Background: KATmt is the first identified cAMP-regulated protein lysine acetylase in mycobacteria.
Results: KATmt acylates fatty acyl CoA ligases in vivo in a cAMP-dependent manner, thus regulating their activity.
Conclusion: Mycobacteria utilize KATmt to regulate the metabolic pool of acetyl and propionyl CoA.
Significance: We provide novel paradigms for linking cAMP signaling and fatty acid metabolism in mycobacteria.
Keywords: Acetyl Coenzyme A, Actinobacteria, Fatty Acid Metabolism, Mass Spectrometry (MS), Mycobacteria, Prokaryotic Signal Transduction, Protein Acylation
Abstract
Acetylation of lysine residues is a posttranslational modification that is used by both eukaryotes and prokaryotes to regulate a variety of biological processes. Here we identify multiple substrates for the cAMP-dependent protein lysine acetyltransferase from Mycobacterium tuberculosis (KATmt). We demonstrate that a catalytically important lysine residue in a number of FadD (fatty acyl CoA synthetase) enzymes is acetylated by KATmt in a cAMP-dependent manner and that acetylation inhibits the activity of FadD enzymes. A sirtuin-like enzyme can deacetylate multiple FadDs, thus completing the regulatory cycle. Using a strain deleted for the KATmt ortholog in Mycobacterium bovis Bacillus Calmette-Guérin (BCG), we show for the first time that acetylation is dependent on intracellular cAMP levels. KATmt can utilize propionyl CoA as a substrate and, therefore, plays a critical role in alleviating propionyl CoA toxicity in mycobacteria by inactivating acyl CoA synthetase (ACS). The precision by which mycobacteria can regulate the metabolism of fatty acids in a cAMP-dependent manner appears to be unparalleled in other biological organisms and is ideally suited to adapt to the complex environment that pathogenic mycobacteria experience in the host.
Introduction
Tuberculosis remains one of the leading causes of death because of an infectious disease. The causative agent of tuberculosis, Mycobacterium tuberculosis, is harbored by almost 30% of the population of the world. However, most infected people do not develop the disease. Thus, a delicate balance is maintained between the pathogen and a healthy host that does not show signs of illness. This balance can tilt toward the pathogen if the host is coinfected with HIV or has a weakened immune response because of dietary or environmental conditions, and understanding the factors that disturb the equilibrium is, therefore, important.
A number of virulence factors in M. tuberculosis allow it to evade the natural immune response of the host, establish infection, and remain persistent over many years in a dormant state. The availability of the genome sequence of M. tuberculosis served as an important molecular tool in dissecting the roles of individual genes in pathogenesis (1). We found some years ago that a number of genes involved in cAMP (3′,5′-cAMP) synthesis (adenylyl cyclases) were encoded in many mycobacterial genomes (2, 3). Cyclic AMP, a universal second messenger, provides a means by which the pathogen can communicate with and hijack host signaling within macrophages during early infection (4, 5). Indeed, it has been reported that deletion of one of the 16 adenylyl cyclase genes in M. tuberculosis results in attenuation of virulence (6), pointing toward an important role for cAMP/adenylyl cyclases in M. tuberculosis pathogenesis.
Elevated cAMP levels are, however, also seen in non-pathogenic strains of mycobacteria, indicating that cAMP has important roles to play in the basic biology of mycobacteria (7). We therefore set out to identify targets of cAMP in mycobacteria and focused on proteins that contained a cyclic nucleotide binding domain identified earlier and characterized in many eukaryotic proteins and the bacterial transcription factor cyclic AMP receptor protein, CRP (8, 9). We identified unique proteins (MSMEG_5458 and Rv0998) from Mycobacterium smegmatis and M. tuberculosis, respectively, that consist of a cyclic nucleotide binding domain fused to an acetyltransferase domain. We demonstrated that these proteins act as cAMP-regulated protein lysine acetyltransferases (10) and showed that a universal stress protein is a substrate in M. smegmatis. Rv0998, which we call KATmt, required the presence of cAMP to show acetyl transferase activity. Recent biophysical and structural analyses revealed the dramatic conformational change that occurs in these proteins on cAMP binding that allows the acetylation of its protein substrates (11, 12).
Orthologs of KATmt are found in all mycobacteria, including Mycobacterium leprae, indicating that this protein must have an important role to play in these organisms. The fusion of a cyclic nucleotide binding domain along with an acetyltransferase domain in a single gene product is not found in any other prokaryotic or eukaryotic genome sequenced thus far, implying that the direct regulation of protein acetylation by intracellular cAMP levels in mycobacteria is unique to this genus. In this study, we have identified substrates of KATmt and show for the first time that a number of mycobacterial fatty acyl Co-A ligases are acylated in vitro and in vivo by KATmt. We thus perhaps uncover some of the first mechanisms downstream of adenylyl cyclases and cAMP within mycobacteria during establishment and, possibly, persistence in the host macrophage.
EXPERIMENTAL PROCEDURES
Bioinformatic Analysis
Six amino acids surrounding the acetylated Lys residue in universal stress proteins were used as a seed sequence for BLASTP analysis against the predicted M. tuberculosis protein database (NCBI). Proteins that were identified were then inspected manually to confirm that the Lys residue was preceded by a small amino acid and followed by a few small hydrophobic residues (13). Proteins that were identified were then subjected to further analysis using Predmod (13).
Multiple sequence alignment of the 34 FadDs2 was done using ClustalW (14, 15), and a phylogenetic tree was generated using Molecular Evolutionary Genetics Analysis software (16).
Cloning, Expression, and Purification of Proteins
The list of primers used for PCR and mutagenesis are provided in supplemental Table S1. All clones were generated and verified by sequencing (Macrogen, South Korea). Cloning strategies are available on request. A clone of FadD13 containing a mutation at Lys-517 was provided by Prof. A. K. Tyagi, University of Delhi, India. Proteins were expressed in the E. coli BL21 endo− strain following induction with 500 μm isopropyl 1-thio-β-d-galactopyranoside for 20 h at 16 °C or for 3 h at 37 °C, as described earlier (10).
For MSMEG_5175 (sirtuin) and MSMEG_5175H104Y, cells were lysed in lysis buffer containing 50 mm Tris-Cl (pH 8.2), 500 mm NaCl, 20% glycerol, 5 mm 2-mercaptoethanol, 2 mm PMSF, and 1 mm benzamidine. Washes were done with buffer containing 50 mm Tris-Cl (pH 8.2), 500 mm NaCl, 20% glycerol, 5 mm 2-mercaptoethanol, and 20 mm imidazole. The proteins were eluted with buffer containing 50 mm Tris-Cl (pH 8.2), 500 mm NaCl, 20% glycerol, 5 mm 2-mercaptoethanol, and 300 mm imidazole. KATmt was expressed in the E. coli SP850 strain on induction using 500 μm 1-thio-β-d-galactopyranoside for 20 h at 16 °C as described earlier (10).
Western Blotting
Samples were electrophoresed on a 12% SDS-polyacrylamide gel and transferred to a PVDF membrane (Immobilon-P, Millipore). FadD13 polyclonal antibody was generated in the laboratory and used at a dilution of 1:5000. Acetyl lysine antibody (Cell Signaling Technology, Inc.) was used at a dilution of 1:2500. Horseradish peroxidase-conjugated secondary antibody (GE Healthcare) or light chain-specific antibody (Jackson ImmunoResearch Laboratories) were used and detected by enhanced chemiluminescence according to the instructions of the manufacturer (GE Healthcare).
Mass Spectrometric Analysis
Recombinant purified protein samples after in vitro acylation were treated with 400 ng of trypsin for 5 h in solution. The peptide mixtures were then analyzed by liquid chromatography and electrospray tandem mass spectrometry (LC-ESI MS/MS). The samples were analyzed on an HCT-Ultra ETD-II ion trap mass spectrometer (Bruker Daltonics, Bremen, Germany), coupled to an Agilent 1100 HPLC. The LC separation was performed on a reverse-phase C18 column at a flow rate of 0.2 ml/min using a H2O-acetonitrile (with 0.1% formic acid) solvent system under a gradient of 90% H2O to 90% acetonitrile over a period of 60 min. The mass spectral analysis was performed under auto-MS/MS conditions, with the three most intense ions from each scan being subjected to CID fragmentation. The fragmentations were carried out inside the ion trap through the collision of He gas with the ions of interest, which were excited kinetically by an increased resonance amplitude of the dipolar field, with a value typically set between 1–2 V. The scan speed was set at 26,000 m/z s-1. The m/z ranges for MS detection and MS/MS precursor ion selection were set from 300–1800 and 500–1400, respectively.
In Vitro Deacylation Assay
FadDs (2 μg) were acetylated or propionylated by KATmt (200 ng). Samples were then directly treated with sirtuinWT or sirtuinH104Y (500 ng) in the presence or absence of 1 mm NAD+. Deacetylation reactions were carried out at 37 °C in buffer containing 25 mm Tris-Cl (pH 8.2), 137 mm NaCl, 2.7 mm KCl, 1 mm MgCl2, and 100 ng/μl BSA for 1 h. Whenever necessary, deacetylation reactions were also performed in the presence of 5 mm nicotinamide, a known sirtuin enzyme inhibitor. Reactions were stopped by boiling in SDS sample buffer and analyzed by Western blotting with acetyl lysine antibody and chemiluminescent HRP substrate (Immobilon, Millipore).
FadD Assays
The activities of acylated or non-acylated FadDs were assessed using an in vitro fatty acyl CoA synthetase assay with 14C-labeled palmitic acid as the substrate. The purified FadD protein (1 μg) was initially incubated with 10 mm DTT for 30 min at 37 °C prior to its use. Subsequently, purified enzymes were incubated with 50 μm acetyl CoA or propionyl CoA, 25 mm Tris-Cl (pH 7.5), 1 mm cAMP, and 300 ng of KATmt for 30 min at 37 °C to acylate the FadDs in a reaction volume of 10 μl.
Fatty acyl CoA synthetase assays were performed in 50 mm Tris-Cl (pH 8.0), 100 μm 14C-palmitic acid, 5 mm ATP, and 2.5 mm MgCl2 in a 10-μl reaction volume and, wherever applicable, CoA (2 mm). After prewarming the reaction mixture at 30 °C for 15 min, purified protein (acetylated or non-acetylated) was added to a final reaction volume of 20 μl and incubated for 5 min at 30 °C. The reactions were quenched with 5% acetic acid and directly spotted on silica gel TLC plates. Products were resolved in n-butanol/acetic acid/water (15:5:8) at 4 °C. The radioactive bands on TLC were detected using a phosphoimager (Bass 1800, Fuji) after 12 h of exposure.
Mycobacterial Cell Culture and Preparation of Cell Lysates
Mycobacterium bovis BCG was grown in Middlebrook's 7H9 broth supplemented with 0.2% glycerol, 10% ODAC (oleic acid, albumin, dextrose and catalase) and 0.03% tyloxapol (Sigma-Aldrich). Initially the cultures were maintained in volume of 5 ml in tissue culture flasks (T-25 cm2, Thermo Scientific) without shaking until the optical density reached 1. This culture was then used as inoculum for large-scale cultures with an initial optical density of 0.1. Whenever necessary, hygromycin (50 μg/ml) was used. For carbon utilization experiments, bacteria were grown in 7H9 medium containing 0.03% tyloxapol and glucose (0.2%). Sodium propionate was used at a concentration of 10 mm. For SDS stress, cells were grown until the optical density was 0.8 and then treated with 0.05% SDS for 1.5 h at 37 °C.
For preparation of whole cell lysates, cells were broken by bead beating in buffer containing 10 mm Tris-Cl (pH 7.5), 100 mm NaCl, 10% glycerol, 5 μg/ml leupeptin, 5 μg/ml aprotinin, 5 μg/ml soybean trypsin inhibitor, 3 mm nicotinamide, and 1 μm trichostatin A. Nonidet P-40 and deoxycholate to final concentrations of 1% and 0.5%, respectively, were added to the lysate and mixed continuously at 4 °C for 2 h, after which samples were centrifuged for 30 min at 4 °C at 13,000 × g. Supernatants were collected, and protein was estimated using a Micro BCA protein assay kit (Invitrogen).
Measurement of cAMP in M. bovis BCG
Cells were centrifuged at 13,000 × g; washed with buffer containing 10 mm Tris-Cl (pH 7.5), 0.89% NaCl, and 0.05% Tween 80 (TBST); and then the pellet was resuspended in 0.1N HCl and the suspension was boiled for 10 min. Aliquots of the lysate were taken for estimation of cAMP by radioimmunoassay.
Immunoprecipitation of FadD13
Antibodies to FadD13 were generated in rabbits. Anti FadD 13 IgG was purified using a T-Gel purification kit (Thermo Scientific Pierce) according to the instructions of the manufacturer. For preclearing, cell lysates (300 μg) were incubated with normal rabbit IgG for 1 h at 4 °C. Protein G beads were added to the lysates and incubated for an additional 30 min. The beads were removed by centrifugation, and the precleared supernatant was interacted with FadD13 IgG overnight at 4 °C. Protein G beads were added to the lysates and incubated for an additional 2 h. The beads were pelleted at 4 °C and washed thrice with buffer containing 10 mm Tris-Cl (pH 7.5), 100 mm NaCl, and 0.1% Triton X-100 and twice with TBS (10 mm Tris-Cl (pH 7.5), 100 mm NaCl). The beads were boiled in SDS sample buffer and subsequently analyzed by SDS-PAGE and Western blotting.
RNA Isolation and Reverse Transcriptase
PCR Cells were harvested and resuspended in 300 μl of Tri reagent (Sigma-Aldrich) prewarmed to 65 °C. Approximately 50 mg of glass beads (with a diameter of 465–600 μm, Sigma-Aldrich) were added, and bead beating was carried out for 90 s with 30-s intervals at 4800 rpm using a BeadBeater (BioSpec Products). The lysate was mixed by pipetting and followed by centrifugation at 12,000 × g for 10 min at 4 °C. The supernatant was treated with chloroform (200 μl) followed by centrifugation at 14,000 × g for 15 min at 4 °C. The RNA was precipitated with 200 μl of isopropyl alcohol, and the RNA pellet was dissolved, quantified, and treated with RNase-free DNase (1–5 units). RNA (2 μg) was used for reverse transcription using 200 units of reverse transcriptase (MBI Fermentas, Canada). The transcript levels of BCG_1055 (KATbcg), FadD13, and sirtuin were assessed using primer pairs Rv0998RTFWD and Rv0998RTRVS, Rv3089RTFWD and Rv3089RTRVS, and Rv1151cRTFWD and Rv1151cRVS, respectively (supplemental Table S1).
Generation of ΔKATbcg strain
M. bovis BCG with a deletion of KATbcg was generated using allelic exchange using a specialized transducing phage, phAE87 (provided by Prof. Sabine Ehrt, Weill Cornell Medical College). A region ∼650 bp upstream of KATbcg was amplified using the Rv0998upFWD and Rv0998upRVS primers (5′ amplicon) (supplemental Table S1). Similarly, a fragment ∼650 bases downstream of KATbcg was amplified using the Rv0998downFWD and Rv0998downRVS primers (3′ amplicon) (supplemental Table S1), and fragments were cloned into the pBKSII (+) vector. Clones were confirmed by sequencing. The AgeI-SphI fragment from pBKS-Rv0998upKO was ligated to similarly digested pJSC284-loxP (provided by Prof. Sabine Ehrt, Weill Cornell Medical College) to generate pJSC284-loxP-Rv0998upKO. The BamHI-HindIII fragment from pBKS-Rv0998downKO was ligated into the similarly cut pJSC284-loxP-Rv0998upKO plasmid to generate the pJSC284-loxP-Rv0998updownKO plasmid. The isolated phage DNA was ligated using 5 Weiss units of DNA ligase at 16 °C for 12 h. After ligation, the phage DNA was subjected to PacI digestion. The digested phage DNA was then ligated to PacI-digested pJSC284-loxP-Rv0998updownKO. The packaging of the ligated phage DNA was carried out in the laboratory of Dr. Apoorva Bhatt, School of Biosciences, University of Birmingham. Approximately 5–20 μg phagemid DNA was electroporated into electrocompetent M. smegmatis cells and plated on 7H10 agar plates. Plaques appeared 2–3 days later at 30 °C. The phages were amplified, and phage DNA was isolated to check the presence of pJSC284-loxP-Rv0998updownKO by PCR. The desired phage was transduced into exponential phase M. bovis BCG, and they were screened for the absence of KATbcg by genomic PCR using the Rv0998FWD and Rv0998RVS primers (supplemental Table S1). They were further confirmed by Southern blotting and RT-PCR.
To construct the ΔKATbcg strain complemented with the KATbcg gene driven by its own promoter, the Rv0998upFWD and Rv0998RVS primers (supplemental Table S1) were used to amplify 650 bp upstream of KATbcg. The PCR product was digested with BamHI and EcoRI and ligated into similarly digested pBKSII (+) to generate pBKS-opromRv0998. The NotI-EcoRI fragment from pBKS-opromRv0998 was cloned into similarly digested pMV306 (a gift from Prof. Sabine Ehrt, Weill Cornell Medical College) to generate pMV306-opromRv0998, which was electroporated in the ΔKATbcg strain. Positive integrants carrying the required insert were screened by colony PCR and validated by Southern hybridization and RT-PCR.
RESULTS
Multiple Substrates for KATmt
A loose consensus for predicting lysine residues that have a propensity for acetylation has been identified (13), and a stretch of small or flexible amino acid directly preceding the acetylated lysine residue is often observed (17, 18). Therefore, we used six amino acids surrounding the acetylated Lys residue in universal stress proteins as a seed sequence for BLASTP analysis against the predicted M. tuberculosis protein database. Proteins that were identified were then inspected manually to confirm that the Lys residue was preceded by a small amino acid and followed by a few small hydrophobic residues, as seen in the universal stress protein sequence. Sixty proteins were thus identified and were then subjected to further analysis using Predmod to determine their propensity for acetylation (13). Eighteen proteins had a significant score (supplemental Fig. 1a), suggesting that these proteins may be acetylated by KATmt.
We selected seven proteins, cloned their genes, and expressed all of them in E. coli (supplemental Fig. 1b). Rv2948c (FadD22), Rv0270 (FadD2), Rv3089 (FadD13), and Rv0166 (FadD5) are acyl CoA synthetases, Rv1007c is predicted to be a methionyl tRNA synthetase, Rv1633 is an enzyme involved in nucleotide excision repair, and Rv0640 is a 50 S ribosomal protein, L11. We also cloned Rv3667 (ACS), which has been shown earlier to be acetylated by the M. smegmatis KAT MSMEG_5458 (19). Purified proteins were treated individually with KATmt in the presence of cAMP and acetyl CoA (Fig. 1a). The Mtb 50 S ribosomal protein L11, Rv0640, was acetylated as purified, probably by an E. coli protein acetyltransferase, and showed no further increase in acetylation in the presence of KATmt. ACS was acetylated by KATmt in a cAMP-dependent manner in agreement with earlier results. Although FadD22 (Rv2948c), FadD2 (Rv0270), FadD5 (Rv0166), and FadD13 (Rv3089) were acetylated in a cAMP-dependent manner, Rv1633 and Rv1007c were not acetylated by KATmt. Therefore, we were able to identify KATmt substrates through sequence analysis with some degree of confidence, and, as shown below, the predictive efficiency was better for the family of FadDs we chose to study in greater detail.
FIGURE 1.
Cyclic AMP-dependent acetylation of FadDs by KATmt. a, two micrograms of purified proteins were incubated in the presence of KATmt (300 ng), cAMP (1 mm), and acetyl CoA (50 μm), followed by Western blotting with acetyl lysine antibody and anti-His antibody. b, CID MS/MS spectrum of the acetylated tryptic peptide 483NPTGKAcILK490 of FadD13. The observed fragment ions (b and y ions) are marked on the spectrum and also summarized schematically. c, FadD13K487A (2 μg) was incubated with KATmt, cAMP, and acetyl CoA, followed by Western blotting with acetyl lysine antibody and anti-His antibody. d, in vitro acetylation of indicated FadDs (500 ng) was performed as indicated, followed by Western blotting with acetyl lysine antibody and anti-His antibody. e, multiple sequence alignment for FadDs that are acetylated by KATmt (upper panel) and those that are not (lower panel). Conserved residues are highlighted by colored boxes. The red box (lower panel) highlights the variation in the highly conserved K/AXP motif. The acetylated lysine is marked with a green dot in both panels. f, dendrogram of full-length sequences of 34 FadDs. The acetylated and non-acetylated FadDs are highlighted with green/red arcs, respectively. Using the same color code and an asterisk, the potential of 12 FadDs to be acetylated by KATmt is predicted.
The biochemical and structural features of FadD13 have been described recently (20–22). We have therefore used it in further studies as a representative substrate for KATmt. We mapped the site of acetylation in FadD13 using LC-MS/MS and identified Lys-487 as the acetylated residue (Fig. 1b). In agreement with the LC-MS/MS data, a mutant FadD13 (K487A) showed no acetylation in the presence of KATmt, cAMP, and acetyl CoA (Fig. 1c and supplemental Fig. 1c), indicating that Lys-487 was the sole site of acetylation.
Strikingly, all tested substrates for KATmt (FadD22, FadD2, FadD5, and FadD13) belonged to the fatty acyl CoA synthetase family of proteins that activate lipids prior to their utilization in metabolic pathways (23). The site of acetylation of FadD22, FadD2, and FadD5 was the Lys residue present at a position equivalent to Lys-487 in FadD13. Because this lysine residue is conserved in FadD enzymes, it was of interest to determine whether all of these proteins were substrates for KATmt. We therefore cloned and expressed all 34 FadD enzymes in E. coli. Twenty-two proteins were purifiable from E. coli, although the remainder were localized to inclusion bodies (supplemental Fig. 1d). We tested all 22 proteins for their ability to be acetylated by KATmt in a cAMP-dependent manner, and, as shown in Fig. 1d, eight enzymes were acetylated. The site of acetylation in these proteins, as analyzed by LC-MS/MS (supplemental Fig. 1e) was the Lys residue present at an equivalent position to Lys-487 in FadD13 (Fig. 1e). LC-MS/MS also confirmed that the other 14 FadD enzymes were not acetylated (because peptides resulting from tryptic cleavage following the C-terminal Lys residue could be detected, supplemental Fig. 1f).
We aligned the sequences of the FadDs that were acetylated as well as those that were not (Fig. 1e). We observed the conservation of motifs GGXNX4EXE/D, E/DX7E/D, K/AXP, and PX4GK in the C-terminal domain sequences of FadDs that were acetylated, whereas non-acetylated FadDs showed conservation in some, but not all, of these motifs together. This implies a concerted influence of a set of conserved residues in the C-terminal domain of FadDs, rather than just residues in the immediate proximity of the acetylated lysine residue that determine substrate specificity. Acetylated FADs clustered together on a dendrogram built on a sequence alignment of the full-length proteins (Fig. 1f), whereas the non-acetylated enzymes branched separately. Inspection of the dendrogram and sequences of the non-purifiable enzymes allowed us to predict whether a protein could be acetylated or not by KATmt (supplemental Fig. 1g). Because FadDs 1, 11, and 15 do not have the PX4GK motif and FadDs 21 and 33 lack the K/AXP, E/DX7E/D, and GGXNX4EXE/D motifs, we predict that they may not be acetylated by KATmt. On the other hand, FadDs 8, 3, 14, and 36 retain most of the important motifs for acetylation and, therefore, could be substrates for KATmt. Manual inspection of the sequences of FadD16 and FadD9 revealed that they lack the PX4GK motif, which includes the acetylated Lys, at the C terminus and are, therefore, unlikely to be acetylated.
Deacetylation of FadDs by a Mycobacterial Sirtuin-like Enzyme
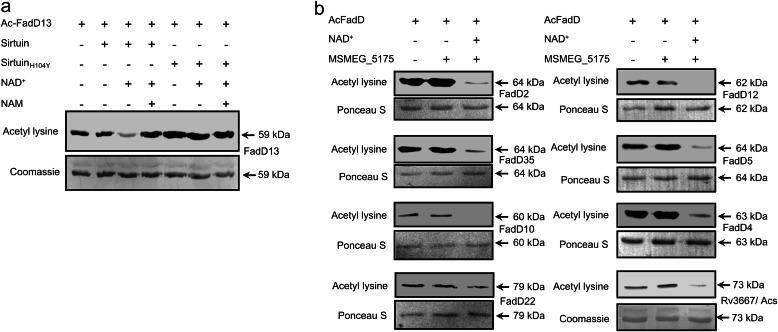
Lysine acetylation is a reversible process (24, 25), and deacetylation can be carried out by the sirtuin class of deacetylases. Sirtuin-like enzymes can be identified in bacterial genomes (25, 26), and one such enzyme, the product of the Rv1151c gene, has been shown to deacetylate ACS (19). We cloned and purified the ortholog of Rv1151c from M. smegmatis, MSMEG_5175 (sirtuin) (the former being more difficult to purify, supplemental Fig. 2, a and b), and monitored its ability to deacetylate FadD13 (supplemental Fig. 2c). Sirtuins are NAD+-dependent enzymes, and a critical His residue is present at the active site, which, on mutation to Tyr, inactivates enzyme activity (27). As seen in Fig. 2a, sirtuin was able to deacetylate FadD13 only in the presence of NAD+, whereas the sirtuinH104Y, lacking the catalytic residue, showed no activity either in the presence or absence of NAD+. Inclusion of nicotinamide, an inhibitor of sirtuin-like enzymes, prevented the deacetylation of FadD13 (28).
FIGURE 2.
Deacetylation of FadDs by sirtuin. a, FadD13 (2 μg) was acetylated by KATmt (200 ng) at 37 °C for 30 min and then incubated with sirtuin (500 ng) or sirtuinH104Y in the presence of NAD+ (1 mm) for 1 h at 37 °C. Nicotinamide (NAM, 2 mm) was added to the reaction mixture as indicated. Western blot analysis was performed with acetyl lysine antibody (upper panel) followed by Coomassie Brilliant Blue staining of the blot (lower panel). b, two micrograms of the indicated FadDs were acetylated using KATmt and then deacetylated using sirtuin in the presence of NAD+ (1 mm), as described previously, followed by Western blotting with acetyl lysine antibody.
We then proceeded to test whether sirtuin could deacetylate all the FadD enzymes that we have shown previously to be acetylated by KATmt. As shown in Fig. 2b, all enzymes could be deacetylated, some more efficiently than others, perhaps being reflective of sirtuin substrate specificity or the presence of additional domains following the adenylation domain, as seen in FadD22 (29). These results, therefore, indicate that the state of FadD acetylation in the cell could be dynamically regulated by KATmt and sirtuin.
Acylation Inhibits the AMP-ligase Activity of FadD Enzymes
Lys-487 in FadD13 has been shown to be critical for the binding of ATP prior to the formation of the adenylated fatty acid, which is subsequently converted to the CoA derivative (20, 22). Therefore, reversible acetylation of this critical catalytic residue and the equivalent Lys residues in the other FadD enzymes could regulate their biochemical activities.
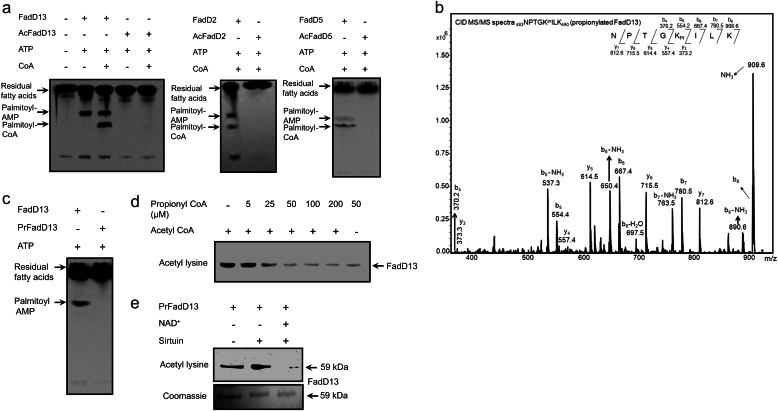
The substrates of all FadDs from mycobacteria have not been identified. However, three of the FadD enzymes (FadD13, FadD5, and FadD2) that were putative substrates for KATmt could charge palmitic acid to form the CoA derivative (Fig. 3a), but to different extents. As shown in Fig. 3a, acetylation of all these enzymes by KATmt prevented the formation of palmitoyl-AMP. Given the conservation of the Lys residue that is acetylated and its critical role in the formation of the fatty acyl-AMP conjugate, we predict that the activities of all the FadD enzymes that serve as substrates for KATmt are inhibited following acetylation.
FIGURE 3.
Inactivation of FadDs by KATmt following acylation. a, FadD or acetylated FadD were incubated with 14C-palmitic acid, ATP (5 mm), and CoA (2 mm), as indicated, at 30 °C for 5 min. Fatty acid migrated on the TLC with a relative mobility of 0.96. Palmitoyl AMP and palmitoyl CoA had Rf values of 0.6 and 0.51, respectively. b, CID MS/MS spectrum of the singly charged species of the propionylated tryptic peptide 483NPTGKPrILK490 of FadD13 (m/z 926.4) The signature b and y ions present in the spectrum are marked, and a schematic summary of all the b and y ions obtained is shown. c, FadD13 or propionylated FadD13 were incubated with 14C-palmitic acid and ATP (5 mm), as indicated, at 30 °C for 5 min. d, assays were performed in the presence of varying concentrations of propionyl CoA with a fixed amount of acetyl CoA (50 μm), and acetylated FadD13 was analyzed using Western blot analysis. e, two micrograms of the indicated FadD13 was propionylated using KATmt (200 ng, 30 °C, 30 min) and then deacetylated using sirtuin (500 ng) in the presence of NAD+ (1 mm) at 37 °C for 1 h, followed by Western blotting with acetyl lysine antibody.
In addition to acetyl CoA, some protein acetyl/propionyl transferases can also utilize propionyl CoA, resulting in the propionylation of proteins (30). Using FadD13 as a substrate, we demonstrated propionylation of the same Lys residue in FadD13 that was acetylated (Fig. 3b), resulting in inhibition of the activity of FadD13 in a manner similar to that seen on acetylation (c). We then tested whether the presence of propionyl CoA affected the utilization of acetyl CoA by KATmt. As shown in Fig. 3d, increasing concentrations of propionyl CoA in the presence of acetyl CoA inhibited the acetylation of FadD13, presumably leading to its propionylation and a reduction in immunoreactivity with the acetyl lysine antibody. Therefore, the relative levels of acetyl CoA or propionyl CoA in the cell could determine whether a substrate of KATmt is acetylated or propionylated.
Propionylated FadD13 showed a low cross-reactivity with the acetyl lysine antibody used for Western blot analysis (Fig. 3d, last lane). We exploited this to determine whether sirtuin was able to depropionylate FadD13. As shown in Fig. 3e, we could detect efficient depropionylation of FadD13 by the sirtuin enzyme, thus providing evidence for a more general mechanism of protein acylation and deacylation in mycobacteria and modulation of activities of substrate proteins.
In Vivo Acetylation of FadD13 in Mb BCG
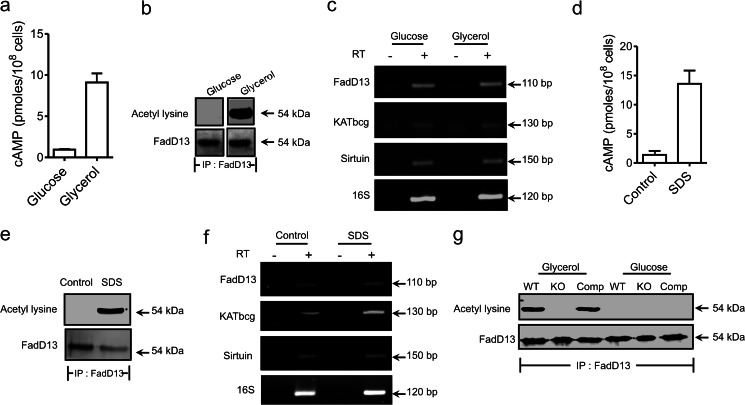
The activity of KATmt is precisely regulated by cAMP, as revealed by biochemical assays (Fig. 1a). We therefore extended these in vitro biochemical studies to monitor cAMP-dependent acetylation and deacetylation of KATmt substrates in vivo. Cyclic AMP levels in mycobacteria are modulated depending on environmental conditions and growth media (7, 31). The sequences of KATmt and its ortholog in M. bovis BCG, BCG_1055 (KATbcg), are identical, as are the sequences of FadD13 and its ortholog BCG_3114. M. bovis BCG grown in glycerol-containing media had 10-fold higher cAMP levels as compared with growth in glucose-containing media (Fig. 4a). We therefore monitored the acetylation status of FadD13 in cells grown under these conditions where intracellular levels of cAMP differed. Cells grown in glycerol showed robust acetylation of FadD13, whereas cells grown in glucose showed no acetylated FadD13 (Fig. 4b), even though levels of the sirtuin ortholog BCG_1212c and KATbcg were equivalent (c). Therefore, there appears to be a correlation between the intracellular levels of cAMP and the activity of KATbcg in vivo, resulting in the subsequent acetylation of its substrates.
FIGURE 4.
FadD13 is acetylated by KATmt in a cAMP-dependent manner. a, intracellular cAMP levels during growth of wild-type M. bovis BCG in medium containing 0.2% glycerol or 0.2% glucose as sole carbon source. Data shown are mean ± S.E. of duplicate determinations, with the experiment being repeated thrice. b, whole cell lysates from M. bovis BCG grown in 7H9 media containing 0.2% glycerol or 0.2% glucose were prepared. FadD13 was immunoprecipitated and subjected to Western blot analysis with acetyl lysine antibody and FadD13 antibody. c, semiquantitative RT-PCR analysis of BCG_3114 (FadD13), BCG_1055 (KATbcg), and BCG_1212c (sirtuin) from RNA prepared from wild-type M. bovis BCG grown in 0.2% glucose or 0.2% glycerol as sole carbon source. 16 S primers were used as a normalization control. d, wild-type M. bovis BCG was treated with 0.05% SDS for 1.5 h, and the intracellular cAMP level was measured. All data shown represent the mean ± S.E. of duplicate determinations, with each assay being performed thrice. e, whole cell lysates was prepared from SDS-treated cells. FadD13 was immunoprecipitated and subjected to Western blot analysis with acetyl lysine antibody and FadD13 antibody. f, semiquantitative RT-PCR analysis of FadD13, KATbcg, and sirtuin from RNA prepared from SDS-treated cells. 16 S was used for normalization. g, whole cell lysates from WT, knockout (KO), and complemented (Comp) strains grown in 7H9 media containing 0.2% glycerol or 0.2% glucose were prepared. FadD13 was immunoprecipitated and subjected to Western blot analysis with acetyl lysine antibody and FadD13 antibody.
We treated cells grown in glucose with 0.05% SDS, a procedure that, as we have shown earlier, results in an increase in intracellular cAMP in M. smegmatis (7). Indeed, cAMP levels were elevated in M. bovis BCG cells more than10-fold following SDS treatment (Fig. 4d). As shown in Fig. 4e, FadD13 was now found to be acetylated following SDS treatment. The levels of both sirtuin and FadD13 remained unaltered in the SDS-treated cells (Fig. 4f), whereas a 4-fold increase in the KATbcg transcript on SDS treatment was observed. This, together with the increase in intracellular cAMP seen on SDS treatment, could now result in activation of KATbcg and increased acetylation of FadD13 (Fig. 4d).
We next generated a strain of M. bovis BCG that was deleted for KATbcg (ΔKATbcg, supplemental Fig. 3a). Approximately 650 bp upstream of KATbcg, which would contain the promoter, were cloned along with the gene and integrated into the knockout strain to generate a complemented strain. The genomic integrity of these strains was confirmed by Southern blot analysis (supplemental Fig. 3b). The transcript for KATbcg was absent in the knockout strain, and transcript levels were restored to wild-type levels in the complemented strain (supplemental Fig. 3c). We grew cells in glycerol and monitored the acetylation status of FadD13 in these strains. As seen in Fig. 4g, FadD13 was not acetylated in the knockout strain grown either in glycerol- or glucose-containing media. Acetylation was restored on complementation of KATbcg in glycerol-containing media to the extent seen in the wild-type strain. These results, therefore, demonstrate that FadD13, and presumably other substrates of KATbcg, are acetylated exclusively by KATbcg and not by any other protein acetyltransferase that may be present in the cell. As a consequence, there is a specific and exquisite regulation of cAMP-dependent protein acetylation in mycobacteria, brought about by this unique acetyltransferase.
Deletion of KATbcg in Mb BCG Leads to Compromised Growth on Propionate-containing Media
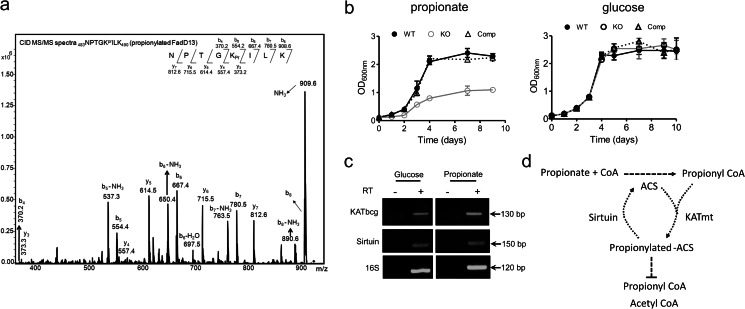
Mycobacteria are sensitive to high levels of intracellular propionyl CoA, presumably because of the formation of toxic propionyl CoA metabolites (32–34). ACS from M. tuberculosis is equally efficient in converting acetate or propionate to their CoA derivatives (35). We hypothesized that inhibition of the activity of ACS by acylation via KATbcg may be an important way of regulating the accumulation of propionyl CoA in the cell. We first confirmed that ACS could be propionylated by KATmt in the presence of cAMP (Fig. 5a). We then grew wild-type, ΔKATbcg and the ΔKATbcg strain complemented with a wild-type copy of KATbcg in media containing either propionate or glucose as the sole carbon source. Growth of the KATbcg deletion strain was not compromised in media containing glucose (where acetyl CoA formation by glycolytic pathways would be predominant), but growth in propionate was reduced dramatically (Fig. 5b). Growth could be restored to levels seen for the wild-type strain following complementation with KATbcg. Although cAMP levels in wild-type cells grown in either propionate (2.7 ± 0.7 pmol/ml/optical density) or glucose (Fig. 4a) were equivalent, the levels of KATbcg mRNA were increased significantly in cells grown in propionate (5.1 ± 0.25-fold as estimated by real-time quantitative RT-PCR, Fig. 5c). However, the mRNA levels of the sirtuin ortholog BCG_1212c remained similar. It is therefore likely that the high levels of intracellular propionyl CoA, which is found in cells grown in propionate as the sole carbon source, necessitate the propionylation of ACS by KATbcg, resulting in its inactivation. This would prevent the further build up of propionyl CoA, thereby alleviating toxicity (Fig. 5d).
FIGURE 5.
Propionylation and inactivation of ACS by KATmt. a, CID MS/MS spectrum of the singly charged species of the propionylated tryptic peptide 614SGKPrIMR620 of ACS (m/z 747.5). The signature b and y ions present in the spectrum are marked, and a schematic summary of all the b and y ions obtained is shown. b, growth analysis of the WT, knockout (KO), and complemented (Comp) strains in 7H9 media containing 0.2% propionate as the sole carbon source. Shown are the A600 values from three independent biological replicates. c, semiquantitative RT-PCR analysis of KATbcg and sirtuin from RNA prepared from wild-type M. bovis BCG grown in 0.2% glucose or 0.2% propionate as the sole carbon source. 16 S primers were used as a normalization control. d, schematic depicting enzymatic inactivation of ACS because of propionylation by KATmt.
DISCUSSION
In this study, we have identified multiple substrates for cAMP-dependent protein acylation in M. tuberculosis and, for the first time, demonstrated the relevance of this posttranslational modification in vivo. Protein acetylation in bacteria occurs widely, but mycobacteria appear to represent the first and so far the only case where acylation is directly regulated by cAMP. The presence of a sirtuin-like enzyme completes the machinery required for modulating protein lysine acylation in vivo, thus allowing mycobacteria to regulate the activities of various proteins on the basis of intracellular levels of cAMP, acetyl CoA, propionyl CoA, and NAD+. Such sophistication in directly regulating protein acylation has not been reported to date.
On the basis of a sequence alignment of acetylated proteins, we were able to recognize conserved amino acids not only in the vicinity of the modified Lys residue but also some distance upstream (Fig. 1e). This appears to be a general feature of all other acetyltransferases (36). A recent structural analysis of FadD13 shows that the Lys residue that is acetylated by KATmt is buried in the catalytic site of the protein (20). Thus, for it to be acetylated, a significant structural change needs to occur. Perhaps the conservation of residues some distance away from the acylated Lys residue may allow an initial recognition of a substrate by KATmt, thus determining the specificity of acylation and also resulting in the required conformational change to expose the Lys residue that is acetylated.
Reversible protein lysine acetylation regulates diverse protein properties, including DNA-protein interactions, protein-protein interaction, subcellular localization, stability, and enzymatic activity (37–43). We show here in mycobacteria that a variety of proteins serve as substrates for KATmt, indicating that multiple processes are likely to be affected by cAMP-dependent protein acetylation in these organisms as well. Of the multiple FadDs encoded in mycobacterial genomes, only a subset of them appears to be substrates for KATmt, emphasizing again the non-redundant properties of the FadD family of enzymes (Fig. 1f). Interestingly, none of the fatty acyl AMP ligases (FAALs), enzymes involved in adenylation of fatty acids prior to their modification by polyketide synthetases during mycobacterial cell wall synthesis (44), appear to be acetylated by KATmt. It is interesting to note that in eukaryotes, a number of mitochondrial metabolic enzymes are modified by acylation, including those involved in lipid β-oxidation (45, 46), reflective of what we see in mycobacteria.
The nutrients available to M. tuberculosis inside the host macrophage are restricted, and a number of studies have shown that fatty acids and cholesterol serve as the primary source of carbon (47–53). However, degradation of cholesterol and odd-chain fatty acids leads to the generation of propionyl CoA, which is toxic to these bacteria (32, 34). There are multiple mechanisms by which the toxicity of propionyl CoA can be alleviated. Isocitrate lyase can utilize propionyl CoA to form succinate and pyruvate (32, 49), propionyl-CoA carboxylase can generate methylmalonyl-CoA, which can enter the methylmalonyl pathway to produce succinyl CoA (33), and methylmalonyl CoA can also be used for the formation of mycobacterial cell wall lipids (54). We show here that propionyl CoA can also be utilized by KATmt for propionylation of proteins and, importantly, that this propionylation can be reversed by the sirtuin enzyme (Fig. 3d). We propose that this allows the selective modification of proteins, depending on the intracellular levels of propionyl CoA, acetyl CoA, cAMP, and NAD+, the latter being required for catalysis by the sirtuin. The precision by which M. tuberculosis can this regulate its metabolism appears to be unparalleled and suited to the complex environment that it is likely to experience in the macrophage during infection and persistence in the granuloma.
Although growth in propionate-containing media did not dramatically increase intracellular levels of cAMP, it appears that the cell is able to enhance protein acetylation by increasing the levels of KATmt mRNA (Fig. 5c). The M. tuberculosis ACS has been biochemically characterized, and in contrast to other species, it can utilize both acetate and propionate equally efficiently to generate their CoA derivatives (19, 35). This appears to be an evolutionary adaptation of this enzyme that would allow the generation of propionyl CoA that can be used for a variety of cellular processes, specifically in mycobacteria. However, under conditions of high concentrations of propionyl CoA, KATmt can propionylate and, thereby, inactivate ACS (Fig. 5d), thus preventing the further increase in propionyl CoA levels, which would prove toxic to the cell.
The promiscuity of the KATmt in utilizing acetyl or propionyl groups for acylation make it particularly suited to be enlisted in homeostatic responses to counter environmental or nutritional conditions that lead to elevated levels of propionyl CoA. This acylation appears obligatory for the alleviation of the toxicity of propionyl CoA metabolites by acting as a feedback loop for regulating propionyl CoA levels in the cell, thereby accounting for the poor growth of the ΔKATbcg strain.
In conclusion, we propose that KATmt influences a number of pathways in mycobacteria. Increases in intracellular cAMP, brought about by the enhanced activity of adenylyl cyclases, would result in the activation of KATmt. Fatty acyl Co-A ligases that degrade lipids (e.g. FadD13) (22) regulate phenolic glycolipid synthesis (FadD22) (29, 55), have a role to play in epithelial invasion (FadD2) (56), recycling of mycolic acids (FadD5) (57), utilization of cholesterol (FadD3) (58), or virulence (FadD10) (59) are regulated by acylation in a cAMP-dependent manner. Enzymes involved in intermediary metabolism are also regulated by acetylation, including ACS. Because ACS generates acetyl CoA and propionyl CoA equally efficiently in mycobacteria, inhibition of its activity following acylation may have a critical role to play in alleviating the toxicity of high levels of propionyl CoA. All of these mechanisms appear to be critically dependent on the levels of intracellular cAMP and/or KATmt, making KATmt an important contributor to mycobacterial physiology and, possibly, pathophysiology.
Acknowledgments
We thank Dr. A. Bhatt from the University of Birmingham for preparing the phage used for generating the knockout strain and for helpful suggestions. We also thank Dr. H. Balaram for useful discussions, Prof. P. Balaram for insights into the analysis of the results, Dr. Avinash R. Shenoy and Dr. Nirmalya Basu for critical reading of the manuscript, and Vani R. Iyer for technical support.
This work was supported by the Department of Biotechnology, Council for Scientific and Industrial Research, by the Department of Science and Technology, Government of India, and by a Research Associate Fellowship from the Indian Institute of Science (to S. N.).

This article contains supplemental Figs. 1–3 and Table 1.
- FadD
- fatty acyl CoA synthetase
- ACS
- acyl CoA synthetase
- BCG
- Bacillus Calmette-Guérin
- CID
- collision-induced dissociation.
REFERENCES
- 1. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 2. McCue L. A., McDonough K. A., Lawrence C. E. (2000) Functional classification of cNMP-binding proteins and nucleotide cyclases with implications for novel regulatory pathways in Mycobacterium tuberculosis. Genome Res. 10, 204–219 [DOI] [PubMed] [Google Scholar]
- 3. Shenoy A. R., Sivakumar K., Krupa A., Srinivasan N., Visweswariah S. S. (2004) A survey of nucleotide cyclases in actinobacteria. Unique domain organization and expansion of the class III cyclase family in Mycobacterium tuberculosis. Comp. Funct. Genomics 5, 17–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shenoy A. R., Visweswariah S. S. (2006) New messages from old messengers. cAMP and mycobacteria. Trends Microbiol. 14, 543–550 [DOI] [PubMed] [Google Scholar]
- 5. McDonough K. A., Rodriguez A. (2012) The myriad roles of cyclic AMP in microbial pathogens. From signal to sword. Nat. Rev. Microbiol. 10, 27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Agarwal N., Lamichhane G., Gupta R., Nolan S., Bishai W. R. (2009) Cyclic AMP intoxication of macrophages by a Mycobacterium tuberculosis adenylate cyclase. Nature 460, 98–102 [DOI] [PubMed] [Google Scholar]
- 7. Dass B. K., Sharma R., Shenoy A. R., Mattoo R., Visweswariah S. S. (2008) Cyclic AMP in mycobacteria. Characterization and functional role of the Rv1647 ortholog in Mycobacterium smegmatis. J. Bacteriol. 190, 3824–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rehmann H., Wittinghofer A., Bos J. L. (2007) Capturing cyclic nucleotides in action. Snapshots from crystallographic studies. Nat. Rev. Mol. Cell Biol. 8, 63–73 [DOI] [PubMed] [Google Scholar]
- 9. Harman J. G. (2001) Allosteric regulation of the cAMP receptor protein. Biochim. Biophys. Acta 1547, 1–17 [DOI] [PubMed] [Google Scholar]
- 10. Nambi S., Basu N., Visweswariah S. S. (2010) cAMP-regulated protein lysine acetylases in mycobacteria. J. Biol. Chem. 285, 24313–24323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nambi S., Badireddy S., Visweswariah S. S., Anand G. S. (2012) Cyclic AMP-induced conformational changes in mycobacterial protein acetyltransferases. J. Biol. Chem. 287, 18115–18129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee H. J., Lang P. T., Fortune S. M., Sassetti C. M., Alber T. (2012) Cyclic AMP regulation of protein lysine acetylation in Mycobacterium tuberculosis. Nat. Struct. Mol. Biol. 19, 811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Basu A., Rose K. L., Zhang J., Beavis R. C., Ueberheide B., Garcia B. A., Chait B., Zhao Y., Hunt D. F., Segal E., Allis C. D., Hake S. B. (2009) Proteome-wide prediction of acetylation substrates. Proc. Natl. Acad. Sci. U.S.A. 106, 13785–13790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 16. Tamura K., Dudley J., Nei M., Kumar S. (2007) MEGA4. Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 17. Lundby A., Lage K., Weinert B. T., Bekker-Jensen D. B., Secher A., Skovgaard T., Kelstrup C. D., Dmytriyev A., Choudhary C., Lundby C., Olsen J. V. (2012) Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dormeyer W., Ott M., Schnölzer M. (2005) Analysis of p300 acetyltransferase substrate specificity by MALDI TOF mass spectrometry. Methods 36, 376–382 [DOI] [PubMed] [Google Scholar]
- 19. Xu H., Hegde S. S., Blanchard J. S. (2011) Reversible acetylation and inactivation of Mycobacterium tuberculosis acetyl-CoA synthetase is dependent on cAMP. Biochemistry 50, 5883–5892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Andersson C. S., Lundgren C. A., Magnúsdóttir A., Ge C., Wieslander A., Martinez Molina D., Högbom M. (2012) The Mycobacterium tuberculosis very-long-chain fatty acyl-CoA synthetase. Structural basis for housing lipid substrates longer than the enzyme. Structure 20, 1062–1070 [DOI] [PubMed] [Google Scholar]
- 21. Jatana N., Jangid S., Khare G., Tyagi A. K., Latha N. (2011) Molecular modeling studies of Fatty acyl-CoA synthetase (FadD13) from Mycobacterium tuberculosis. A potential target for the development of antitubercular drugs. J. Mol. Model 17, 301–313 [DOI] [PubMed] [Google Scholar]
- 22. Khare G., Gupta V., Gupta R. K., Gupta R., Bhat R., Tyagi A. K. (2009) Dissecting the role of critical residues and substrate preference of a Fatty Acyl-CoA synthetase (FadD13) of Mycobacterium tuberculosis. PLoS One 4, e8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Duckworth B. P., Nelson K. M., Aldrich C. C. (2012) Adenylating enzymes in Mycobacterium tuberculosis as drug targets. Curr. Top Med. Chem. 12, 766–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ficner R. (2009) Novel structural insights into class I and II histone deacetylases. Curr. Top Med. Chem. 9, 235–240 [DOI] [PubMed] [Google Scholar]
- 25. Thao S., Escalante-Semerena J. C. (2011) Biochemical and thermodynamic analyses of Salmonella enterica Pat, a multidomain, multimeric N(ϵ)-lysine acetyltransferase involved in carbon and energy metabolism. mBio 2, e00216–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Starai V. J., Escalante-Semerena J. C. (2004) Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol. 340, 1005–1012 [DOI] [PubMed] [Google Scholar]
- 27. Tanny J. C., Moazed D. (2001) Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2. Evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. U.S.A. 98, 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Longo V. D., Kennedy B. K. (2006) Sirtuins in aging and age-related disease. Cell 126, 257–268 [DOI] [PubMed] [Google Scholar]
- 29. Ferreras J. A., Stirrett K. L., Lu X., Ryu J. S., Soll C. E., Tan D. S., Quadri L. E. (2008) Mycobacterial phenolic glycolipid virulence factor biosynthesis. Mechanism and small-molecule inhibition of polyketide chain initiation. Chem. Biol. 15, 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen Y., Sprung R., Tang Y., Ball H., Sangras B., Kim S. C., Falck J. R., Peng J., Gu W., Zhao Y. (2007) Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell Proteomics 6, 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bai G., Knapp G. S., McDonough K. A. (2011) Cyclic AMP signalling in mycobacteria. Redirecting the conversation with a common currency. Cell Microbiol. 13, 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muñoz-Elías E. J., Upton A. M., Cherian J., McKinney J. D. (2006) Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulence. Mol. Microbiol. 60, 1109–1122 [DOI] [PubMed] [Google Scholar]
- 33. Savvi S., Warner D. F., Kana B. D., McKinney J. D., Mizrahi V., Dawes S. S. (2008) Functional characterization of a vitamin B12-dependent methylmalonyl pathway in Mycobacterium tuberculosis. Implications for propionate metabolism during growth on fatty acids. J. Bacteriol. 190, 3886–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Upton A. M., McKinney J. D. (2007) Role of the methylcitrate cycle in propionate metabolism and detoxification in Mycobacterium smegmatis. Microbiology 153, 3973–3982 [DOI] [PubMed] [Google Scholar]
- 35. Li R., Gu J., Chen P., Zhang Z., Deng J., Zhang X. (2011) Purification and characterization of the acetyl-CoA synthetase from Mycobacterium tuberculosis. Acta Biochim. Biophys. Sin. 43, 891–899 [DOI] [PubMed] [Google Scholar]
- 36. Crosby H. A., Rank K. C., Rayment I., Escalante-Semerena J. C. (2012) Structural insights into the substrate specificity of the Rhodopseudomonas palustris protein acetyltransferase RpPat. Identification of a loop critical for recognition by RpPat. J. Biol. Chem. 287, 41392–41404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kouzarides T. (2000) Acetylation. A regulatory modification to rival phosphorylation? EMBO J. 19, 1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu L., Scolnick D. M., Trievel R. C., Zhang H. B., Marmorstein R., Halazonetis T. D., Berger S. L. (1999) p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell Biol. 19, 1202–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Takemura R., Okabe S., Umeyama T., Kanai Y., Cowan N. J., Hirokawa N. (1992) Increased microtubule stability and α tubulin acetylation in cells transfected with microtubule-associated proteins MAP1B, MAP2 or tau. J. Cell Sci. 103, 953–964 [DOI] [PubMed] [Google Scholar]
- 40. Sadoul K., Boyault C., Pabion M., Khochbin S. (2008) Regulation of protein turnover by acetyltransferases and deacetylases. Biochimie 90, 306–312 [DOI] [PubMed] [Google Scholar]
- 41. Sadoul K., Wang J., Diagouraga B., Khochbin S. (2011) The tale of protein lysine acetylation in the cytoplasm. J. Biomed. Biotechnol. 2011, 970382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schwer B., Bunkenborg J., Verdin R. O., Andersen J. S., Verdin E. (2006) Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. U.S.A. 103, 10224–10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 44. Trivedi O. A., Arora P., Sridharan V., Tickoo R., Mohanty D., Gokhale R. S. (2004) Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature 428, 441–445 [DOI] [PubMed] [Google Scholar]
- 45. Hirschey M. D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D. B., Grueter C. A., Harris C., Biddinger S., Ilkayeva O. R., Stevens R. D., Li Y., Saha A. K., Ruderman N. B., Bain J. R., Newgard C. B., Farese R. V., Jr., Alt F. W., Kahn C. R., Verdin E. (2010) SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. He W., Newman J. C., Wang M. Z., Ho L., Verdin E. (2012) Mitochondrial sirtuins. Regulators of protein acylation and metabolism. Trends Endocrinol. Metab. 23, 467–476 [DOI] [PubMed] [Google Scholar]
- 47. Liu K., Yu J., Russell D. G. (2003) pckA-deficient Mycobacterium bovis BCG shows attenuated virulence in mice and in macrophages. Microbiology 149, 1829–1835 [DOI] [PubMed] [Google Scholar]
- 48. Marrero J., Rhee K. Y., Schnappinger D., Pethe K., Ehrt S. (2010) Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc. Natl. Acad. Sci. U.S.A. 107, 9819–9824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McKinney J. D., Höner zu Bentrup K., Muñoz-Elías E. J., Miczak A., Chen B., Chan W. T., Swenson D., Sacchettini J. C., Jacobs W. R., Jr., Russell D. G. (2000) Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406, 735–738 [DOI] [PubMed] [Google Scholar]
- 50. Muñoz-Elías E. J., McKinney J. D. (2005) Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 11, 638–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pandey A. K., Sassetti C. M. (2008) Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U.S.A. 105, 4376–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chang J. C., Miner M. D., Pandey A. K., Gill W. P., Harik N. S., Sassetti C. M., Sherman D. R. (2009) igr Genes and Mycobacterium tuberculosis cholesterol metabolism. J. Bacteriol. 191, 5232–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Griffin J. E., Pandey A. K., Gilmore S. A., Mizrahi V., McKinney J. D., Bertozzi C. R., Sassetti C. M. (2012) Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem. Biol. 19, 218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Russell D. G., VanderVen B. C., Lee W., Abramovitch R. B., Kim M. J., Homolka S., Niemann S., Rohde K. H. (2010) Mycobacterium tuberculosis wears what it eats. Cell Host Microbe 8, 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Siméone R., Léger M., Constant P., Malaga W., Marrakchi H., Daffé M., Guilhot C., Chalut C. (2010) Delineation of the roles of FadD22, FadD26 and FadD29 in the biosynthesis of phthiocerol dimycocerosates and related compounds in Mycobacterium tuberculosis. FEBS J. 277, 2715–2725 [DOI] [PubMed] [Google Scholar]
- 56. Dam T., Danelishvili L., Wu M., Bermudez L. E. (2006) The fadD2 gene is required for efficient Mycobacterium avium invasion of mucosal epithelial cells. J. Infect. Dis. 193, 1135–1142 [DOI] [PubMed] [Google Scholar]
- 57. Dunphy K. Y., Senaratne R. H., Masuzawa M., Kendall L. V., Riley L. W. (2010) Attenuation of Mycobacterium tuberculosis functionally disrupted in a fatty acyl-coenzyme A synthetase gene fadD5. J. Infect. Dis. 201, 1232–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Casabon I., Crowe A. M., Liu J., Eltis L. D. (2013) FadD3 is an acyl-CoA synthetase that initiates catabolism of cholesterol rings C and D in actinobacteria. Mol. Microbiol. 87, 269–283 [DOI] [PubMed] [Google Scholar]
- 59. Hotter G. S., Wards B. J., Mouat P., Besra G. S., Gomes J., Singh M., Bassett S., Kawakami P., Wheeler P. R., de Lisle G. W., Collins D. M. (2005) Transposon mutagenesis of Mb0100 at the ppe1-nrp locus in Mycobacterium bovis disrupts phthiocerol dimycocerosate (PDIM) and glycosylphenol-PDIM biosynthesis, producing an avirulent strain with vaccine properties at least equal to those of M. bovis BCG. J. Bacteriol. 187, 2267–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]