5′-taa tac gac tca cta tag gg acc aaa caa aac aaa taa aac aaa gcc acc atg-3′) as described previously (33). The 3′-primer determined the length of the particular nascent chain. In vitro transcription was performed using the AmpliScribe T7 High Yield Transcription kit. After transcription, mRNA was purified using the RNeasy Mini kit (Qiagen). In vitro translation and targeting of truncated mRNA were carried out using a rabbit reticulocyte lysate system, canine pancreatic microsomal membranes (RM; Promega), and l-[35S]methionine (Hartmann Analytics; >37 TBq/mmol) as described (34). For each 25-μl translation reaction, 1 μg of RNA was added. Truncated constructs were translated for 30 min at 32 °C (TRH) or 20 min at 26 °C (Som). For targeting analysis, translation reactions containing RM were centrifuged for 3 min at 46,000 rpm and 4 °C in a TLA55 rotor over a 50-μl sucrose cushion of 0.5 m sucrose, 0.1 mm KOAc, 50 mm Hepes, pH 7.4, and 5 mm MgOAc. Sedimented RM were washed by adding 50 μl of sucrose cushion and repeating the centrifugation step. Targeted nascent chains were resuspended in microsomal resuspension buffer and analyzed by SDS-PAGE and autoradiography. For all experiments, translation reactions without RM were used as internal sedimentation controls and treated equally as samples containing RM. As quantification controls, polysomes were sedimented for 30 min at 46,000 rpm and 4 °C over a 100-μl sucrose cushion (total). Signals were visualized by autoradiography. For quantification, signals were visualized using a phosphorimaging system (Typhoon) and quantified using the quantification software Image Quant TL.
Recombinant Protein Expression, Purification, and Circular Dichroism (CD) Analysis
cDNA encoding amino acid residues 25–88 of the human somatostatin protein were inserted into a pTXB1 vector (New England Biolabs) modified with a His6 tag introduced into the AgeI site and expressed in Escherichia coli Bl21(DE3) cells (Stratagene) by induction with 1 mm isopropyl 1-thio-β-d-galactopyranoside at an A600 of 0.6 for 3 h. Following induction, cells were lysed by sonification in lysis buffer (50 mm NaH2PO4, 300 mm NaCl, 10 mm imidazole, lysozyme (1 mg/ml), and 5 μg/ml DNase, pH 8.0) supplemented with proteinase inhibitor (1:500; Roche Applied Science). The lysate was cleared by centrifugation, subjected to Ni2+ affinity beads (Qiagen), and agitated overnight at 4 °C. Washing and elution steps were performed on a column according to the manufacturer's instructions. The eluted protein fraction was equilibrated with cleavage buffer (20 mm Tris/HCl, pH 8.0, 50 mm NaCl, and 0.1 mm EDTA) using a Vivaspin 6 Centricon (Amicon; 5-kDa cutoff). The intein tag cleavage reaction was performed overnight at room temperature by addition of 30 mm DTT. The cleaved protein fraction was precipitated with ammonium sulfate for 1 h on ice, sedimented by centrifugation in a Ti50 rotor for 30 min at 31,100 rpm and 4 °C, and resuspended in NPI buffer (50 mm NaH2PO4 and 300 mm NaCl, pH 8.0) supplemented with 5% glycerol. In the next step, the protein sample was again subjected to Ni2+ affinity chromatography to clear the cleaved prodomain from the remaining intein tag. The binding step of protein to nickel-nitrilotriacetic acid (Ni-NTA) beads was conducted for 20 min at 4 °C. The flow-through was collected, concentrated, and equilibrated in NPI buffer without glycerol. The sample was again precipitated with ammonium sulfate for 1 h on ice. After centrifugation in a Ti50 rotor for 30 min at 31,100 rpm and 4 °C the pellet was resuspended in CD buffer (20 mm NaPO4, pH 7.2 and 100 mm KCl) containing 5% glycerol. Protein fractions were analyzed by SDS-PAGE and Coomassie Blue staining. The purified protein band was excised from the gel and verified by mass spectrometry using Sequest. Far-UV circular dichroism spectra of the recombinant somatostatin prodomain (125 μg/ml) in CD buffer containing 5% glycerol were recorded in 50% trifluoroethanol as solvent that approximates the hydrophobic surroundings of the protein interior of the isolated peptide (35). Data were collected at 26 °C using a Jasco J-810 spectropolarimeter. The spectrum was obtained using a 2-mm path length, and data were collected at 1-nm intervals from 260 to 190 nm.
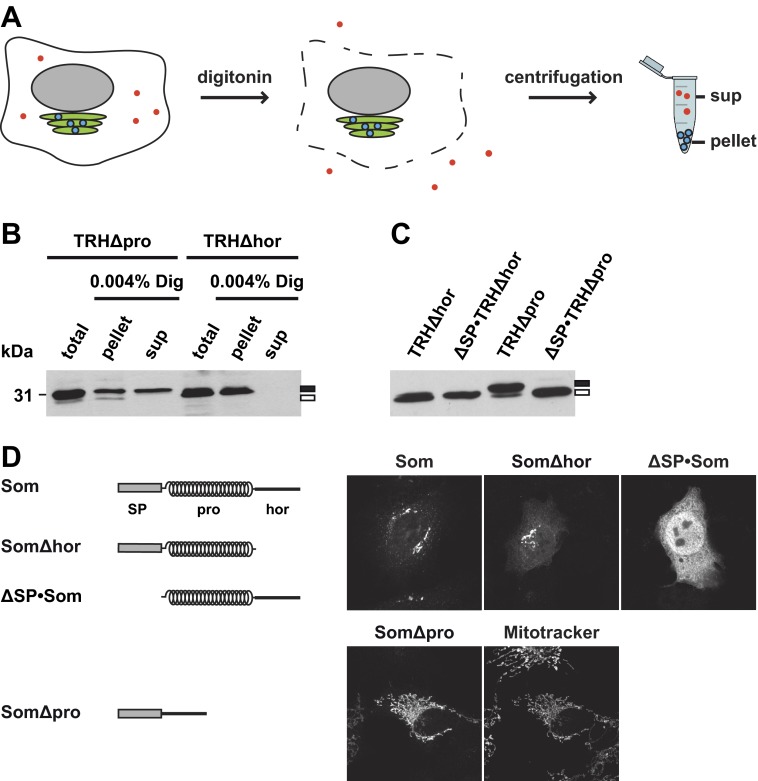
Digitonin Solubilization Assay
Digitonin was used to selectively permeabilize the plasma membrane of cells, leaving the organelle membranes intact (37). Transiently transfected N2a cells were rinsed twice with ice-cold PBS, scraped off the plate, and pelleted by centrifugation. The cells were resuspended in transport buffer (20 mm Hepes, pH 7.4, 110 mm KOAc, 2 mm Mg(OAc)2, and 0.5 mm EGTA) supplemented with proteinase inhibitor (1:500; Roche Applied Science), and the plasma membrane was permeabilized by adding 0.004% digitonin for 5 min on ice. After permeabilization, samples were centrifuged at 100,000 × g for 30 min. Following centrifugation, the supernatant (cytosolic) and pellet (membrane) fraction were harvested. The pellet was resuspended in PBS containing 0.5% Tx/DOC (0.5% Triton X-100 and 0.5% sodium deoxycholate in PBS) and proteinase inhibitor (1:500). For total lysates as control, cells were lysed in PBS containing 0.5% Tx/DOC supplemented with proteinase inhibitor (1:500). Equal volumes of the supernatant, pellet, and total lysate fraction were analyzed by SDS-PAGE and immunoblotting using the mouse monoclonal anti-V5 antibody (Invitrogen).
Immunofluorescence Analysis
Transiently transfected cells were grown on glass coverslips. To analyze mitochondrial morphology and function, cells were incubated with MitoTracker Red CMXRos (50 nm) 30 min prior to fixation. 24 h after transfection, cells were fixed with 3.7% paraformaldehyde for 15 min, permeabilized with 0.2% Triton X-100 in PBS for 10 min, washed with PBS, blocked with PBS containing 1% BSA for 1 h at room temperature, incubated with primary antibody in PBS containing 1% BSA for 1 h at 37 °C, washed again with PBS, and incubated with a fluorescently labeled secondary Alexa Fluor antibody in PBS containing 1% BSA for 30 min at 37 °C. Cells were mounted onto glass slides and examined by fluorescence microscopy using a Zeiss Axiovert 200M microscope.
Bioinformatics Analysis
For the prediction of disordered secretory proteins, SignalP 3.0 was used as a classifier for the prediction of signal peptides in proteins (38). SignalP 3.0 was designed by two different methods, namely hidden Markov models and neural networks. In our analysis, we considered the presence of a signal peptide in a protein if both methods predicted a signal peptide. Mostly disordered proteins (>70% of the amino acid sequence) were identified by the disorder prediction program POODLE-W (39), IUPred (40), and RONN 3.1 (41). In total, 20,331 human protein sequences listed in the Swiss-Prot database (release August 2009) were analyzed. Secondary structure prediction of the prodomains was performed using the prediction servers JPred 3 (42) and PSIPRED (43).
Statistical Analysis
Quantifications were based on at least three independent experiments. Data are shown as means ± S.E. Statistical analysis was performed using Student's t test. p values are as follows: *, p < 0.05; **, p < 0.005; ***, p < 0.0005.
RESULTS
Intrinsically Disordered Proteins Are Underrepresented in the Secretome
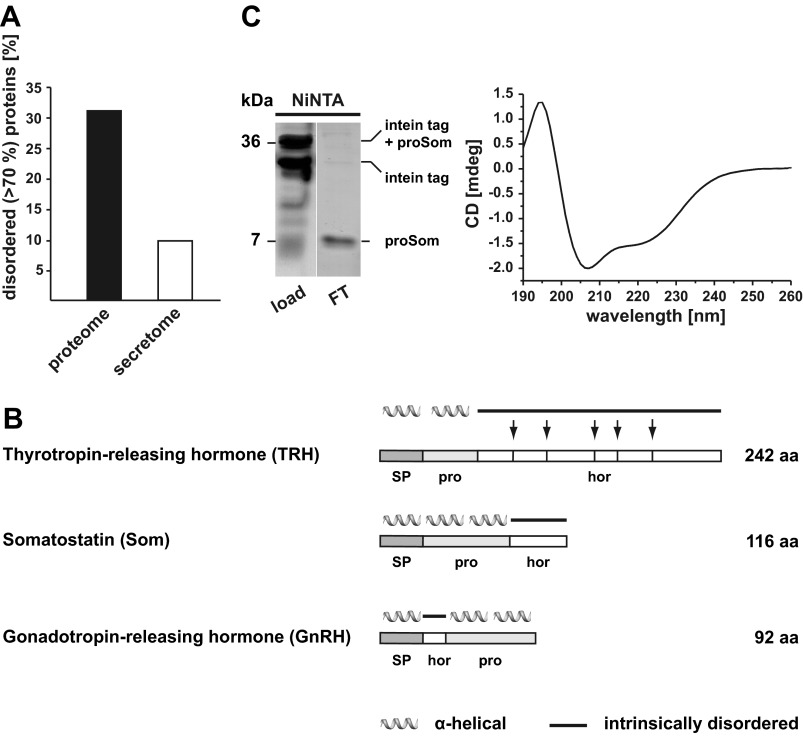
Previous studies on the biogenesis of pathogenic mutants of the prion protein and model substrates revealed that IDPs equipped with an N-terminal signal peptide are inefficiently imported into the ER (18, 19, 44). Based on these findings, we performed an in silico screen for the identification of secretory proteins that are dominated by intrinsically disordered domains (>70% of the amino acid sequence). First, we determined the fraction of IDPs in the total human proteome. Of the 20,331 proteins listed in the Swiss-Prot database (August 2009), 31% were predicted to be mostly disordered (POODLE-W), corroborating previously published data (39, 45–47). Next, we used SignalP 3.0 as a classifier for the prediction of signal peptides in proteins (38). Interestingly, only 10% of the proteins containing an N-terminal ER signal peptide were predicted to be mostly disordered. Using other algorithms for the identification of disordered proteins, for example IUPred and RONN, a similar trend was observed. Based on these analyses, we concluded that IDPs are significantly less abundant in the secretome than in the total proteome (Fig. 1A).
FIGURE 1.
Intrinsically disordered neuropeptide hormones contain an α-helical prodomain. A, intrinsically disordered proteins are underrepresented in the secretome. POODLE-W was used to predict mostly disordered proteins (>70% of the amino acid sequence) in the human proteome. Whereas disordered proteins account for 30% of the total proteome, only 10% of disordered proteins were predicted to contain an ER signal peptide (secretome) using the signal peptide prediction server SignalP 3.0. B, intrinsically disordered neuropeptide hormones are synthesized as larger precursors with α-helical prodomains. Shown is a schematic representation of the neuropeptide hormones analyzed in this study. The precursors contain an N-terminal ER signal peptide (SP), a prodomain (pro), and the hormone domain (hor). In the case of TRH, the hormone domain is further processed to generate five biologically active TRH peptides (arrows). Secondary structure prediction of preprohormones was performed using the prediction server JPred. α-Helical structure is indicated by helices; intrinsic disorder is indicated by a straight line. C, the prodomain of Som has the intrinsic ability to form an α-helical structure. The isolated proSom was expressed as an intein fusion protein in E. coli. The recombinant protein was purified using Ni-NTA-agarose beads. The intein tag of the purified protein was cleaved by incubation with DTT for 16 h and subsequently loaded onto a Ni-NTA column (left panel, load). By collecting the flow-through (FT), the somatostatin prodomain was purified from the His tag-containing intein tag bound to the Ni-NTA column. Protein samples were analyzed by SDS-PAGE and Coomassie Blue staining (left panel). The secondary structure of the purified prodomain of somatostatin was analyzed by CD (right panel). The spectral trace is consistent with an α-helical conformation. x axis, wavelength in nanometer (nm); y axis, ellipticity in millidegrees (mdeg).
Intrinsically Disordered Neuropeptide Hormones Contain an α-Helical Prodomain
When performing the screen described above, we noticed that the mature forms of several neuropeptide hormones are either too small to adopt a stable conformation or are predicted to be intrinsically disordered. Among this class of neuropeptide hormones, we identified TRH, Som, and GnRH (Fig. 1B). Interestingly, these neuropeptide hormones are synthesized as larger precursors (preproproteins) with a prodomain in addition to the N-terminal ER signal peptide. Similarly to the signal peptide, which is removed during translocation into the ER lumen by the signal peptidase, the prodomain is not part of the mature hormone. Processing of the propeptide is mediated by specific protein convertases mainly in the trans-Golgi network or secretory granules (48–50).
A computational approach using two different prediction methods indicated that the prodomains of the intrinsically disordered neuropeptide hormones listed above have the propensity to adopt an α-helical conformation (Fig. 1B). To provide experimental evidence for this prediction, we cloned the isolated prodomain of Som (proSom; aa 25–88) lacking the ER signal peptide into a bacterial expression vector. Because we could not efficiently express the isolated prodomain in E. coli, we fused the prodomain to a C-terminal precursor consisting of an intein tag followed by a His tag and a chitin-binding domain. The fusion protein was purified from bacterial lysates by Ni2+ affinity chromatography under native conditions, and self-cleavage of the intein tag was induced by DTT to release the target protein from the intein tag. Analysis by CD spectroscopy indicated that the isolated prodomain has the propensity to adopt an α-helical conformation (Fig. 1C). Notably, it was shown previously that the isolated prodomain of GnRH also shows significant α-helical propensity in circular dichroism spectra (51).
Deletion of the Prodomain Interferes with Secretion of the Hormone
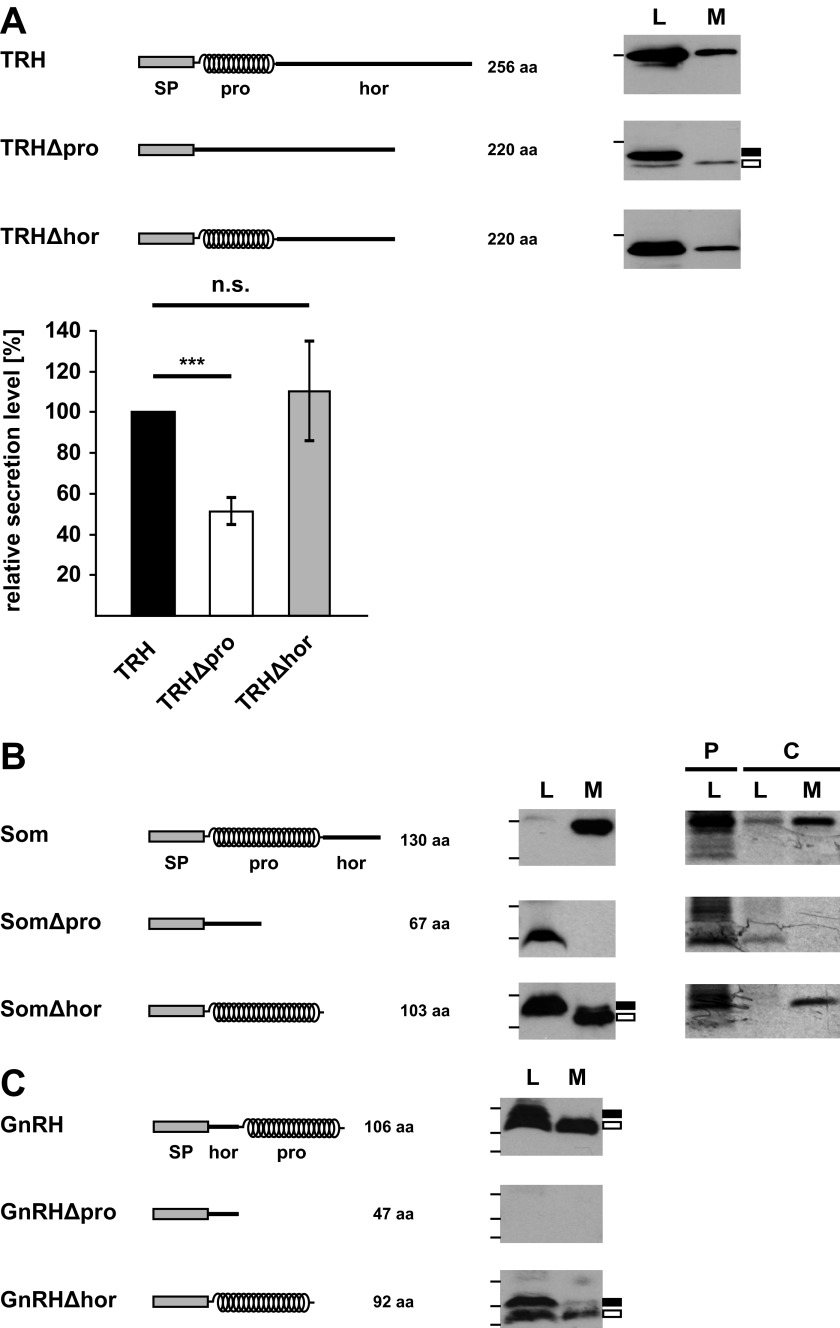
To study a possible role of the prodomains in the biogenesis of the mature hormones, we cloned wild type TRH, Som, and GnRH and deletion mutants thereof lacking either the prodomain (Δpro) or the hormone domain (Δhor). All mutants contained the authentic N-terminal signal peptide of the respective hormone (Fig. 2, A, B, and C). Please note that there are no significant sequence homologies between either the different prodomains or the hormone domains. To monitor secretion in transiently transfected N2a cells, conditioned medium was analyzed by metabolic labeling and Western blotting. All wild type constructs were secreted as unprocessed propeptides because N2a cells do not express the protein convertases required for processing of the prodomain. Similarly, the TRH, Som, and GnRH mutants lacking the hormone domains (Δhor) were secreted (Fig. 2, A, B, and C). However, secretion of mutants lacking the prodomain was significantly impaired or not detectable. Conditioned medium contained significantly reduced levels of TRHΔpro when compared with TRH or TRHΔhor (Fig. 2A), whereas the mutants lacking the prodomains were not detectable in conditioned medium of SomΔpro- or GnRHΔpro-expressing cells (Fig. 2, B and C). TRHΔpro and SomΔpro were present in total cell lysates, indicating that the impaired secretion was not due to an impaired protein synthesis. GnRHΔpro could not be detected in total cell lysates or under conditions of proteasomal inhibition (data not shown).
FIGURE 2.
Deletion of the prodomain interferes with secretion of the hormone domain. A–C, left panels, schematic representation of the constructs used. α-Helical structure is indicated by helices; intrinsically disorder is indicated by a straight line. SP, ER signal peptide; pro, prodomain; hor, hormone domain. A, deletion of the prodomain significantly reduces secretion of the TRH hormone domain. N2a cells were transiently transfected with the constructs indicated. To examine secretion of proteins into the cell culture supernatant, cells were cultivated in cell culture medium without supplements for 3 h at 37 °C. TRH present in the medium (M) and total cell lysates (L) were analyzed by Western blotting. The bar on the left side of the panel represents a molecular size of 36 kDa. Filled rectangle, uncleaved signal peptide; unfilled rectangle, cleaved signal peptide. Right panel, secretion efficiency was determined by quantifying the band intensities of the respective constructs in the medium relative to the amount present in total lysates. Data represent mean ± S.E. (error bars) of at least three independent experiments. The secretion of wild type TRH was set as 100%. ***, p < 0.0005; n.s., not significant. B, the hormone domain of Som is not secreted in the absence of the prodomain. Wild type Som and the indicated mutants present in the media and lysates of transiently transfected N2a cells were analyzed by Western blotting (middle panel) as described in A. In addition, expression and secretion of the different constructs were analyzed by immunoprecipitation (right panel). Transiently transfected cells were pulse-labeled for 15 min with l-[35S]methionine. Cells were either harvested immediately (pulse (P)) or after a further incubation in complete medium for 1 h (chase (C)). The Som constructs present in the lysate or in the medium after the chase were immunoprecipitated. Immunopellets were analyzed by SDS-PAGE and autoradiography. Molecular sizes of 16 (upper bar) and 7 kDa (lower bar) are indicated on the left side of the panel. Filled rectangle, uncleaved signal peptide; unfilled rectangle, cleaved signal peptide. C, the hormone domain of GnRH is not expressed in the absence of the prodomain. Total cell lysates (L) and medium (M) of transiently transfected N2a cells were analyzed by Western blotting as described in A. The GnRH construct lacking the prodomain (GnRHΔpro) could not be detected in lysates or in medium of transfected cells. Molecular size markers are indicated as bars on the left side of the panel and represent 16, 7, and 4 kDa. Filled rectangle, uncleaved signal peptide; unfilled rectangle, cleaved signal peptide.
In sum, these experiments revealed that deletion of the prodomain significantly impairs or prevents secretion of peptide hormones. In contrast, the respective prodomains could be expressed and were secreted in the absence of the hormone domain.
Heterologous α-Helical Domains Restore Secretion of Mutants Devoid of the Prodomain
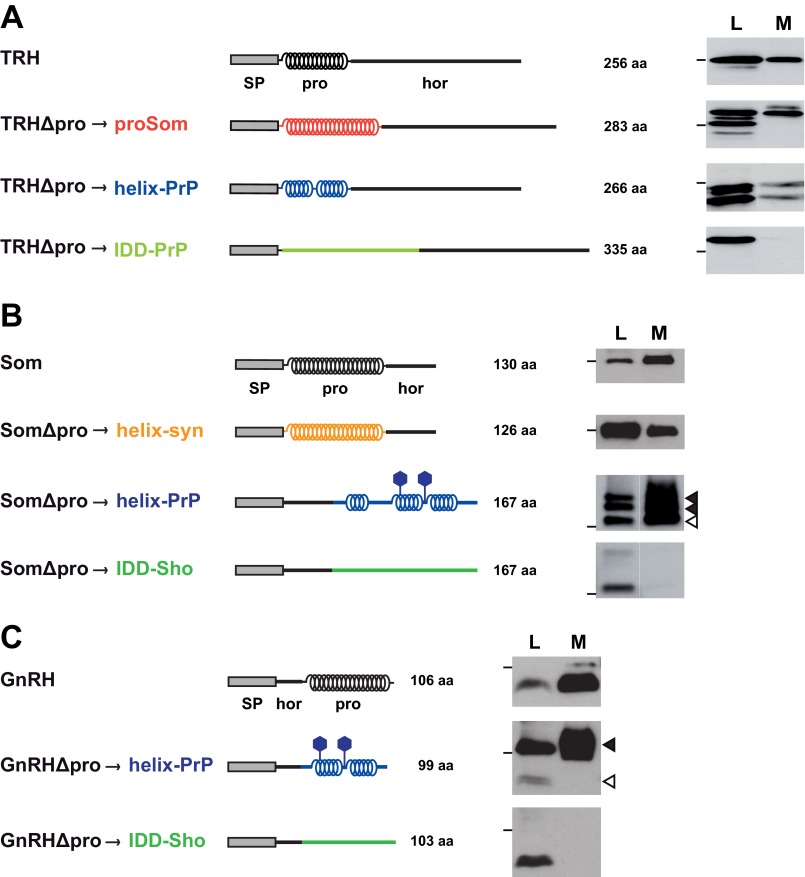
It was shown previously that impaired ER import of IDDs could be restored by α-helical domains (18, 19). To address the possibility that the activity to promote secretion of the hormone domains does not reside in the amino acid sequence of the prodomains but rather in their ability to adopt an α-helical conformation, we performed domain-swapping experiments. The prodomains of TRH, Som, and GnRH were exchanged by heterologous domains of known secondary structure. As examples for α-helical domains, we used (i) the prodomain of Som (aa 25–88), (ii) the C-terminal domain of the prion protein (aa 173–219 or aa 130–230) (52–54), and (iii) a synthetically designed α-helix (25). To test the effect of intrinsically disordered domains, we used part of the N-terminal domain of the prion protein (aa 23–115) (52) or shadoo (aa 25–125) (55, 56). N2a cells were transiently transfected with the different constructs, and conditioned medium was analyzed by Western blotting. This approach revealed that irrespective of the amino acid sequence heterologous domains promoted secretion of the hormone domain provided that they can adopt an α-helical structure. In contrast, when the prodomain was replaced by an intrinsically disordered domain, the respective TRH, Som, and GnRH constructs were not secreted. Notably, the intrinsically disordered proteins were detectable in cell lysates, indicating that lack of secretion was not due to impaired protein synthesis (Fig. 3, A, B, and C). This analysis supports the concept that efficient secretion of the hormone precursors is dependent on the ability of the prodomain to adopt an α-helical conformation and is not linked to a specific amino acid sequence.
FIGURE 3.
Heterologous α-helical domains can restore secretion of the hormone domain. A–C, the prodomains of TRH (A), Som (B), and GnRH (C) were replaced by heterologous domains of defined structures. Schematic representations of the different constructs are shown on the left side. Different amino acid sequences are indicated by different colors. proSom, α-helical prodomain of somatostatin; helix-PrP, α-helical part of the structured C-terminal domain of the prion protein; IDD-PrP, part of the intrinsically disordered N-terminal domain of the prion protein; helix-syn, synthetic α-helix; IDD-Sho, part of the disordered domain of shadoo. α-Helical structure is indicated by helices; intrinsic disorder is indicated by a straight line. SP, ER signal peptide; pro, prodomain; hor, hormone domain. Filled hexagon, N-linked glycosylation site. N2a cells were transiently transfected, and the lysate (L) and medium (M) fractions were analyzed by Western blotting as described in Fig. 2A. Molecular sizes of 36 (A) and 16 kDa (B and C) are indicated as bars on the left side of the panel. Filled arrowhead, glycosylated fraction; unfilled arrowhead, unglycosylated fraction.
The Prodomain Promotes Productive Translocation of the Precursor into the ER Lumen
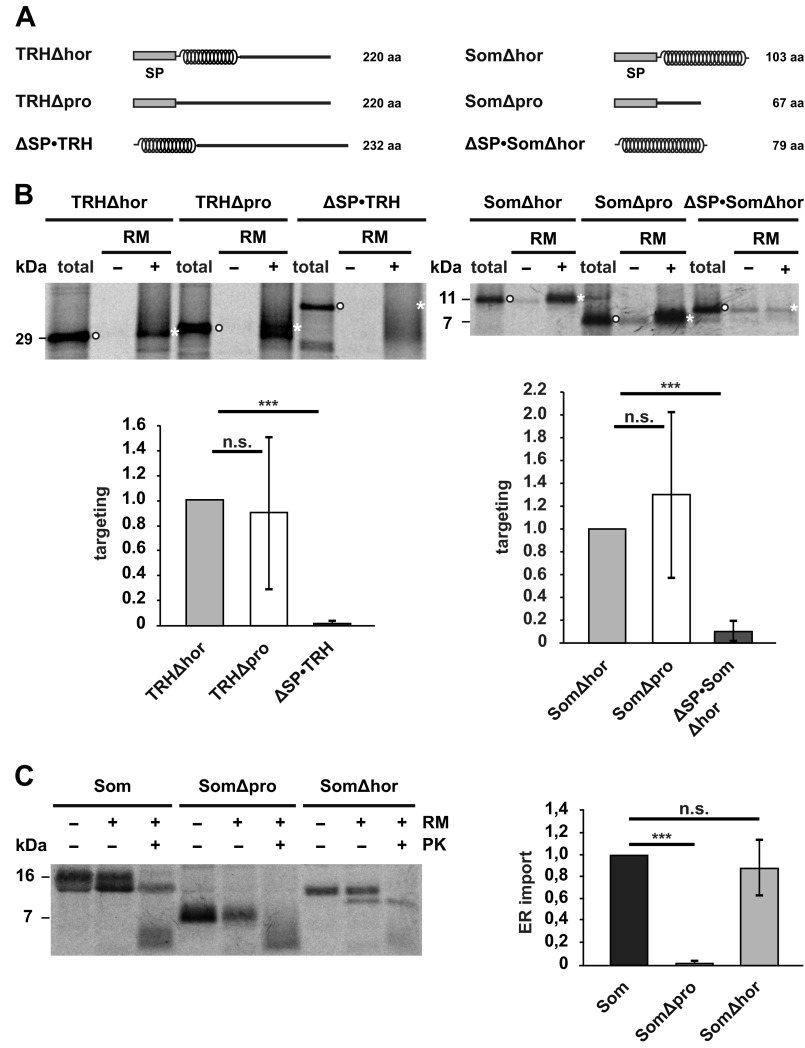
Next, we sought to investigate which step in the biogenesis is dependent on the α-helical prodomains. Because TRHΔpro and SomΔpro are detectable by Western blotting in cell lysates (Fig. 2, A and B), we analyzed whether targeting of these mutants to the ER membrane/Sec61 translocon is impaired. Co-translational targeting to the ER membrane was investigated in vitro. Stalled ribosome-nascent chain complexes were generated by translating truncated mRNAs lacking a stop codon in reticulocyte lysate supplemented with [35S]methionine. After synthesis in the presence of canine pancreatic microsomal membranes, ribosome-nascent chain complexes bound to ER membranes were separated from untargeted nascent chains by sedimentation. The percentage of nascent chains recovered in this way was quantified by SDS-PAGE and autoradiography. To analyze whether structural features of the nascent chain modulate this step, we compared co-translational targeting of TRHΔhor with that of TRHΔpro and SomΔhor with that of SomΔpro. As a negative control, we included constructs lacking the N-terminal signal peptide (ΔSP). This comparative analysis (Fig. 4, A and B) revealed that intrinsically disordered nascent chains (TRHΔpro and SomΔpro) were as efficiently targeted to ER membranes as nascent chains composed of an α-helix (TRHΔhor and SomΔhor). Thus, the structure of the nascent chain does not seem to regulate co-translational targeting of ribosome-nascent chain complexes to the ER membrane at least under the in vitro conditions tested.
FIGURE 4.
α-Helical domains promote translocation of the nascent chain into the ER lumen. A, schematic representation of the constructs used. α-Helical structure is indicated by helices; intrinsic disorder is indicated by a straight line. SP, signal peptide. B, intrinsically disordered nascent chains are co-translationally targeted to ER membranes in vitro. Truncated mRNAs lacking a stop codon were translated in vitro using rabbit reticulocyte lysate supplemented with l-[35S]methionine in the presence (+) or absence (−) of rough RM. Targeted nascent chains were pelleted by centrifugation over a sucrose cushion for 3 min at 50,000 × g. In parallel, polysomes of translation reactions without microsomes were sedimented by centrifugation for 30 min at 50,000 × g (total). Samples were analyzed by SDS-PAGE and autoradiography. Targeting efficiency was determined by quantifying the band intensities of the respective constructs in the +RM fraction (white asterisks) relative to the amount present in the total fraction (white dots). Data represent mean ± S.E. (error bars) of at least three independent experiments. Targeting efficiency of the Δhor mutants was set as 1. ***, p < 0.0005; n.s., not significant. C, an intrinsically disordered protein is not imported into microsomes in vitro. Left panel, indicated constructs were translated in vitro using a rabbit reticulocyte lysate supplemented with l-[35S]methionine in the presence (+) or absence (−) of RM. After translation, samples were treated with proteinase K (+PK) or left untreated (−PK) and analyzed by SDS-PAGE and autoradiography. Right panel, import into microsomes was determined by quantifying the ratio between the signal of the proteinase K-protected fraction (+RM, +PK) to the total protein signal in the presence of microsomes (+RM, −PK). The relative amount of proteinase K-resistant wild type somatostatin was set as 1. Data represent mean ± S.E. (error bars) of at least three independent experiments. ***, p < 0.0005; n.s., not significant. The lower molecular smear (<7 kDa) present in the fractions treated with proteinase K (+PK) represents nonspecific degradation products that also appear after proteinase K digestion in the absence of microsomal membranes (data not shown).
To analyze translocation of the polypeptide chains into the ER lumen, we used two different approaches. First, we studied ER import in vitro. Similarly to the approach described above, the proteins were synthesized in reticulocyte lysate in the presence of canine pancreatic microsomal membranes. Import efficiency was measured by treating the samples with proteases that degrade all proteins that have not been imported. The proteins were then analyzed by SDS-PAGE and phosphorimaging to quantify the fraction of proteins that resisted proteolytic digestion, i.e. have been imported into the ER vesicles. Another marker for ER import is the appearance of a second, faster migrating band on SDS-PAGE that is indicative of cleavage of the ER signal peptide. This approach revealed clear differences between SomΔpro and SomΔhor. In the presence of microsomes SomΔhor became resistant to proteolytic digestion, whereas SomΔpro was degraded under these conditions. In addition, cleavage of the signal peptide was only observed for wild type Som and SomΔhor but not for SomΔpro (Fig. 4C).
In a second approach, we analyzed ER import of the TRH mutants in transiently transfected cells. The cells were treated with low concentrations of digitonin (0.004% for 5 min on ice) to selectively disrupt the plasma membrane but leave the cellular organelles like the ER and trans-Golgi network intact (37). After centrifugation, proteins present in the ER can be found in the pellet fraction, whereas cytosolic proteins remain in the supernatant (Fig. 5A). The relative amount of the constructs present in the cytosolic or ER fraction was then determined by Western blotting. Consistent with the finding that TRHΔhor is efficiently secreted (Fig. 2A), the construct was exclusively detected in the pellet (ER) fraction. However, a significant proportion of the construct lacking the prodomain was found in the cytosolic fraction (Fig. 5B). To specifically address processing of the ER signal peptide, we deleted the signal peptide sequence to generate ΔSP·TRHΔhor and ΔSP·TRHΔpro. After transient expression in N2a cells, the constructs were analyzed by Western blotting. Migration of TRHΔhor and ΔSP·TRHΔhor in SDS-PAGE was similar, indicating that the signal peptide of TRHΔhor was efficiently processed upon expression in cells. However, the majority of TRHΔpro appeared to be slightly larger than ΔSP·TRHΔpro, suggesting that the signal peptide was not removed from the mutant lacking the prodomain (Fig. 5C).
FIGURE 5.
The non-secreted fractions of TRHΔpro or SomΔpro are located in the cytosol or mitochondria and contain an uncleaved signal peptide. A, schematic illustration of the digitonin solubilization assay. Cells were treated with low concentrations of digitonin to selectively disrupt the plasma membrane (indicated by dashed line) but not membranes of intracellular organelles like the ER (green) or nucleus (gray). After centrifugation, secretory proteins (blue dots) are found in the pellet, whereas cytosolic proteins (red dots) remain in the supernatant (sup). B, TRHΔpro is partially present in the cytosol. Transiently transfected N2a cells were solubilized in buffer containing 0.004% digitonin (Dig) for 5 min on ice. Samples were centrifuged for 30 min at 100,000 × g. The supernatant (sup) and the pellet fraction (pellet) were analyzed by Western blotting. Total cell lysates were analyzed in parallel (total). C, TRHΔpro contains an uncleaved signal peptide. Transiently transfected cells expressing the constructs indicated were analyzed by Western blotting. B and C, filled rectangle, uncleaved signal peptide; unfilled rectangle, cleaved signal peptide. D, non-translocated SomΔpro is forwarded to mitochondria. Transiently transfected SH-SY5Y cells expressing the constructs indicated were analyzed by indirect immunofluorescence. Mitochondria were labeled with MitoTracker Red CMXRos.
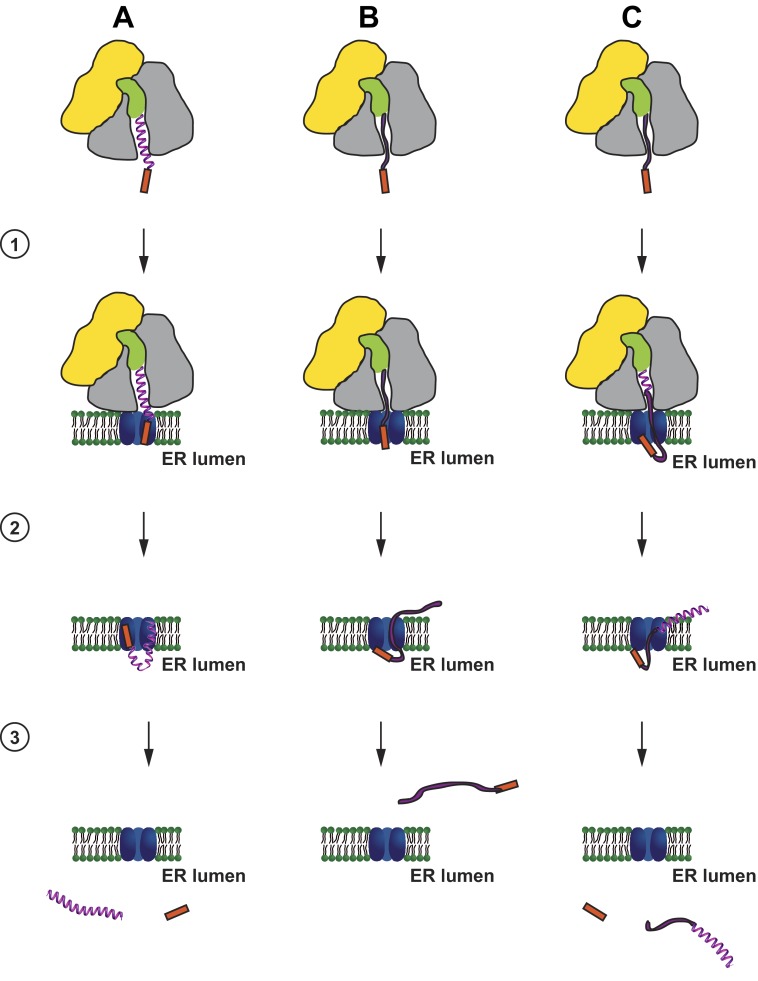
When we used the same approach to analyze SomΔpro, we were surprised to see that this mutant, which was not secreted from transiently transfected cells (Fig. 2B) and not imported into microsomal membranes in vitro (Fig. 3C), was found in the pellet fraction similarly to wild type Som (data not shown). To investigate this finding in more detail, we performed an indirect immunofluorescence analysis of transiently transfected cells. As expected for secretory proteins, in cells expressing wild type Som, the protein was predominantly found in the Golgi complex (Fig. 5D). As a marker for cytosolic localization, we included ΔSP·Som, a mutant that is not imported into the ER because it lacks the N-terminal signal peptide. Unexpectedly, the immunofluorescence pattern of SomΔpro was reminiscent of mitochondrial localization. Indeed, co-staining with MitoTracker Red CMXRos confirmed that SomΔpro predominantly colocalizes with mitochondria (Fig. 5D). The mitochondrial targeting of SomΔpro explains why it is present in the pellet fraction of digitonin-solubilized cells because mitochondria remain intact under the conditions used and sediment after centrifugation similarly to the ER. Mitochondrial targeting mediated by the ER signal peptide of Som has been addressed in a recent study (57). In sum, the combined in vitro and cell culture approaches support a model in which α-helical prodomains can specifically promote productive translocation of intrinsically disordered proteins into the ER lumen (Fig. 6).
FIGURE 6.
Putative model of the activity of α-helical domains in promoting productive ER import of secreted proteins. Nascent chains composed of α-helical (A) or intrinsically disordered domains (B) or both (C) are co-translationally targeted to the Sec61 translocon (1). A, proteins dominated by α-helical domains translocate through the translocon (2) and are released into the ER lumen with a cleaved ER signal peptide (red) (3). B, in contrast, intrinsically disordered proteins might partially translocate (2) but are finally released into the cytosol with an uncleaved signal peptide (3). C, ER import of intrinsically disordered proteins can be rescued by α-helical domains even if they are located at the C terminus. Cytosolic and translocon-associated proteins that are possibly involved in the different steps are not shown. Yellow, small ribosomal subunit; gray, large ribosomal subunit; green, tRNA; blue, translocon. The model is based on the experiments described in this study and previous work (19, 57).
DISCUSSION
In this study, we analyzed the role of prodomains in the biogenesis of different intrinsically disordered neuropeptide hormones. Using deletion mutagenesis and domain-swapping experiments, our in vitro and cell culture experiments revealed that the prodomain is required for a productive translocation of the hormone into the ER lumen. Notably, the activity of the prodomain to promote ER import is based on its propensity to adopt an α-helical conformation.
Intrinsically Disordered Proteins Are Underrepresented in the Secretome
Natively unstructured or IDPs lack a stable secondary/tertiary structure under physiological conditions in vitro. Despite the absence of a well defined structure, IDPs fulfill a variety of physiological functions; moreover, it appears that some activities require the flexibility of disordered proteins (for reviews, see Refs. 27–32 and 58). To specifically concentrate on IDPs in the secretome, we used computational approaches to identify IDPs containing an N-terminal ER signal peptide. Our finding that only 10% of the secretory proteins are predicted to be intrinsically disordered whereas IDPs account for about 30% of the total proteome strongly suggests that IDPs are underrepresented in the secretome. So far, one can only speculate about the reasons underlying this phenomenon. It is conceivable that the physiological activities of secretory proteins or exposure to the extracellular environment favors stable secondary structures. Alternatively, expanded stretches of intrinsically disordered domains might be disadvantageous for proteins that are subjected to the complex folding and quality control pathways in the ER lumen (for reviews, see Refs. 59–61). Specifically, failure to adopt a native conformation can prevent further trafficking of secretory and membrane proteins to the plasma membrane and result in retrograde transport to the cytosol where proteasomal degradation occurs (for reviews, see Refs. 62–64). It is therefore possible that intrinsic disorder is mistaken for misfolding and that IDPs erroneously are subjected to the ER-associated degradation pathway. We observed that the non-secreted fraction of the IDPs is found in the cytosol, which might support such a scenario. However, cytosolic IDPs contained an uncleaved ER signal peptide, a feature that is not expected for the cytosolic fraction of ER-associated degradation substrates. Rather an uncleaved ER signal peptide would be indicative for an abortive ER import and/or an alternative pre-emptive, co-translocational quality control pathway that operates before translocation of the protein into the ER is completed (13, 14, 65).
Prodomains Promote Productive ER Import of Intrinsically Disordered Neuropeptide Hormones
Prodomains are found in a variety of proteins. As a unifying feature, they are proteolytically removed during maturation and are not part of the physiologically active protein. Various functions have been described for prodomains, for example to facilitate folding and processing, to repress the enzymatic activity of the mature protein, and to promote trafficking. In addition, it was shown that the unprocessed propeptide or the liberated prodomain can have physiological activities different from that of the mature hormone (66–74).
Our study revealed a novel activity of prodomains: to promote ER import of neuropeptide hormones that are intrinsically disordered. The first hint for such an activity was the observation that deletion of the prodomains significantly interfered with secretion of the hormone. In contrast, mutants lacking the hormone domain were present in the conditioned medium of transiently transfected cells. Notably, a similar finding was described previously for TRH constructs lacking the prodomain (75).
Some of the constructs lacking the prodomain are rather short, raising the possibility that impaired secretion is linked to the reduced length of the polypeptide chain. To address this question, we performed domain-swapping experiments to generate constructs that are of similar length but of different structure. Specifically, we analyzed constructs that are either entirely unstructured or contain an α-helical domain in addition to the unstructured hormone domain. This analysis revealed two important findings. First, impaired secretion of the Δpro constructs can be restored by heterologous domains provided that they have the propensity to adopt an α-helical conformation. Second, increasing the length of an IDP is not sufficient to restore its secretion at least in the size range tested (335 aa for TRHΔpro→IDD-PrP). Similarly, ER import of IDPs is not significantly promoted by more efficient ER signal peptides (19). Because all chimeric constructs were efficiently secreted as unprocessed propeptides, it appears that secretion of the hormone domain is not dependent on processing of the propeptide.
Our experimental approaches clearly demonstrate that secretion of different intrinsically disordered neuropeptide hormones is critically dependent on α-helical prodomains. However, these findings do not imply that this is their only physiological function. For example, the prodomain of somatostatin itself comprises another peptide hormone named neuronostatin, which regulates different neuronal functions (66). Similarly, part of the prodomain of GnRH known as GnRH-associated peptide is present in GnRH-containing secretory granules and inhibits prolactin secretion (51).
Intrinsically Disordered Proteins Are Not Translocated into the ER
Another important aspect of this study was to define which step between translation and secretion is impaired in the absence of α-helical prodomains. In transiently transfected cells, the non-secreted fraction of the proteins expressed without a prodomain was found in the cytosol. Different scenarios are plausible to explain such a finding. First, targeting of the ribosome-nascent chain complex to the Sec61 translocon might be impaired. Our in vitro targeting assays revealed that the Δpro and Δhor constructs were co-translationally targeted to ER membranes with similar efficiencies, arguing against this possibility. Second, after targeting to the Sec61 translocon, translocation into the ER lumen might be aborted so that the protein is finally released into the cytosol. One marker for such a pathway is an uncleaved ER signal peptide. Cleavage of the ER signal peptide normally occurs during import before the protein is fully translocated into the ER lumen. Indeed, the results obtained with our in vitro and cell culture assays are indicative of an abortive ER import. In vitro, SomΔpro, which was co-translationally targeted to microsomal membranes, was not imported into the microsomes and contained an unprocessed ER signal peptide. Similarly, in transiently transfected cells, TRHΔpro and SomΔpro were present in the cytosol with an uncleaved ER signal peptide. Remarkably, the non-translocated fraction of SomΔpro was forwarded to mitochondria. A detailed analysis of the activity of the ER signal peptide of Som to mediate mitochondrial targeting is described elsewhere (57). Third, secretory proteins could be found in the cytosol after they had been subjected to the ER-associated degradation pathway. In this pathway, the protein is completely translocated into the ER lumen before it is subjected to quality control mechanisms and retrogradely transported to the cytosol. As mentioned above, the non-secreted fraction of the IDPs in the cytosol contained an uncleaved signal peptide, suggesting that this fraction was never completely released into the ER lumen.
On the basis of these findings, we suggest a model in which the productive translocation of IDPs through the Sec61 translocon is impaired (Fig. 6). This concept does not rule out that a part of the chain is transiently exposed to the luminal side or interacts with proteins in the ER lumen. For example, overexpression of p58IPK significantly impairs ER import of proteins dominated by IDDs (19). p58IPK is an Hsp70 co-chaperone present predominantly in the ER lumen. Because p58IPK can interact with both the nascent chain and BiP (65, 76), such interactions could impair productive translocation of IDPs into the ER lumen. Other translocon-associated proteins, such as Sec63 or Sec62, might be involved in such a pathway as well. Finally, it appears plausible that α-helical structures have an intrinsic activity to promote the translocation process, for example via interactions with the inner side of the translocation pore and/or with translocon-associated proteins or just by supporting the linear force of the translating ribosome. In this view, IDPs are not actively expelled but are rather too “weak” to enter the ER lumen.
Our study emphasizes the notion that structural features of the protein can significantly modulate ER import efficiency. It will now be of interest to uncover how this feature is used by the cell to regulate ER import of secretory proteins and to identify cellular proteins involved in this process.
Acknowledgments
We thank Veronika Müller for excellent technical assistance and Roland Beckmann and Shashi Bhushan for providing the synthetic α-helical construct.
The work was supported by Deutsche Forschungsgemeinschaft Grant SFB 596 and by the Max Planck Society.
- ER
- endoplasmic reticulum
- IDP
- intrinsically disordered protein
- IDD
- intrinsically disordered domain
- Som
- somatostatin
- TRH
- thyrotropin-releasing hormone
- GnRH
- gonadotropin-releasing hormone
- aa
- amino acids
- PrP
- prion protein
- Sho
- shadoo
- RM
- microsomal membranes
- Ni-NTA
- nickel-nitrilotriacetic acid
- proSom
- prodomain of Som.
REFERENCES
- 1. Gilmore R., Blobel G., Walter P. (1982) Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J. Cell Biol. 95, 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gilmore R., Walter P., Blobel G. (1982) Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J. Cell Biol. 95, 470–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meyer D. I., Dobberstein B. (1980) Identification and characterization of a membrane component essential for the translocation of nascent proteins across the membrane of the endoplasmic reticulum. J. Cell Biol. 87, 503–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walter P., Blobel G. (1980) Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 77, 7112–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Walter P., Blobel G. (1982) Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature 299, 691–698 [DOI] [PubMed] [Google Scholar]
- 6. Walter P., Gilmore R., Blobel G. (1984) Protein translocation across the endoplasmic reticulum. Cell 38, 5–8 [DOI] [PubMed] [Google Scholar]
- 7. Zimmermann R., Eyrisch S., Ahmad M., Helms V. (2011) Protein translocation across the ER membrane. Biochim. Biophys. Acta 1808, 912–924 [DOI] [PubMed] [Google Scholar]
- 8. Rapoport T. A. (2007) Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450, 663–669 [DOI] [PubMed] [Google Scholar]
- 9. von Heijne G. (1985) Signal sequences. The limits of variation. J. Mol. Biol. 184, 99–105 [DOI] [PubMed] [Google Scholar]
- 10. Martoglio B., Dobberstein B. (1998) Signal sequences: more than just greasy peptides. Trends Cell Biol. 8, 410–415 [DOI] [PubMed] [Google Scholar]
- 11. Hegde R. S., Bernstein H. D. (2006) The surprising complexity of signal sequences. Trends Biochem. Sci. 31, 563–571 [DOI] [PubMed] [Google Scholar]
- 12. Hegde R. S., Kang S. W. (2008) The concept of translocational regulation. J. Cell Biol. 182, 225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oyadomari S., Yun C., Fisher E. A., Kreglinger N., Kreibich G., Oyadomari M., Harding H. P., Goodman A. G., Harant H., Garrison J. L., Taunton J., Katze M. G., Ron D. (2006) Cotranslocational degradation protects the stressed endoplasmic reticulum from protein overload. Cell 126, 727–739 [DOI] [PubMed] [Google Scholar]
- 14. Kang S. W., Rane N. S., Kim S. J., Garrison J. L., Taunton J., Hegde R. S. (2006) Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell 127, 999–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hegde R. S., Ploegh H. L. (2010) Quality and quantity control at the endoplasmic reticulum. Curr. Opin. Cell Biol. 22, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yogev O., Naamati A., Pines O. (2011) Fumarase: a paradigm of dual targeting and dual localized functions. FEBS J. 278, 4230–4242 [DOI] [PubMed] [Google Scholar]
- 17. Shaffer K. L., Sharma A., Snapp E. L., Hegde R. S. (2005) Regulation of protein compartmentalization expands the diversity of protein function. Dev. Cell 9, 545–554 [DOI] [PubMed] [Google Scholar]
- 18. Heske J., Heller U., Winklhofer K. F., Tatzelt J. (2004) The C-terminal domain of the prion protein is necessary and sufficient for import into the endoplasmic reticulum. J. Biol. Chem. 279, 5435–5443 [DOI] [PubMed] [Google Scholar]
- 19. Miesbauer M., Pfeiffer N. V., Rambold A. S., Müller V., Kiachopoulos S., Winklhofer K. F., Tatzelt J. (2009) α-Helical domains promote translocation of intrinsically disordered polypeptides into the endoplasmic reticulum. J. Biol. Chem. 284, 24384–24393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Whitley P., Nilsson I. M., von Heijne G. (1996) A nascent secretory protein may traverse the ribosome endoplasmic reticulum translocase complex as an extended chain. J. Biol. Chem. 271, 6241–6244 [DOI] [PubMed] [Google Scholar]
- 21. Woolhead C. A., McCormick P. J., Johnson A. E. (2004) Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell 116, 725–736 [DOI] [PubMed] [Google Scholar]
- 22. Lu J., Deutsch C. (2005) Secondary structure formation of a transmembrane segment in Kv channels. Biochemistry 44, 8230–8243 [DOI] [PubMed] [Google Scholar]
- 23. Mingarro I., Nilsson I., Whitley P., von Heijne G. (2000) Different conformations of nascent polypeptides during translocation across the ER membrane. BMC Cell Biol. 1, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Woolhead C. A., Johnson A. E., Bernstein H. D. (2006) Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Mol. Cell 22, 587–598 [DOI] [PubMed] [Google Scholar]
- 25. Bhushan S., Gartmann M., Halic M., Armache J. P., Jarasch A., Mielke T., Berninghausen O., Wilson D. N., Beckmann R. (2010) α-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 17, 313–317 [DOI] [PubMed] [Google Scholar]
- 26. Daniel C. J., Conti B., Johnson A. E., Skach W. R. (2008) Control of translocation through the Sec61 translocon by nascent polypeptide structure within the ribosome. J. Biol. Chem. 283, 20864–20873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tompa P., Fuxreiter M., Oldfield C. J., Simon I., Dunker A. K., Uversky V. N. (2009) Close encounters of the third kind: disordered domains and the interactions of proteins. BioEssays 31, 328–335 [DOI] [PubMed] [Google Scholar]
- 28. Fuxreiter M., Tompa P. (2012) Fuzzy complexes: a more stochastic view of protein function. Adv. Exp. Med. Biol. 725, 1–14 [DOI] [PubMed] [Google Scholar]
- 29. Fuxreiter M., Tompa P., Simon I., Uversky V. N., Hansen J. C., Asturias F. J. (2008) Malleable machines take shape in eukaryotic transcriptional regulation. Nat. Chem. Biol. 4, 728–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Uversky V. N., Dunker A. K. (2010) Understanding protein non-folding. Biochim. Biophys. Acta 1804, 1231–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Longhi S. (2012) The measles virus N(TAIL)-XD complex: an illustrative example of fuzziness. Adv. Exp. Med. Biol. 725, 126–141 [DOI] [PubMed] [Google Scholar]
- 32. Dyson H. J., Wright P. E. (2005) Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 33. Beckmann R., Spahn C. M., Eswar N., Helmers J., Penczek P. A., Sali A., Frank J., Blobel G. (2001) Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107, 361–372 [DOI] [PubMed] [Google Scholar]
- 34. Rutkowski D. T., Ott C. M., Polansky J. R., Lingappa V. R. (2003) Signal sequences initiate the pathway of maturation in the endoplasmic reticulum lumen. J. Biol. Chem. 278, 30365–30372 [DOI] [PubMed] [Google Scholar]
- 35. Tsai F. C., Sherman J. C. (1993) Circular dichroism analysis of a synthetic peptide corresponding to the α,α-corner motif of hemoglobin. Biochem. Biophys. Res. Commun. 196, 435–439 [DOI] [PubMed] [Google Scholar]
- 36.Deleted in proof
- 37. Plutner H., Davidson H. W., Saraste J., Balch W. E. (1992) Morphological analysis of protein transport from the ER to Golgi membranes in digitonin-permeabilized cells: role of the P58 containing compartment. J. Cell Biol. 119, 1097–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 39. Shimizu K., Muraoka Y., Hirose S., Tomii K., Noguchi T. (2007) Predicting mostly disordered proteins by using structure-unknown protein data. BMC Bioinformatics 8, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dosztányi Z., Csizmok V., Tompa P., Simon I. (2005) IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434 [DOI] [PubMed] [Google Scholar]
- 41. Yang Z. R., Thomson R., McNeil P., Esnouf R. M. (2005) RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics 21, 3369–3376 [DOI] [PubMed] [Google Scholar]
- 42. Cole C., Barber J. D., Barton G. J. (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36, W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 44. Zanusso G., Petersen R. B., Jin T., Jing Y., Kanoush R., Ferrari S., Gambetti P., Singh N. (1999) Proteasomal degradation and N-terminal protease resistance of the codon 145 mutant prion protein. J. Biol. Chem. 274, 23396–23404 [DOI] [PubMed] [Google Scholar]
- 45. Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F., Jones D. T. (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 337, 635–645 [DOI] [PubMed] [Google Scholar]
- 46. Dunker A. K., Obradovic Z., Romero P., Garner E. C., Brown C. J. (2000) Intrinsic protein disorder in complete genomes. Genome Inform. Ser. Workshop Genome Inform. 11, 161–171 [PubMed] [Google Scholar]
- 47. Oldfield C. J., Cheng Y., Cortese M. S., Brown C. J., Uversky V. N., Dunker A. K. (2005) Comparing and combining predictors of mostly disordered proteins. Biochemistry 44, 1989–2000 [DOI] [PubMed] [Google Scholar]
- 48. Patel Y. C., Galanopoulou A. (1995) Processing and intracellular targeting of prosomatostatin-derived peptides: the role of mammalian endoproteases. Ciba Found. Symp. 190, 26–40; discussion 40–50 [DOI] [PubMed] [Google Scholar]
- 49. Mulcahy L. R., Vaslet C. A., Nillni E. A. (2005) Prohormone-convertase 1 processing enhances post-Golgi sorting of prothyrotropin-releasing hormone-derived peptides. J. Biol. Chem. 280, 39818–39826 [DOI] [PubMed] [Google Scholar]
- 50. Bundgaard J. R., Rehfeld J. F. (2008) Distinct linkage between post-translational processing and differential secretion of progastrin derivatives in endocrine cells. J. Biol. Chem. 283, 4014–4021 [DOI] [PubMed] [Google Scholar]
- 51. Chavali G. B., Nagpal S., Majumdar S. S., Singh O., Salunke D. M. (1997) Helix-loop-helix motif in GnRH associated peptide is critical for negative regulation of prolactin secretion. J. Mol. Biol. 272, 731–740 [DOI] [PubMed] [Google Scholar]
- 52. Donne D. G., Viles J. H., Groth D., Mehlhorn I., James T. L., Cohen F. E., Prusiner S. B., Wright P. E., Dyson H. J. (1997) Structure of the recombinant full-length hamster prion protein PrP(29–231): the N terminus is highly flexible. Proc. Natl. Acad. Sci. U.S.A. 94, 13452–13457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Riek R., Hornemann S., Wider G., Billeter M., Glockshuber R., Wüthrich K. (1996) NMR structure of the mouse prion protein domain PrP(121–321). Nature 382, 180–182 [DOI] [PubMed] [Google Scholar]
- 54. Riek R., Hornemann S., Wider G., Glockshuber R., Wüthrich K. (1997) NMR characterization of the full-length recombinant murine prion protein, mPrP(23–231). FEBS Lett. 413, 282–288 [DOI] [PubMed] [Google Scholar]
- 55. Daude N., Ng V., Watts J. C., Genovesi S., Glaves J. P., Wohlgemuth S., Schmitt-Ulms G., Young H., McLaurin J., Fraser P. E., Westaway D. (2010) Wild-type Shadoo proteins convert to amyloid-like forms under native conditions. J. Neurochem. 113, 92–104 [DOI] [PubMed] [Google Scholar]
- 56. Watts J. C., Drisaldi B., Ng V., Yang J., Strome B., Horne P., Sy M. S., Yoong L., Young R., Mastrangelo P., Bergeron C., Fraser P. E., Carlson G. A., Mount H. T., Schmitt-Ulms G., Westaway D. (2007) The CNS glycoprotein Shadoo has PrP(C)-like protective properties and displays reduced levels in prion infections. EMBO J. 26, 4038–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pfeiffer N. V., Dirndorfer D., Lang S., Resenberger U. K., Restelli L. M., Hemion C., Miesbauer M., Frank S., Neutzner A., Zimmermann R., Winklhofer K. F., Tatzelt J. (2013) Structural features within the nascent chain regulate alternative targeting of secretory proteins to mitochondria. EMBO J. 32, 1036–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dunker A. K., Silman I., Uversky V. N., Sussman J. L. (2008) Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 18, 756–764 [DOI] [PubMed] [Google Scholar]
- 59. Walter P., Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 [DOI] [PubMed] [Google Scholar]
- 60. Claessen J. H., Kundrat L., Ploegh H. L. (2012) Protein quality control in the ER: balancing the ubiquitin checkbook. Trends Cell Biol. 22, 22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Braakman I., Bulleid N. J. (2011) Protein folding and modification in the mammalian endoplasmic reticulum. Annu. Rev. Biochem. 80, 71–99 [DOI] [PubMed] [Google Scholar]
- 62. Nakatsukasa K., Brodsky J. L. (2008) The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic 9, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ellgaard L., Helenius A. (2003) Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4, 181–191 [DOI] [PubMed] [Google Scholar]
- 64. Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) ERAD: the long road to destruction. Nat. Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- 65. Rutkowski D. T., Kang S. W., Goodman A. G., Garrison J. L., Taunton J., Katze M. G., Kaufman R. J., Hegde R. S. (2007) The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol. Biol. Cell 18, 3681–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Samson W. K., Zhang J. V., Avsian-Kretchmer O., Cui K., Yosten G. L., Klein C., Lyu R. M., Wang Y. X., Chen X. Q., Yang J., Price C. J., Hoyda T. D., Ferguson A. V., Yuan X. B., Chang J. K., Hsueh A. J. (2008) Neuronostatin encoded by the somatostatin gene regulates neuronal, cardiovascular, and metabolic functions. J. Biol. Chem. 283, 31949–31959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Suter U., Heymach J. V., Jr., Shooter E. M. (1991) Two conserved domains in the NGF propeptide are necessary and sufficient for the biosynthesis of correctly processed and biologically active NGF. EMBO J. 10, 2395–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen Y. G., Danoff A., Shields D. (1995) The propeptide of anglerfish preprosomatostatin-I rescues prosomatostatin-II from intracellular degradation. J. Biol. Chem. 270, 18598–18605 [DOI] [PubMed] [Google Scholar]
- 69. Kjeldsen T., Andersen A. S., Hach M., Diers I., Nikolajsen J., Markussen J. (1998) α-Factor pro-peptide N-linked oligosaccharides facilitate secretion of the insulin precursor in Saccharomyces cerevisiae. Biotechnol. Appl. Biochem. 27, 109–115 [DOI] [PubMed] [Google Scholar]
- 70. Conticello S. G., Kowalsman N. D., Jacobsen C., Yudkovsky G., Sato K., Elazar Z., Petersen C. M., Aronheim A., Fainzilber M. (2003) The prodomain of a secreted hydrophobic mini-protein facilitates its export from the endoplasmic reticulum by hitchhiking on sorting receptors. J. Biol. Chem. 278, 26311–26314 [DOI] [PubMed] [Google Scholar]
- 71. Salvesen G. S., Dixit V. M. (1999) Caspase activation: the induced-proximity model. Proc. Natl. Acad. Sci. U.S.A. 96, 10964–10967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Khan A. R., James M. N. (1998) Molecular mechanisms for the conversion of zymogens to active proteolytic enzymes. Protein Sci. 7, 815–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ibáñez C. F. (2002) Jekyll-Hyde neurotrophins: the story of proNGF. Trends Neurosci. 25, 284–286 [DOI] [PubMed] [Google Scholar]
- 74. Stoller T. J., Shields D. (1989) The propeptide of preprosomatostatin mediates intracellular transport and secretion of α-globin from mammalian cells. J. Cell Biol. 108, 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Romero A., Cakir I., Vaslet C. A., Stuart R. C., Lansari O., Lucero H. A., Nillni E. A. (2008) Role of a pro-sequence in the secretory pathway of prothyrotropin-releasing hormone. J. Biol. Chem. 283, 31438–31448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Petrova K., Oyadomari S., Hendershot L. M., Ron D. (2008) Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 27, 2862–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]