Background: Curli fibers are recognized by the TLR2/TLR1 complex.
Results: CD14 binds curli fibers and enhances TLR2/TLR1-dependent NF-κB activation as well as cytokine and nitrite production.
Conclusion: CD14 is an adaptor protein for the TLR2/TLR1 complex binding the fibrillar structure of curli fibers.
Significance: Understanding the immune receptor complexes that recognize amyloids will have implications for amyloid-associated diseases.
Keywords: Amyloid, Bacteria, Bacterial Pathogenesis, Biofilm, Toll-like Receptors (TLR), CD14, Salmonella, TLR2, Curli
Abstract
Amyloids, protein aggregates with a cross β-sheet structure, contribute to inflammation in debilitating disorders, including Alzheimer's disease. Enteric bacteria also produce amyloids, termed curli, contributing to inflammation during infection. It has been demonstrated that curli and β-amyloid are recognized by the immune system via the Toll-like receptor (TLR) 2/TLR1 complex. Here we investigated the role of CD14 in the immune recognition of bacterial amyloids. We used HeLa 57A cells, a human cervical cancer cell line containing a luciferase reporter gene under the control of an NF-κB promoter. When HeLa 57A cells were transiently transfected with combinations of human expression vectors containing genes for TLR2, TLR1, and CD14, membrane-bound CD14 enhanced NF-κB activation through the TLR2/TLR1 complex stimulated with curli fibers or recombinant CsgA, the curli major subunit. Similarly, soluble CD14 augmented the TLR2/TLR1 response to curli fibers in the absence of membrane-bound CD14. We further revealed that IL-6 and nitric oxide production were significantly higher by wild-type (C57BL/6) bone marrow-derived macrophages compared with TLR2-deficient or CD14-deficient bone marrow-derived macrophages when stimulated with curli fibers, recombinant CsgA, or synthetic CsgA peptide, CsgA-R4–5. Binding assays demonstrated that recombinant TLR2, TLR1, and CD14 bound purified curli fibers. Interestingly, CD14-curli interaction was specific to the fibrillar form of the amyloid, as demonstrated by using synthetic CsgA peptides proficient and deficient in fiber formation, respectively. Activation of the TLR2/TLR1/CD14 trimolecular complex by amyloids provides novel insights for innate immunity with implications for amyloid-associated diseases.
Introduction
Amyloids are protein aggregates with a conserved cross β-sheet structure where the axis of the β-sheets lies perpendicular to the fiber axis (1). More than 60 different amyloidogenic proteins are produced in humans (2). Several of these amyloid proteins have been shown to serve a functional role. For instance, Pmel17, involved in melanin production, forms amyloids that sequester toxic melanin precursors inside the melanosomes, protecting the melanocyte from cytotoxicity (3–5). Furthermore, peptide hormones take on an amyloid-like structure during their storage in secretory granules (6). Despite the apparent functionality of amyloids, accumulation of many fibrillar amyloids in various organs have been associated with many debilitating human diseases, including Alzheimer's disease, prion disease, Type II Diabetes Mellitus, and Creutzfeldt-Jakob disease (7–10). These complex amyloid-associated diseases still lack satisfactory treatment options for afflicted patients.
Many bacteria, including important pathogens such as Escherichia coli, Salmonella enterica serovar Typhimurium, Staphylococcus aureus, and Mycobacterium tuberculosis, as well as certain genera of non-pathogenic bacteria, have evolved to produce amyloids as a component of their extracellular matrix in biofilms (11–16). Because of their conserved quaternary structure, both human and bacterial amyloids share unique physical properties, including binding to fluorescent dyes such as Thioflavin T (ThT)2 and Congo red. When bound to fibrillar amyloids, Congo red displays an apple-green birefringence under a polarized light microscope (17–19). Furthermore, both human and bacterial amyloids show similar functional features. For instance, both β-amyloid 1–42, the amyloid found in the plaques of Alzheimer's disease patients, and curli fibers, produced by enteric bacteria, bind to fibronectin (20–22) and laminin (23) and trigger fibrinogen and plasminogen activation (24, 25).
Curli fibers, produced by members of the Enterobacteriaceae family, including E. coli and Salmonella typhimurium, represent the best-characterized bacterial amyloid to date. Curli fibers aid in the formation of enteric biofilms mediating bacterial adherence to self as well as to biotic and abiotic surfaces (26–28). The proteins necessary for curli fiber formation are encoded by two operons, csgBAC and csgDEFG (20, 29). CsgD acts as a positive transcriptional regulator for both operons, csgBAC and csgDEFG, and initiates transcription under stress-related conditions, including low osmolarity and low temperature (30). CsgA, the major subunit of the curli fibers, contains five repeats, R1-R5, which fold into the β-sheet necessary for fiber formation. Of these repeats, R1 and R5 seem to be the most important for E. coli CsgA fiber formation (31). It has been long understood that CsgB is the nucleator protein necessary for CsgA polymerization to occur efficiently, but only recently has the exact mechanism of this nucleation come to light (32). Studies showed that CsgB forms short fibers attached to the outer membrane that form a template for CsgA folding into the β-sheets necessary for polymerization (33). Nonetheless, CsgA still forms fibers at a slower rate, even in the absence of the nucleator protein CsgB when CsgA concentrations are high (34). CsgE acts as a chaperone for the major subunit of the fibers, CsgA, and prevents CsgA polymerization within the bacterial cell, as this could be toxic to the cell (35). CsgG forms an outer membrane pore that allows for CsgA to cross from the periplasm into the extracellular space (36). Finally, CsgF helps localization of CsgB and aids in the anchoring of the fibers to the outer membrane (37).
Toll-like Receptors (TLRs) are a member of the pattern recognition receptor family of innate immune receptors, which recognize conserved molecular patterns of microbes as well as endogenous danger molecules initiating inflammatory immune responses (reviewed in Ref. 38). TLR signaling cascades lead to the translocation of NF-κB into the nucleus through a signaling cascade involving an intracellular adaptor protein, termed myeloid differentiation primary response protein 88 (MyD88), leading to the transcription of various cytokines and chemokines (reviewed in Ref. 39). TLR4, together with adaptor molecules MD2 and CD14, recognizes hexa-acylated lipid A of LPS found in the outer membrane of most Gram-negative bacterial species (40, 41). TLR5 recognizes flagellin proteins that form the flagella of motile bacteria (42, 43). By forming heterodimers with either TLR1 or TLR6, TLR2 recognizes a wide range of molecules, including triacylated or diacylated lipopeptides, lipoteichoic acids, and zymosan (44, 45). Recently, we have shown that TLR2 recognizes curli fibers of enteric bacteria as well as the Alzheimer's disease-associated amyloid fiber, β-amyloid (46–48). Furthermore, we demonstrated that TLR2 requires the formation of a heterocomplex with TLR1 to recognize curli fibers (46). Consistent with our findings, a recent study revealed the involvement of TLR1 in the recognition of β-amyloid 1–42 by TLR2 (49). In addition, TLR2-TLR1 complex activation by serum amyloid A, an acute phase protein, has been demonstrated (50).
CD14, a Glycosylphosphatidylinositol (GPI)-anchored membrane protein lacking a transmembrane domain, is present in both a membrane-bound and a soluble form in the human body (51, 52). Soluble CD14 is primarily produced by hepatocytes in the liver, and its levels increase in inflammatory states, a characteristic of all acute phase proteins in the body, including serum amyloid A and C-reactive protein (53). Membrane-bound CD14 is primarily expressed by monocyte cell lineages (54, 55). Interestingly, CD14 has been implicated in the inflammatory milieu in the brain that occurs as a result of amyloid deposition (56, 57). In addition, CD14 has been shown to act as an adaptor molecule for various TLRs, including TLR4, TLR2, TLR9, and TLR7 (58–61). Although the adaptor function of CD14 for the TLR2-TLR1 complex has been demonstrated for a synthetic ligand, Pam3CSK4, the role of CD14 in the recognition of amyloids remains unknown.
Here we explored the function of CD14 in immune recognition of amyloids. We used curli fibers of S. typhimurium and investigated whether CD14 could act as an adaptor protein for the TLR2-TLR1 complex.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
The HeLa 57A cell line stably transfected with an NF-κB luciferase reporter was provided by Dr. R. T. Hay (The Wellcome Trust Centre for Gene Regulation and Expression, College of Life Sciences, University of Dundee, UK). HeLa 57A cells were grown in DMEM (Invitrogen) supplemented with 10% heat-inactivated FBS. HeLa 57A cells were grown to confluence in a 75-cm2 flask (Nunc) in a 5% CO2 incubator at 37 °C. Pam3CSK4, the synthetic triacylated lipoprotein used as a synthetic ligand for the TLR2-TLR1 complex, was purchased from InvivoGen (catalog no. tlrl-pms). Recombinant human TLR2 and mouse TLR1 (catalog nos. 2616-TR-050 and 1476-TR-050, respectively) were purchased from R&D Biosystems. Recombinant human soluble CD14 was purchased from Sigma-Aldrich (catalog no. SRP3149). The CsgA synthetic peptides CsgA R4–5 and CsgA R4–5N122A were described previously (48) and were purchased from Biosynthesis, Inc. ExGen 500 reagent for the transfection of HeLa 57A cells was purchased from Fermentas (catalog no. RO511).
Purification of Curli Fibers and Recombinant CsgA
All purified curli fibers were purified from the S. typhimurium msbB mutant (RPW3) (62) utilizing the established protocol described previously (63). The plasmid pSW5–50 contained the gene csgA cloned in GST fusion protein vector pGEX-4T-2. GST-CsgA was purified from E. coli DH5α (pSW5–50) as described previously (64). GST was purified from E. coli DH5α (pGEX-4T-2) utilizing the same protocol.
Plasmid Isolations and Transfections
The pTRACER (Invitrogen) human expression plasmid containing human TLR2, human TLR1, and human CD14 were maintained in E. coli DH5α as described previously (65). Overnight cultures were grown at 37 °C in Luria Bertani (LB) broth supplemented with 100 μg/ml of carbenicillin. The Qiagen Midi prep kit was utilized according to the instructions of the manufacturer to isolate the plasmids from these bacteria for transfection experiments. Transfections were conducted as described previously (46). Briefly, trypsinized cells were seeded at 5 × 105 cells/well in 500 μl of DMEM supplemented with 10% FBS. 24 h later, ExGen 500 reagent (Fermentas) was added according to the instructions of the manufacturer. Vectors carrying the hTLR2, hTLR1, hCD14, and lacZ genes were added in various combinations to a total amount of 250 ng of transfected plasmid DNA. In the transfections, the lacZ vector was used to normalize the transfection efficiency. The 48-well plate was then spun in a centrifuge for 5 min at 700 rpm. Cells were then incubated for 48 h before the experiment.
Luciferase Assay
48 h post-transfection, the cells were washed in 500 μl of cold PBS twice, and 500 μl of DMEM supplemented with 2% FBS was added into the wells. HeLa 57A cells were treated with 10 μg of purified curli fibers, GST-tagged curli monomer (GST-CsgA), or 50 ng of Pam3CSK4 synthetic TLR2/1 ligand. The same amount of GST purified in the same manner as the GST-CsgA was utilized as a negative control. The plates were centrifuged for 5 min at 700 rpm. The cells were then incubated for 8 h in a 5% CO2 incubator at 37 °C. After the 8-hour incubation, the medium was removed, and cells were washed with 500 μl of cold PBS twice. Cells were lysed by adding 100 μl of 1× reporter lysis buffer (Promega) to the cells, and they were flash-frozen at −80 °C overnight to lyse the cells. Following lysis, the lysates were assayed using the Promega luciferase assay kit according to the instructions of the manufacturer. A β-galactosidase assay was also run on the lysates to determine the transfection efficiency and normalize the luciferase assay results.
For determining the effect of soluble CD14, the above protocol was followed, but in this case purified curli samples were incubated overnight with 10 μg/ml of soluble CD14 before stimulation of the transfected HeLa 57A cells. After overnight incubation, fibers were spun down at 15,000 × g for 5 min, and unbound soluble CD14 was removed. These experiments were conducted in triplicate and repeated two times.
Bone Marrow-derived Macrophages (BMDMs)
Six- to eight-week-old female C57BL/6, TLR2-deficient mice (B6.129-TLR2tm1Kir/J) and CD14-deficient mice (B6.129-Cd14tm1Frm/J) were obtained from The Jackson Laboratories. BMDMs were differentiated as described previously (47). Briefly, femurs from three individual C57BL/6 TLR2-deficient and CD14-deficient mice were flushed, and a single cell suspension of the bone marrow was prepared in RPMI. The suspension was centrifuged at 1000 rpm for 10 min to pellet the cells. The cells were then resuspended in bone marrow macrophage medium. On the seventh day of culture, cells were seeded into the wells of a 24-well plate at a density of 500,000 cells/well. 24 h later, cells were stimulated with 7 μg of curli, GST-CsgA, 50 ng Pam3CSK4, or polymerized synthetic CsgA R4-R5 peptides. Supernatants were collected 6 h after stimulation. An IL-6 ELISA was conducted according to the instructions of the manufacturer (eBiosciences). A Griess reagent assay for nitrite in the supernatant was also conducted according to the instructions of the manufacturer (Acros Organics).
Protein Binding Assay
A protein binding assay described previously was modified (50). Briefly, wells of Maxisorp ELISA plates (Nunc) were coated with 5 μg of recombinant CD14, TLR2, and TLR1 in PBS overnight at 4 °C. The plate was then washed five times with PBS containing 0.5% Tween 20 and incubated for 2 h with 1% BSA in PBS. Next, the plate was incubated with increasing concentrations of curli (10 μg/ml, 20 μg/ml, 30 μg/ml, 50 μg/ml, 75 μg/ml, and 100 μg/ml) in 1% BSA in PBS for 2 h and then washed five times with PBS containing 0.5% Tween 20. The wells were then incubated with a 1:500 dilution of rabbit α-CsgA antibody in 1% BSA in PBS for 1 h and then washed five times with PBS containing 0.5% Tween 20. The plate was then incubated with an alkaline phosphatase-conjugated goat anti-rabbit IgG antibody diluted 1:1000 in 1% BSA in PBS for 1 h. The plate was washed eight times to ensure elimination of all excess secondary antibody. Wells were then incubated overnight with pNpp solution, and absorbance was read at 405 nm in a BMG Labtech POLARstar Omega microplate reader.
Inhibition of Binding
Wells of Maxisorp ELISA (Nunc) plates were coated with 2.5 μg of recombinant TLR2, TLR1, and CD14 in PBS overnight at 4 °C. Plates were then blocked with 1% BSA in PBS for 2 h. Wells were washed three times with PBS containing 0.5% TWEEN 20. Wells were incubated with 5 μg of GST-CsgA combined with 0.1 μg/ml, 0.5 μg/ml, 1.0 μg/ml, 5.0 μg/ml, 10.0 μg/ml 20 μg/ml, and 50 μg/ml of curli for 2 h. Wells were washed three times with PBS containing 0.5% Tween 20. Wells were then incubated with α-GST antibody (Sigma, catalog no. G7781) diluted 1:2000 in 1% BSA in PBS for 1 h. Wells were washed three times with PBS containing 0.5% Tween 20, and then wells were incubated with alkaline phosphatase-conjugated goat anti-rabbit IgG diluted 1:5000 for 1 h. Finally, plates were washed five times with PBS containing 0.5% Tween 20 and then incubated with 1 mg/ml p-Nitrophenyl Phosphate (pNPP) solution. Plates were incubated for 1 h at 37 °C, and absorbance was read using BMG Labtech Polarstar plate reader at 405-nm wavelength.
Synthetic Peptides (CsgA R4–5 and CsgA R4–5N122A)
Polymerization of the synthetic peptides CsgA R4–5 and CsgA R4–5N122A in to fibers was conducted as described previously, with some modifications (66). Briefly, 1 mg of lyophilized peptide was dissolved in 500 μl of hexafluoroisopropanol (HFIP), vortexed for 1 min, and incubated with shaking for 1 h at room temperature. HFIP was evaporated using a SpeedVac concentrator system (Savant) at room temperature. Dried peptide was then dissolved in 500 μl of dimethyl sulfoxide and vortexed until fully dissolved. The sample was injected into a HiTrap desalting column (GE Healthcare, catalog no. 17-1407-01) that had been previously equilibrated with 25 ml of equilibration buffer (50 mm Tris-HCl, 1 mm EDTA (pH 7.4)). 1 ml of equilibration buffer was injected, and that flow-through was discarded. Peptides were then eluted and collected twice using 1 ml of equilibration buffer.
Thioflavin T Assay
Equal volumes of 50 μm synthetic peptide and 10 μm ThT (Sigma, catalog no. T3516-5G) were combined and distributed into a black Nunc 96 MicroWelltm plate (catalog no. 237108) and sealed to prevent evaporation. ThT fluorescence intensity was monitored using a BMG Labtech POLARstar Omega microplate reader with 440/490-nm excitation/emission filters set for 36 h at 37 °C.
Dot Blot
5 μg of rabbit α-CsgA, CsgA R4–5, CsgA R4–5N122A, BSA, and purified curli were spotted and incubated on a PVDF membrane for 1 h. The membrane was then blocked in blocking buffer (5% dry milk in TBS (50 mm Tris, 0.5 m NaCl (pH 7.4))) for 1 h at room temperature. The blocking buffer was removed, and the membrane was incubated for 1 h at room temperature with a 1:500 dilution of rabbit α-CsgA serum in blocking buffer plus 0.05% Tween 20. The membrane was then washed with TBS plus 0.05% Tween 20 three times for 10 min each time with rocking. Next, the membrane was incubated with a 1:5000 dilution of goat anti-rabbit IgG antibody conjugated to IRDye® 800CW (Li-Cor Biosciences, catalog no. 926-32211) in blocking buffer for 1 h at room temperature and protected from light. The membrane was then washed three times in TBS plus 0.05% Tween 20. The membrane was visualized using an Odyssey infrared imager.
Statistical Analysis
One-way analysis of variance with Bonferroni post-test was utilized to calculate statistically significant differences in all experiments. Values of p < 0.05 were considered statistically significant.
RESULTS
CD14 Is Not Required but, Instead, Increases the Activation of TLR2/TLR1 by Curli Fibers
Both curli fibers and β-amyloid are recognized by TLR2 in cooperation with TLR1 (46, 56). Adaptor protein CD14 has been demonstrated previously to increase the activation of the TLR2-TLR1 heterocomplex by the triacylated lipopeptide Pam3CSK4 by binding and increasing the proximity of the ligand to the receptor complex (59). Interestingly, CD14 was implicated in the pathogenesis of Alzheimer's disease (56, 57). Although mice deficient in CD14 showed a slower progression of Alzheimer's disease, primary microglia deficient in CD14 expression had decreased phagocytosis and decreased levels of reactive oxygen species production in response to β-amyloid (56, 57). To determine whether CD14 acts as an adaptor for the TLR2-TLR1 complex in the recognition of amyloids, we utilized the HeLa 57A cell line stably transfected with a NF-κB luciferase reporter (67). Because HeLa cells do not express any TLRs, they represent a convenient model to investigate the contribution of TLRs to host responses. Similarly, HeLA cells do not express membrane-bound CD14 (52). HeLa 57A cells were transiently transfected with a combination of human expression vectors containing the genes for human CD14, human TLR2, and human TLR1. Cells were then stimulated with purified curli fibers, the recombinant curli major subunit GST-CsgA, and the synthetic TLR2-TLR1 ligand Pam3CSK4 as a positive control, or GST as a negative control. Cell lysates were collected and assayed for luciferase activity 8 h after stimulation. Consistent with the previous findings, cells transfected with both TLR2 and TLR1 had increased levels of luciferase activity, the indicator of NF-κB activation, upon stimulation with curli fibers, Pam3CSK4, and GST-CsgA. GST-CsgA polymerizes following the purification process (data not shown). HeLa 57A cells that were transfected with CD14 together with TLR2 and TLR1 showed significantly higher levels of luciferase activity after treatment with curli, Pam3CSK4, and GST-CsgA compared with cells transfected with only TLR2 and TLR1. As expected, cells stimulated with the GST protein purified with a similar method to GST-CsgA did not induce NF-κB activation. Similarly, cells transfected with CD14 alone lacked any significant NF-κB activation (Fig. 1). Overall, these data suggested that CD14 was not required for the activation of the TLR2-TLR1 heterocomplex by curli fibers. Nonetheless, the presence of CD14 enhanced the TLR2-TLR1 heterocomplex activation by curli fibers as well as its major subunit, CsgA.
FIGURE 1.
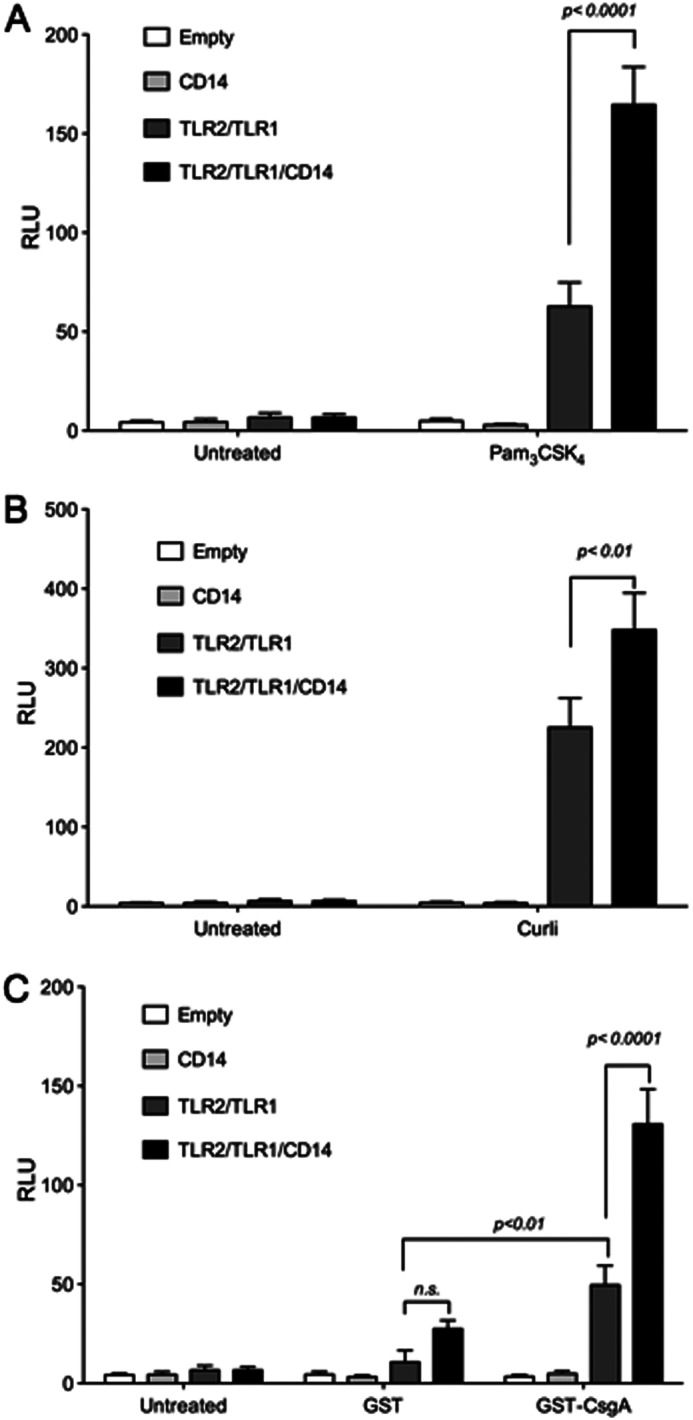
CD14 enhances NF-κB activation by the TLR2-TLR1 complex upon curli stimulation. HeLa 57A cells were transfected with an empty human expression vector or human expression vector carrying the indicated TLRs and CD14. Cells were stimulated with 50 ng of Pam3CSK4 (A), 10 μg of purified curli fibers (B), or GST or GST-CsgA fusion peptide (C) and lysed 8 h after the start of stimulation. NF-κB activation was monitored by measuring the luciferase activity as relative luminescence units (RLU). Bars represent averages mean ± S.E. from three independent experiments.
Both Membrane-bound and Soluble CD14 Increases the Activity of the TLR2-TLR1 Complex in Response to Curli Fibers
The adaptor molecule CD14 is expressed by monocytes and epithelial cells in its membrane-bound form. In addition, soluble CD14 is released into the serum by hepatocytes (51). In the absence of membrane bound CD14, soluble CD14 has been shown to aid the TLR4-MD2 complex to recognize lipid A (60). Although membrane-bound CD14 was identified as an adaptor molecule in the recognition of a synthetic ligand, triacylated lipopeptide Pam3CSK4, no studies have yet addressed whether soluble CD14 could replace the function of the membrane-bound CD14 molecule in aiding the TLR2-TLR1 complex. To determine the roles of membrane-bound and soluble CD14 in the immune recognition of curli fibers, HeLa 57A cells were transfected with vectors for TLR2 and TLR1 alone or in combination with the vector for CD14. Transfection of the cells with a vector carrying the gene for CD14 results in the expression of membrane-bound CD14. All groups of HeLa 57A cells were then treated with curli fibers alone or curli fibers that were pretreated with 10 μg of soluble CD14. Both the presence of membrane-bound CD14 and the pretreatment of curli fibers with soluble CD14 resulted in increased NF-κB activation through the TLR2-TLR1 heterocomplex. Additionally, when both membrane-bound and soluble CD14 were present during stimulation, no change in NF-κB was seen when compared with cells with just one form of CD14 present during stimulation (Fig. 2). These data suggest that the presence of either membrane-bound or soluble CD14 enhances the TLR2/TLR1 activation by curli fibers.
FIGURE 2.
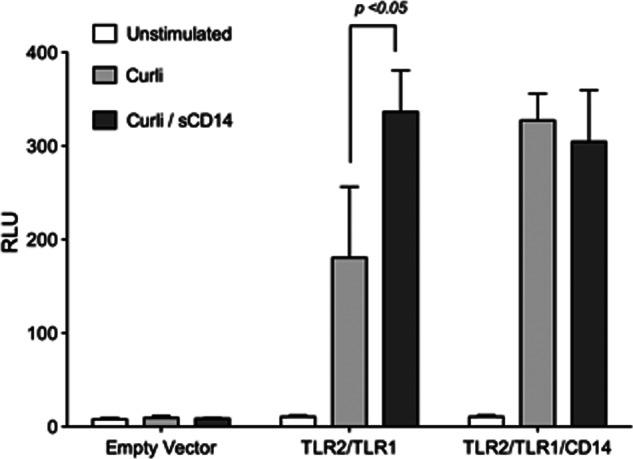
Soluble CD14 enhances recognition of curli fibers by the TLR2-TLR1 complex. HeLa 57A cells were transfected with an empty human expression vector or a human expression vector carrying the indicated TLRs and CD14. Cells were stimulated with 10 μg of curli fibers or 10 μg of curli fibers that had been incubated overnight with 10 μg/ml of soluble CD14. Cells were lysed 8 h after the start of stimulation, and NF-κB activation was monitored by measuring relative luminescence units (RLU). Bars represent mean ± S.E. from three independent experiments.
CD14 Leads to Increased IL-6 and Nitric Oxide Production By Macrophages Stimulated with Curli
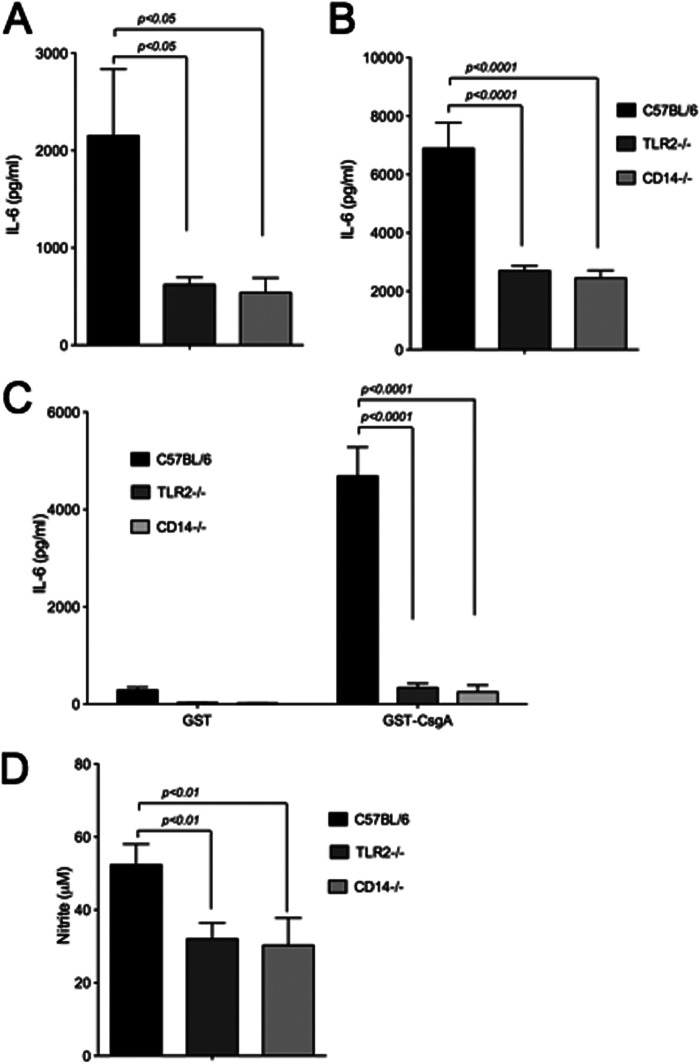
Among the monocyte populations, macrophages express high levels of CD14 (68). Curli fibers elicit cytokine and nitric oxide production in macrophages, which leads to hypotension and increased plasma nitrite/nitrate levels in a mouse model of E. coli sepsis in a TLR2-dependent manner (48, 69, 70). To determine the role of CD14 during activation of immune cells by curli fibers, we used BMDMs from wild-type C57BL/6, TLR2-deficient, and CD14-deficient mice. BMDMs were stimulated with curli, GST-CsgA, and Pam3CSK4, and supernatants were assayed for IL-6 production and the nitric oxide breakdown product nitrite using ELISA and a Griess reagent assay, respectively. To eliminate the effects of soluble CD14, which could be present in the fetal bovine serum, serum levels were decreased to 2% 24 h before stimulation of BMDMs.
Consistent with previous reports, BMDMs from C57BL/6 mice produced IL-6 in response to the synthetic TLR2-TLR1 complex ligand Pam3CSK4, whereas BMDMs deficient in CD14 or TLR2 showed significantly decreased IL-6 production (Fig. 3A). Similarly, CD14-deficient or TLR2-deficient BMDMs stimulated with curli fibers (Fig. 3B) or GST-CsgA (C) produced significantly lower levels of IL-6 compared with wild-type C57BL/6 macrophages. As expected, the GST protein purified with a similar method as the GST-CsgA did not induce IL-6 production in BMDMs (Fig. 3C).
FIGURE 3.
Membrane-bound CD14 contributes to IL-6 and nitrite production by BMDMs. BMDMs from C57BL/6, TLR2-deficient (TLR2−/−), and CD14-deficient (CD14−/− mice) were stimulated with 50 ng of Pam3CSK4 (A), 7 μg of purified curli fibers (B), or GST or GST-CsgA fusion peptide (C) for 6 h. Supernatants were collected at 6 h, and ELISA was conducted to measure IL-6 production. D, BMDMs were also stimulated with 10 μg of purified curli fibers for 24 h, and nitrite levels in the supernatant were measured. Bars represent mean ± S.E. from three independent experiments.
In addition, nitrite levels were significantly lower in the supernatants of TLR2- and CD14-deficient BMDMs compared with wild-type C57BL/6 BMDMs supernatants after stimulation with curli fibers (Fig. 3D). Together, this data suggested that CD14 contributed to the immune recognition of curli fibers as well as its major subunit, CsgA, by macrophages.
Curli Fibers bind TLR2, TLR1, and CD14
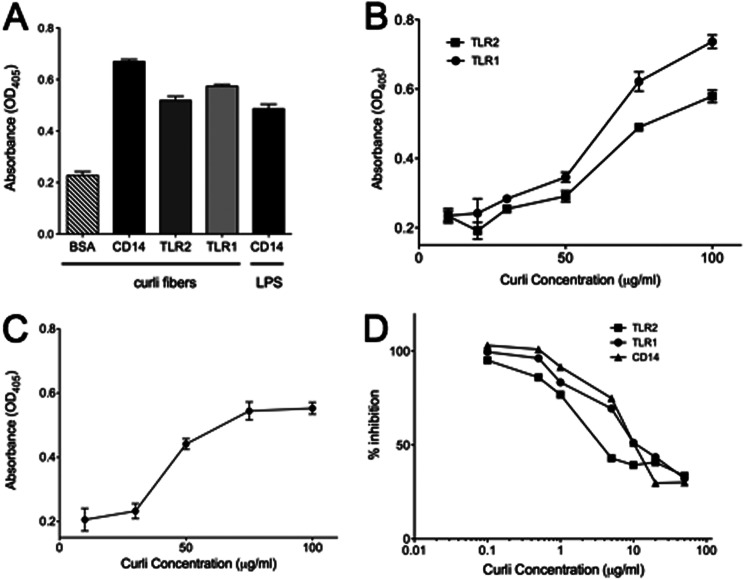
Structural and biochemical studies have demonstrated direct binding of LPS to CD14 and elucidated the hydrophobic binding pocket to which LPS binds on CD14 (71, 72). Although studies indicated a possible interaction between β-amyloid and CD14 in eliciting immune responses (56, 57, 68, 73), no studies have yet addressed whether CD14 could directly bind to amyloid fibers. To investigate the ability of curli fibers to bind to the individual components of the TLR2-TLR1-CD14 complex, we coated the wells of a 96-well MaxSorp ELISA plate with recombinant CD14, TLR2, TLR1, or BSA and investigated whether curli fibers or LPS bound to these molecules using α-CsgA antibodies by an ELISA. The results showed that although curli fibers did not bind to BSA, it bound to all the components of the proposed trimolecular receptor complex: CD14, TLR2, and TLR1. The validity of the assay was confirmed with binding of LPS by immobilized CD14 (Fig. 4A). Next, increasing amounts of curli fibers were added to wells coated with recombinant CD14, TLR2, TLR1, or BSA. The amount of curli fibers bound to the components of the trimolecular receptor complex was determined using α-CsgA antibodies. Curli fibers bound to immobilized TLR2 or TLR1 increased with increasing concentrations of curli fibers (Fig. 4B). Similar results were obtained for curli fibers binding to CD14 (Fig. 4C).
FIGURE 4.
CD14, TLR2, and TLR1 bind purified curli fibers. A, ELISA plates coated with 5 μg/ml of recombinant CD14, TLR2, or TLR1 were incubated with 50 μg/ml of purified curli fibers for 2 h. α-CsgA antibody was used as primary antibody, and alkaline phosphatase-conjugated goat anti-rabbit IgG antibody was used for secondary antibody for detection and visualization of binding of curli to each receptor. LPS binding to CD14 was utilized as a positive control. B, an ELISA plate coated with 5 μg/ml of recombinant TLR2 or TLR1 was incubated with an increasing concentration of purified curli fibers (10 μg/ml, 30 μg/ml, 50 μg/ml, 75 μg/ml, and 100 μg/ml). α-CsgA antibody was used as a primary antibody, and alkaline phosphatase-conjugated goat anti-rabbit IgG antibody was used for secondary antibody for detection. C, an ELISA plate coated with 5 μg/ml of recombinant CD14 was incubated with increasing concentrations of purified curli fibers (10 μg/ml, 30 μg/ml, 50 μg/ml, 75 μg/ml, and 100 μg/ml). α-CsgA antibody was used as a primary antibody, and alkaline phosphatase-conjugated goat anti-rabbit IgG antibody was used for secondary antibody for detection. Curve data points and bars represent mean ± S.E. from at least four independent experiments. D, ELISA plates coated with 2.5 μg/ml of recombinant CD14, TLR2, or TLR1 were incubated with 5.0 μg/ml of recombinant GST-CsgA mixed with increasing concentrations of purified curli fibers (0.1 μg/ml, 0.5 μg/ml, 1 μg/ml, 5 μg/ml, 10 μg/ml, 20 μg/ml, and 50 μg/ml) for 2 h. α-CsgA antibody was used as a primary antibody and alkaline phosphatase-conjugated goat anti-rabbit IgG antibody was used for secondary antibody for detection and visualization of binding of curli to each receptor.
To further investigate the interactions of curli with CD14, TLR2, and TLR1, curli was tested as an inhibitor of CD14, TLR2, or TLR1 binding to GST-CsgA. Wells coated with CD14, TLR2, or TLR1 were incubated with 5 μg of GST-CsgA in the presence of increasing concentrations of purified curli fibers. 50% inhibition of GST-CsgA binding to all three receptors was achieved at approximately equal amounts of purified curli fibers, suggesting a similar avidity of curli for each member of the trimolecular complex (Fig. 4D). Together, these data suggest that curli fibers bind to all the components of the TLR2-TLR1-CD14 receptor complex.
Amyloid Fibrillar Structure of Curli Is Required for CD14 Binding
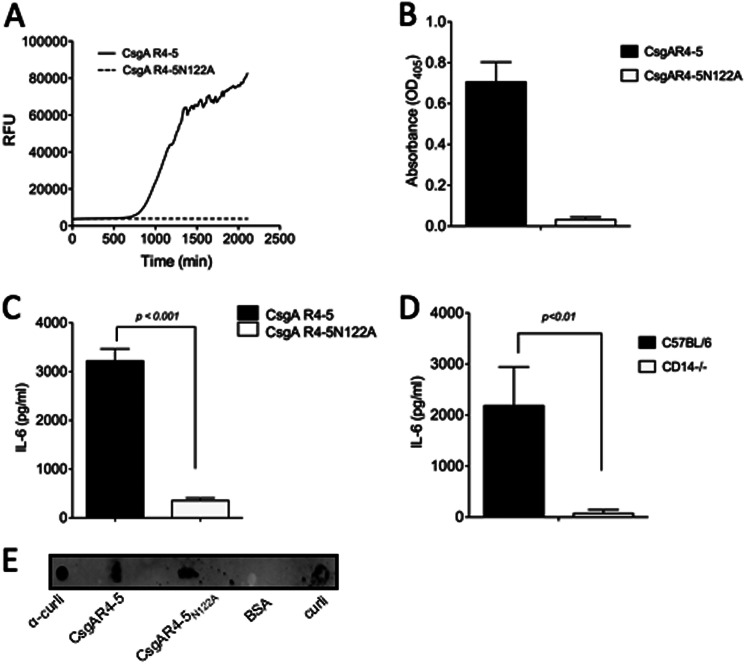
To eliminate any possibility of contamination that may have been present in the curli fibers or the recombinant GST-CsgA preparation, two synthetic peptides, described in an earlier study, corresponding to the fourth and fifth repeat regions of CsgA, were utilized (46, 48). Although the CsgAR-4–5 peptide represents the native CsgA, CsgAR-4–5N122A contains an amino acid substitution at the 122nd residue of CsgA that is critical for the β-sheet structure of amyloid fibers. Synthetic peptides were incubated for 36 h to allow polymerization. A ThT binding assay was performed to confirm the fibrillar structure. Although CsgA R4–5 showed robust polymerization, as evidenced by the sigmoidal nature of the ThT fluorescence, the mutated CsgA R4–5N122A peptide demonstrated no increase in fluorescence intensity, indicating that this peptide does not form amyloid fibers (Fig. 5A). After polymerization, the two synthetic fibers were used in binding assays to determine whether the fibrillar nature of CsgA was required to bind CD14. As shown, although CD14 bound to fibrillar CsgAR4–5, it did not bind CsgA R4–5N122A (Fig. 5B). To confirm that the α-CsgA antibodies bound to both CsgA R4–5 and CsgA R4–5N122A, we performed a dot blot, demonstrating that the α-CsgA primary antibody used for protein binding assays bound to both CsgA R4–5 and CsgA R4–5N122A (Fig. 5E).
FIGURE 5.
CD14 binds synthetic curli fibers and enhances IL-6 production by BMDMs. Polymerization of 50 μm CsgA R4–5 (A) and CsgA R4–5N122A was monitored by measuring fluorescence intensity (excitation, 440; emission, 490) in a BMG Omega Polar Star plate reader. Equal volumes of peptides and 10 μm ThT were mixed and incubated at 37 °C. B, ELISA plates coated with CD14 were incubated with 50 μg/ml of synthetic peptides, CsgA R4–5, and CsgA R4–5N122A, and binding of the synthetic peptides to CD14 was detected using α-CsgA antibody and the secondary antibody, alkaline phosphatase-conjugated goat anti-rabbit IgG antibody. Absorbance was read at 405 nm. C, BMDMss from C57BL/6 were stimulated with 90 μg/ml of polymerized CsgA R4–5 and CsgA R4–5N122A for 6 h, and IL-6 in the supernatant was measured using ELISA. Bars represent mean ± S.E. from at least three independent experiments. D, differentiated BMDMs from C57BL/6 mice and CD14-deficient (CD14−/−) mice were stimulated with polymerized CsgA R4–5 (90 μg/ml) for 6 h, and IL-6 in the supernatant was measured using ELISA. Bars represent mean ± S.E. from at least three independent experiments. E, dot blot of 5 μg of α-CsgA antibody, CsgA R4–5, CsgA R4–5N122A, BSA, and purified curli fibers. The primary antibody used for detection was α-CsgA antibody. Goat anti-rabbit IgG antibody conjugated to IRDye® 800CW was used as secondary antibody.
To further elucidate that the fibrillar structure of curli was required for recognition and cytokine production, we stimulated wild-type C57BL/6 BMDMs with fibrillar CsgA R4–5 peptide and non-fibrillar CsgA R4–5N122A. Six hours after stimulation, supernatants were removed, and IL-6 production was determined by ELISA. There was significantly greater IL-6 production by BMDMs stimulated with the fibrillar peptide CsgA R4–5 as compared with the non-fibrillar CsgA R4–5N122A (Fig. 5C). Lastly, BMDMs from wild-type C57BL/6 and CD14-deficient mice were stimulated by fibrillar CsgA R4–5. CsgA R4–5 caused significantly higher IL-6 production by wild-type BMDMs as compared with CD14-deficient BMDMs at 6 h, as determined by ELISA (Fig. 5D). Overall, these data suggest that CD14 is a key player for immune responses to amyloid fibers and that the fibrillar structure of the amyloid fibers is required for CD14 binding.
DISCUSSION
Bacteria form multicellular communities, termed biofilms, that provide protection from environmental insults and nutrient deprivation (74). Common factors, including exopolysaccharides and extracellular DNA, provide structural support to the bacterial biofilms. Amyloids, proteins resistant to enzymatic digestion and chemical treatments, have also been identified as another common component of bacterial biofilms (11, 12, 75). Curli fibers produced in enterobacterial biofilms are the best characterized bacterial amyloid to date. Production of curli fibers by enteric bacteria results in the activation of the immune system during enteric infection (70). We recently determined that curli fibers of E. coli and S. typhimurium are recognized by the immune system through the activation of the TLR2-TLR1 complex (46–48). It is interesting to note that several amyloids with a similar quaternary structure rich in β-sheets are also produced in humans, leading to inflammatory responses through the activation of TLRs. In the case of Alzheimer's disease, β-amyloid that forms plaques in the brain of the patients elicits nitric oxide and cytokine activation (49, 57, 76, 77). Although various TLRs have been implicated in this inflammatory process, caused by the accumulation of β-amyloid (56, 57, 78), TLR2 has been demonstrated convincingly to recognize the quaternary fibrillar structure of β-amyloid as well as curli fibers (46, 48). Furthermore, consistent with the data showing the activation of the TLR2-TLR1 complex by curli fibers, β-amyloid has also been shown to activate the TLR2-TLR1 complex (49), suggesting that the immune activation mechanisms by bacterial and eukaryotic amyloids may be similar because of their conserved structure.
Adaptor molecule CD14 has been shown to aid several receptor complexes, including TLR4-MD2, TLR7, and TLR9 in recognition of lipid A, single-stranded RNA, and double-stranded bacterial CpG DNA, respectively (58, 61, 79). Interestingly, blocking CD14 by neutralizing antibodies or genetic deficiency for this molecule on microglia results in reduced levels of immune activation by β-amyloid (76). Furthermore, deletion of CD14 attenuates the progression of Alzheimer's disease pathology in the brain in mouse models of Alzheimer's disease (57). Although several studies suggested that CD14 aids the TLR2-TLR1 complex to recognize bacterial lipopeptides (59, 80), no studies have yet addressed a conclusive role for this molecule in response to amyloids. In this study, we investigated the potential role of CD14 as an adaptor molecule for the TLR2-TLR1 complex to recognize the amyloids. Our results suggest that the TLR2-TLR1/CD14 receptor complex recognizes the curli fibers of S. typhimurium. Interestingly, CD14 was neither able to signal alone nor essential for the activity of TLR2/TLR1 on epithelial cells. However, the presence of CD14 significantly increased the activation of NF-κB by the TLR2-TLR1 complex upon recognition of curli fibers or its major subunit, GST-CsgA (Fig. 1). Consistent with these results, a recent study showed that curli fibers induce TLR2 activation on a colon carcinoma cell line, T-84, that is known to lack CD14 expression, leading to cytokine expression and modulation of epithelial permeability (81, 82). In contrast, CD14 deficiency on macrophages resulted in blunted IL-6 and nitric oxide production upon activation by curli fibers or GST-CsgA to the levels observed by TLR2 deficiency, suggesting that CD14 may be essential for macrophages in responding to amyloids (Fig. 3).
To address the need for membrane-bound CD14 versus soluble CD14 for maximal TLR2-TLR1 activation, HeLa 57A epithelial cells were grown under serum starvation conditions to limit the amount of soluble CD14 that is present in fetal calf serum. When membrane-bound CD14 was lacking, the addition of soluble CD14 compensated for the CD14 deficiency in the epithelial cells. These data suggest that, in the cells that are not expressing CD14, soluble CD14 may act as an adaptor molecule for the TLR2-TLR1 complex (Fig. 2).
Curli fibers bind to TLR2, TLR1, and CD14 in vitro (Fig. 4, A–C). Inhibition studies demonstrated little to no difference in the avidity of curli to TLR1, CD14, or TLR2 as demonstrated by the fact that 1–10 μg/ml curli was enough to inhibit 50% of the binding of GST-CsgA to all three receptors. Furthermore, the kd value was ∼10−7 m for the interaction between curli and each member of the receptor complex (Fig. 4D). It is important to point out that the in vitro binding assays used here may not recapitulate the actual binding interactions between curli and the TLR2-TLR1-CD14 receptor complex because the structure and accessible residues for binding could be different when all three components of the receptor complex assemble as compared with individual receptors. Experiments using two synthetic peptides, CsgAR4–5 and CsgAR4–5N122A, of which the native peptide forms fibers or the mutated peptide cannot form fibers, respectively, demonstrated that the amyloid fibrillar structure is required for the ability of curli fibers to bind CD14 and activate macrophages (Fig. 5). Previous studies indicated that TLR2 and CD14 are involved in the clearance of amyloid-β. Although TLR2 and CD14 activation induces the phagocytosis and clearance of amyloid-β, CD14-deficient macrophages fail to clear β-amyloid (57, 83, 84). CD14 was also implicated to control the endocytotic processes of LPS independent of TLR4 (85). Nonetheless, the role of the TLR2-TLR1-CD14 complex in the internalization of amyloid-expressing bacteria remains unknown. We are currently investigating this process, and our preliminary data indicate that expression of curli fibers does not affect the uptake of S. typhimurium by macrophages (unpublished data).3
Together these data lead us to a possible model for the function of CD14 as an adaptor protein for the TLR2-TLR1 complex. Curli fibers, released from the biofilm or still attached to bacteria, are recognized by TLR2-TLR1, allowing for activation of NF-κB, which leads to cytokine production such as IL-6. However, when soluble CD14 is present in the environment, we think that CD14 binds to the curli fibers, probably eliciting a conformational change in the receptor complex, exposing the binding/activation site. This would, in turn, lead to an increased NF-κB activation, eventually resulting in increased cytokine production. Similarly, membrane-bound CD14 may bind to curli fibers, allowing receptor complex formation and better exposure of the TLR2-TLR1 activation site. Again, this would allow for increased NF-κB activation, resulting in greater cytokine production.
Overall, our data demonstrate that CD14 acts as an adaptor molecule for the activation of the TLR2-TLR1 complex in response to S. typhimurium curli fibers. The requirement for the fibrillar nature of curli to bind to CD14, activating NF-κB and inducing cytokine production, suggests that CD14 may be a common receptor for other amyloid-forming proteins. Although there is evidence that the adaptor function of CD14 may be common across the spectrum of TLR2/TLR1 ligands, as demonstrated for mycobacterial lipopeptides, ara-lipoarabinomannan, synthetic triacylated lipopeptide Pam3CSK4, and OspA of Borrelia burgdorferi (59, 86–89), there is more information needed in regards to additional TLR2 ligands, including amyloids, and the role of CD14.
Acknowledgments
We thank Dr. R. T. Hay (The Wellcome Trust Centre for Gene Regulation and Expression, College of Life Sciences, University of Dundee, UK) for providing the HeLa 57A cell line.
This work was supported by American Heart Association Scientist Development Grant 0835248N and by a grant from the Pennsylvania Department of Health (to G. J. R.).
G. J. Rapsinski, T. N. Newman, G. O. Oppong, J. P. M. van Putten, and Ç. Tükel, unpublished data.
- ThT
- Thioflavin T
- TLR
- Toll-like receptor
- BMDM
- bone marrow-derived macrophage.
REFERENCES
- 1. Toyama B. H., Weissman J. S. (2011) Amyloid structure. Conformational diversity and consequences. Annu. Rev. Biochem. 80, 557–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mok K. H., Pettersson J., Orrenius S., Svanborg C. (2007) HAMLET, protein folding, and tumor cell death. Biochem. Biophys. Res. Commun. 354, 1–7 [DOI] [PubMed] [Google Scholar]
- 3. Theos A. C., Truschel S. T., Raposo G., Marks M. S. (2005) The Silver locus product Pmel17/gp100/Silv/ME20. Controversial in name and in function. Pigment Cell Res. 18, 322–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leonhardt R. M., Vigneron N., Rahner C., Van den Eynde B. J., Cresswell P. (2010) Endoplasmic reticulum export, subcellular distribution, and fibril formation by Pmel17 require an intact N-terminal domain junction. J. Biol. Chem. 285, 16166–16183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pfefferkorn C. M., McGlinchey R. P., Lee J. C. (2010) Effects of pH on aggregation kinetics of the repeat domain of a functional amyloid, Pmel17. Proc. Natl. Acad. Sci. U.S.A. 107, 21447–21452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maji S. K., Perrin M. H., Sawaya M. R., Jessberger S., Vadodaria K., Rissman R. A., Singru P. S., Nilsson K. P., Simon R., Schubert D., Eisenberg D., Rivier J., Sawchenko P., Vale W., Riek R. (2009) Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325, 328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aigelsreiter A., Janig E., Stumptner C., Fuchsbichler A., Zatloukal K., Denk H. (2007) How a cell deals with abnormal proteins. Pathogenetic mechanisms in protein aggregation diseases. Pathobiology 74, 145–158 [DOI] [PubMed] [Google Scholar]
- 8. Bucciantini M., Giannoni E., Chiti F., Baroni F., Formigli L., Zurdo J., Taddei N., Ramponi G., Dobson C. M., Stefani M. (2002) Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416, 507–511 [DOI] [PubMed] [Google Scholar]
- 9. Hull R. L., Westermark G. T., Westermark P., Kahn S. E. (2004) Islet amyloid. A critical entity in the pathogenesis of type 2 diabetes. J. Clin. Endocr. Metab. 89, 3629–3643 [DOI] [PubMed] [Google Scholar]
- 10. Ross C. A., Poirier M. A. (2004) Protein aggregation and neurodegenerative disease. Nat. Med. 10, S10–17 [DOI] [PubMed] [Google Scholar]
- 11. Alteri C. J., Xicohténcatl-Cortes J., Hess S., Caballero-Olín G., Girón J. A., Friedman R. L. (2007) Mycobacterium tuberculosis produces pili during human infection. Proc. Natl. Acad. Sci. U.S.A. 104, 5145–5150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barnhart M. M., Chapman M. R. (2006) Curli biogenesis and function. Annu. Rev. Microbiol. 60, 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chapman M. R., Robinson L. S., Pinkner J. S., Roth R., Heuser J., Hammar M., Normark S., Hultgren S. J. (2002) Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295, 851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Romero D., Aguilar C., Losick R., Kolter R. (2010) Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U.S.A. 107, 2230–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Römling U., Bokranz W., Rabsch W., Zogaj X., Nimtz M., Tschäpe H. (2003) Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. Int. J. Med. Microbiol. 293, 273–285 [DOI] [PubMed] [Google Scholar]
- 16. Schwartz K., Syed A. K., Stephenson R. E., Rickard A. H., Boles B. R. (2012) Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog. 8, e1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khurana R., Coleman C., Ionescu-Zanetti C., Carter S. A., Krishna V., Grover R. K., Roy R., Singh S. (2005) Mechanism of thioflavin T binding to amyloid fibrils. J. Struct. Biol. 151, 229–238 [DOI] [PubMed] [Google Scholar]
- 18. Miura T., Yamamiya C., Sasaki M., Suzuki K., Takeuchi H. (2002) Binding mode of Congo red to Alzheimer's amyloid β-peptide studied by UV Raman spectroscopy. J. Raman Spectrosc. 33, 530–535 [Google Scholar]
- 19. Klunk W. E., Pettegrew J. W., Abraham D. J. (1989) Quantitative evaluation of Congo red binding to amyloid-like proteins with a β-pleated sheet conformation. J. Histochem. Cytochem. 37, 1273–1281 [DOI] [PubMed] [Google Scholar]
- 20. Collinson S. K., Clouthier S. C., Doran J. L., Banser P. A., Kay W. W. (1996) Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J. Bacteriol. 178, 662–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Olsén A., Jonsson A., Normark S. (1989) Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338, 652–655 [DOI] [PubMed] [Google Scholar]
- 22. Olsén A., Arnqvist A., Hammar M., Sukupolvi S., Normark S. (1993) The RpoS σ factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol. Microbiol. 7, 523–536 [DOI] [PubMed] [Google Scholar]
- 23. Castillo G. M., Lukito W., Peskind E., Raskind M., Kirschner D. A., Yee A. G., Snow A. D. (2000) Laminin inhibition of β-amyloid protein (Aβ) fibrillogenesis and identification of an Aβ binding site localized to the globular domain repeats on the laminin a chain. J. Neurosci. Res. 62, 451–462 [DOI] [PubMed] [Google Scholar]
- 24. Sjöbring U., Pohl G., Olsén A. (1994) Plasminogen, absorbed by Escherichia coli expressing curli or by Salmonella enteritidis expressing thin aggregative fimbriae, can be activated by simultaneously captured tissue-type plasminogen activator (t-PA). Mol. Microbiol. 14, 443–452 [DOI] [PubMed] [Google Scholar]
- 25. Tucker H. M., Kihiko M., Caldwell J. N., Wright S., Kawarabayashi T., Price D., Walker D., Scheff S., McGillis J. P., Rydel R. E., Estus S. (2000) The plasmin system is induced by and degrades amyloid-β aggregates. J. Neurosci. 20, 3937–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goulter-Thorsen R. M., Taran E., Gentle I. R., Gobius K. S., Dykes G. A. (2011) CsgA production by Escherichia coli O157:H7 alters attachment to abiotic surfaces in some growth environments. Appl. Environ. Microbiol. 77, 7339–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mauclaire L., Brombacher E., Bunger J. D., Zinn M. (2010) Factors controlling bacterial attachment and biofilm formation on medium-chain-length polyhydroxyalkanoates (mcl-PHAs). Colloids Surf. 76, 104–111 [DOI] [PubMed] [Google Scholar]
- 28. Antão E. M., Wieler L. H., Ewers C. (2009) Adhesive threads of extraintestinal pathogenic Escherichia coli. Gut Pathog. 1, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hammar M., Arnqvist A., Bian Z., Olsén A., Normark S. (1995) Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18, 661–670 [DOI] [PubMed] [Google Scholar]
- 30. Gerstel U., Römling U. (2003) The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res. Microbiol. 154, 659–667 [DOI] [PubMed] [Google Scholar]
- 31. Wang X., Hammer N. D., Chapman M. R. (2008) The molecular basis of functional bacterial amyloid polymerization and nucleation. J. Biol. Chem. 283, 21530–21539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hammer N. D., McGuffie B. A., Zhou Y., Badtke M. P., Reinke A. A., Brännström K., Gestwicki J. E., Olofsson A., Almqvist F., Chapman M. R. (2012) The C-terminal repeating units of CsgB direct bacterial functional amyloid nucleation. J. Mol. Biol. 422, 376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shu Q., Crick S. L., Pinkner J. S., Ford B., Hultgren S. J., Frieden C. (2012) The E. coli CsgB nucleator of curli assembles to β-sheet oligomers that alter the CsgA fibrillization mechanism. Proc. Natl. Acad. Sci. U.S.A. 109, 6502–6507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dueholm M. S., Nielsen S. B., Hein K. L., Nissen P., Chapman M., Christiansen G., Nielsen P. H., Otzen D. E. (2011) Fibrillation of the major curli subunit CsgA under a wide range of conditions implies a robust design of aggregation. Biochemistry 50, 8281–8290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nenninger A. A., Robinson L. S., Hammer N. D., Epstein E. A., Badtke M. P., Hultgren S. J., Chapman M. R. (2011) CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. Mol. Microbiol. 81, 486–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robinson L. S., Ashman E. M., Hultgren S. J., Chapman M. R. (2006) Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 59, 870–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nenninger A. A., Robinson L. S., Hultgren S. J. (2009) Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc. Natl. Acad. Sci. U.S.A. 106, 900–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takeuchi O., Akira S. (2010) Pattern recognition receptors and inflammation. Cell 140, 805–820 [DOI] [PubMed] [Google Scholar]
- 39. Takeda K., Akira S. (2004) TLR signaling pathways. Semin. Immunol. 16, 3–9 [DOI] [PubMed] [Google Scholar]
- 40. Lien E., Means T. K., Heine H., Yoshimura A., Kusumoto S., Fukase K., Fenton M. J., Oikawa M., Qureshi N., Monks B., Finberg R. W., Ingalls R. R., Golenbock D. T. (2000) Toll-like receptor 4 imparts ligand-specific recognition of bacterial lipopolysaccharide. J. Clin. Invest. 105, 497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Poltorak A. (1998) Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice. Mutations in Tlr4 gene. Science 282, 2085–2088 [DOI] [PubMed] [Google Scholar]
- 42. Hayashi F., Smith K. D., Ozinsky A., Hawn T. R., Yi E. C., Goodlett D. R., Eng J. K., Akira S., Underhill D. M., Aderem A. (2001) The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103 [DOI] [PubMed] [Google Scholar]
- 43. Gewirtz A. T., Navas T. A., Lyons S., Godowski P. J., Madara J. L. (2001) Cutting edge. Bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J. Immunol. 167, 1882–1885 [DOI] [PubMed] [Google Scholar]
- 44. Aliprantis A. O. (1999) Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science 285, 736–739 [DOI] [PubMed] [Google Scholar]
- 45. Schwandner R. (1999) Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J. Biol. Chem. 274, 17406–17409 [DOI] [PubMed] [Google Scholar]
- 46. Tükel C., Nishimori J. H., Wilson R. P., Winter M. G., Keestra A. M., van Putten J. P., Bäumler A. J. (2010) Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cell. Microbiol. 12, 1495–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tükel C., Raffatellu M., Humphries A. D., Wilson R. P., Andrews-Polymenis H. L., Gull T., Figueiredo J. F., Wong M. H., Michelsen K. S., Akçelik M., Adams L. G., Bäumler A. J. (2005) CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol. Microbiol. 58, 289–304 [DOI] [PubMed] [Google Scholar]
- 48. Tükel C., Wilson R. P., Nishimori J. H., Pezeshki M., Chromy B. A., Bäumler A. J. (2009) Responses to amyloids of microbial and host origin are mediated through toll-like receptor 2. Cell Host Microbe 6, 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu S., Liu Y., Hao W., Wolf L., Kiliaan A. J., Penke B., Rübe C. E., Walter J., Heneka M. T., Hartmann T., Menger M. D., Fassbender K. (2012) TLR2 is a primary receptor for Alzheimer's amyloid β peptide to trigger neuroinflammatory activation. J. Immunol. 188, 1098–1107 [DOI] [PubMed] [Google Scholar]
- 50. Cheng N., He R., Tian J., Ye P. P., Ye R.D. (2008) Cutting edge. TLR2 is a functional receptor for acute-phase serum amyloid A. J. Immunol. 181, 22–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pan Z., Zhou L., Hetherington C. J., Zhang D. E. (2000) Hepatocytes contribute to soluble CD14 production, and CD14 expression is differentially regulated in hepatocytes and monocytes. J. Biol. Chemistry 275, 36430–36435 [DOI] [PubMed] [Google Scholar]
- 52. Hetherington C. J., Kingsley P. D., Crocicchio F., Zhang P., Rabin M. S., Palis J., Zhang D. E. (1999) Characterization of human endotoxin lipopolysaccharide receptor CD14 expression in transgenic mice. J. Immunol. 162, 503–509 [PubMed] [Google Scholar]
- 53. Morrow J. F., Stearman R. S., Peltzman C. G., Potter D. A. (1981) Induction of hepatic synthesis of serum amyloid A protein and actin. Proc. Natl. Acad. Sci. U.S.A. 78, 4718–4722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ziegler-Heitbrock H. W., Ulevitch R. J. (1993) CD14. Cell surface receptor and differentiation marker. Immunol. Today 14, 121–125 [DOI] [PubMed] [Google Scholar]
- 55. Haziot A., Chen S., Ferrero E., Low M. G., Silber R., Goyert S. M. (1988) The monocyte differentiation antigen, CD14, is anchored to the cell membrane by a phosphatidylinositol linkage. J. Immunol. 141, 547–552 [PubMed] [Google Scholar]
- 56. Reed-Geaghan E. G., Savage J. C., Hise A. G., Landreth G. E. (2009) CD14 and toll-like receptors 2 and 4 are required for fibrillar Aβ-stimulated microglial activation. J. Neurosci. 29, 11982–11992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Reed-Geaghan E. G., Reed Q. W., Cramer P. E., Landreth G. E. (2010) Deletion of CD14 attenuates Alzheimer's disease pathology by influencing the brain's inflammatory milieu. J. Neurosci. 30, 15369–15373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Baumann C. L., Aspalter I. M., Sharif O., Pichlmair A., Blüml S., Grebien F., Bruckner M., Pasierbek P., Aumayr K., Planyavsky M., Bennett K. L., Colinge J., Knapp S., Superti-Furga G. (2010) CD14 is a coreceptor of Toll-like receptors 7 and 9. J. Exp. Med. 207, 2689–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Manukyan M., Triantafilou K., Triantafilou M., Mackie A., Nilsen N., Espevik T., Wiesmüller K. H., Ulmer A. J., Heine H. (2005) Binding of lipopeptide to CD14 induces physical proximity of CD14, TLR2 and TLR1. Eur. J. Immunol. 35, 911–921 [DOI] [PubMed] [Google Scholar]
- 60. Moreno C., Merino J., Ramírez N., Echeverría A., Pastor F., Sánchez-Ibarrola A. (2004) Lipopolysaccharide needs soluble CD14 to interact with TLR4 in human monocytes depleted of membrane CD14. Microbes Infect. 6, 990–995 [DOI] [PubMed] [Google Scholar]
- 61. Jiang Z., Georgel P., Du X., Shamel L., Sovath S., Mudd S., Huber M., Kalis C., Keck S., Galanos C., Freudenberg M., Beutler B. (2005) CD14 is required for MyD88-independent LPS signaling. Nat. Immunol. 6, 565–570 [DOI] [PubMed] [Google Scholar]
- 62. Low K. B., Ittensohn M., Le T., Platt J., Sodi S., Amoss M., Ash O., Carmichael E., Chakraborty A., Fischer J., Lin S. L., Luo X., Miller S. I., Zheng L., King I., Pawelek J. M., Bermudes D. (1999) Lipid A mutant Salmonella with suppressed virulence and TNFα induction retain tumor-targeting in vivo. Nat. Biotechnol. 17, 37–41 [DOI] [PubMed] [Google Scholar]
- 63. Collinson S. K., Emödy L., Müller K. H., Trust T. J., Kay W. W. (1991) Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 173, 4773–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Humphries A. D., Raffatellu M., Winter S., Weening E. H., Kingsley R. A., Droleskey R., Zhang S., Figueiredo J., Khare S., Nunes J., Adams L. G., Tsolis R. M., Bäumler A. J. (2003) The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol. Microbiol. 48, 1357–1376 [DOI] [PubMed] [Google Scholar]
- 65. Keestra A. M., de Zoete M. R., van Aubel R. A., van Putten J. P. (2007) The central leucine-rich repeat region of chicken TLR16 dictates unique ligand specificity and species-specific interaction with TLR2. J. Immunol. 178, 7110–7119 [DOI] [PubMed] [Google Scholar]
- 66. Broersen K., Jonckheere W., Rozenski J., Vandersteen A., Pauwels K., Pastore A., Rousseau F., Schymkowitz J. (2011) A standardized and biocompatible preparation of aggregate-free amyloid beta peptide for biophysical and biological studies of Alzheimer's disease. Protein Eng. Des. Sel. 24, 743–750 [DOI] [PubMed] [Google Scholar]
- 67. Rodriguez M. S. (1999) Nuclear retention of IκBα protects it from signal-induced degradation and inhibits nuclear factor κB transcriptional activation. J. Biol. Chem. 274, 9108–9115 [DOI] [PubMed] [Google Scholar]
- 68. Anas A., van der Poll T., de Vos A. F. (2010) Role of CD14 in lung inflammation and infection. Crit. Care 14, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bian Z., Brauner A., Li Y., Normark S. (2000) Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J. Infect. Dis. 181, 602–612 [DOI] [PubMed] [Google Scholar]
- 70. Bian Z., Yan Z. Q., Hansson G. K., Thorén P., Normark S. (2001) Activation of inducible nitric oxide synthase/nitric oxide by curli fibers leads to a fall in blood pressure during systemic Escherichia coli infection in mice. J. Infect. Dis. 183, 612–619 [DOI] [PubMed] [Google Scholar]
- 71. Kim J. I., Lee C. J., Jin M. S., Lee C. H., Paik S. G., Lee H., Lee J. O. (2005) Crystal structure of CD14 and its implications for lipopolysaccharide signaling. J. Biol. Chem. 280, 11347–11351 [DOI] [PubMed] [Google Scholar]
- 72. Jerala R. (2007) Structural biology of the LPS recognition. Int. J. Med. Microbiol. 297, 353–363 [DOI] [PubMed] [Google Scholar]
- 73. Bate C., Veerhuis R., Eikelenboom P., Williams A. (2004) Microglia kill amyloid-β1–42 damaged neurons by a CD14-dependent process. Neuroreport 15, 1427–1430 [DOI] [PubMed] [Google Scholar]
- 74. Donlan R. M., Costerton J. W. (2002) Biofilms. Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Larsen P., Nielsen J. L., Dueholm M. S., Wetzel R., Otzen D., Nielsen P. H. (2007) Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 9, 3077–3090 [DOI] [PubMed] [Google Scholar]
- 76. Fassbender K., Walter S., Kühl S., Landmann R., Ishii K., Bertsch T., Stalder A. K., Muehlhauser F., Liu Y., Ulmer A. J., Rivest S., Lentschat A., Gulbins E., Jucker M., Staufenbiel M., Brechtel K., Walter J., Multhaup G., Penke B., Adachi Y., Hartmann T., Beyreuther K. (2004) The LPS receptor (CD14) links innate immunity with Alzheimer's disease. FASEB J. 18, 203–205 [DOI] [PubMed] [Google Scholar]
- 77. Heneka M. T., O'Banion M. K. (2007) Inflammatory processes in Alzheimer's disease. J. Neuroimmunol. 184, 69–91 [DOI] [PubMed] [Google Scholar]
- 78. Udan M. L., Ajit D., Crouse N. R., Nichols M. R. (2008) Toll-like receptors 2 and 4 mediate Aβ(1–42) activation of the innate immune response in a human monocytic cell line. J. Neurochem. 104, 524–533 [DOI] [PubMed] [Google Scholar]
- 79. Wright S. D., Ramos R. A., Tobias P. S., Ulevitch R. J., Mathison J. C. (1990) CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science 249, 1431–1433 [DOI] [PubMed] [Google Scholar]
- 80. Shin D. M., Yuk J. M., Lee H. M., Lee S. H., Son J. W., Harding C. V., Kim J. M., Modlin R. L., Jo E. K. (2010) Mycobacterial lipoprotein activates autophagy via TLR2/1/CD14 and a functional vitamin D receptor signalling. Cell. Microbiol. 12, 1648–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Oppong G. O., Rapsinski G. J., Newman T. N., Nishimori J. H., Biesecker S. G., Tukel C. (2012) Epithelial cells augment barrier function via activation of the Toll-like receptor 2/phosphatidylinositol 3-kinase pathway upon recognition of Salmonella enterica serovar Typhimurium curli fibrils in the gut. Infect. Immun. 81, 478–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Cario E., Rosenberg I. M., Brandwein S. L., Beck P. L., Reinecker H. C., Podolsky D. K. (2000) Lipopolysaccharide activates distinct signaling pathways in intestinal epithelial cell lines expressing Toll-like receptors. J. Immunol. 164, 966–972 [DOI] [PubMed] [Google Scholar]
- 83. Letiembre M., Liu Y., Walter S., Hao W., Pfander T., Wrede A., Schulz-Schaeffer W., Fassbender K. (2009) Screening of innate immune receptors in neurodegenerative diseases. A similar pattern. Neurobiol. Aging 30, 759–768 [DOI] [PubMed] [Google Scholar]
- 84. Liu Y., Walter S., Stagi M., Cherny D., Letiembre M., Schulz-Schaeffer W., Heine H., Penke B., Neumann H., Fassbender K. (2005) LPS receptor (CD14). A receptor for phagocytosis of Alzheimer's amyloid peptide. Brain 128, 1778–1789 [DOI] [PubMed] [Google Scholar]
- 85. Zanoni I., Ostuni R., Marek L. R., Barresi S., Barbalat R., Barton G. M., Granucci F., Kagan J. C. (2011) CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell 147, 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yu W., Soprana E., Cosentino G., Volta M., Lichenstein H. S., Viale G., Vercelli D. (1998) Soluble CD14(1–152) confers responsiveness to both lipoarabinomannan and lipopolysaccharide in a novel HL-60 cell bioassay. J. Immunol. 161, 4244–4251 [PubMed] [Google Scholar]
- 87. Schromm A. B., Reiling N., Howe J., Wiesmüller K. H., Roessle M., Brandenburg K. (2010) Influence of serum on the immune recognition of a synthetic lipopeptide mimetic of the 19-kDa lipoprotein from Mycobacterium tuberculosis. Innate. Immun. 16, 213–225 [DOI] [PubMed] [Google Scholar]
- 88. Ranoa D. R., Kelley S. L., Tapping R. I. (2013) Human LBP and CD14 independently deliver triacylated lipoproteins to TLR1 and TLR2 and enhance formation of the ternary signaling complex. J. Biol. Chem. 288, 9729–9741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Nakata T., Yasuda M., Fujita M., Kataoka H., Kiura K., Sano H., Shibata K. (2006) CD14 directly binds to triacylated lipopeptides and facilitates recognition of the lipopeptides by the receptor complex of Toll-like receptors 2 and 1 without binding to the complex. Cell Microbiol. 8, 1899–1909 [DOI] [PubMed] [Google Scholar]