Background: LaeA, a putative methyltransferase in Aspergillus nidulans, is a master regulator of secondary metabolism.
Results: LaeA automethylates at a methionine residue near the AdoMet-binding site. This modification is not required for in vivo function.
Conclusion: Automethylation of LaeA reveals a novel protein methionine methyltransferase activity.
Significance: Elucidating the substrate(s) of LaeA will provide insights into the physiological function of LaeA in modulating gene expression.
Keywords: Aspergillus, Gene Regulation, Histone Methylation, Methionine, Protein Methylation, S-Adenosylmethionine, Secondary Metabolism, LaeA, S-Methylmethionine, Automethylation
Abstract
The filamentous fungi in the genus Aspergillus are opportunistic plant and animal pathogens that can adapt to their environment by producing various secondary metabolites, including lovastatin, penicillin, and aflatoxin. The synthesis of these small molecules is dependent on gene clusters that are globally regulated by the LaeA protein. Null mutants of LaeA in all pathogenic fungi examined to date show decreased virulence coupled with reduced secondary metabolism. Although the amino acid sequence of LaeA contains the motifs characteristic of seven-β-strand methyltransferases, a methyl-accepting substrate of LaeA has not been identified. In this work we did not find a methyl-accepting substrate in Aspergillus nidulans with various assays, including in vivo S-adenosyl-[methyl-3H]methionine labeling, targeted in vitro methylation experiments using putative protein substrates, or in vitro methylation assays using whole cell extracts grown under different conditions. However, in each experiment LaeA was shown to self-methylate. Amino acid hydrolysis of radioactively labeled LaeA followed by cation exchange and reverse phase chromatography identified methionine as the modified residue. Point mutations show that the major site of modification of LaeA is on methionine 207. However, in vivo complementation showed that methionine 207 is not required for the biological function of LaeA. LaeA is the first protein to exhibit automethylation at a methionine residue. These findings not only indicate LaeA may perform novel chemistry with S-adenosylmethionine but also provide new insights into the physiological function of LaeA.
Introduction
The Ascomycete genus Aspergillus represents a diverse collection of filamentous fungi. In human health, Aspergillus is an opportunistic pathogen and is one of the leading causes of infection-related deaths in immunocompromised patients (1, 2). Furthermore, Aspergillus is of significant biological, commercial, and medical importance because of the small molecule secondary metabolites it synthesizes. These include beneficial molecules such as the β-lactam antibiotic penicillin (3) and anticholesterol hypolipidemic agent lovastatin (4), as well as the carcinogenic aflatoxins (5) and the cytotoxic gliotoxin (6).
The fungal genes required for the biosynthesis of a secondary metabolite are organized into gene clusters composed of hallmark enzymes (5, 7), including polyketide synthases, nonribosomal peptide synthetases, terpene cyclases, dehydrogenases, esterases, and methyltransferases (8). A cluster-specific transcription factor is often required to recognize palindromic sequences and promote the transcription of an entire biosynthetic gene cluster. For example, the sterigmatocystin and aflatoxin gene clusters are controlled by the binuclear zinc finger protein AflR (9). Although gene clusters are of considerable biological and economic interest, much remains unknown about their structure, function, and regulation. In fact, of the 55 clusters predicted by the software SMURF in Aspergillus flavus, only six clusters have actually been characterized (10, 11).
Recent work has focused on elucidating the regulation of a secondary metabolism. In a mutagenesis screen to identify Aspergillus nidulans mutants with reduced sterigmatocystin, a protein was found to regulate AflR and designated LaeA4 (loss of aflR expression A) (12). LaeA is a nuclear protein that when absent results in a decrease in secondary metabolism and virulence (13, 14). Further studies have revealed that LaeA is a master regulator, controlling the transcription of not only the genes in 13 of the 22 secondary metabolite clusters in Aspergillus fumigatus but also 10% of the genome itself (15).
LaeA is a highly conserved protein in filamentous fungi and a conserved virulence factor in all pathogenic fungi examined to date (13, 14, 16–19). An analysis of the 374 amino acids of LaeA shows the presence of S-adenosylmethionine (AdoMet)-binding motifs common to seven-β-strand methyltransferases (12, 20). LaeA is known to be a nuclear protein that interacts with the VeA and VelB proteins in a heterotrimeric complex, called the velvet complex, which functions to coordinate development and secondary metabolism in response to light (21). Because LaeA has similarities to methyltransferases and is localized to the nucleus, it has been suggested that it regulates transcription by protein lysine or protein arginine methyltransferase functions (12, 22, 23). Although no direct biochemical studies on the methyl-accepting specificity of LaeA have been done, this protein has been linked to changes in chromatin structure (24).
Because of the current enigmatic nature of the molecular function of LaeA and the velvet complex, determining the methylated substrates of LaeA would be important to elucidate the global regulatory mechanism of secondary metabolism. Here, we show that the putative methyltransferase LaeA does not methylate histones or the velvet complex proteins in vitro. Additionally, both in vitro and in vivo biochemical analyses were unsuccessful in identifying targets of LaeA. However, in these experiments LaeA was consistently found to be automethylated at a methionine residue. In this work, we have characterized this automethylation reaction.
EXPERIMENTAL PROCEDURES
Construction of Plasmids
Standard techniques were used for nucleic acid manipulations according to Sambrook and Russell (25). All plasmids used in this study are listed in Table 1 and primers used in this study are listed in Table 2. Unless noted otherwise, all plasmids were constructed using a PCR-mediated cloning approach (26). Point mutations and truncations were made using the QuikChange mutagenesis kit (Stratagene). For heterologous expression of LaeA in Escherichia coli, laeA cDNA was introduced into the 6× histidine and maltose-binding protein (MBP) expression vector pKLD116 (27). This resulted in the construction of the full-length MPB-LaeA expression vector (pGSG01). N-terminal truncations of LaeA were made, including MBP-LaeAΔ1–86 (pGSG02), MBP-LaeAΔ1–30 (pGSG04), and MBP-LaeAΔ1–42 (pGSG06). Subsequent methionine to alanine point mutations were constructed in MBP-LaeAΔ1–86 (pGSG02) creating pJMP141, pJMP143, pJMP144, and pJMP145.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pKLD116 | His6-MBP | Ref. 27 |
| pGSG01 | His6-MBP-LaeA | This study |
| pGSG02 | His6-MBP-LaeAΔ1–86 | This study |
| pGSG04 | His6-MBP-LaeAΔ1–30 | This study |
| pGSG06 | His6-MBP-LaeAΔ1–42 | This study |
| pJMP141 | His6-MBP-LaeAΔ1–86, M101A, M195A | This study |
| pJMP143 | His6-MBP-LaeAΔ1–86, M207A | This study |
| pJMP144 | His6-MBP-LaeAΔ1–86, M213A | This study |
| pJMP145 | His6-MBP-LaeAΔ1–86, M268A | This study |
| pJW53 | ¾ pyroA in SspI site of pBluescript II SK− | Ref. 73 |
| pFNO3 | GA5-GFP-A. fumigatus pyrG | Ref. 28 |
| pJW45–4 | laeA(p)::laeA::laeA(t) in ½ trpC | Ref. 12 |
| pJMP148 | laeA(p)::laeA::GFP::laeA(t) in ½ trpC | This study |
| pJMP150 | laeA(p)::laeAM207A::GFP::laeA(t) in ½ trpC | This study |
| pJMP151 | laeA(p)::laeAΔ1–86::GFP::laeA(t) in ½ trpC | This study |
| pJMP152 | laeA(p)::laeA::laeA(t) in ¾ pyroA | This study |
| pJMP154 | laeA(p)::laeA::GFP::laeA(t) in ¾ pyroA | This study |
| pJMP156 | laeA(p)::laeAM207A::GFP::laeA(t) in ¾ pyroA | This study |
| pJMP157 | laeA(p)::laeAΔ1–86::GFP::laeA(t) in ¾ pyroA | This study |
| pGST-velB | GST-VelB | Ref. 31 |
| pJMP134 | His6-veA-GST-Stag | Ref. 31 |
TABLE 2.
Oligonucleotides used in this study
| Primers | Sequence (5′ to 3′) | Purpose |
|---|---|---|
| LaeA_N_116 | GCAGGTGCATGTGGACGTCCCGGGCTAGCATGTTTGAGATGGGCCCGGTGGGAACTCG | pGSG01 |
| LaeA_C_116 | GCGGCCGCAAGCTTGCCTGCAGGCCATGTTATCTTAATGGTTTCCTAGCCTGG | pGSG01 |
| AnLAEAdel86F | CCACtagtGAGAATCTCTACTTCCAAGGCTCGGATCATGAAGAGAACGGACGCACCTACC | pGSG02 |
| AnLaeAdel30F | GAGAATCTCTACTTCCAAGGCCTTAGCGACAGAGGCCGGTCAAGGC | pGSG04 |
| AnLaeAdel30R | GCCTTGACCGGCCTCTGTCGCTAAGGCCTTGGAAGTAGAGATTCTC | pGSG04 |
| AnLaeAdel42F | GAGAATCTCTACTTCCAAGGCCTTAGCGACATCCAGTCCATCACTGAACG | pGSG06 |
| AnLaeAdel42R | CGTTCAGTGATGGACTGGATGTCGCTAAGGCCTTGGAAGTAGAGATTCTC | pGSG06 |
| JP AnLaeA M101A For | ACCATGGCTTTCGCAGGGGAgcGTATTTTCTTCCGTGCGATG | LaeAM101A |
| JP AnLaeA M101A Rev | CATCGCACGGAAGAAAATACgcTCCCCTGCGAAAGCCATGGT | LaeAM101A |
| JP AnLaeA M195A For | GACTTCGAAGCGCCATGGGCCgcGGGGGAGGATTCCTGGGATC | LaeAM195A |
| JP AnLaeA M195A Rev | GATCCCAGGAATCCTCCCCCgcGGCCCATGGCGCTTCGAAGTC | LaeAM195A |
| JP AnLaeA M207A For | GGGATCTAATCCATCTGCAGgcGGGTTGCGGTAGTGTCATGG | LaeAM207A |
| JP AnLaeA M207A Rev | CCATGACACTACCGCAACCCgcCTGCAGATGGATTAGATCCC | LaeAM207A |
| JP AnLaeA M213A For | CAGATGGGTTGCGGTAGTGTCgcGGGCTGGCCAAACTTGTATC | LaeAM213A |
| JP AnLaeA M213A Rev | GATACAAGTTTGGCCAGCCCgcGACACTACCGCAACCCATCTG | LaeAM213A |
| JP AnLaeA M268A For | CTTAAACAGGCGACAGCAGAGACCgcGCGGCCAATCGCCCATAGCTC | LaeAM268A |
| JP AnLaeA M268A Rev | GAGCTATGGGCGATTGGCCGCgcGGTCTCTGCTGTCGCCTGTTTAAG | LaeAM268A |
In vivo complementation vectors were based off of the vector pJW45-4 (12) harboring the native laeA gene from A. nidulans. First, GA5-GFP (glycine-alanine linker fused to GFP) was amplified from pFNO3 (28) and inserted in-frame at the C terminus of laeA yielding pJMP148. Truncation of the N-terminal 86 amino acids of laeA and the methionine 207 to alanine point mutant was constructed in pJMP148 to make pJMP151 and pJMP150, respectively. Finally, HindIII fragments from pJW45-4, pJMP148, pJMP150, and pJMP151 were subsequently cloned into the HindIII site of pJW53 (29) yielding the pyroA targeting complementation vectors of pJMP152, pJMP154, pJMP156, and pJMP157.
Expression and Purification of Proteins
MBP-LaeA proteins were expressed in Rosetta DE3 (Novagen) and subsequently purified as described previously (27) with the modifications indicated below. Briefly, proteins were first purified using nickel-nitrilotriacetic acid resin (Qiagen) followed by a second round of purification on amylose resin (New England Biolabs). Removal of the N-terminal 6× histidine and MBP tag was achieved by proteolytic cleavage using recombinant tobacco etch virus (30) and subsequent removal of the recombinant tobacco etch virus and MBP tags with nickel-nitrilotriacetic acid resin. Purification of VeA-GST and GST-VelB was done according to Palmer et al. (31).
Creation of Mutant Fungal Strains and Fungal Physiology Experiments
Fungal strains used in this study are listed in Table 3. All strains were maintained on glucose minimal medium (GMM) (32) at 37 °C and when appropriate were supplemented with 0.5 mm pyridoxine-HCl, 0.01 μg/ml riboflavin, 5 mm uridine, and 5 mm uracil. In vivo complementation of ΔlaeA strains was achieved by targeted integration of laeA complementation vectors at the pyroA locus in RJMP155.16 (riboB2, pyroA4, and ΔlaeA). Transformation was conducted as described previously in Szewczyk et al. (33). Transformants were screened and verified through a combination of diagnostic PCR and Southern blots (data not shown). Strains harboring a single integration of the complementation vector at the pyroA locus were subsequently crossed to RJW130.1 (pyroA4 and ΔlaeA) and/or RJMP260.28 (pyroA4, hhoA-mCherry, and ΔlaeA) to generate prototrophic strains. Analysis of sterigmatocystin production was done from point-inoculated cultures grown for 4 days in dark and light conditions, and sterigmatocystin was extracted and visualized via thin layer chromatography (31). Analysis of asexual and sexual sporulation was conducted as described previously (34) with the minor modification of using Champe's media for sexual developmental induction (35). Visualizing the cellular localization of LaeA-GFP fusion proteins was conducted as described previously in Palmer et al. (31) using a Zeiss AxioVision A10 microscope.
TABLE 3.
Strains of A. nidulans used in this study
| Strain | Genotype | Source |
|---|---|---|
| RJMP103.5 | Wild type | Ref. 74 |
| RJW41A | metG1, ΔlaeA::metG | Ref. 21 |
| RJW152.1 | ΔlaeA::metG, pyroA::gpdA(p)::laeA | J. W. Bok, unpublished data |
| RJMP155.16 | riboB2, pyroA4, metG1, ΔlaeA::metG | This study |
| TJMP154.5 | riboB2, metG1, ΔlaeA::metG, pyroA::laeA | This study |
| TJMP156.1 | riboB2, metG1, ΔlaeA::metG, pyroA::laeA-GFP | This study |
| TJMP158.7 | riboB2, metG1, ΔlaeA::metG, pyroA::laeAM207A-GFP | This study |
| TJMP159.3 | riboB2, metG1, ΔlaeA::metG, pyroA::laeAΔ1–86-GFP | This study |
| RJW130.1 | metG1, pyroA4, ΔlaeA::metG | J. W. Bok, unpublished data |
| RJMP254.1 | ΔlaeA::metG, pyroA::laeA | This study |
| RJMP256.3 | ΔlaeA::metG, pyroA::laeA-GFP | This study |
| RJMP258.1 | ΔlaeA::metG, pyroA::laeAM207A-GFP | This study |
| RJMP259.1 | ΔlaeA::metG, pyroA::laeAΔ1–86-GFP | This study |
| TJMP125.1 | riboB2, pyroA4, pyrG89, hhoA::mCherry::riboB, ΔnkuA, veA1 | J. M. Palmer, unpublished data |
| RJMP260.28 | pyroA4, ΔlaeA::metG, hhoA::mCherry::riboB | This study |
| RJMP260.1 | ΔlaeA::metG, hhoA::mCherry::riboB | This study |
| RJMP261.26 | ΔlaeA::metG, hhoA::mCherry::riboB, pyroA::laeA-GFP | This study |
| RJMP263.3 | ΔlaeA::metG, hhoA::mCherry::riboB, pyroA::laeAM207A-GFP | This study |
| RJMP264.5 | ΔlaeA::metG, hhoA::mCherry::riboB, pyroA::laeAΔ1–86-GFP | This study |
Protein Extraction from Aspergillus
Extracts were prepared from cells in vegetative, asexual, and sexual growth stages. Vegetative growth was achieved by incubating A. nidulans in liquid shaking cultures (GMM, 230 rpm, 37 °C) for 24 h with an inoculum density of 1 × 106 spores/ml. Asexual development was induced by growing mycelia as described above for vegetative growth, filtering the mycelia through sterile Miracloth (Calbiochem), and then transferring the mycelia to solid media (GMM) under constant room light for an additional 24 h. Sexual developmental was induced as described above for asexual induction except the solid medium plates were wrapped in parafilm and aluminum foil and incubated for a further 48 h in the dark. Mycelia were collected from all three growth stages and immediately frozen in liquid nitrogen. Whole cell extracts were prepared by grinding mycelia to a fine powder under liquid nitrogen, followed by vortexing in extraction buffer (100 mm Tris-HCl, pH 7.5, 250 mm NaCl, 10% glycerol, 0.1% Nonidet P-40, 1 mm EDTA, and one Roche Complete Protease inhibitor tablet without EDTA per 50 ml), and removing cellular debris by centrifugation at 20,000 × g for 20 min.
Crude nuclear extracts were prepared essentially as described previously (36). Briefly, 250 ml of liquid GMM medium was inoculated with 1 × 106 spores/ml and incubated at 37 °C for 48 h at 250 rpm. Mycelia were collected by filtering through sterile Miracloth, frozen in liquid nitrogen, and ground to a fine powder using a mortar and pestle. Approximately 5 g of ground mycelia was vortexed with cold nuclear isolation buffer (10 mm Tris-HCl, 1 m sorbitol, 10 mm EDTA, 0.15 mm spermine, 0.5 mm spermidine, 2.5 mm PMSF, pH 7.5). Cell debris was removed by centrifugation at 1000 × g for 10 min at 4 °C, and the supernatant was filtered through Miracloth. The supernatant was centrifuged at 10,000 × g for 15 min at 4 °C, and the pellet was subsequently washed in cold resuspension buffer (10 mm Tris-HCl, 1 m sorbitol, 1 mm EDTA, 0.15 mm spermine, 0.5 mm spermidine, 2.5 mm PMSF, pH 7.5). Crude nuclei were then pelleted by centrifugation at 10,000 × g for 15 min at 4 °C. Finally, crude nuclei were resuspended in cold ST buffer (10 mm Tris-HCl, 1 m sorbitol, protease inhibitor mixture for fungi (Sigma P8215)). All protein extracts were quantified using the protein assay according to manufacturer's recommendations (Bio-Rad).
In Vitro Methylation Reactions and Fluorography
Methylation reactions were performed in a final volume of 100 μl and included [3H]AdoMet diluted from a 7.1 μm stock solution (S-adenosyl-l-[methyl-3H]methionine; PerkinElmer Life Sciences, 75–85 Ci/mmol, 0.55 mCi/ml in 10 mm H2SO4/EtOH (9:1, v/v)). Recombinant proteins, whole cell extracts, nuclear extracts, or human recombinant histones H2A, H2B, H3.3, and H4 (New England Biolabs, 1 mg/ml stock) were added and incubated in a buffer of 50 mm Tris-HCl, pH 7.5, 300 mm NaCl, 5% glycerol, and 1 mm DTT. Reactions were allowed to proceed at 37 °C for 18–20 h. The reactions were terminated by adding SDS-PAGE loading buffer (final concentration of 250 mm Tris-HCl, pH 6.8, 10% (w/v) SDS, 50% (v/v) glycerol, 5% (v/v) β-mercaptoethanol, and 0.05% (w/v) bromphenol blue) and heated at 100 °C for 3 min. The entire sample was loaded onto 10% Tris glycine or 10% BisTris-HCl polyacrylamide slab gels as described in Frankel and Clarke (37) and Kinoshita and Kinoshita-Kikuta (38), respectively. The samples were separated by electrophoresis for ∼5 h at a constant current of 60–80 mA. The gel was stained with Coomassie (0.1% (w/v) Brilliant Blue R-250, 10% (v/v) glacial acetic acid, and 50% (v/v) methanol) for 1 h, destained in 10% (v/v) acetic acid and 5% (v/v) methanol, and imaged using an Alpha Imager 2200 (Alpha Innotech Corp.) to visualize protein bands. To identify methylated species, fluorography was performed by soaking the gels for 1 h in EN3HANCE (PerkinElmer Life Sciences) followed by a 30-min water treatment. The gel was subsequently dried for 3 h at 80 °C before being exposed to film (Denville Scientific, 5 × 7-inch HyBlot CL). Gels were exposed to film for 3 days to 9 months at −80 °C.
High Resolution Cation Exchange Chromatography Amino Acid Analysis
Radioactive bands of interest from dried gels were cut out and rehydrated in 1 ml of water for 1 h. The gel slice was diced into smaller pieces and placed into several 6 × 50-mm glass tubes with 100 μl of 6 m HCl. Acid hydrolysis was carried out in a Waters Pico-Tag Vapor-Phase apparatus in a vacuum vial with an additional 500 μl of 6 m HCl for 18 h at 110 °C. After the hydrolysates were vacuum dried, they were resuspended in 100 μl of water, and centrifuged at 10,600 × g for 10 min to remove any debris. The sample was added to 360 μl of citrate buffer (0.2 m Na+, pH 2.2) and 1 μmol each of the following standards: dl-methionine S-methylsulfonium chloride (S-methylmethionine, Sigma M0501); l-ω-NG-monomethylarginine acetate salt (Sigma M7033); l-ω-NG,NG-asymmetric dimethylarginine dihydrochloride salt (Sigma D4268); l-1-(π)-methylhistidine (Sigma M9005); ϵ-monomethyl-lysine hydrochloride salt (Sigma M6004); ϵ-N-dimethyl-lysine (Sigma 19773), and ϵ-N-trimethyl-lysine hydrochloride salt (Sigma M1660).
All 500 μl of sample was loaded onto a 0.9 × 8-cm column of PA-35 sulfonated polystyrene beads (6–12 μm, Benson Polymeric Inc., Sparks, NV). The column was equilibrated and eluted with citrate buffer (0.35 m Na+, pH 5.27) at 55 °C and a flow rate of 1 ml/min, and 1-ml fractions were collected. The elution times of amino acids were determined by ninhydrin assay. Into a 96-well plate, 30 μl of cation exchange fraction was added to 200 μl of water and 100 μl of ninhydrin reagent (20 mg/ml ninhydrin and 3 mg/ml hydrindantin in 75% (v/v) dimethyl sulfoxide and 25% (v/v) 4 m lithium acetate, pH 4.2) and heated at 100 °C for 15 min. Absorbance was measured at 570 nm using a SpectraMax M5 microplate reader with a path length of 1 cm. The radioactivity in each column fraction was quantified using a Beckman LS6500 scintillation counter and expressed as an average of three 5-min counting cycles after mixing 200 μl of sample, 400 μl of water, and 5 ml of scintillation reagent (Safety Solve, Research Products International, 111177).
o-Phthalaldehyde (OPA) Amino Acid Analysis
Amino acid standards or cation exchange column fractions were fluorescently labeled with OPA, separated with an Agilent Eclipse AAA HPLC column (5 μm, 4.6-mm inner diameter, and 150-mm length) at 1.5 ml/min, and detected by a Gilson Model 121 fluorometer (excitation and emission filters of 305–395 and 430–470 nm, respectively, and a setting of 0.1 relative fluorescence units) as described previously (39). During the HPLC analysis, 0.5-ml column fractions were collected and mixed with 5 ml of scintillation reagent to quantify radioactivity. The radioactivity was expressed as an average of three 10-min counting cycles.
Multiple Sequence Alignment and Protein Modeling of LaeA
A multiple-sequence alignment of LaeA proteins from different fungi was generated using Clustal Omega (40) and Jalview (41). Three-dimensional structure prediction of LaeA based on the primary sequence was performed using Phyre2 (42). The prediction of ligands and their respective binding sites in LaeA was calculated by 3DLigandSite (43).
In Vivo Methylation Assay
Mycelia were grown overnight in 50 ml of GMM. Approximately 1 g of mycelia was collected by vacuum filtration, transferred to 2 ml of fresh GMM with 125 μl of [3H]AdoMet (7.1 μm, 75–85 Ci/mmol) to a final concentration of 0.4 μm, and incubated at 37 °C in a gyrorotary shaker at 230 rpm. After 6 h, the samples were centrifuged, and the supernatant was removed. The tissue was lyophilized overnight and ground to a powder. The sample was resuspended in extraction buffer (100 mm Tris-HCl, pH 7.5, 250 mm NaCl, 10% glycerol, 0.1% Nonidet P-40, 1 mm EDTA, and 1 Roche Complete Protease inhibitor without EDTA per 50 ml). The mycelia were vortexed, stored on ice for 10 min, and centrifuged at 20,000 × g for 20 min. The supernatant (“Soluble Fraction”) was removed. Both the soluble fraction and the pellet (“Insoluble Fraction”) were added to SDS-PAGE loading buffer, heated at 100 °C for 3 min, and loaded on a gel as described above.
RESULTS
Soluble N-terminal Truncation of LaeA Demonstrates Activity in Vivo
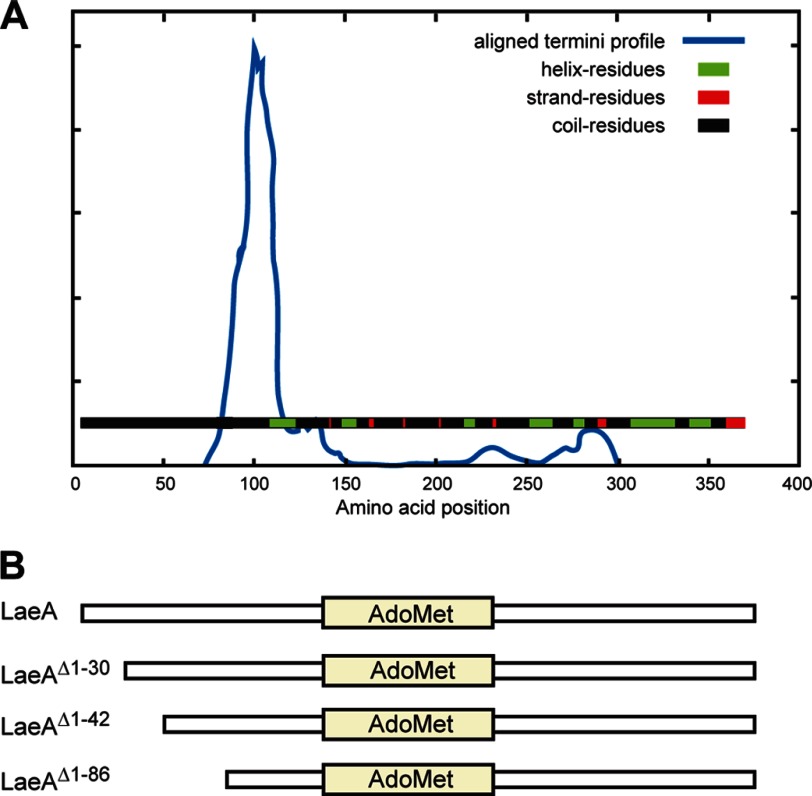
To elucidate the function of LaeA, we heterologously expressed LaeA in E. coli. Our initial expression trial of LaeA with a 6× histidine N-terminal tag proved to be insoluble. To improve solubility, we constructed an N-terminal maltose-binding protein fusion protein (MBP-LaeA), which proved to be soluble and eluted as a monomer in gel filtration analysis. Although the MBP-LaeA full-length protein was soluble, upon cleavage of the N-terminal MBP tag, LaeA rapidly became insoluble. To facilitate the in vitro functional characterization of LaeA, partial proteolysis experiments were conducted on MBP-LaeA (data not shown). These data indicated that portions of the protein were indeed soluble. DomPred domain prediction (44) of LaeA revealed a putative domain boundary at about residue 100 (Fig. 1), and multiple sequence alignments of LaeA homologs indicated that the N terminus of LaeA was variable up to residue 88 (Fig. 2). We thus made several N-terminal truncations of LaeA, and we found that truncation mutants of the N-terminal 30, 42, and 86 amino acids all resulted in soluble protein (Fig. 1; data not shown).
FIGURE 1.
DomPred predicts a domain boundary of LaeA near amino acid 100. A, full-length LaeA amino acid sequence was inputted to the DomPred domain prediction server. The output “aligned termini profile” predicts that LaeA has two domains split near amino acid residue 100. B, because the first motif of the AdoMet-binding site starts at amino acid 141, we chose to make N-terminal truncation mutants by removing 30, 42, or 86 residues.
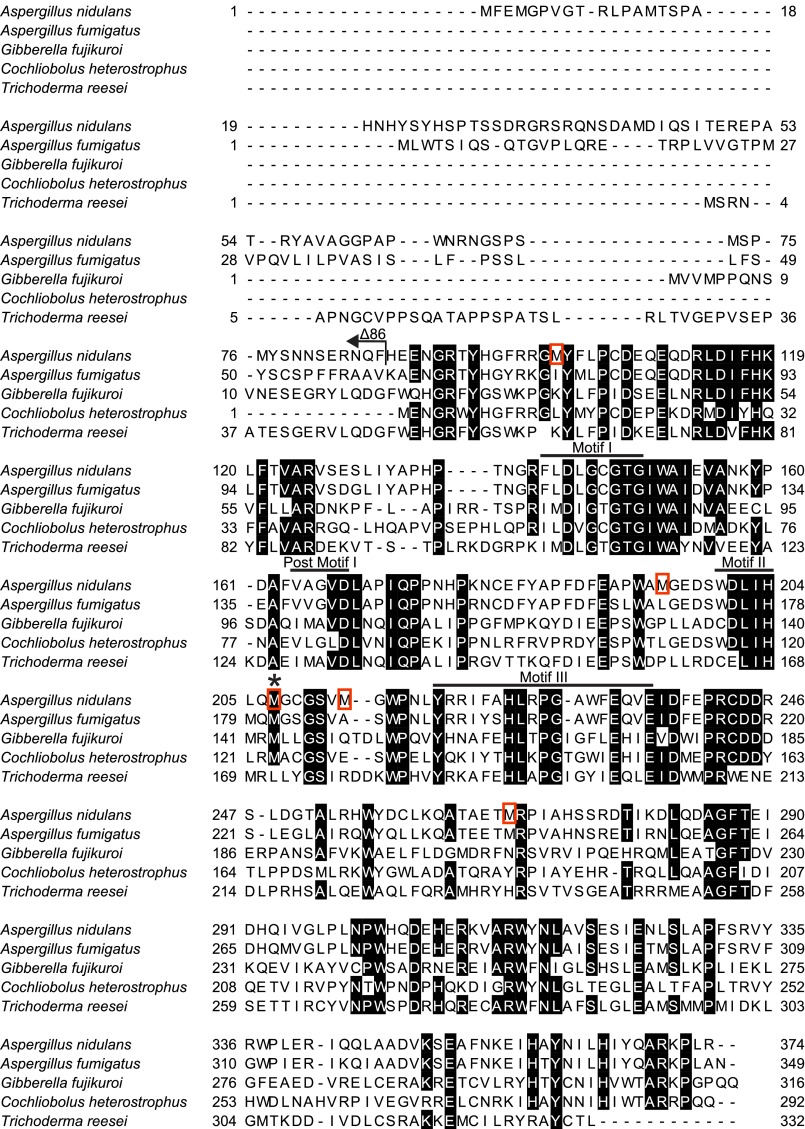
FIGURE 2.
Sequence alignment of LaeA from A. nidulans with homologs in other fungal species. Amino acids with 80% or greater conservation are indicated in black. The arrow indicates the amino acid sequence missing at the N terminus of the LaeAΔ1–86 protein. Bars are used to denote the amino acids comprising the four common methyltransferase motifs of seven-β-strand methyltransferases. Red boxes highlight methionine residues mutated in this study. Amino acid sequences are taken from UniProt accession numbers C8VQG9, Q6TFC7, E0WDF6, G4XKY9, and I3RU94.
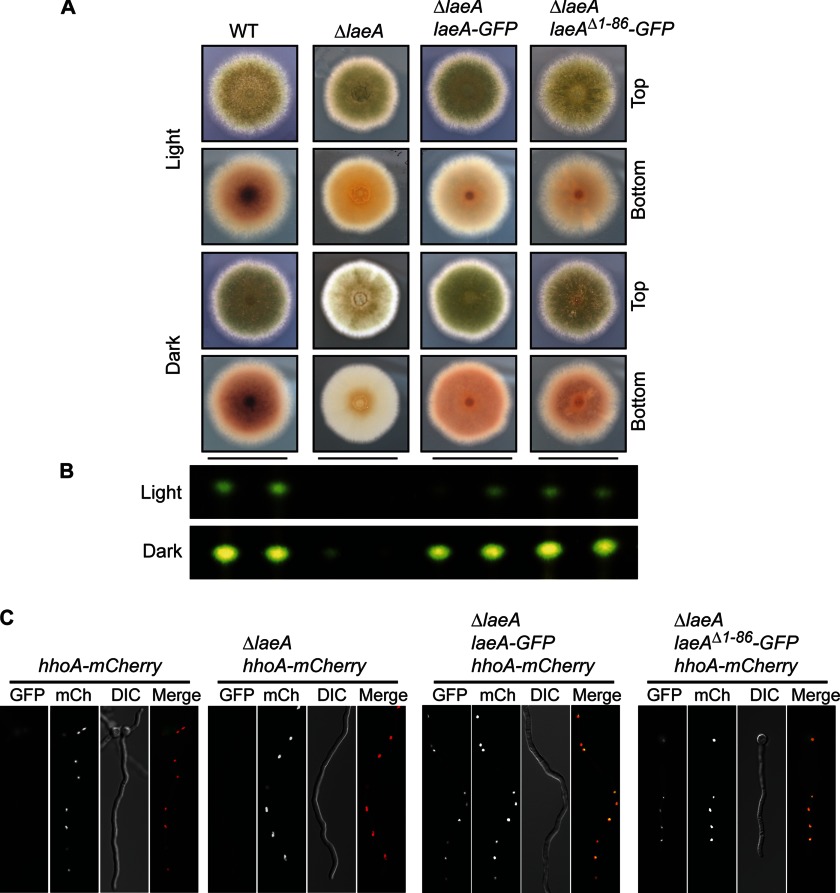
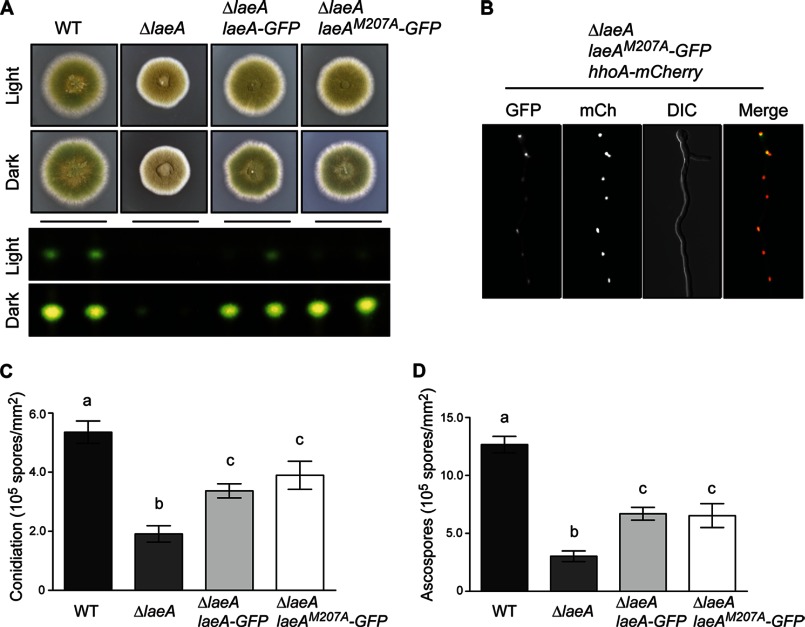
We next determined whether the largest truncation mutant (LaeAΔ1–86) was biologically active and able to complement ΔlaeA in vivo. To simultaneously test the function of the LaeAΔ1–86 mutant and determine cellular localization, we constructed several laeA complementation vectors in the 3/4 pyroA targeting vector that are driven by the native laeA promoter (p) as well as terminator (t): laeA(p)::laeA::laeA(t), laeA(p)::laeA-GFP::laeA(t), and laeA(p)::laeAΔ1–86-GFP::laeA(t). These constructs were then targeted to the pyroA locus in a ΔlaeA background. Transformants were obtained and crossed to prototrophy. Importantly, LaeAΔ1–86-GFP was capable of complementing the ΔlaeA phenotype as sterigmatocystin production and sexual development returned to levels similar to wild type (Fig. 3, A and B). Moreover, LaeAΔ1–86-GFP localized to the nucleus (Fig. 3C). These data suggest that the N terminus of LaeA is dispensable for function and validated the use of the LaeAΔ1–86 protein for further in vitro methylation assays.
FIGURE 3.
N-terminal 86 amino acids of LaeA are dispensable for function. Complementation of ΔlaeA was accomplished by targeted integration of constructs at the pyroA locus. A, strains of A. nidulans were grown for 4 days under light or dark conditions on glucose minimal media where null mutants of laeA produce fewer conidia and reduced colony pigmentation. As expected, a carboxyl GFP tag of laeA complements these gross phenotypes. Additionally, truncation of the N-terminal 86 amino acids (LaeAΔ1–86-GFP) also results in restoration of the gross phenotype of ΔlaeA strains. B, production of the mycotoxin sterigmatocystin was extracted and visualized via thin layer chromatography from cultures shown in A. Sterigmatocystin is nearly absent in ΔlaeA strains, and this phenotype is restored in both the LaeA-GFP and LaeAΔ1–86-GFP complementation strains. C, LaeA-GFP localized constitutively in the nucleus as expected, and the N-terminal truncation mutant LaeAΔ1–86-GFP also localizes exclusively in the nucleus. Histone H1 fused to mCherry (hhoA-mCherry) serves as a marker for nuclei. Strains used are as follows: WT (RJMP260.12), ΔlaeA (RJMP260.1), ΔlaeA laeA-GFP (RJMP261.26), and ΔlaeA laeAΔ1–86-GFP (RJMP264.5). DIC, differential interference contrast.
Is LaeA a Protein Methyltransferase?
From the presence of the four appropriately spaced sequence motifs (I, post-I, II, and III) associated with the AdoMet binding domain of seven-β-strand methyltransferases present in the amino acid sequence of LaeA, a catalytic function in methyl transfer is indicated (Fig. 2) (12, 20). Such a function has been supported by the loss of LaeA function in vivo in a double mutation in motif I (23). Furthermore, it has been shown that LaeA can bind AdoMet in vitro (31). These data all suggest that LaeA could be a bona fide methyltransferase. Although BLAST searches of the LaeA sequence reveal it contains common motifs shared by seven-β-strand methyltransferases (Fig. 2), this group of enzymes is involved in modifying such a variety of DNA, RNA, protein, lipid, and small molecule substrates that it is impossible to infer potential LaeA methyl-accepting substrates.
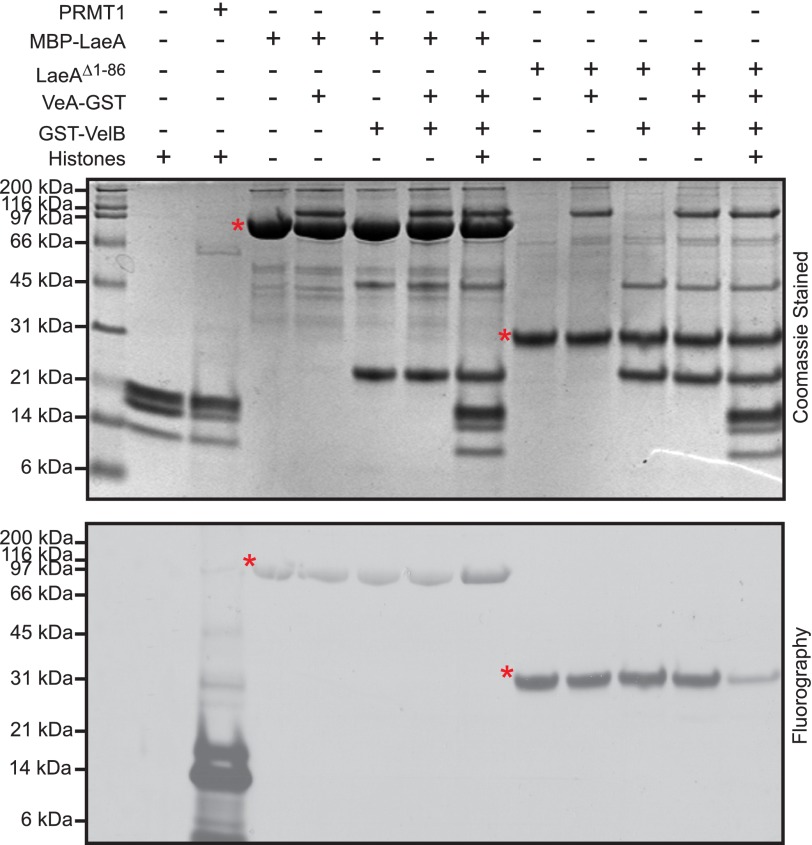
However, the function of LaeA in controlling gene expression suggested the possibility that LaeA could be a protein methyltransferase similar to those histone methyltransferases involved in transcriptional activation and repression (45–47). LaeA is known to transcriptionally regulate as much as 10% of the transcriptome (15) and subsequently has been linked to changes in chromatin structure (24). However, it is unknown whether this is a direct or indirect effect. To specifically address the possibility that LaeA methylates histone proteins or is required to be in a protein complex to methylate a substrate, we incubated MBP-LaeA or LaeAΔ1–86 with human recombinant histone proteins (H2A, H2B, H3.3, and H4) or with proteins associated with the velvet complex (VeA and VelB). The reaction components were separated by gel electrophoresis and analyzed by fluorography (Fig. 4). As a control, we showed that the protein arginine methyltransferase PRMT1 robustly methylates histones (48). Although we did not observe any methylation of recombinant histones or the velvet complex in these experiments, we were quite surprised to find that both MBP-LaeA and LaeAΔ1–86 automethylated (Fig. 4). The extent of automethylation appeared to be ∼10-fold greater for LaeAΔ1–86 than for MBP-LaeA, suggesting a potential regulatory role of the N terminus.
FIGURE 4.
Recombinant LaeAΔ1–86 or MBP-LaeA does not methylate purified VeA, VelB, or histone proteins. In vitro methylation reactions were prepared with 1.7 μm [3H]AdoMet and incubated for 20 h at 37 °C with methyltransferases and potential methyl-accepting substrates, including 5 μg each of recombinant human histone H2A, H2B, H3.3, and H4, 1 μg of purified PRMT1 (positive control), 27.4 μg of MBP-LaeA, ∼5 μg of VeA, ∼5 μg of VelB, and 8.8 μg of LaeAΔ1–86. The samples were separated on a 10% BisTris gel with MES running buffer and stained with Coomassie. Polypeptide molecular weight markers (Bio-Rad broad range, ∼3 μg of each protein, catalogue number 161-0317) were electrophoresed in a parallel lane and shown on the left, and include myosin (200 kDa), β-galactosidase (116 kDa), phosphorylase b (97 kDa), serum albumin (66 kDa), ovalbumin (45 kDa), carbonic anhydrase (31 kDa), trypsin inhibitor (21 kDa), lysozyme (14 kDa), and aprotinin (6 kDa). Fluorography was performed by treating the gel with EN3HANCE and exposing the dried gels to film for 5 days at −80 °C as described under “Experimental Procedures.” Red asterisks indicate the position of MBP-LaeA and LaeAΔ1–86.
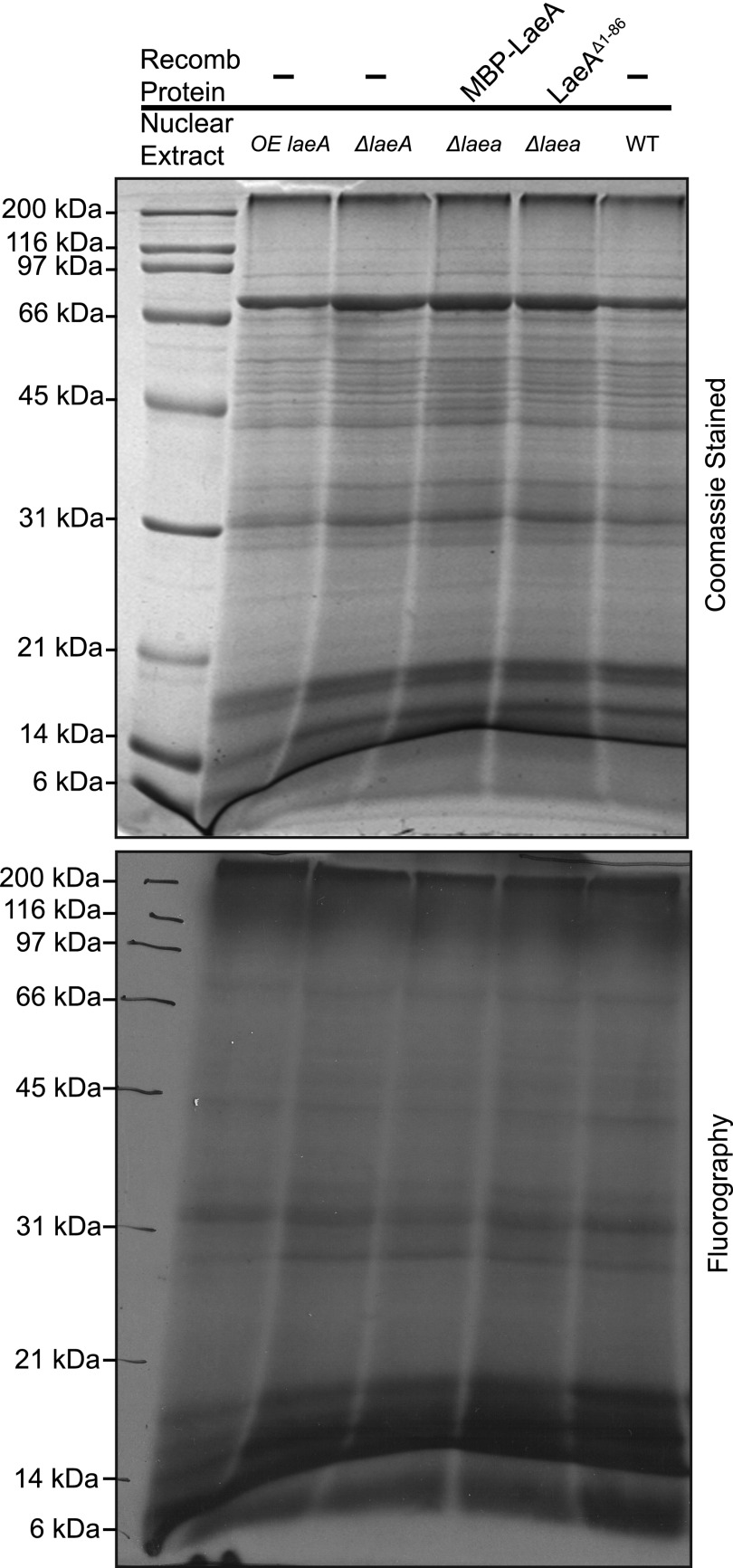
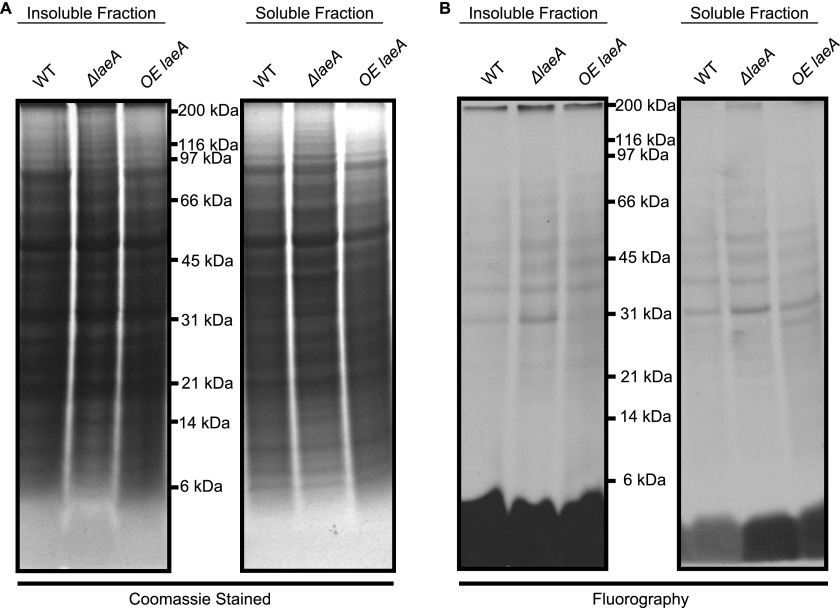
Because LaeA can translocate into the nucleus, we looked for methylated substrates in wild type, ΔlaeA, and overexpressing laeA crude nuclear extracts incubated with [3H]AdoMet. Analysis of the polypeptides separated by SDS-PAGE indicated no differences in the pattern of Coomassie staining or in the fluorography of 3H-methylated proteins (Fig. 5). We then asked whether we could find potential LaeA substrates in whole cell extracts from wild type and ΔlaeA strains derived from vegetative, asexual, and sexual growth stages. Coomassie staining and fluorography of extracts incubated with [3H]AdoMet and separated by SDS-PAGE showed no significant differences in the pattern of polypeptides or methylated species (Fig. 6). Finally, we in vivo labeled wild type, ΔlaeA, and OE laeA strains with [3H]AdoMet. Gel electrophoresis and fluorography revealed no significant protein or methylation differences between the three different strains (Fig. 7). Interestingly, out of all the potential methylation substrates, LaeA automethylation represented the predominant methylated species when the enzyme was incubated with any of the extracts.
FIGURE 5.
In vitro methylation assay using wild type, laeA overexpressing, and ΔlaeA nuclear extracts. Approximately 110 μg of crude nuclear protein from each strain was isolated as described under “Experimental Procedures” and incubated with 100 nm [3H]AdoMet for 4 h at 37 °C. When added, MBP-LaeA and LaeAΔ1–86 were present at 4 μg of protein. The samples were electrophoresed on a 10% Tris glycine polyacrylamide gel with Tris glycine running buffer and Coomassie-stained. Fluorography was performed by treating the gel with EN3HANCE and exposing the dried gel to film for 9 months at −80 °C. The positions of Bio-Rad broad range marker proteins are shown on the left. Recomb, recombinant.
FIGURE 6.
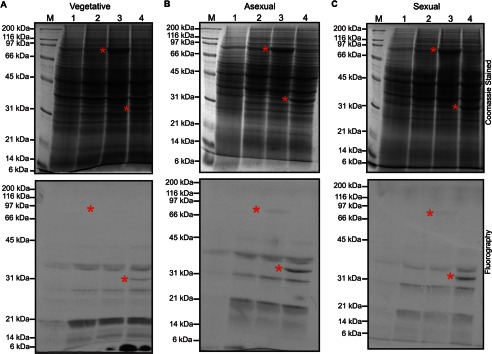
In vitro methylation of wild type and ΔlaeA extracts of A. nidulans at vegetative, asexual, and sexual growth phases do not reveal LaeA methyl-accepting substrates. Whole cell extracts from wild type or ΔlaeA strains in vegetative (A), asexual (B), or sexual (C) growth stages were prepared as described under “Experimental Procedures.” Samples analyzed were as follows: lane 1, 300 μg of wild type whole cell extract; lane 2, 300 μg of ΔlaeA whole cell extract; lane 3, 300 μg of ΔlaeA whole cell extract mixed with 10 μg of purified MBP-LaeA, and lane 4, 300 μg of ΔlaeA whole cell extract mixed with 10 μg of purified LaeAΔ1–86. Each sample was incubated with 100 nm [3H]AdoMet for 3 h at 37 °C, separated by 10% Tris glycine polyacrylamide gels with Tris glycine running buffer, and stained with Coomassie. The gels were treated with EN3HANCE, and the dried gels were exposed to film for 1 month at −80 °C. The positions of Bio-Rad broad range marker proteins (M) are shown on the left. Red asterisks denote the position of MBP-LaeA or LaeAΔ1–86 in the Coomassie-stained gel and in the fluorograph.
FIGURE 7.
In vivo protein methylation of wild type, laeA-overexpressing, and ΔlaeA A. nidulans strains separated into insoluble and soluble fractions. Aspergillus mycelia was labeled with [3H]AdoMet as described under “Experimental Procedures.” Lyophilized mycelia were ground to a powder and resuspended in 100 mm Tris-HCl, pH 7.5, 250 mm NaCl, 10% glycerol, 0.1% Nonidet P-40, and 1 mm EDTA. The resulting lysate was vortexed, kept at 0 °C for 10 min, and centrifuged at 20,000 × g for 20 min at 4 °C. The supernatant Soluble Fraction and the pellet Insoluble Fraction were fractionated by SDS-PAGE as described under “Experimental Procedures” on a 10% BisTris polyacrylamide gel with MES-SDS running buffer. A, Coomassie-stained gel. B, fluorograph of EN3HANCE-treated gels after 1 month at −80 °C. The positions of Bio-Rad broad range marker proteins are indicated.
LaeAΔ1–86 and MBP-LaeA Are Automethylated at Methionine 207
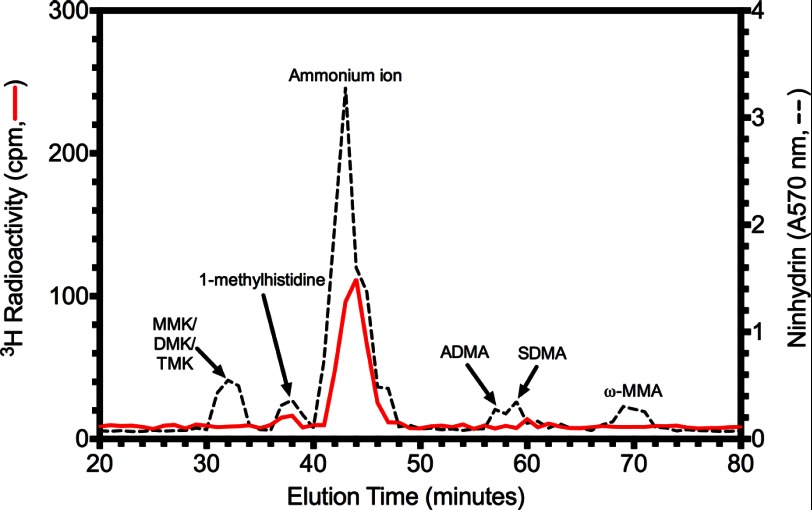
Because a number of protein arginine and lysine methyltransferases have been shown to be automethylated at the same types of residues as their substrates (49–52), we decided to identify the automethylation sites of LaeA in the expectation that it might lead us to the residue of methylation in its substrate. To determine the modified residue, the radioactive LaeAΔ1–86 species was excised from the gel, acid-hydrolyzed, and characterized for possible neutral and basic 3H-methylated amino acid products by high resolution cation exchange chromatography. We found that there was no incorporation of 3H radioactivity in peaks co-migrating with added basic amino acid standards of mono-, di-, and tri-methylated lysine, mono-methylated histidine, and mono- and di-methylated arginine (Fig. 8). However, we did note a significant radioactive peak migrating between the methylated histidine and arginine standards near the peak of ammonium ion, which results from the hydrolysis of the polyacrylamide gel. We thus considered the possibility that the radioactivity was associated with S-methylmethionine, another basic amino acid derivative.
FIGURE 8.
A novel LaeA automethylated amino acid residue. 30 μg of purified LaeAΔ1–86 was incubated with 2.8 μm [3H]AdoMet for ∼20 h at 37 °C as described under “Experimental Procedures.” Reaction mixtures separated by 10% BisTris PAGE were stained, destained, and treated with EN3HANCE. After fluorography, the [3H]AdoMet-labeled LaeA protein band was excised, rehydrated with water, and acid-hydrolyzed as described under “Experimental Procedures.” After mixing with standards of methylated amino acids (1 μmol each) including 1-(π)-methylhistidine, ω-NG-monomethylarginine (ω-MMA), ω-NG,NG-dimethylarginine (ADMA), ω-NG,NG′-dimethylarginine (SDMA), ϵ-N-monomethyllysine hydrochloride (MMK), ϵ-N-dimethyllysine (DMK), and ϵ-N-trimethyllysine (TMK), high resolution cation exchange chromatography was performed as described under “Experimental Procedures.” The positions of the standard amino acids were determined by ninhydrin assay as described under “Experimental Procedures” and indicated by a dashed line. Radioactivity was determined in 200 μl of each fraction and is shown by a red solid line.
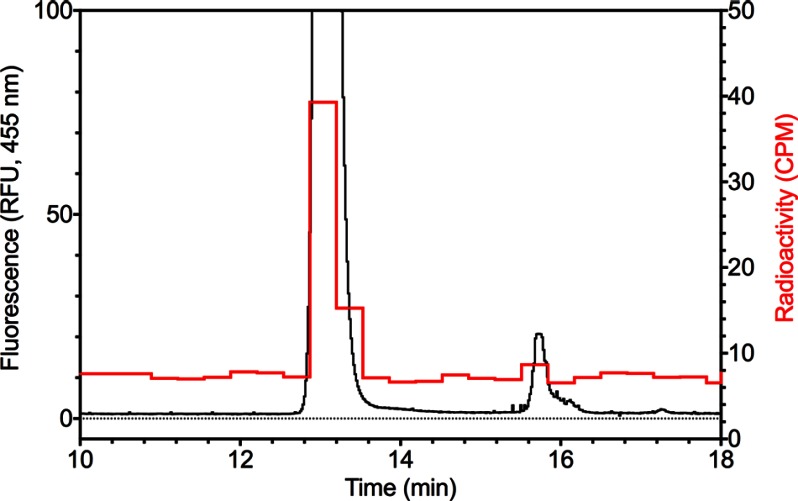
We then repeated the high resolution chromatographic analysis with the hydrolysate of 3H-automethylated MBP-LaeAΔ1–86 and a standard of S-methylmethionine greatly in excess of the amount of ammonium ion. We found that the radioactivity eluted just prior to the standard, in a position that would be expected from the tritium isotope effect previously observed with methyl derivatives of lysine, histidine, and arginine (Fig. 9B) (53–56). To confirm the identification of S-methyl[3H]methionine, we fluorescently labeled the cation exchange chromatography fractions with OPA and analyzed the components by reverse phase HPLC and fluorometry. Analysis of the cation exchange fraction containing S-methylmethionine identified the fluorescently modified amino acid and confirmed its co-elution with the radioactivity (Fig. 10). These results show that LaeA is automethylated on a methionine residue and that the function of the enzyme may be as a protein methionine methyltransferase.
FIGURE 9.
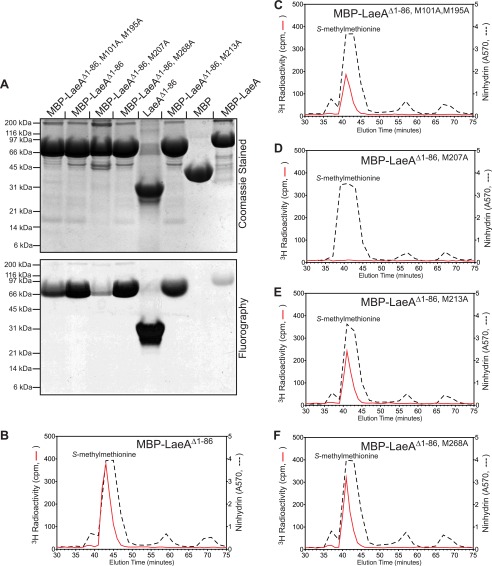
Direct demonstration of LaeA automethylation at methionine 207 by gel electrophoresis and high resolution cation exchange chromatography. Constructs were made to express His-tagged MBP fusion proteins of LaeAΔ1–86 where each methionine residue in the LaeAΔ1–86 sequence was replaced individually by an alanine residue. 150 μg of each of the purified LaeA proteins, with the exception of MBP-LaeA (lane 8, 82 μg), were incubated with 1.4 μm [3H]AdoMet for 20 h at 37 °C, and the reaction products were separated by 10% BisTris PAGE with MES running buffer as described under “Experimental Procedures.” A, gel was stained with Coomassie. The position of Bio-Rad broad range marker proteins are shown on the left. Controls included the unmutated MBP-LaeAΔ1–86 (2nd lane), unmutated LaeAΔ1–86 (5th lane), MBP alone (7th lane), and unmutated MBP-full-length LaeA (8th lane). Fluorography was performed by treating the gel with EN3HANCE and exposing the dried gel to film for 4 days at −80 °C. The portion of the EN3HANCE-treated gel containing the radiolabeled LaeA protein was excised, rehydrated with water, and acid-hydrolyzed as described under “Experimental Procedures.” B–F, after mixing with standards (1 μmol each, with the exception of S-methylmethionine with 5 μmol) of methylated amino acids, including 1-(π)-methylhistidine, S-methylmethionine, ω-NG-monomethylarginine, and ω-NG,NG-dimethylarginine, high resolution cation exchange chromatography was performed as described under “Experimental Procedures.” The positions of the standard amino acids were determined by ninhydrin assay as described under “Experimental Procedures” and indicated by a dashed line. Radioactivity was determined in 200 μl of each fraction and is shown in solid red lines.
FIGURE 10.
OPA amino acid analysis verifies S-methylmethionine is the radioactive product of LaeA automethylation. Cation exchange chromatography fractions from the experiment shown in Fig. 9B were fluorescently labeled with OPA reagent and analyzed by reverse phase HPLC coupled to a fluorometer as described under “Experimental Procedures.” Fractions of 0.5 ml were collected, and radioactivity was quantified as described under “Experimental Procedures” (red line). The fluorescence HPLC trace (black line) represents relative fluorescence units (RFU); the OPA-derivative of S-methylmethionine elutes at 13.1 min.
We then prepared MBP-LaeAΔ1–86, where each of the five methionine residues was replaced by an alanine residue. Fluorography was used to determine the automethylation state (Fig. 9A). Compared with the control of MBP-LaeAΔ1–86, purified protein with substitutions at methionine 213 or methionine 268 did not have a significant reduction in automethylation. When methionines 101 and 195 were substituted to alanine, automethylation was slightly reduced. However, most of the automethylation was abolished with the methionine 207 substitution (Fig. 9A).
To confirm the site of methylation, the [3H]AdoMet-labeled LaeA protein species from the fluorograph in Fig. 9A were acid-hydrolyzed and fractionated by high resolution cation exchange chromatography. We found that the 3H peak at the position of S-methylmethionine did not decrease significantly, or only slightly decreased, in the methionine to alanine substitutions at positions 101, 195, 213, and 268 (Fig. 9, B, C, E, and F). However, complete abolishment of methionine methylation only occurs when methionine 207 is substituted with alanine (Fig. 9D). These results suggest that the methionine 207 is the only site of automethylation. The small amount of automethylation seen in the methionine 207 mutants after gel electrophoresis (Fig. 9A) may be explained by automethylation of an alternative nucleophile in the protein or by residual binding of [3H]AdoMet.
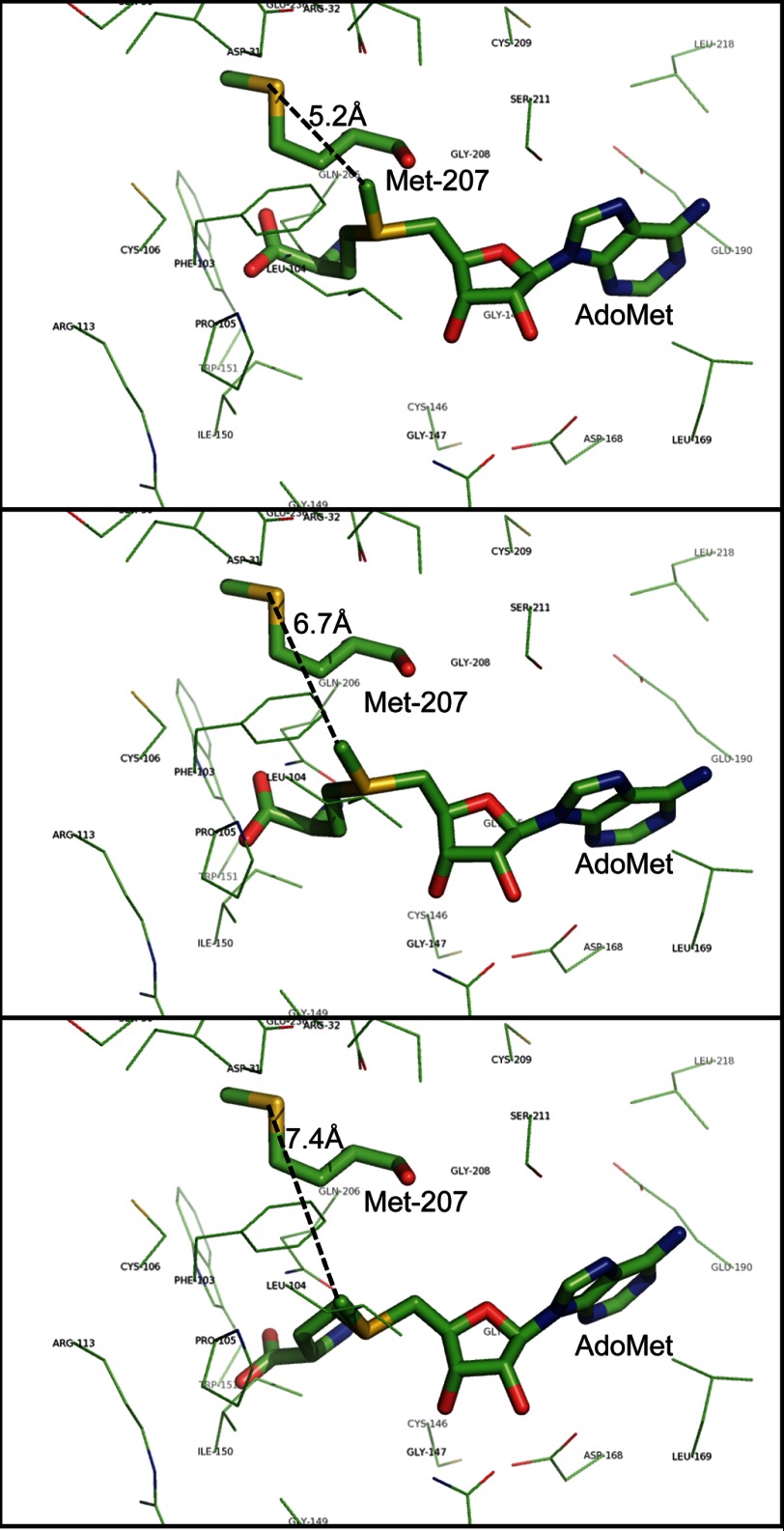
Methionine 207 is located in the amino acid sequence of LaeA immediately after motif II (Fig. 2). In the three-dimensional structures of a number of seven-β-strand family members, residues in this region interact with methyl-accepting substrates and are close to the methyl group of AdoMet (57). Because no crystal structure exists for LaeA, the three-dimensional structure was modeled using Phyre2 (42). The ligand prediction server 3DLigandSite was used to identify and to place a binding site of AdoMet into LaeA (43). Eight different AdoMet binding conformations were predicted in LaeA. The transferable methyl group of AdoMet ranged from 5.2 to 7.4 Å away from the sulfur group of methionine 207 in these structures, and no other residues were found between them (Fig. 11). This result is consistent with the reaction of the methionine thioether and the methyl group of AdoMet to produce S-methylmethionine.
FIGURE 11.
Modeling of the LaeA structure suggests that the sulfur atom of methionine 207 is close to the position of the transferable methyl group of bound AdoMet. The three-dimensional structure of full-length LaeA was based on the primary sequence (UniProt accession number C8VQG9) and derived from Phyre2 modeling; the position of the ligand AdoMet was modeled by 3DLigandSite. The positions of three possible conformations of AdoMet are shown that encompass the extent of the eight different ligand structures obtained. LaeA side chain residues within 4 Å of AdoMet are shown as thin lines with the exception of methionine 207, which is shown as a thick line with the sulfur atom in yellow. The bound AdoMet is also shown as a thick line. The distance of this sulfur atom to the transferable methyl group of AdoMet is 5.2, 6.7, and 7.4 Å in these three simulated conformations and is represented by black dashed lines.
Methionine 207 Is Not Required for Function in Vivo
To determine the functional role of methionine 207, we constructed a LaeAM207A point mutant in vivo. We found that the M207A mutant was capable of complementing the ΔlaeA allele based on initial colony morphology (Fig. 12A). We also found that LaeAM207A could restore production of sterigmatocystin (Fig. 12A) and that LaeAM207A is localized in the nucleus (Fig. 12B). These results indicate that methionine 207 is not required for these functions of LaeA. Finally, because null mutants of laeA are deficient in the production of both asexual spores (conidia) and sexual spores (ascospores), we measured the sporulation capacity of the complementation strains. We found that LaeAM207A restores sporulation to the same extent as the complemented control strain (Fig. 12, C and D). We noted, however, that neither the wild type nor the M207A constructs restored sporulation levels back to wild type. This observation is common as rarely does complementation at an alternative genetic locus result in the full restoration of the mutant phenotype. Taken together, these data suggest that methylation of methionine 207 is not required for LaeA function in vivo.
FIGURE 12.
Methionine 207 of LaeA is not required for function in vivo. A methionine 207 to alanine point mutation was constructed and integrated at the pyroA locus in a ΔlaeA strain of A. nidulans. A, LaeAM207A-GFP strains restored the gross phenotype of ΔlaeA strains as well as production of sterigmatocystin in both light and dark growth conditions. B, using fluorescent microscopy, LaeAM207A-GFP was found to be located in nuclei. C, quantification of asexual sporulation (conidiation) was measured on plates grown for 4 days under light conditions. Null mutants of laeA produce fewer conidia under these conditions. Complementation strains partially restore the production of conidia, although not to wild type levels. LaeA-GFP and LaeAM207A-GFP produce similar amounts of conidia. D, production of sexual spores (ascospores) is also reduced in ΔlaeA strains compared with wild type. This phenotype is partially restored in both LaeA-GFP and LaeAM207A-GFP complementation strains. Statistical differences were measured by analysis of variance using Prism 5.0 (GraphPad) and different letters represent statistical differences p < 0.001. Strains used in A, C, and D were WT (RJMP103.5), ΔlaeA (RJW41A), ΔlaeA laeA-GFP (RJMP256.3), and ΔlaeA laeAM207A-GFP (RJMP258.1). RJMP263.3 (ΔlaeA, hhoA-mCherry, pyroA::laeAM207A-GFP) was used in B.
DISCUSSION
Based on its primary amino acid sequence that resembles seven-β-strand methyltransferases, its nuclear localization, and its large scale effects on gene expression, LaeA has been postulated to be a histone methyltransferase. However, no direct biochemical studies on the possible substrates of LaeA have been described. In this study, we attempted to identify substrates of LaeA using in vitro and in vivo biochemical and molecular biology techniques. Because LaeA automethylation is readily observed in in vitro experiments with [3H]AdoMet (Figs. 4 and 6), we expected that methyl group transfer to other substrates would also be readily detected. However, this was not the case in our in vitro (Figs. 5 and 6) and in vivo (Fig. 7) labeling experiments. Taken together, these results suggest that either LaeA is not a protein methyltransferase, that it modifies substrates at levels undetectable by our methods, or that specific conditions are required for target methylation.
Importantly, we consistently observed LaeA automethylation at a methionine residue located close to the AdoMet-binding site. This finding is significant because it demonstrates a novel type of automethylation.
Although methyltransferases are generally characterized by their action on exogenous methyl-accepting substrates, some have been previously shown to self-methylate. The first such enzyme was the bovine protein repair l-isoaspartyl-(d-aspartyl)methyltransferase, which self-methylates at a modified asparaginyl and at an aspartyl residue (58, 59). Several protein arginine methyltransferases, including mammalian PRMT1 (60, 61), mammalian and trypanosomal PRMT6 (51, 62), and Arabidopsis PRMT10 (63) also demonstrate automethylation. In these cases, automethylation does not appear to have an effect on enzymatic activity. However, there are several cases in which the automethylation of an enzyme may have an influence on function. In the SET domain family, the automethylation of Chlamydia nuclear effector methyltransferase and human histone-lysine-N-methyltransferase (SETMAR or Metnase) is hypothesized to increase methylation activity toward histone substrates (64) or to repress methyltransferase-associated TopoIIα decatenation (65), respectively. Finally, automethylation roles involving transcription and pre-mRNA splicing (50), the inactivation of unused methyltransferases (66), and auto-regulation (49) have been suggested for CARM1/PRMT4, the DNA methyltransferase Dnmt3a, and PRMT8, respectively.
The site of automethylation can also provide insights into the substrates of methyltransferases. Although the SETMAR protein lysine methyltransferase automethylates both lysine and arginine residues (65), PRMT1 (61), CARM1/PRMT4 (50), and PRMT8 (49) automethylation is specific for arginine residues. Furthermore, in the lysine methyltransferase G9a, the sequence context of an automethylated lysine residue (an ARKT motif) is identical to the site of G9a modification on lysine 9 of histone H3 (52). Therefore, the presence of the automethylated methionine residue in LaeA may indicate that other potential substrate(s) could be methylated at a methionine residue, perhaps in a similar sequence context. The only prior evidence for methionine protein methylation is from a single report to a still unidentified methyltransferase that modifies cytochrome c in Euglena gracilis (67). A BLAST search of LaeA against the proteins encoded by the E. gracilis genome reveals no homologs.
Although this study showed the LaeA-dependent methylation of a methionine residue in a protein, free S-methylmethionine has been extensively studied for its role in plant stress tolerance (68) and as a methyl donor for a select few methyltransferases, such as YagD in E. coli (69), the human betaine-homocysteine S-methyltransferase 2 (70), and the yeast Mht1 methyltransferase (71). It is thus possible that the methyl group may be transferred from the methylmethionine residue in LaeA to another acceptor.
As illustrated by our three-dimensional modeling of LaeA (Fig. 11), the sulfur group of methionine 207 is close to the transferable methyl group of AdoMet, ranging between ∼5.2 and 7.4 Å. This is similar to the 6-Å distance between the sulfhydryl group of cysteine and the methyl group of AdoMet in the previously characterized automethylation of Dnmt3a (66). The close positioning of methionine 207 to the AdoMet-binding site appears to be important as this residue in LaeA is conserved in other filamentous fungi, with the notable exception of Trichoderma reesei LAE1, which interestingly does not complement ΔlaeA strains of A. nidulans (Fig. 2) (72). Despite the conservation of methionine 207, the LaeAM207A point mutant fully complements ΔlaeA in vivo. These data suggest that automethylation of methionine 207 could be occurring in the absence of substrate. However, it is also possible that LaeA automethylation could be serving as a self-regulator, as evident by how mutating the automethylation site of CARM1 does not affect enzymatic activity but does impair CARM1-activated transcription and pre-mRNA splicing (50).
Acknowledgment
We thank Xiaoyu Liang for contributions to the characterization of the automethylated species.
This work was supported, in whole or in part, by National Institutes of Health Grants GM026020 (to S. G. C.), 1 R01 Al065728-01 (to N. P. K.), AI55397 from NRSA (to J. M. P.), and GM007185 from NRSA (Ruth L. Kirschstein award) (to A. N. P.).
- LaeA
- Loss of AflR Expression A
- AdoMet
- S-adenosylmethionine
- [3H]AdoMet
- S-adenosyl-l-[methyl-3H]methionine
- OPA
- o-phthalaldehyde
- MBP
- maltose-binding protein
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Latgé J. P. (1999) Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12, 310–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Latgé J. P. (2001) The pathobiology of Aspergillus fumigatus. Trends Microbiol. 9, 382–389 [DOI] [PubMed] [Google Scholar]
- 3. Brakhage A. A. (1998) Molecular regulation of β-lactam biosynthesis in filamentous fungi. Microbiol. Mol. Biol. Rev. 62, 547–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alberts A. W., Chen J., Kuron G., Hunt V., Huff J., Hoffman C., Rothrock J., Lopez M., Joshua H., Harris E., Patchett A., Monaghan R., Currie S., Stapley E., Albers-Schonberg G., Hensens O., Hirshfield J., Hoogsteen K., Liesch J., Springer J. (1980) Mevinolin: a highly potent competitive inhibitor of hydroxymethylglutaryl-coenzyme A reductase and a cholesterol-lowering agent. Proc. Natl. Acad. Sci. U.S.A. 77, 3957–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Keller N. P., Turner G., Bennett J. W. (2005) Fungal secondary metabolism–from biochemistry to genomics. Nat. Rev. Microbiol. 3, 937–947 [DOI] [PubMed] [Google Scholar]
- 6. Garvey G. S., Keller N. P. (2010) Fungal secondary metabolites and their fundamental roles in human mycoses. Curr. Fungal Infect. Rep. 4, 256–265 [Google Scholar]
- 7. Keller N. P., Hohn T. M. (1997) Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 21, 17–29 [PubMed] [Google Scholar]
- 8. Brakhage A. A. (2013) Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 11, 21–32 [DOI] [PubMed] [Google Scholar]
- 9. Fernandes M., Keller N. P., Adams T. H. (1998) Sequence-specific binding by Aspergillus nidulans AflR, a C6 zinc cluster protein regulating mycotoxin biosynthesis. Mol. Microbiol. 28, 1355–1365 [DOI] [PubMed] [Google Scholar]
- 10. Amaike S., Keller N. P. (2011) Aspergillus flavus. Annu. Rev. Phytopathol. 49, 107–133 [DOI] [PubMed] [Google Scholar]
- 11. Forseth R. R., Amaike S., Schwenk D., Affeldt K. J., Hoffmeister D., Schroeder F. C., Keller N. P. (2013) Homologous NRPS-like gene clusters mediate redundant small-molecule biosynthesis in Aspergillus flavus. Angew. Chem. Int. Ed. Engl. 52, 1590–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bok J. W., Keller N. P. (2004) LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3, 527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bok J. W., Balajee S. A., Marr K. A., Andes D., Nielsen K. F., Frisvad J. C., Keller N. P. (2005) LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot. Cell 4, 1574–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sugui J. A., Pardo J., Chang Y. C., Müllbacher A., Zarember K. A., Galvez E. M., Brinster L., Zerfas P., Gallin J. I., Simon M. M., Kwon-Chung K. J. (2007) Role of laeA in the regulation of alb1, gliP, conidial morphology, and virulence in Aspergillus fumigatus. Eukaryot. Cell 6, 1552–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Perrin R. M., Fedorova N. D., Bok J. W., Cramer R. A., Wortman J. R., Kim H. S., Nierman W. C., Keller N. P. (2007) Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 3, e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Amaike S., Keller N. P. (2009) Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus. Eukaryot. Cell 8, 1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. López-Berges M. S., Hera C., Sulyok M., Schäfer K., Capilla J., Guarro J., Di Pietro A. (2013) The velvet complex governs mycotoxin production and virulence of Fusarium oxysporum on plant and mammalian hosts. Mol. Microbiol. 87, 49–65 [DOI] [PubMed] [Google Scholar]
- 18. Wu D., Oide S., Zhang N., Choi M. Y., Turgeon B. G. (2012) ChLae1 and ChVel1 regulate T-toxin production, virulence, oxidative stress response, and development of the maize pathogen Cochliobolus heterostrophus. PLoS Pathog. 8, e1002542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wiemann P., Brown D. W., Kleigrewe K., Bok J. W., Keller N. P., Humpf H.-U., Tudzynski B. (2010) FfVel1 and FfLae1, components of a velvet-like complex in Fusarium fujikuroi, affect differentiation, secondary metabolism, and virulence. Mol. Microbiol. 77, 972–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Petrossian T. C., Clarke S. G. (2009) Multiple motif scanning to identify methyltransferases from the yeast proteome. Mol. Cell. Proteomics 8, 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bayram O., Krappmann S., Ni M., Bok J. W., Helmstaedt K., Valerius O., Braus-Stromeyer S., Kwon N. J., Keller N. P., Yu J. H., Braus G. H. (2008) VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320, 1504–1506 [DOI] [PubMed] [Google Scholar]
- 22. Fox E. M., Howlett B. J. (2008) Secondary metabolism: regulation and role in fungal biology. Curr. Opin. Microbiol. 11, 481–487 [DOI] [PubMed] [Google Scholar]
- 23. Bok J. W., Noordermeer D., Kale S. P., Keller N. P. (2006) Secondary metabolic gene cluster silencing in Aspergillus nidulans. Mol. Microbiol. 61, 1636–1645 [DOI] [PubMed] [Google Scholar]
- 24. Reyes-Dominguez Y., Bok J. W., Berger H., Shwab E. K., Basheer A., Gallmetzer A., Scazzocchio C., Keller N., Strauss J. (2010) Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol. Microbiol. 76, 1376–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26. Bok J. W., Keller N. P. (2012) Fast and easy method for construction of plasmid vectors using modified quick-change mutagenesis. Methods Mol. Biol. 944, 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rocco C. J., Dennison K. L., Klenchin V. A., Rayment I., Escalante-Semerena J. C. (2008) Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid 59, 231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang L., Ukil L., Osmani A., Nahm F., Davies J., De Souza C. P., Dou X., Perez-Balaguer A., Osmani S. A. (2004) Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 3, 1359–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tsitsigiannis D. I., Kowieski T. M., Zarnowski R., Keller N. P. (2004) Endogenous lipogenic regulators of spore balance in Aspergillus nidulans. Eukaryot. Cell 3, 1398–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Blommel P. G., Fox B. G. (2007) A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr. Purif. 55, 53–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Palmer J. M., Theisen J. M., Duran R. M., Grayburn W. S., Calvo A. M., Keller N. P. (2013) Secondary metabolism and development is mediated by LlmF control of VeA subcellular localization in Aspergillus nidulans. PLoS Genet. 9, e1003193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shimizu K., Keller N. P. (2001) Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157, 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Szewczyk E., Nayak T., Oakley C. E., Edgerton H., Xiong Y., Taheri-Talesh N., Osmani S. A., Oakley B. R. (2006) Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1, 3111–3120 [DOI] [PubMed] [Google Scholar]
- 34. Palmer J. M., Mallaredy S., Perry D. W., Sanchez J. F., Theisen J. M., Szewczyk E., Oakley B. R., Wang C. C., Keller N. P., Mirabito P. M. (2010) Telomere position effect is regulated by heterochromatin-associated proteins and NkuA in Aspergillus nidulans. Microbiology 156, 3522–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Champe S. P., el-Zayat A. A. (1989) Isolation of a sexual sporulation hormone from Aspergillus nidulans. J. Bacteriol. 171, 3982–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Palmer J. M., Perrin R. M., Dagenais T. R., Keller N. P. (2008) H3K9 methylation regulates growth and development in Aspergillus fumigatus. Eukaryot. Cell 7, 2052–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Frankel A., Clarke S. (2000) PRMT3 is a distinct member of the protein arginine N-methyltransferase family. J. Biol. Chem. 275, 32974–32982 [DOI] [PubMed] [Google Scholar]
- 38. Kinoshita E., Kinoshita-Kikuta E. (2011) Improved Phos-tag SDS-PAGE under neutral pH conditions for advanced protein phosphorylation profiling. Proteomics 11, 319–323 [DOI] [PubMed] [Google Scholar]
- 39. Dhar S., Vemulapalli V., Patananan A. N., Huang G. L., Di Lorenzo A., Richard S., Comb M. J., Guo A., Clarke S. G., Bedford M. T. (2013) Loss of the major type I arginine methyltransferase PRMT1 causes substrate scavenging by other PRMTs. Sci. Rep. 3, 1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009) Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 43. Wass M. N., Kelley L. A., Sternberg M. J. (2010) 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res. 38, W469–W473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marsden R. L., McGuffin L. J., Jones D. T. (2002) Rapid protein domain assignment from amino acid sequence using predicted secondary structure. Protein Sci. 11, 2814–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Palmer J. M., Keller N. P. (2010) Secondary metabolism in fungi: does chromosomal location matter? Curr. Opin. Microbiol. 13, 431–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Strauss J., Reyes-Dominguez Y. (2011) Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genet. Biol. 48, 62–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jenuwein T., Allis C. D. (2001) Translating the histone code. Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 48. Wang H., Huang Z. Q., Xia L., Feng Q., Erdjument-Bromage H., Strahl B. D., Briggs S. D., Allis C. D., Wong J., Tempst P., Zhang Y. (2001) Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293, 853–857 [DOI] [PubMed] [Google Scholar]
- 49. Sayegh J., Webb K., Cheng D., Bedford M. T., Clarke S. G. (2007) Regulation of protein arginine methyltransferase 8 (PRMT8) activity by its N-terminal domain. J. Biol. Chem. 282, 36444–36453 [DOI] [PubMed] [Google Scholar]
- 50. Kuhn P., Chumanov R., Wang Y., Ge Y., Burgess R. R., Xu W. (2011) Automethylation of CARM1 allows coupling of transcription and mRNA splicing. Nucleic Acids Res. 39, 2717–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Frankel A., Yadav N., Lee J., Branscombe T. L., Clarke S., Bedford M. T. (2002) The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J. Biol. Chem. 277, 3537–3543 [DOI] [PubMed] [Google Scholar]
- 52. Chin H. G., Estève P.-O., Pradhan M., Benner J., Patnaik D., Carey M. F., Pradhan S. (2007) Automethylation of G9a and its implication in wider substrate specificity and HP1 binding. Nucleic Acids Res. 35, 7313–7323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gottschling H., Freese E. (1962) A tritium isotope effect on ion exchange chromatography. Nature 196, 829–831 [DOI] [PubMed] [Google Scholar]
- 54. Webb K. J., Zurita-Lopez C. I., Al-Hadid Q., Laganowsky A., Young B. D., Lipson R. S., Souda P., Faull K. F., Whitelegge J. P., Clarke S. G. (2010) A novel 3-methylhistidine modification of yeast ribosomal protein Rpl3 is dependent upon the YIL110W methyltransferase. J. Biol. Chem. 285, 37598–37606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Webb K. J., Al-Hadid Q., Zurita-Lopez C. I., Young B. D., Lipson R. S., Clarke S. G. (2011) The ribosomal L1 protuberance in yeast is methylated on a lysine residue catalyzed by a seven-β-strand methyltransferase. J. Biol. Chem. 286, 18405–18413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zurita-Lopez C. I., Sandberg T., Kelly R., Clarke S. G. (2012) Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming ω-NG-monomethylated arginine residues. J. Biol. Chem. 287, 7859–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Martin J. L., McMillan F. M. (2002) SAM (dependent) I AM: the S-adenosylmethionine dependent methyltransferase fold. Curr. Opin. Struct. Biol. 12, 783–793 [DOI] [PubMed] [Google Scholar]
- 58. Lindquist J. A., Barofsky E., McFadden P. N. (1996) Determination of two sites of automethylation in bovine erythrocyte protein (d-aspartyl/l-isoaspartyl) carboxyl methyltransferase. J. Protein Chem. 15, 115–122 [DOI] [PubMed] [Google Scholar]
- 59. Lindquist J. A., McFadden P. N. (1994) Automethylation of protein (d-aspartyl/l-isoaspartyl) carboxyl methyltransferase, a response to enzyme aging. J. Protein Chem. 13, 23–30 [DOI] [PubMed] [Google Scholar]
- 60. Lakowski T. M., 't Hart P., Ahern C. A., Martin N. I., Frankel A. (2010) Nη-substituted arginyl peptide inhibitors of protein arginine N-methyltransferases. ACS Chem. Biol. 5, 1053–1063 [DOI] [PubMed] [Google Scholar]
- 61. Gui S., Wooderchak W. L., Daly M. P., Porter P. J., Johnson S. J., Hevel J. M. (2011) Investigation of the molecular origins of protein-arginine methyltransferase I (PRMT1) product specificity reveals a role for two conserved methionine residues. J. Biol. Chem. 286, 29118–29126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fisk J. C., Zurita-Lopez C., Sayegh J., Tomasello D. L., Clarke S. G., Read L. K. (2010) TbPRMT6 is a type I protein arginine methyltransferase that contributes to cytokinesis in Trypanosoma brucei. Eukaryot. Cell 9, 866–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Niu L., Lu F., Zhao T., Liu C., Cao X. (2012) The enzymatic activity of Arabidopsis protein arginine methyltransferase 10 is essential for flowering time regulation. Protein Cell 3, 450–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pennini M. E., Perrinet S., Dautry-Varsat A., Subtil A. (2010) Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen Chlamydia trachomatis. PLoS Pathog. 6, e1000995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Williamson E. A., Rasila K. K., Corwin L. K., Wray J., Beck B. D., Severns V., Mobarak C., Lee S.-H., Nickoloff J. A., Hromas R. (2008) The SET and transposase domain protein Metnase enhances chromosome decatenation: regulation by automethylation. Nucleic Acids Res. 36, 5822–5831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Siddique A. N., Jurkowska R. Z., Jurkowski T. P., Jeltsch A. (2011) Auto-methylation of the mouse DNA-(cytosine C5)-methyltransferase Dnmt3a at its active site cysteine residue. FEBS J. 278, 2055–2063 [DOI] [PubMed] [Google Scholar]
- 67. Farooqui J. Z., Tuck M., Paik W. K. (1985) Purification and characterization of enzymes from Euglena gracilis that methylate methionine and arginine residues of cytochrome c. J. Biol. Chem. 260, 537–545 [PubMed] [Google Scholar]
- 68. Ogawa S., Mitsuya S. (2012) S-Methylmethionine is involved in the salinity tolerance of Arabidopsis thaliana plants at germination and early growth stages. Physiol. Plant. 144, 13–19 [DOI] [PubMed] [Google Scholar]
- 69. Thanbichler M., Neuhierl B., Böck A. (1999) S-Methylmethionine metabolism in Escherichia coli. J. Bacteriol. 181, 662–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Szegedi S. S., Castro C. C., Koutmos M., Garrow T. A. (2008) Betaine-homocysteine S-methyltransferase-2 is an S-methylmethionine homocysteine methyltransferase. J. Biol. Chem. 283, 8939–8945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vinci C. R., Clarke S. G. (2007) Recognition of age-damaged (R,S)-adenosyl-l-methionine by two methyltransferases in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 282, 8604–8612 [DOI] [PubMed] [Google Scholar]
- 72. Karimi-Aghcheh R., Bok J. W., Phatale P. A., Smith K. M., Baker S. E., Lichius A., Omann M., Zeilinger S., Seiboth B., Rhee C., Keller N. P., Freitag M., Kubicek C. P. (2013) Functional analyses of Trichoderma reesei LAE1 reveal conserved and contrasting roles of this regulator. G3 3, 369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tsitsigiannis D. I., Zarnowski R., Keller N. P. (2004) The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulans. J. Biol. Chem. 279, 11344–11353 [DOI] [PubMed] [Google Scholar]
- 74. Shaaban M., Palmer J. M., El-Naggar W. A., El-Sokkary M. A., Habib el-S. E., Keller N. P. (2010) Involvement of transposon-like elements in penicillin gene cluster regulation. Fungal Genet. Biol. 47, 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]