Background: The Abi1 scaffold protein affects cell migration in vitro by regulating actin cytoskeletal dynamics.
Results: Knockdown of Abi1 or its binding partner, Wasp, disrupts eye development and retinal progenitor cell movement in Xenopus embryos.
Conclusion: Abi1 and Wasp are essential for eye morphogenesis in Xenopus.
Significance: Cytoskeletal regulation by Abi1 is critical in vivo for morphogenesis during embryonic development.
Keywords: Cell Migration, Development, Eye, Signal Transduction, Xenopus
Abstract
Abl interactor 1 (Abi1) is a scaffold protein that plays a central role in the regulation of actin cytoskeleton dynamics as a constituent of several key protein complexes, and homozygous loss of this protein leads to embryonic lethality in mice. Because this scaffold protein has been shown in cultured cells to be a critical component of pathways controlling cell migration and actin regulation at cell-cell contacts, we were interested to investigate the in vivo role of Abi1 in morphogenesis during the development of Xenopus embryos. Using morpholino-mediated translation inhibition, we demonstrate that knockdown of Abi1 in the whole embryo, or specifically in eye field progenitor cells, leads to disruption of eye morphogenesis. Moreover, signaling through the Src homology 3 domain of Abi1 is critical for proper movement of retinal progenitor cells into the eye field and their appropriate differentiation, and this process is dependent upon an interaction with the nucleation-promoting factor Wasp (Wiskott-Aldrich syndrome protein). Collectively, our data demonstrate that the Abi1 scaffold protein is an essential regulator of cell movement processes required for normal eye development in Xenopus embryos and specifically requires an Src homology 3 domain-dependent interaction with Wasp to regulate this complex morphogenetic process.
Introduction
Abl interactor 1 (Abi1) is a regulator of actin cytoskeleton dynamics through its role as a scaffold protein for several multiprotein complexes. One such complex associates with actin filaments through spectrin (1) and another through Eps8 (epidermal growth factor receptor kinase substrate 8) (2). Abi1 and Esp8 form a trimeric complex with the guanine nucleotide exchange factor Sos-1 to control actin polymerization by Rho family GTPases, by transducing activating signals from Ras to Rac (3). Abi1 also has a second role in this pathway as a critical component of the Wave complex, which includes Nap1, Sra1, HSPC300, and a Wasp family verprolin-homologous (Wave) protein. This complex links Rac to Wave, allowing Wave to stimulate actin polymerization by the Arp2/3 complex in response to Rac activation (4).
Abi1 also binds directly to Wasp and N-Wasp and cooperates with Cdc42 to stimulate them to induce Arp2/3-dependent actin assembly. Using cell culture systems, it was demonstrated that Abi1 and Wave, but not N-Wasp, are critical for Rac-dependent membrane protrusion and macropinocytosis (5). In contrast, Abi1 and N-Wasp, but not Wave, regulate epidermal growth factor receptor endocytosis and cell-surface distribution (5). Thus, Abi1 regulates both Wave and N-Wasp activities in specific actin-dependent processes.
Abi is not only capable of interacting with Wave and Wasp proteins to activate the Arp2/3 complex but can also stimulate actin nucleation by interacting with the Diaphanous (Dia)-related formins in the absence of Wave (6). In cell culture, knockdown of Abi1 or Dia1 severely disrupted cell-cell junctions, indicating that Abi1 may be important for junction formation and maintenance though recruitment and stimulation of Dia1 (6).
In mammals, Abi1 has two paralogs as follows: Abi2, which is over 90% identical to Abi1 in its functional domains, and Abi3/NESH, which is more divergent. Abi1 and Abi2 are expressed in the embryo, and recent studies have begun to address their roles in development. Abi2-KO mice survive to adulthood but exhibit a severe memory deficit and defective lens fiber orientation and migration, probably due to defects in dendritic spine morphogenesis and adherens junction formation (7). In contrast, epiblast-specific disruption of Abi1 in mice caused lethality at embryonic day 11.5, and embryos displayed morphogenetic defects in the developing heart and brain because of localized impairment of actin polymerization and reduced cell migration (8). Another report noted that mice lacking Abi1 or α4 integrin exhibited similar phenotypes, namely mid-gestational lethality with abnormalities in placental and cardiovascular development (9). Moreover, Abi1 protein bound to phosphorylated α4 integrin, and this interaction was required for cell spreading in vitro, suggesting a role for Abi1 in linking α4 integrin to the regulation of actin dynamics to control cell movements during development (9).
The Xenopus laevis Abi1 protein has high similarity to its mammalian homolog. Xenopus Abi1 was shown to interact with Esp8 and co-localize with N-Wasp in apical surface epithelial cells and to affect actin cables in a Rac-independent manner (10). An amino-truncated homolog of Abi2, Xlan-4, is also found in Xenopus and is expressed during CNS development (11).
Abi1 is also a substrate and binding partner of c-Abl1, a cytoplasmic and nuclear nonreceptor tyrosine kinase from the Src family that is implicated in processes of cell differentiation, cell division, and cell adhesion (12). The oncogenic fusion protein, Bcr-Abl, induces tyrosine phosphorylation of Abi1 and translocation of Abi1/Wave2 to the plasma membrane, where actin polymerization occurs (13). Abi1 can also regulate the activity of c-Abl1 in a complex manner. Overexpression of Abi1 inhibits both transformation of NIH3T3 cells by v-Abl (14, 15) and serum-induced proliferation of NIH3T3/EGF receptor cells (2). However, binding to Abi1 has also been found to increase the catalytic activity of c-Abl1 in vitro and enhance the phosphorylation of several of its substrates, including Wave2 (16–18).
One morphogenetic event that requires the precise regulation of actin dynamics is the formation of the eye. In the first stages of eye development, specific cells in the animal hemisphere of the blastula-stage embryo receive signals that make them competent to contribute to the retina. During late gastrulation, cell movements are required to position these cells in the anterior neural plate where they are able to express a retinal fate (19). Morphogenetic movements are also required for the evagination of the optic vesicles from the ventral forebrain as the neural tube forms and for the subsequent invagination of both the distal optic vesicle and lens ectoderm to form the optic cup and lens vesicle, respectively (20). All these processes require coordinated regulation of actin dynamics and the assistance of the Rho family of small GTPases. Because Abi1 has the potential to regulate actin dynamics via multiple pathways, in this study we have examined the role of Abi1 in cell movement during eye field formation in X. laevis.
EXPERIMENTAL PROCEDURES
cDNAs and Plasmids
The abi1 gene, including 5′UTR, was amplified from an IMAGE clone (BC081178) obtained from Open Biosystems and cloned into pCS2+. abi1 RNA resistant to the Abi1 morpholino was constructed by deleting the 5′UTR region targeted by the MO.3 Further mutants of abi1, including Δ18–145 and ΔSH3(447–499), were created from the pCS2+ Abi1-FLAG/HA plasmid using QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA). wasp cDNA was amplified from an IMAGE clone (BC129739) from Open Biosystems using primers to tag with either FLAG or HA epitopes and clone into pCS107. Subsequent mutants, including wasp6pt mut (MO-resistant), or wasp Δproline were derived from pCS107-wasp-Flag using QuikChange site-directed mutagenesis. To isolate X. laevis c-abl1a and -b, 5′-RACE-ready cDNA (SMARTer RACE cDNA amplification kit, Clontech) was generated from total RNA isolated from stage 32 embryos using TRIzol reagent (Invitrogen). Using reverse primers designed against X. laevis c-abl1a or Xenopus tropicalis c-abl1b 5′-ORF sequences (obtained from the NCBI database), 5′-RACE reactions were carried out. The resulting products were cloned into the TOPO cloning vector and sequenced. Full-length versions of c-abl1a and -b (including 5′UTR MO target sites) were then obtained by RT-PCR using forward primers based on the RACE-derived sequences and a reverse primer based on X. laevis EST sequence and subcloned into pCS108 with insertion of two C-terminal FLAG tags. The dominant-negative kinase-dead mutants, c-abl1a (K270R) and c-abl1b (K290R), were derived from the wild-type constructs using QuikChange site-directed mutagenesis.
Whole Mount in Situ Hybridization
Xenopus embryos were collected at stage 34 for abi1, wasp, or c-abl1 mRNA expression. For analysis of eye field transcription factor expression, MO- or RNA-injected embryos were harvested at stage 16. The embryos were fixed in MEMFA (0.1 m MOPS, pH 7.4, 2 mm EGTA, 1 mm MgSO4, 3.7% formaldehyde) for 2 h and processed for whole mount in situ hybridization using standard methods (21) with the following probes: abi1 (ORF), wasp (ORF), c-abl1a (ORF), rx1 (22), and pax6 (23). Before analysis, embryos were bleached in formamide/hydrogen peroxide solution (3% H2O2, 5% formamide, in 0.5× SSC).
Blastomere Injections
Xenopus embryos were injected either at the two-cell stage into both blastomeres with Abi1 MO (37.5 ng) or at the 32-cell stage into one D1.1.1 blastomere with the following MOs: Abi1 MO (10 ng), Wasp MO (10 ng), or c-Abl1b MO (10 ng); or mRNAs: abi1 (150 pg), truncation mutants of abi1 (150 pg), wasp (200 pg), deletion mutants of wasp (200 pg), or dominant-negative mutants of c-abl1a/b (1 ng). The MOs used were 25 nucleotides long with the base sequences Abi1 MO (5′-GCGCATCGCTTCCTCCTTGTACACT-3′), Wasp MO (5′-CCATTTTAGGCCCCCCTCGGCTCAT-3′), and c-Abl1b MO (5′-CAGGCTGCTGCCCCATAGGATGAAC-3′). For rescue of MO effects, mRNAs were prepared from cDNAs encoding Abi1ΔUTR (lacking the MO-targeted 5′UTR) or Wasp6pt mut (containing mutations in wobble codons in the ORF, rendering the RNA resistant to the MO).
Immunoprecipitation and Western Blot Analysis
For Western blots to test MO efficiency on exogenously expressed proteins, oocytes were injected with Abi1, Wasp, or c-Abl1b MOs with or without abi1, abi1ΔUTR, wasp, wasp6pt mut, or c-abl1b mRNAs, as indicated. Oocytes were cultured overnight, and lysates were then prepared with TNSG (20 mm Tris, pH 8.0, 137 mm NaCl, 10% glycerol, 1% Nonidet P-40) lysis buffer with protease and phosphatase inhibitors, as described previously (24). Immunoblotting was performed as described previously using HRP-conjugated anti-FLAG (1:2000; Sigma) or anti-HA (1:2000 Roche Applied Science) antibodies. To determine the effect of the c-Abl1b MO on endogenous c-Abl1 protein, embryos were injected at the two-cell stage with 10–40 ng of c-Abl1b MO or 40 ng of standard control MO, collected at stage 12.5, lysed, and subjected to immunoblotting with anti-c-Abl antibody (1:1000, catalog no. MABT203, Millipore, Billerica, MA), followed by HRP-conjugated anti-mouse antibody (1:2000, catalog no. 12-349, Millipore). As a loading control, blots were reprobed with anti-Erk2 antibody (1:5000, catalog no. sc-154, Santa Cruz Biotechnology, Dallas, TX), followed by HRP-conjugated anti-rabbit antibody (1:2000, catalog no. 12-348, Millipore). For immunoprecipitations, embryos at the two-cell stage were injected with the following mRNAs: abi1 or abi1 mutants, including abi1Δ18–145 and abi1 ΔSH3(447–499) with or without wasp or wasp Δproline(Δ301–391) or with or without c-abl1a/b or dominant-negative c-abl1 mutants. Embryos were grown to stage 12.5 and then lysed as above. Immunoprecipitations were carried out in 15 embryo equivalent lysates with monoclonal antibodies raised against FLAG (G191, ABM) or HA (3F10, Roche Applied Science) for 2 h and protein-A/G-agarose (Santa Cruz Biotechnology) for 2 h followed by four washes with TNSG buffer. Beads were then incubated with 35 μl of SDS sample buffer for 5 min at 95 °C and subjected to immunoblotting using HRP-conjugated anti-FLAG or anti-HA antibodies, as above, or HRP-conjugated anti-phosphotyrosine (Tyr(P)) antibody (1:6000, catalog no. 16-105, Millipore). For anti-Tyr(P), the blocking buffer was 1% fish gelatin, 1% BSA, 2% goat serum in TBST. For all other antibodies, 5% milk in TBST was used.
Cell Fate Analysis
Embryos were injected at the 32-cell stage in one D1.1.1 blastomere with various MOs along with 200 pg of GFP RNA or 3.75 ng of Alexa 488-conjugated dextran (Invitrogen) as a lineage tracer, collected at stage 32, and fixed in MEMFA for 2 h. For sectioning, fixed samples were washed with PBS before transferring to 15% sucrose solution containing 15% fish gelatin (Sigma) for overnight equilibration. Sections of 15 μm thickness were obtained using standard cryo-sectioning techniques. Subsequently, the slides were dried at room temperature for 30 min and mounted with Vectashield mounting medium. Sections were examined under the fluorescence microscope Zeiss Observer .Z1, and images were acquired with a Carl Zeiss AxioCam MRM camera using Axiovision software.
Tracing of Cell Positions during Gastrulation
Embryos were injected in one D1.1.1 blastomere with various MOs, or resistant RNAs along with β-galactosidase mRNA (200 pg), grown to stage 12.5, and fixed for 1 h in MEMFA. Immediately after fixing, embryos were washed several times in PBST and incubated in 1.5 mg/ml Red-Gal (5-bromo-6-chloro-3-indolyl β-d-galactopyranoside; Sigma) in 1 ml of LacZ staining solution (20 mm K3Fe(CN)6, 20 mm K4Fe(CN)6, 2 mm MgCl2, 0.01% deoxycholate, 0.02% Nonidet P-40) at 37 °C for 45 min to 2 h until red color developed. Subsequently, embryos were washed several times in PBST, fixed in Bouin's fixative substitute (IMEB Inc., San Marcos, CA) for 1 h, washed several times in 70% EtOH in PBS, and bleached as described earlier. Cell position analysis was performed using ImageJ version 1.44o software (National Institutes of Health). RGB images were split into their constituent channels using the “RGB split” command, and the green channel, which shows the maximum contrast between Red-Gal staining and background, was used for analysis. The scale was set using the plastic mesh base of the dish used to photograph embryos (1 mm2). To calculate the area of cell spread, the freehand selection tool was used to draw around the stained region, and its area was determined using the “Analyze>Measure” command.
Whole Mount TUNEL Assay
Terminal deoxynucleotidyltransferase-mediated dUTP-X nick end labeling (TUNEL) assays to detect double-stranded breaks in DNA due to cell apoptosis were performed as described (25). Briefly, embryos injected in one D1.1.1 blastomere with Abi1 MO (10 ng), Wasp MO (10 ng), or STD MO (10 ng) were grown to stage 12.5, fixed in MEMFA, and washed sequentially with PBST (PBS containing 0.01% Tween 20), and terminal deoxynucleotidyltransferase buffer. Subsequently, embryos were incubated overnight at room temperature in terminal deoxynucleotidyltransferase buffer containing 150 units/μl terminal deoxynucleotide transferase and 0.5 μm digoxygenin-dUTP. Embryos were then subjected to multiple 1-h washes as follows: twice with PBS containing 1 mm EDTA at 65 °C, four times with PBS at 23 °C, once with PBT (PBS containing 2 mg/ml BSA and 0.1% Triton X-100), and once with PBT containing 20% normal goat serum. Subsequently, embryos were incubated with alkaline phosphatase-conjugated anti-digoxygenin Fab-fragments (1:2000; Roche Applied Science) in PBT containing 20% normal goat serum. Embryos were then washed five times, for 1 h each, with PBT and 10 min with alkaline phosphatase buffer (100 mm Tris, pH 9.5, 50 mm MgCl2, 100 mm NaCl, 0.1% Tween 20), and the chromogenic reaction was performed in BM purple (Roche Applied Science). Subsequently, embryos were washed briefly in alkaline phosphatase buffer, re-fixed in MEMFA, washed a few times in 70% ethanol in PBS, and bleached as described previously.
RESULTS
Knockdown of Abi1 in X. laevis Affects Eye Development
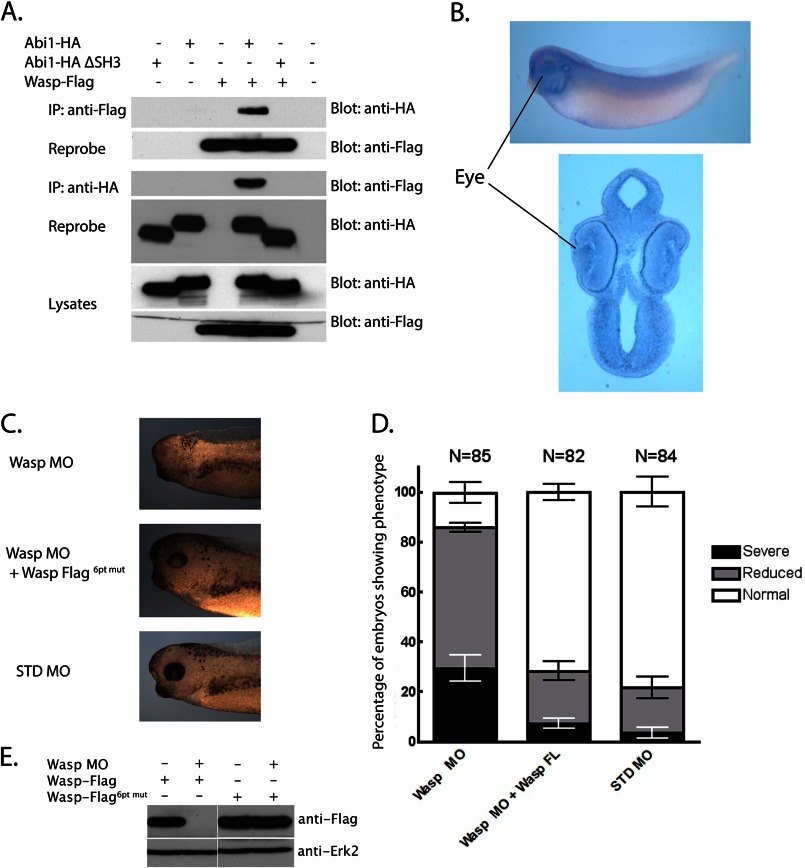
Because cell culture studies have suggested an important role for Abi1 in cell movement (26), and Abi1 null mice are embryonic lethal, we used X. laevis embryos to study the role of Abi1 during the major cell movements that shape tissue and organ development. Xenopus Abi1 protein is 86% identical to its murine ortholog. To determine the role of Xenopus Abi1 in embryogenesis, we used an Abi1 antisense MO to block Abi1 translation. Injection of the Abi1 MO into both blastomeres of two-cell embryos results in a shortened body, reduced head size, and defects in melanocyte distribution, as well as abnormal eye development (Fig. 1A). Embryos injected with standard control MO (STD MO) developed normally. Swimming tadpole stage embryos (37–38) showed eye defects ranging from absence of eye formation to more than 50% reduction in eye size (Fig. 1A). To determine whether the phenotype observed is specific to Abi1 knockdown, we designed an HA-tagged construct of Abi1 that lacks the 5′-untranslated region of the mRNA and is thus resistant to the MO (abi1-HAΔ5′UTR RNA). Co-injection of abi1-HAΔ5′UTR RNA along with the Abi1 MO markedly rescued the eye defect observed with Abi1 MO alone (Fig. 1A). Western blot analyses show that Abi1 MO inhibits exogenously expressed Abi1 protein, whereas the Abi1 HAΔ5′UTR is resistant (Fig. 1B).
FIGURE 1.
Knockdown of Abi1 in X. laevis affects eye development. A, knockdown of Abi1 by injection of Abi1 MO (37.5 ng) into both blastomeres of two-cell stage embryos results in defects in eye development; injecting STD MO (37.5 ng) does not. Co-injection of abi1Δ5′UTR RNA (150 pg) largely restores normal eye development. B, Western blot to assess Abi1 MO efficacy and specificity. Xenopus oocytes were injected with abi1-HA RNA (10 ng) or abi1Δ5′UTR RNA (10 ng), alone or with Abi1 MO (10 ng). Wild-type abi1 RNA is susceptible to the MO, whereas abi1Δ5′UTR is resistant. C, whole mount in situ hybridization of stage 34 embryos using an abi1 antisense probe reveals expression in eye (lateral view and sagittal section). D, injection of Abi1 MO (10 ng) into the D1.1.1 blastomere of 32-cell stage embryos results in defects in eye development; injecting STD MO (10 ng) does not. Co-injection of abi1Δ5′UTR RNA (150 pg) largely restores eye development. E, summary of effects of 32-cell stage injections in D on eye development. Error bars represent mean ± S.E. F, sagittal sections of stage 37 embryos injected with Abi1 MO or STD MO along with Alexa 488-dextran (3.75 ng) into D1.1.1 blastomere. Note that the fluorescent cells containing Abi1 MO are found at the midline but not in the presumptive eye area.
Because the developing eye is a very tractable system for exploring how signaling molecules of interest may be involved in cell movement and morphogenesis (27–29), we focused our study on this aspect of the Abi1 morphant phenotype. In support of the Abi1 MO effect directly causing the small eye phenotype, whole mount in situ hybridization analyses at stage 34 showed that abi1 is widely expressed but that there are particularly high transcript levels in the eye, as well as brain, branchial arches, and otic vesicles. Tissue sections through the head region show expression in the retina and lens (Fig. 1C).
In X. laevis, the D1.1.1 blastomere of 32-cell stage embryos is a major progenitor of the cells that populate the retina later in development (30). To determine whether loss of Abi1 causes eye defects by affecting the ability of these progenitors to populate the retina, rather than a secondary morphogenetic effect, we injected Abi1 MO either alone or with abi1-HAΔ5′UTR RNA into the D1.1.1 blastomere. Examination of embryos at stage 37–38 shows that more than 78% of Abi1 MO-injected embryos show a defect in eye development (27.2 ± 7.1% S.E., eyes absent; 51.7 ± 4.8% S.E., eyes reduced), which is rescued by injecting Abi1 HAΔ5′UTR RNA (9.2 ± 2% S.E., eyes absent; 19.6 ± 2.3% S.E., eyes reduced), indicating that the eye developmental defect is specific to Abi1 (Fig. 1, D and E). Cross-sections of stage 37–38 embryos injected with either Abi1 MO or control MO along with dextran-Alexa 488 as a lineage tracer show that loss of Abi1 in D1.1.1 progenitors prevents the majority of D1.1.1 progeny cells from populating the retina. Instead, the cells populate the brain, head mesoderm, and pharynx (Fig. 1F). As expected, control MO containing D1.1.1 progeny populate the brain, both retinas, epidermis, and cement gland (Fig. 1F). These data indicate that Abi1 is necessary for proper contribution of progenitor cells to the retina.
Abi1 SH3 Domain Is Required for Rescuing Abi1 MO-induced Eye Developmental Defects
To gain further insight into possible signaling partners and pathways required by Abi1 to regulate eye morphogenesis, we created abi1-HAΔ5′UTR expression constructs lacking known functional domains (Fig. 2A) as follows: the N-terminal domain (residues 18–145), which notably binds Wave proteins, α4 integrin, the formin proteins Diaphanous1/2, and Nap1; or the C-terminal SH3 domain, which binds N-Wasp and c-Abl1 (5, 9, 18, 31–33). These constructs (wild-type abi1, abi1(Δ18–145), or abi1 ΔSH3) were introduced along with Abi1 MO into the D1.1.1 blastomere of embryos to test the ability of these mutants to rescue the eye defects caused by loss of Abi1. Analyses of stage 37–38 embryos show that abi1(Δ18–145) and wild-type abi1 constructs were able to markedly rescue (60 ± 7% normal eye for Δ18–145; 67 ± 3.5% for wild-type) the Abi1 MO-induced eye phenotype; however, abi1 ΔSH3 failed to do so (16.5 ± 3.3% normal; Fig. 2, B and C). These data indicate that the Abi1 SH3 domain is critical for eye development in X. laevis and that the N-terminal Wave-binding, SNARE, and homeodomain homologous region domains are not critical. From this we can conclude that the N-terminal binding partners (i.e. α4 integrin, Wave, Diaphanous1/2, and Nap1) are not necessary for Abi1's role in eye formation, but c-Abl and/or Wasp may be required.
FIGURE 2.
Abi1 SH3 domain is required for rescuing Abi1 MO-mediated defects in eye development. A, schematic depicting the full-length and mutant Abi1 protein and interaction domains along with binding partners. HHR, homeodomain homologous region; P/PP, proline-rich regions; SNARE, soluble NSF attachment protein receptor domain. B, embryos were injected at the 32-cell stage into the D1.1.1 blastomere with Abi1 MO (10 ng) and 150 pg of MO-resistant wild-type or mutant (Δ18–145 or ΔSH3 (447–499)) abi1Δ5′UTR RNAs, and examined at stage 37 for defects in eye development. C, summary of effects of injections in B on eye development. Error bars represent mean ± S.E.
Xenopus c-Abl Is Expressed in the Developing Eye and Can Interact with Abi1 but Is Not a Major Contributor to Abi1-mediated Eye Development
It was previously reported that in addition to binding N-Wasp, the Abi1 SH3 domain also binds the c-Abl tyrosine kinase (14, 34). c-Abl1 has two isoforms that both contain one SH3 and one SH2 domain, a tyrosine kinase domain, a DNA binding domain, and an F-actin binding region (supplemental Fig. 1). The two isoforms only differ in the N terminus, where c-Abl1b contains a myristoylation sequence that is absent from c-Abl1a. This difference may affect the activity and localization of the proteins (12, 35).
To determine whether c-Abl may play a role in Abi1-mediated signaling that affects eye development in Xenopus, we first determined that c-abl is expressed appropriately in the eye. Whole mount in situ hybridization analyses showed that c-abl transcripts are localized in the eye, as well as the branchial arches, otic vesicle, forebrain, and pronephros, at stage 34 (Fig. 3A). To confirm that c-Abl and Abi1 can interact in our system, X. laevis embryos were co-injected with RNA encoding HA-tagged Abi1 and FLAG-tagged c-Abl1, and co-immunoprecipitation analysis was performed (Fig. 3B and supplemental Fig. 3). Full-length Abi1 was co-immunoprecipitated with both isoforms of c-Abl, indicating that c-Abl can interact with Abi1. In addition, detection of phosphotyrosine in the anti-HA immunoprecipitate in the presence of c-Abl1 indicates that c-Abl1 proteins can phosphorylate Abi1 in Xenopus, as in humans.
FIGURE 3.
c-Abl1 interacts with Abi1 but does not affect eye development. A, whole mount in situ hybridization of stage 34 embryos using a c-abl1 antisense probe, showing expression in eye, among other tissues. ba, branchial arches; ov, otic vesicle; fb, forebrain; p, pronephros. B, Xenopus embryos were injected with HA-tagged abi1 RNA (500 pg) and/or FLAG-tagged c-abl1b RNA (1 ng). Extracts were co-immunoprecipitated (IP) with anti-FLAG or -HA antibodies and probed with anti-FLAG, -HA, or -Tyr(P) (p-Tyr) antibodies as indicated. Abi1 is detected in the c-Abl1b immune complexes and vice versa, and Abi1 is phosphorylated in the presence of Abl1b. C, top, Western blot of Xenopus oocyte lysates probed with anti-FLAG antibody, showing that c-Abl1b MO (10 ng) effectively blocks translation of exogenous c-Abl1b-FLAG protein (10 ng). Center, Western blot to show effect of c-Abl1b MO on endogenous protein. Xenopus embryos were injected into both blastomeres at the two-cell stage with 10–40 ng of c-Abl1b MO or 40 ng of STD MO. Embryos were analyzed at stage 12.5, using a c-Abl1 antibody. Bottom, 10 ng of c-Abl1b MO or STD MO was co-injected with GFP mRNA into the D1.1.1 blastomere at the 32-cell stage, and eye development was analyzed at stage 37–38. The fluorescent cells containing c-Abl1b MO can enter the eye field, and the eye forms normally. D, top, embryos were injected into the D1.1.1 blastomere at the 32-cell stage with GFP mRNA alone or with c-abl1a (K270R) or c-abl1b (K290R) RNAs, and eye development was analyzed at stage 37. The fluorescent progeny cells can enter the eye field, and the eye forms normally. Bottom, Western blot performed on siblings of the embryos shown above, showing that the dominant-negative c-Abl1 proteins are expressed above endogenous c-Abl1 levels.
To test whether c-Abl plays a role in Abi1-mediated signaling that affects eye development, we designed antisense morpholino oligonucleotides to block the translation of c-Abl1a and -1b, and these MOs were tested for effectiveness against exogenously expressed proteins and endogenous expression. Unfortunately, we were unable to generate an MO against the c-Abl1a isoform that could visibly reduce endogenous c-Abl1 expression in embryos. However, the c-Abl1b MO effectively blocks exogenous translation of the c-Abl1b isoform and can block most of the endogenous c-Abl1 expression detected in embryos at stage 12.5 (Fig. 3C), suggesting that the c-Abl1b isoform is the major component of the total c-Abl protein in embryos during early eye development. Moreover, there does not appear to be any effect of the c-Abl1b MO on population of the retina by D1.1.1 progeny cells at stage 37, and eye morphogenesis is normal (Fig. 3C), as is retinal progenitor movement at the late gastrula stage (supplemental Fig. 2, A and B).
As an additional test of whether c-Abl1 may play a role in eye development, we generated kinase-dead versions of both c-Abl1 isoforms (equivalent to the human c-Abl1b K290R mutant) and expressed them at levels 10–50-fold above endogenous levels in the D1.1.1 blastomere (Fig. 3D). Eye morphogenesis and progenitor cell population of the retina were unaffected by these dominant-negative mutants (Fig. 3D). As expected, these mutant proteins were co-immunoprecipitated with wild-type Abi1 from embryos exogenously expressing the mutants along with wild-type c-Abl1 (supplemental Fig. 3). Also as anticipated, the mutant c-Abl1 isoforms failed to phosphorylate Abi1, as evidenced by the absence of phosphotyrosine in the anti-HA immunoprecipitates, although the wild-type versions did phosphorylate Abi1 (supplemental Fig. 3). Taken together, these data strongly suggest that loss of c-Abl does not disrupt eye formation during Xenopus embryogenesis, and importantly, c-Abl is not a major contributor to Abi1-mediated signaling that regulates eye formation. Therefore, we tested whether another protein that is associated with the Abi1 SH3 domain (5), Wasp, affects Abi1-mediated signaling during eye formation.
Abi1 SH3 Domain Interacting Protein, Wasp, Is Required for Eye Development
To confirm that an Abi1/Wasp (X-Wasp) interaction can occur in X. laevis, Wasp and either Abi1 or an Abi1 mutant lacking the SH3 domain were exogenously expressed in oocytes, followed by co-immunoprecipitation analysis. Wasp was co-immunoprecipitated with full-length Abi1 but not with Abi1 lacking the SH3 domain (Fig. 4A). Having confirmed an Abi1 SH3 domain-dependent interaction between Wasp and Abi1, we investigated whether wasp is expressed in a location consistent with a role in eye development. In situ hybridization analyses demonstrate that wasp is expressed in multiple tissues, including the lens and retina of the eye, and the brain and branchial arches (Fig. 4B). To determine whether Wasp plays a role in eye development, and thus may mediate Abi1 function in this process, the D1.1.1 blastomere of 32-cell stage embryos was injected with an MO targeted against Wasp either alone or along with MO-resistant FLAG-tagged wasp (wasp-FLAG6pt mut) RNA (Fig. 4C). Analysis of stage 37–38 embryos showed that the Wasp MO causes eye defects with a frequency >85% (29.4 ± 5.1% S.E. absent eyes; 56.4 ± 1.9% S.E. reduced eyes) that are quite similar to those observed with an Abi1 knockdown (Fig. 4, C and D). This phenotype is specific to Wasp as it is rescued (71.8 ± 3.2% S.E. normal eyes) by co-injecting wasp MO-resistant RNA (7.2 ± 1.9% S.E. absent eyes; 21 ± 3.8% S.E. reduced eyes; Fig. 4, C and D). Western blot analysis confirmed that the Wasp MO inhibits expression of exogenously expressed FLAG-tagged Wasp in oocytes, although the Wasp FLAG6pt mut is resistant to the MO (Fig. 4E). These data indicate that knockdown of Wasp, like Abi1, leads to reduced or absent eyes in embryos.
FIGURE 4.
Abi1 SH3-interacting protein, Wasp, is required for eye development. A, Xenopus embryos were injected at the two-cell stage in both blastomeres with RNA encoding FLAG-tagged Wasp along with HA-tagged Abi1 or Abi1 ΔSH3 (1 ng each). At stage 12.5, extracts were prepared, co-immunoprecipitated (IP) with anti-FLAG or -HA antibodies, and immunoblotted as indicated. Abi1 is found in Wasp immune complexes and vice versa. B, whole mount in situ hybridization of stage 34 embryos using a wasp antisense probe, showing expression in eye, among other tissues (lateral view and sagittal section). C, Wasp MO (10 ng) was injected in the D1.1.1 blastomere either alone or with MO-resistant wasp-FLAG6pt mut RNA (200 pg), and embryos were examined at stage 37. Eye development is disrupted in the presence of Wasp MO but is rescued by co-injection of MO-resistant wasp RNA. D, histogram showing percentages of embryos with the indicated defects in eye development. Error bars represent mean ± S.E. E, Western blot analysis of lysates from Xenopus oocytes injected with wild-type or MO-resistant wasp RNA along with Wasp MO.
Wasp Proline-rich Domain Is Essential for Normal Eye Development
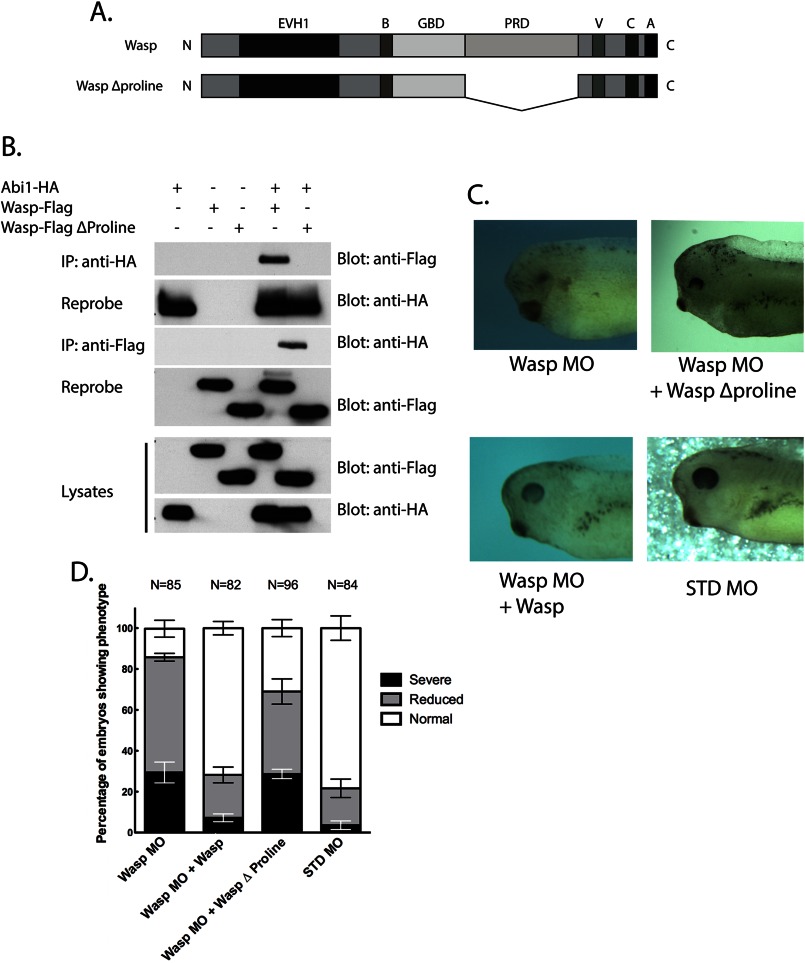
Similar to other SH3 domain interacting proteins, N-Wasp has been shown to bind the Abi1 SH3 domain via a proline-rich domain (5). To confirm this observation in X. laevis, we performed co-expression and co-immunoprecipitation assays in oocytes expressing wild-type FLAG-tagged Wasp or a FLAG-tagged Wasp protein lacking the proline-rich domain (Wasp-FLAG Δproline, Fig. 5A) along with an HA-tagged Abi1 protein. Wasp-FLAG Δproline failed to co-immunoprecipitate with Abi1, although the wild-type Wasp did associate with Abi1 (Fig. 5B).
FIGURE 5.
Wasp proline-rich domain is required for rescuing Wasp MO-mediated defects in eye development. A, schematic depicting full-length Wasp protein and the WaspΔproline mutant. A, acidic region; B, basic region; C, central hydrophobic region; EVH1, Ena/VASP homology domain 1; GBD, GTPase-binding domain; PRD, proline-rich domain; V, verprolin homology domain. B, embryos were injected with mRNA encoding HA-tagged Abi1 along with FLAG-tagged Wasp6pt mut or Wasp6pt mutΔproline (Δ301–391) (1 ng each). At stage 12.5, extracts were prepared, co-immunoprecipitated (IP) with antibodies against HA or FLAG, and immunoblotted with the indicated antibodies. The interaction with Abi1 is lost when wild-type Wasp is replaced with the Δproline mutant. C, Wasp MO (10 ng) was injected into the D1.1.1 blastomere along with 200 pg of RNA encoding Wasp-FLAG6pt mut or Wasp-FLAG6pt mutΔproline, and eye phenotypes were assessed at stage 37. Wasp-FLAG6pt mut rescues eye development, whereas the proline deletion mutant does not. D, histogram summarizing eye development phenotypes observed in the Wasp MO rescue experiment in C. Error bars represent mean ± S.E.
We have established that the SH3 domain of Abi1 is essential for eye development and that one known binding partner, c-Abl, is not critical in this process, although Wasp is necessary. However, to further test whether a Wasp and Abi1 interaction may be required for a role in eye development, we co-injected the D1.1.1 blastomere with Wasp MO and either full-length wasp-FLAG6pt mut RNA or wasp-FLAG6pt mut Δproline (a wasp mutant RNA lacking the proline-rich domain and resistant to the MO). Whereas full-length wasp RNA rescued the MO-induced eye defect (Figs. 5C and 4C), the mutant wasp RNA lacking the proline-rich domain was unable to do so (Fig. 5, C and D), resulting in 69% of embryos harboring eye defects (28.7 ± 2.3% S.E. absent eyes; 40.3 ± 6.2% S.E. reduced eyes; Fig. 5, C and D). Taken together, these data indicate that an interaction between Abi1 and Wasp is required for normal eye development.
Loss of Interaction between Abi1 and Wasp Disrupts Movement of Retinal Progenitor Cells
Because we demonstrated that an interaction between Abi1 and Wasp plays a critical role in proper eye development, we tested whether these signaling molecules that regulate actin polymerization (5) were responsible for controlling cell movement into the eye field. Abi1 MO or Wasp MO was co-injected with β-galactosidase RNA (as a cell lineage tracer, with Red-Gal as the substrate) into one D1.1.1 blastomere. The positions of the labeled cells were determined during late gastrulation (stage 12.5) when retinal progenitor cells populate the presumptive eye field. Analyses of the embryos show that in control MO-injected clones, D1.1.1 progeny were dispersed broadly across the dorsal animal quadrant (Fig. 6A). However, in Abi1 MO- or Wasp MO-injected embryos, the D1.1.1 clones were restricted to the midline (Fig. 6A). The area of Red-Gal staining (in mm2) used as a measure of the distance traversed during the lateral spread of progeny clones and Wasp MO-containing progeny was revealed to spread less (62 ± 5% S.E.) than controls (Fig. 6B). MO-mediated loss of Abi1 also caused a restricted spread of cells from the midline (60 ± 4% S.E. that of controls) when compared with controls (Fig. 6, A and B). Co-injection of resistant RNA with either the Abi1 MO or the Wasp MO substantially rescued the normal cell movement of these cells (92 ± 4% S.E. of standard control MO area for Abi1 HAΔ5′UTR; 83 ± 4% S.E. of standard control MO area for Wasp FLAG6pt mut) into the presumptive eye field (Fig. 6, A and B). To assess whether an interaction between Abi1 and Wasp is important for proper movement of these progenitors into the eye field, we performed similar MO rescue experiments with an MO-resistant Abi1 mutant that does not interact with Wasp (Abi1Δ5′UTRΔSH3) and an MO-resistant Wasp mutant that does not interact with Abi1 (Wasp-FLAG6pt mut Δproline). Expression of the Abi1Δ5′UTRΔSH3 mutant in the presence of the Abi1 MO did not rescue progenitor cell movement into the eye field; these cells remained restricted to the midline (53 ± 6% S.E. that of the area of controls; Fig. 6, A and B). In the reciprocal experiment, expressing the Wasp-FLAG6pt mut Δ proline mutant in the presence of the Wasp MO resulted in restricted cell movement as well (55 ± 5% S.E. of the area of controls; Fig. 6, A and B).
FIGURE 6.
Knockdown of Abi1 or Wasp in the D1.1.1 blastomere disrupts movement of progenitor cells into the eye field. A, D1.1.1 blastomere of 32-cell stage embryos was injected with 10 ng of Abi1 MO or Wasp MO and 200 pg of RNA encoding β-galactosidase, with or without abi1-HAΔ5′UTR (150 pg) or wasp-FLAG6pt mut (200 pg) RNA. Embryos were collected at stage 12.5 and stained for β-galactosidase (using Red-Gal as the substrate). Progenitor cells (red) are restricted to the midline when harboring the Abi1 MO or the Wasp MO, and this is rescued by co-expression of Abi1-HAΔ5′UTR or Wasp-FLAG6pt mut, respectively. In contrast, an Abi1 mutant lacking the SH3 domain or a Wasp mutant lacking the proline-rich domain cannot rescue the MO phenotypes. B, histogram depicts mean area of β-galactosidase spread (in mm2). Error bars represent mean ± S.E. C, Abi1, Wasp, or STD MOs were injected into the D1.1.1 blastomere, and stage 12.5 embryos were subjected to TUNEL assay (see “Experimental Procedures”). Only the positive control embryos are stained (dark blue).
To confirm that the reduction in the area occupied by Abi1 MO and Wasp MO containing progeny was not a result of cell death, we performed TUNEL assays on the MO-injected embryos at stage 12.5. We did not detect significant cell death in Abi1 MO or Wasp MO-injected embryos (Fig. 6C), suggesting that inhibition of cell movement, rather than cell death, is primarily responsible for the restricted spread of cells lacking Abi1 or Wasp during late gastrulation. Collectively, these data indicate that both Abi1 and Wasp are necessary for progenitor cells to properly move into the eye field and strongly suggest that the interaction between Abi1 and Wasp is required for this process.
Expression of rx1 and pax6 Is Repressed in Abi1 or Wasp Knockdown Embryos
During Xenopus embryogenesis, as early as stage 12.5, retinal progenitor cells occupy the presumptive eye field and express eye field transcription factors, including rx1 and pax6. Having established that retinal progenitor cell movement was restricted as early as the late gastrula stage, we tested whether the knockdown of Abi1 or Wasp affected eye field fate at this stage. We injected Abi1 MO or Wasp MO into one D1.1.1 blastomere and examined embryos at stage 12.5 by whole mount in situ hybridization. Expression of the early eye field markers rx1 and pax6 were significantly repressed on the injected side of the embryos, suggesting that early formation of the presumptive eye field was perturbed (Fig. 7, A and B for rx1; and Fig. 7, C and D for pax6). The repression of eye field markers was relieved by co-injection of either MO-resistant HA-tagged wild-type abi1 RNA (abi1-HAΔ5′UTR) or FLAG-tagged wild-type wasp RNA (wasp-FLAG6pt mut) along with the Abi1 MO or Wasp MO, respectively (Fig. 7, A–D). Of note, the Abi1 and Wasp interaction mutants (Abi1-HAΔ5′UTRΔSH3 and Wasp-FLAG6pt mutΔproline, respectively) failed to rescue the expression of these eye field markers (Fig. 7, A–D). Together, the data indicate that blocking either Abi1 or Wasp expression affects eye field progenitor cell movement and leads to reduced expression of markers of eye field fate. Moreover, these critical processes for proper eye formation require an interaction between Abi1 and Wasp.
FIGURE 7.
Early markers of eye development are reduced in Abi1 or Wasp knockdown embryos. A and C, embryos were injected unilaterally into one D1.1.1 blastomere, either with 10 ng of Abi1 MO with or without 150 pg of abi1-HAΔ5′UTR or abi1-HAΔ5′UTR ΔSH3 RNA, or with 10 ng of Wasp MO with or without 200 pg of wasp-FLAG6pt mut or wasp-FLAG6pt mut Δproline. Embryos were collected at stage 16 and subjected to whole mount in situ hybridization with antisense RNA probes against the eye-specific transcription factors, rx1 and pax6. Asterisk indicates injected side, and bars on embryos probed for pax6 mark expression in the putative eye field, with the midline indicated. Expression of both rx1 and pax6 is reduced in embryos containing the Abi1 MO or Wasp MO alone or with mutants lacking the interaction domains for these proteins. In contrast, wild-type abi1 or wasp RNAs rescue eye marker expression in the presence of their respective MOs. B and D, histograms depicting percentage of embryos showing reduced eye marker (rx1 or pax6 as indicated) expression on the injected side compared with the uninjected side.
DISCUSSION
Abi1 is an adaptor protein that interacts with several complexes that regulate actin dynamics, and Abi1-deficient mouse embryos die at mid-gestation with defects in the cardiovascular system, brain, and placenta, consistent with deregulation of the actin cytoskeleton and cell/cell interactions (8, 9). Because Abi1, through its effects on actin dynamics, has the potential to regulate multiple complex morphogenetic events, we examined the effect of Abi1 knockdown during Xenopus embryogenesis. In common with Abi1 knock-out mice, these embryos displayed several abnormalities, including reduced head size, abnormal pigmentation, and severe eye defects, which may be related to defects in cell movement or adhesion as a result of cytoskeletal disruption.
Because the development of the eye, although complex, represents a very tractable system for examining signaling events affecting morphogenesis, we chose to examine the role of Abi1 in eye development. During the specification of the eye field, retinal progenitor cells need to be properly positioned within the anterior neural plate to receive the local environmental signals that will direct them to undergo the subsequent steps of retinal morphogenesis, cellular lamination, and cell type specification (19, 36, 37). The observed dramatic loss of eye formation due to morpholino-mediated knockdown of Abi1 was not the result of major perturbations in the whole embryo, because restricting the MO injection to the D1.1.1 retinal progenitor cells had the same effect. GFP or β-galactosidase tracing of these cells showed that migration into the eye field was restricted, although a TUNEL assay indicated that apoptosis was not increased.
Abi1 interacts with several binding partners and complexes, and it was not inherently apparent which of these may be critical for proper eye formation. Because the binding sites for several of these proteins are known, we made mutants of Abi1 and expressed them in eye field progenitor cells where the endogenous Abi1 protein expression was inhibited by the MO. The Abi1(Δ18–145) mutant removes the SNARE domain, Wave-binding region, and most of the homeodomain homologous region, which are responsible for α4 integrin, Diaphanous, and Nap1 binding (6, 9, 26). Expression of this mutant along with an Abi1 MO in eye field progenitor cells effectively rescued eye formation, indicating that these motifs were not critical for this function of Abi1. In stark contrast, loss of the C-terminal SH3 domain completely prevented the rescue of eye formation.
The Abi1 SH3 domain can interact with the C-terminal end of the nonreceptor tyrosine kinase, c-Abl1. c-Abl1 is a known regulator of actin dynamics via multiple pathways; for example, it contains both F- and G-actin binding domains that mediate actin bundling, (38) and also phosphorylates and promotes the activity of the nucleation-promoting factor Wave2 (17). Abi1 is phosphorylated by c-Abl1 and can also modulate its activity against other substrates, suggesting c-Abl as a possible mediator of Abi1's role in eye development. However, eye formation was not visibly affected by injection of a morpholino targeting c-Abl1b or by overexpression of dominant-negative mutants of either c-Abl1 isoform. Therefore, another Abi1-associated protein, Wasp, was examined as a possible Abi1 signaling mediator in eye development.
The Wasp/N-Wasp proteins are central players in the signaling networks controlling actin dynamics through activation of the Arp2/3 complex. Native Wasp exists in an auto-inhibited complex, but it is activated upon attaining an open conformation, which is stimulated by binding of Cdc42 to its GTPase binding domain (4). Wasp contains a proline-rich region that binds the SH3 domain of Abi1, which promotes its activity by assisting in its release from auto-inhibition or promoting its oligomerization (5, 39).
MO-mediated knockdown of Wasp in the D1.1.1 blastomeres results in the disruption of eye formation and restriction of eye field progenitor cells to the midline, similar to the Abi1 MO. In the presence of Wasp MO, eye formation and normal eye progenitor cell movement were restored by an MO-resistant form of wild-type wasp RNA; however, a wasp mutant RNA lacking the proline-rich region that is necessary for an interaction with Abi1 failed to rescue these processes. Moreover, a similar result was observed in neurula stage embryos regarding the rescue of eye cell fate marker (rx1 and pax6) expression in D1.1.1 progeny harboring either Abi1 or Wasp MOs. Again, only the abi1/wasp interaction mutant RNAs failed to relieve the MO-mediated reduction in rx1 and pax6 expression, although wild-type versions rescued eye cell fate. Collectively, these data show that Abi1 and Wasp interaction mutants cannot substitute for their respective wild-type proteins in the context of eye development, strongly suggesting that the Abi1/Wasp interaction plays a critical role in proper eye field formation.
Although the mechanisms regulating retinal progenitor cell movement during eye field formation are poorly defined, previous studies by our laboratory and others have demonstrated an important role for reverse signaling by the Eph ligand, ephrinB1. Loss of ephrinB1 in retinal progenitors inhibits the movement of these cells into the eye field and reduces their contribution to the retina (29). The Xenopus Dishevelled protein (Xdsh) interacts with ephrinB1 and transduces its signals through the planar cell polarity pathway to regulate retinal progenitor movement (27). The noncanonical Wnt, Wnt11, also promotes eye field formation by acting through Frizzled 5 to cause local antagonism of canonical Wnt signaling and by regulating the cohesion of eye field cells (40). Thus, Wnt11 and ephrinB1 have overlapping functions and may collaborate in modulating retinal progenitor cell movement into the eye field; however, it appears that ephrinB1 reverse signaling through Xdsh can act independently of Wnt11 in this function. Furthermore, the fibroblast growth factor receptor 2 (FGFR2) can modulate this process by phosphorylating the intracellular domain of ephrinB1, which blocks the interaction with Xdsh (28).
Interestingly, phosphorylation of the ephrinB1 intracellular domain also allows it to associate with the Grb4 adaptor protein (41), and Grb4, in turn, has been identified as a binding partner of Abi1 (42). Thus, an additional mechanism for the modulation of ephrinB1-induced retinal progenitor cell movement by FGFR2 could be proposed, whereby the interaction of FGFR2-phosphorylated ephrinB1 with Abi1, via Grb4, may impair the ability to Abi1 to stimulate cell movement, perhaps by altering its localization or inhibiting its interaction with Wasp. Conversely, in the absence of FGFR2, unphosphorylated ephrinB1 would be unable to form a complex with Abi1, which therefore would be free to stimulate cell movement. Thus, interactions between the ephrinB1 and FGFR2 signaling pathways may play a central role in the accurate positioning of retinal progenitor cells within the anterior neural plate, and thus in normal eye development. However, many alternative or additional mechanisms for regulation of Abi1 and Wasp activities in retinal progenitors are also possible, and the elucidation of these would be an interesting topic of further study.
Acknowledgments
We thank Dr. Sally Moody for providing the Rx1 plasmid and Dr. Takaya Nakayama for the Pax6 plasmid. We thank Kathleen Mood for technical support and Dr. Hee Jun Cho and other members of the Cancer Genetics and Signaling Group at NCI-Frederick for helpful discussions and suggestions. We also thank Dr. Sally Moody and Dr. Harry Isaacs for critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Intramural Research Program of the NCI.

This article contains supplemental Figs. 1–3.
- MO
- morpholino oligonucleotide
- SH3
- Src homology 3
- RACE
- rapid amplification of cDNA ends
- STD MO
- standard control MO.
REFERENCES
- 1. Ziemnicka-Kotula D., Xu J., Gu H., Potempska A., Kim K. S., Jenkins E. C., Trenkner E., Kotula L. (1998) Identification of a candidate human spectrin Src homology 3 domain-binding protein suggests a general mechanism of association of tyrosine kinases with the spectrin-based membrane skeleton. J. Biol. Chem. 273, 13681–13692 [DOI] [PubMed] [Google Scholar]
- 2. Biesova Z., Piccoli C., Wong W. T. (1997) Isolation and characterization of e3B1, an eps8-binding protein that regulates cell growth. Oncogene 14, 233–241 [DOI] [PubMed] [Google Scholar]
- 3. Scita G., Nordstrom J., Carbone R., Tenca P., Giardina G., Gutkind S., Bjarnegård M., Betsholtz C., Di Fiore P. P. (1999) EPS8 and E3B1 transduce signals from Ras to Rac. Nature 401, 290–293 [DOI] [PubMed] [Google Scholar]
- 4. Derivery E., Gautreau A. (2010) Generation of branched actin networks: assembly and regulation of the N-WASP and WAVE molecular machines. BioEssays 32, 119–131 [DOI] [PubMed] [Google Scholar]
- 5. Innocenti M., Gerboth S., Rottner K., Lai F. P., Hertzog M., Stradal T. E., Frittoli E., Didry D., Polo S., Disanza A., Benesch S., Di Fiore P. P., Carlier M. F., Scita G. (2005) Abi1 regulates the activity of N-WASP and WAVE in distinct actin-based processes. Nat. Cell Biol. 7, 969–976 [DOI] [PubMed] [Google Scholar]
- 6. Ryu J. R., Echarri A., Li R., Pendergast A. M. (2009) Regulation of cell-cell adhesion by Abi/Diaphanous complexes. Mol. Cell. Biol. 29, 1735–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grove M., Demyanenko G., Echarri A., Zipfel P. A., Quiroz M. E., Rodriguiz R. M., Playford M., Martensen S. A., Robinson M. R., Wetsel W. C., Maness P. F., Pendergast A. M. (2004) ABI2-deficient mice exhibit defective cell migration, aberrant dendritic spine morphogenesis, and deficits in learning and memory. Mol. Cell. Biol. 24, 10905–10922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dubielecka P. M., Ladwein K. I., Xiong X., Migeotte I., Chorzalska A., Anderson K. V., Sawicki J. A., Rottner K., Stradal T. E., Kotula L. (2011) Essential role for Abi1 in embryonic survival and WAVE2 complex integrity. Proc. Natl. Acad. Sci. U.S.A. 108, 7022–7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ring C., Ginsberg M. H., Haling J., Pendergast A. M. (2011) Abl-interactor-1 (Abi1) has a role in cardiovascular and placental development and is a binding partner of the α4 integrin. Proc. Natl. Acad. Sci. U.S.A. 108, 149–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roffers-Agarwal J., Xanthos J. B., Miller J. R. (2005) Regulation of actin cytoskeleton architecture by Eps8 and Abi1. BMC Cell Biol. 6, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reddy B. A., Kloc M., Etkin L. D. (1992) The cloning and characterization of a localized maternal transcript in Xenopus laevis whose zygotic counterpart is detected in the CNS. Mech. Dev. 39, 143–150 [DOI] [PubMed] [Google Scholar]
- 12. Colicelli J. (2010) ABL tyrosine kinases: evolution of function, regulation, and specificity. Sci. Signal. 3, re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Y., Clough N., Sun X., Yu W., Abbott B. L., Hogan C. J., Dai Z. (2007) Bcr-Abl induces abnormal cytoskeleton remodeling, β1 integrin clustering, and increased cell adhesion to fibronectin through the Abl interactor 1 pathway. J. Cell Sci. 120, 1436–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi Y., Alin K., Goff S. P. (1995) Abl-interactor-1, a novel SH3 protein binding to the carboxyl-terminal portion of the Abl protein, suppresses v-abl transforming activity. Genes Dev. 9, 2583–2597 [DOI] [PubMed] [Google Scholar]
- 15. Ikeguchi A., Yang H. Y., Gao G., Goff S. P. (2001) Inhibition of v-Abl transformation in 3T3 cells overexpressing different forms of the Abelson interactor protein Abi1. Oncogene 20, 4926–4934 [DOI] [PubMed] [Google Scholar]
- 16. Tani K., Sato S., Sukezane T., Kojima H., Hirose H., Hanafusa H., Shishido T. (2003) Abl interactor 1 promotes tyrosine 296 phosphorylation of mammalian enabled (Mena) by c-Abl kinase. J. Biol. Chem. 278, 21685–21692 [DOI] [PubMed] [Google Scholar]
- 17. Leng Y., Zhang J., Badour K., Arpaia E., Freeman S., Cheung P., Siu M., Siminovitch K. (2005) Abelson-interactor-1 promotes WAVE2 membrane translocation and Abelson-mediated tyrosine phosphorylation required for WAVE2 activation. Proc. Natl. Acad. Sci. U.S.A. 102, 1098–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sato M., Maruoka M., Yokota N., Kuwano M., Matsui A., Inada M., Ogawa T., Ishida-Kitagawa N., Takeya T. (2011) Identification and functional analysis of a new phosphorylation site (Y398) in the SH3 domain of Abi1. FEBS Lett. 585, 834–840 [DOI] [PubMed] [Google Scholar]
- 19. Kenyon K. L., Zaghloul N., Moody S. A. (2001) Transcription factors of the anterior neural plate alter cell movements of epidermal progenitors to specify a retinal fate. Dev. Biol. 240, 77–91 [DOI] [PubMed] [Google Scholar]
- 20. Fuhrmann S. (2010) Eye morphogenesis and patterning of the optic vesicle. Curr. Top. Dev. Biol. 93, 61–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sive H., Grainger R. M., Harland RM. (2000) Early Development of Xenopus laevis, pp. 231–247, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 22. Mathers P. H., Grinberg A., Mahon K. A., Jamrich M. (1997) The Rx homeobox gene is essential for vertebrate eye development. Nature 387, 603–607 [DOI] [PubMed] [Google Scholar]
- 23. Hirsch N., Harris W. A. (1997) Xenopus Pax-6 and retinal development. J. Neurobiol. 32, 45–61 [PubMed] [Google Scholar]
- 24. Chong L. D., Park E. K., Latimer E., Friesel R., Daar I. O. (2000) Fibroblast growth factor receptor-mediated rescue of x-ephrin B1-induced cell dissociation in Xenopus embryos. Mol. Cell. Biol. 20, 724–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sible J. C., Anderson J. A., Lewellyn A. L., Maller J. L. (1997) Zygotic transcription is required to block a maternal program of apoptosis in Xenopus embryos. Dev. Biol. 189, 335–346 [DOI] [PubMed] [Google Scholar]
- 26. Steffen A., Rottner K., Ehinger J., Innocenti M., Scita G., Wehland J., Stradal T. E. (2004) Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 23, 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee H. S., Bong Y. S., Moore K. B., Soria K., Moody S. A., Daar I. O. (2006) Dishevelled mediates ephrinB1 signalling in the eye field through the planar cell polarity pathway. Nat. Cell Biol. 8, 55–63 [DOI] [PubMed] [Google Scholar]
- 28. Lee H. S., Mood K., Battu G., Ji Y. J., Singh A., Daar I. O. (2009) Fibroblast growth factor receptor-induced phosphorylation of ephrinB1 modulates its interaction with Dishevelled. Mol. Biol. Cell 20, 124–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moore K. B., Mood K., Daar I. O., Moody S. A. (2004) Morphogenetic movements underlying eye field formation require interactions between the FGF and ephrinB1 signaling pathways. Dev. Cell 6, 55–67 [DOI] [PubMed] [Google Scholar]
- 30. Huang S., Moody S. A. (1993) The retinal fate of Xenopus cleavage stage progenitors is dependent upon blastomere position and competence: studies of normal and regulated clones. J. Neurosci. 13, 3193–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Innocenti M., Frittoli E., Ponzanelli I., Falck J. R., Brachmann S. M., Di Fiore P. P., Scita G. (2003) Phosphoinositide 3-kinase activates Rac by entering in a complex with Eps8, Abi1, and Sos-1. J. Cell Biol. 160, 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Courtney K. D., Grove M., Vandongen H., Vandongen A., LaMantia A. S., Pendergast A. M. (2000) Localization and phosphorylation of Abl-interactor proteins, Abi1 and Abi-2, in the developing nervous system. Mol. Cell. Neurosci. 16, 244–257 [DOI] [PubMed] [Google Scholar]
- 33. Juang J. L., Hoffmann F. M. (1999) Drosophila Abelson interacting protein (dAbi) is a positive regulator of abelson tyrosine kinase activity. Oncogene 18, 5138–5147 [DOI] [PubMed] [Google Scholar]
- 34. Dai Z., Pendergast A. M. (1995) Abi-2, a novel SH3-containing protein, interacts with the c-Abl tyrosine kinase and modulates c-Abl transforming activity. Genes Dev. 9, 2569–2582 [DOI] [PubMed] [Google Scholar]
- 35. Koleske A. J., Gifford A. M., Scott M. L., Nee M., Bronson R. T., Miczek K. A., Baltimore D. (1998) Essential roles for the Abl and Arg tyrosine kinases in neurulation. Neuron 21, 1259–1272 [DOI] [PubMed] [Google Scholar]
- 36. Saha M. S., Servetnick M., Grainger R. M. (1992) Vertebrate eye development. Curr. Opin. Genet. Dev. 2, 582–588 [DOI] [PubMed] [Google Scholar]
- 37. Li H., Tierney C., Wen L., Wu J. Y., Rao Y. (1997) A single morphogenetic field gives rise to two retina primordia under the influence of the prechordal plate. Development 124, 603–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Van Etten R. A., Jackson P. K., Baltimore D., Sanders M. C., Matsudaira P. T., Janmey P. A. (1994) The COOH terminus of the c-Abl tyrosine kinase contains distinct F- and G-actin binding domains with bundling activity. J. Cell Biol. 124, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Padrick S. B., Rosen M. K. (2010) Physical mechanisms of signal integration by WASP family proteins. Annu. Rev. Biochem. 79, 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cavodeassi F., Carreira-Barbosa F., Young R. M., Concha M. L., Allende M. L., Houart C., Tada M., Wilson S. W. (2005) Early stages of zebrafish eye formation require the coordinated activity of Wnt11, Fz5, and the Wnt/β-catenin pathway. Neuron 47, 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bong Y. S., Park Y. H., Lee H. S., Mood K., Ishimura A., Daar I. O. (2004) Tyr-298 in ephrinB1 is critical for an interaction with the Grb4 adaptor protein. Biochem. J. 377, 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cowan C. A., Henkemeyer M. (2001) The SH2/SH3 adaptor Grb4 transduces B-ephrin reverse signals. Nature 413, 174–179 [DOI] [PubMed] [Google Scholar]