Abstract
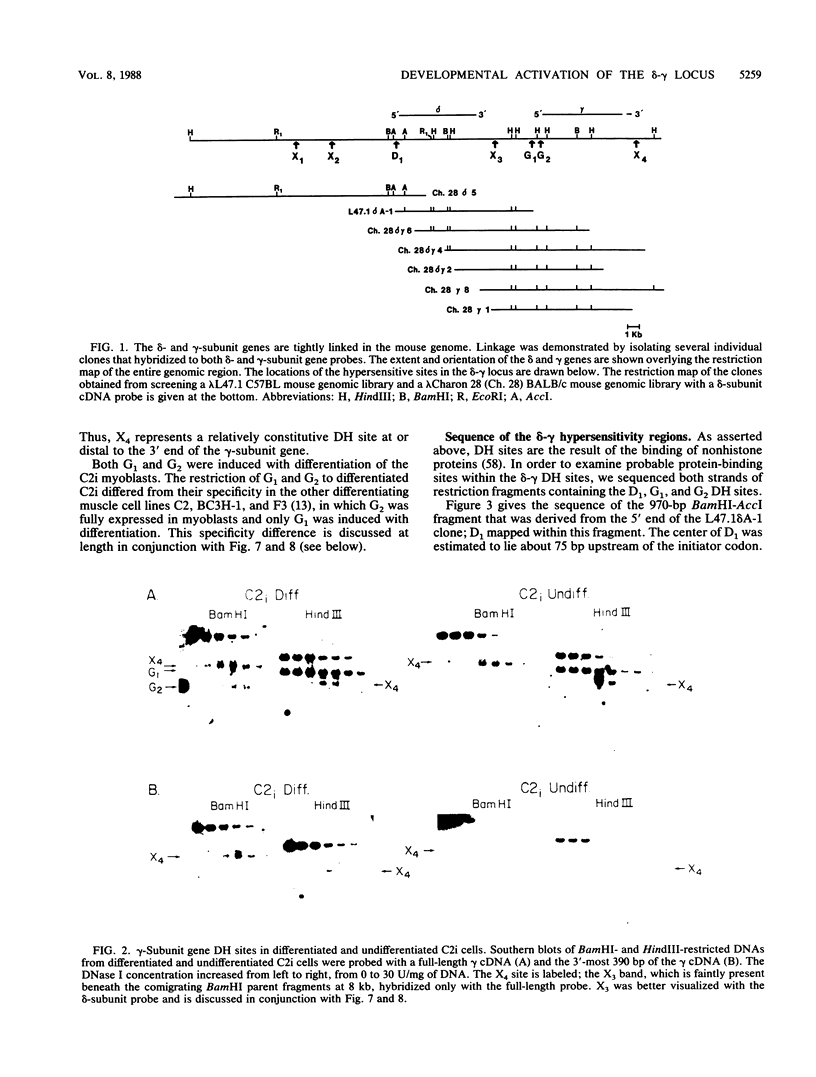
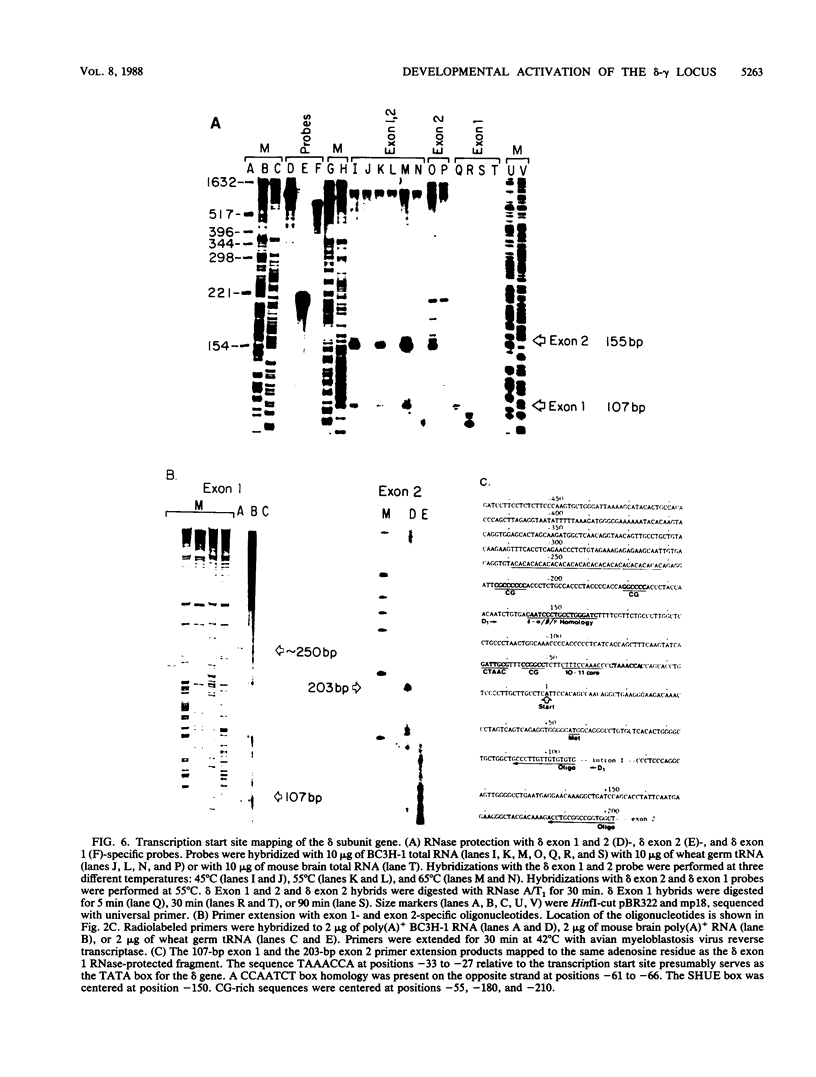
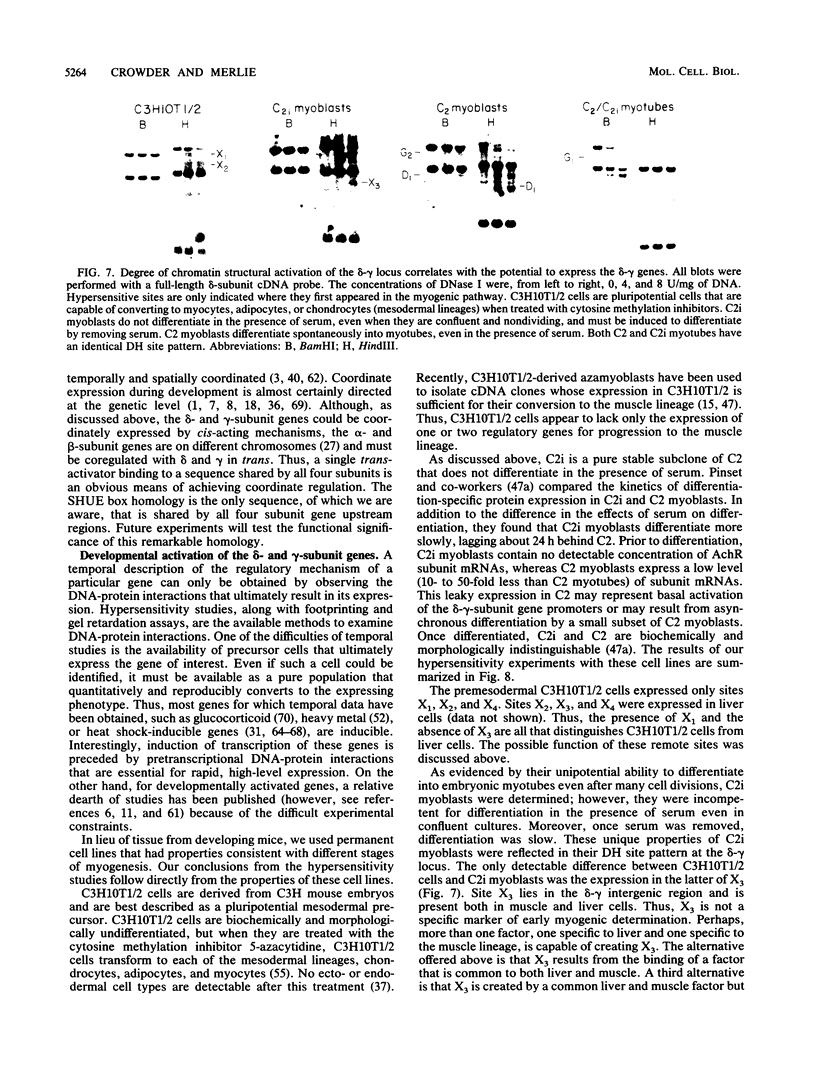
We used the DNase I-hypersensitive sites around the mouse acetylcholine receptor delta-subunit gene as a guide toward the cloning and sequencing of delta and gamma transcriptional regulatory regions and as a means to assess chromatin structural activation of the delta- and gamma-subunit genes during myogenesis. Genomic cloning of hypersensitive sites downstream of the delta-subunit gene revealed the presence of the gamma-subunit gene approximately 5 kilobases away; the hypersensitive sites mapped to the 5' end of the gamma-subunit gene. Sequence comparison of restriction fragments containing hypersensitive sites in analogous locations at the 5' ends of the delta- and gamma-subunit genes uncovered little overall homology between the two genomic fragments; however, an 11- of 13-base-pair match between the two sequences was found. Homologs to this sequence were also found to be present in the upstream regions of the chicken alpha- and mouse beta-subunit genes. By RNase protection and primer extension analyses, the delta-subunit gene transcription start site was mapped to 56 base pairs upstream of the initiator ATG codon. Clonal cell lines with various potentials to differentiate to the skeletal muscle phenotype were examined for their hypersensitive site pattern within the delta-gamma locus. Only remote hypersensitive sites flanking the locus appear in pluripotential mesodermal cells. A cell line of determined but inducible myoblasts expressed only one more intergenic site, while in permissively differentiating myoblasts hypersensitive sites were already present at the 5' ends of the delta and gamma genes prior to differentiation. Terminal differentiation resulted in an identical pattern of hypersensitive sites in all muscle cell lines examined so far, with an intergenic site near the gamma-subunit gene being the only site specific to the differentiated muscle phenotype.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin T. J., Yoshihara C. M., Blackmer K., Kintner C. R., Burden S. J. Regulation of acetylcholine receptor transcript expression during development in Xenopus laevis. J Cell Biol. 1988 Feb;106(2):469–478. doi: 10.1083/jcb.106.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom K. S., Amaya E., Carbon J., Clarke L., Hill A., Yeh E. Chromatin conformation of yeast centromeres. J Cell Biol. 1984 Nov;99(5):1559–1568. doi: 10.1083/jcb.99.5.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount P., Merlie J. P. Native folding of an acetylcholine receptor alpha subunit expressed in the absence of other receptor subunits. J Biol Chem. 1988 Jan 15;263(2):1072–1080. [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Briggs M. R., Kadonaga J. T., Bell S. P., Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986 Oct 3;234(4772):47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- Bryan P. N., Olah J., Birnstiel M. L. Major changes in the 5' and 3' chromatin structure of sea urchin histone genes accompany their activation and inactivation in development. Cell. 1983 Jul;33(3):843–848. doi: 10.1016/0092-8674(83)90026-0. [DOI] [PubMed] [Google Scholar]
- Buonanno A., Merlie J. P. Transcriptional regulation of nicotinic acetylcholine receptor genes during muscle development. J Biol Chem. 1986 Sep 5;261(25):11452–11455. [PubMed] [Google Scholar]
- Buonanno A., Mudd J., Shah V., Merlie J. P. A universal oligonucleotide probe for acetylcholine receptor genes. Selection and sequencing of cDNA clones for the mouse muscle beta subunit. J Biol Chem. 1986 Dec 15;261(35):16451–16458. [PubMed] [Google Scholar]
- Burch J. B., Weintraub H. Temporal order of chromatin structural changes associated with activation of the major chicken vitellogenin gene. Cell. 1983 May;33(1):65–76. doi: 10.1016/0092-8674(83)90335-5. [DOI] [PubMed] [Google Scholar]
- Cartwright I. L., Abmayr S. M., Fleischmann G., Lowenhaupt K., Elgin S. C., Keene M. A., Howard G. C. Chromatin structure and gene activity: the role of nonhistone chromosomal proteins. CRC Crit Rev Biochem. 1982;13(1):1–86. doi: 10.3109/10409238209108709. [DOI] [PubMed] [Google Scholar]
- Choi O. R., Engel J. D. A 3' enhancer is required for temporal and tissue-specific transcriptional activation of the chicken adult beta-globin gene. Nature. 1986 Oct 23;323(6090):731–734. doi: 10.1038/323731a0. [DOI] [PubMed] [Google Scholar]
- Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980 Sep 19;209(4463):1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- Crowder C. M., Merlie J. P. DNase I-hypersensitive sites surround the mouse acetylcholine receptor delta-subunit gene. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8405–8409. doi: 10.1073/pnas.83.21.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H., Lassar A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987 Dec 24;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. 1985 Aug 29-Sep 4Nature. 316(6031):774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Emerson B. M., Nickol J. M., Jackson P. D., Felsenfeld G. Analysis of the tissue-specific enhancer at the 3' end of the chicken adult beta-globin gene. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4786–4790. doi: 10.1073/pnas.84.14.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine B., Sassoon D., Buckingham M., Changeux J. P. Detection of the nicotinic acetylcholine receptor alpha-subunit mRNA by in situ hybridization at neuromuscular junctions of 15-day-old chick striated muscles. EMBO J. 1988 Mar;7(3):603–609. doi: 10.1002/j.1460-2075.1988.tb02853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritton H. P., Igo-Kemenes T., Nowock J., Strech-Jurk U., Theisen M., Sippel A. E. Alternative sets of DNase I-hypersensitive sites characterize the various functional states of the chicken lysozyme gene. Nature. 1984 Sep 13;311(5982):163–165. doi: 10.1038/311163a0. [DOI] [PubMed] [Google Scholar]
- Goldman D., Boulter J., Heinemann S., Patrick J. Muscle denervation increases the levels of two mRNAs coding for the acetylcholine receptor alpha-subunit. J Neurosci. 1985 Sep;5(9):2553–2558. doi: 10.1523/JNEUROSCI.05-09-02553.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D., Brenner H. R., Heinemann S. Acetylcholine receptor alpha-, beta-, gamma-, and delta-subunit mRNA levels are regulated by muscle activity. Neuron. 1988 Jun;1(4):329–333. doi: 10.1016/0896-6273(88)90081-5. [DOI] [PubMed] [Google Scholar]
- Gross D. S., Huang S. Y., Garrard W. T. Chromatin structure of the potential Z-forming sequence (dT-dG)n X (dC-dA)n. Evidence for an "alternating-B" conformation. J Mol Biol. 1985 May 25;183(2):251–265. doi: 10.1016/0022-2836(85)90218-9. [DOI] [PubMed] [Google Scholar]
- Grosveld F., van Assendelft G. B., Greaves D. R., Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987 Dec 24;51(6):975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- Hamada H., Seidman M., Howard B. H., Gorman C. M. Enhanced gene expression by the poly(dT-dG).poly(dC-dA) sequence. Mol Cell Biol. 1984 Dec;4(12):2622–2630. doi: 10.1128/mcb.4.12.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris D. A., Falls D. L., Dill-Devor R. M., Fischbach G. D. Acetylcholine receptor-inducing factor from chicken brain increases the level of mRNA encoding the receptor alpha subunit. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1983–1987. doi: 10.1073/pnas.85.6.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidmann O., Buonanno A., Geoffroy B., Robert B., Guénet J. L., Merlie J. P., Changeux J. P. Chromosomal localization of muscle nicotinic acetylcholine receptor genes in the mouse. Science. 1986 Nov 14;234(4778):866–868. doi: 10.1126/science.3022377. [DOI] [PubMed] [Google Scholar]
- Igo-Kemenes T., Hörz W., Zachau H. G. Chromatin. Annu Rev Biochem. 1982;51:89–121. doi: 10.1146/annurev.bi.51.070182.000513. [DOI] [PubMed] [Google Scholar]
- Ishii S., Merlino G. T., Pastan I. Promoter region of the human Harvey ras proto-oncogene: similarity to the EGF receptor proto-oncogene promoter. Science. 1985 Dec 20;230(4732):1378–1381. doi: 10.1126/science.2999983. [DOI] [PubMed] [Google Scholar]
- Keene M. A., Corces V., Lowenhaupt K., Elgin S. C. DNase I hypersensitive sites in Drosophila chromatin occur at the 5' ends of regions of transcription. Proc Natl Acad Sci U S A. 1981 Jan;78(1):143–146. doi: 10.1073/pnas.78.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Klarsfeld A., Changeux J. P. Activity regulates the levels of acetylcholine receptor alpha-subunit mRNA in cultured chicken myotubes. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4558–4562. doi: 10.1073/pnas.82.13.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarsfeld A., Daubas P., Bourachot B., Changeux J. P. A 5'-flanking region of the chicken acetylcholine receptor alpha-subunit gene confers tissue specificity and developmental control of expression in transfected cells. Mol Cell Biol. 1987 Feb;7(2):951–955. doi: 10.1128/mcb.7.2.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny S. F., Emerson C. P., Jr 5-Azacytidine induction of stable mesodermal stem cell lineages from 10T1/2 cells: evidence for regulatory genes controlling determination. Cell. 1984 Oct;38(3):791–800. doi: 10.1016/0092-8674(84)90274-5. [DOI] [PubMed] [Google Scholar]
- LaPolla R. J., Mayne K. M., Davidson N. Isolation and characterization of a cDNA clone for the complete protein coding region of the delta subunit of the mouse acetylcholine receptor. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7970–7974. doi: 10.1073/pnas.81.24.7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar A. B., Paterson B. M., Weintraub H. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell. 1986 Dec 5;47(5):649–656. doi: 10.1016/0092-8674(86)90507-6. [DOI] [PubMed] [Google Scholar]
- Merlie J. P., Isenberg K. E., Russell S. D., Sanes J. R. Denervation supersensitivity in skeletal muscle: analysis with a cloned cDNA probe. J Cell Biol. 1984 Jul;99(1 Pt 1):332–335. doi: 10.1083/jcb.99.1.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlie J. P., Sanes J. R. Concentration of acetylcholine receptor mRNA in synaptic regions of adult muscle fibres. Nature. 1985 Sep 5;317(6032):66–68. doi: 10.1038/317066a0. [DOI] [PubMed] [Google Scholar]
- Mishina M., Kurosaki T., Tobimatsu T., Morimoto Y., Noda M., Yamamoto T., Terao M., Lindstrom J., Takahashi T., Kuno M. Expression of functional acetylcholine receptor from cloned cDNAs. Nature. 1984 Feb 16;307(5952):604–608. doi: 10.1038/307604a0. [DOI] [PubMed] [Google Scholar]
- Mishina M., Takai T., Imoto K., Noda M., Takahashi T., Numa S., Methfessel C., Sakmann B. Molecular distinction between fetal and adult forms of muscle acetylcholine receptor. Nature. 1986 May 22;321(6068):406–411. doi: 10.1038/321406a0. [DOI] [PubMed] [Google Scholar]
- Nandi A., Das G., Salzman N. P. Characterization of a surrogate TATA box promoter that regulates in vitro transcription of the simian virus 40 major late gene. Mol Cell Biol. 1985 Mar;5(3):591–594. doi: 10.1128/mcb.5.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A. The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell. 1982 Sep;30(2):567–578. doi: 10.1016/0092-8674(82)90253-7. [DOI] [PubMed] [Google Scholar]
- Nef P., Mauron A., Stalder R., Alliod C., Ballivet M. Structure linkage, and sequence of the two genes encoding the delta and gamma subunits of the nicotinic acetylcholine receptor. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7975–7979. doi: 10.1073/pnas.81.24.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda M., Takahashi H., Tanabe T., Toyosato M., Kikyotani S., Furutani Y., Hirose T., Takashima H., Inayama S., Miyata T. Structural homology of Torpedo californica acetylcholine receptor subunits. Nature. 1983 Apr 7;302(5908):528–532. doi: 10.1038/302528a0. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Rich A. The sequence (dC-dA)n X (dG-dT)n forms left-handed Z-DNA in negatively supercoiled plasmids. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1821–1825. doi: 10.1073/pnas.80.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinney D. F., Pearson-White S. H., Konieczny S. F., Latham K. E., Emerson C. P., Jr Myogenic lineage determination and differentiation: evidence for a regulatory gene pathway. Cell. 1988 Jun 3;53(5):781–793. doi: 10.1016/0092-8674(88)90095-5. [DOI] [PubMed] [Google Scholar]
- Pinset C., Montarras D., Chenevert J., Minty A., Barton P., Laurent C., Gros F. Control of myogenesis in the mouse myogenic C2 cell line by medium composition and by insulin: characterization of permissive and inducible C2 myoblasts. Differentiation. 1988 Jun;38(1):28–34. doi: 10.1111/j.1432-0436.1988.tb00588.x. [DOI] [PubMed] [Google Scholar]
- Reznikoff C. A., Brankow D. W., Heidelberger C. Establishment and characterization of a cloned line of C3H mouse embryo cells sensitive to postconfluence inhibition of division. Cancer Res. 1973 Dec;33(12):3231–3238. [PubMed] [Google Scholar]
- Roesler W. J., Vandenbark G. R., Hanson R. W. Cyclic AMP and the induction of eukaryotic gene transcription. J Biol Chem. 1988 Jul 5;263(19):9063–9066. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidereit C., Geisse S., Westphal H. M., Beato M. The glucocorticoid receptor binds to defined nucleotide sequences near the promoter of mouse mammary tumour virus. Nature. 1983 Aug 25;304(5928):749–752. doi: 10.1038/304749a0. [DOI] [PubMed] [Google Scholar]
- Senear A. W., Palmiter R. D. Expression of the mouse metallothionein-I gene alters the nuclease hypersensitivity of its 5' regulatory region. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):539–547. doi: 10.1101/sqb.1983.047.01.063. [DOI] [PubMed] [Google Scholar]
- Shermoen A. W., Beckendorf S. K. A complex of interacting DNAase I-hypersensitive sites near the Drosophila glue protein gene, Sgs4. Cell. 1982 Jun;29(2):601–607. doi: 10.1016/0092-8674(82)90176-3. [DOI] [PubMed] [Google Scholar]
- Shibahara S., Kubo T., Perski H. J., Takahashi H., Noda M., Numa S. Cloning and sequence analysis of human genomic DNA encoding gamma subunit precursor of muscle acetylcholine receptor. Eur J Biochem. 1985 Jan 2;146(1):15–22. doi: 10.1111/j.1432-1033.1985.tb08614.x. [DOI] [PubMed] [Google Scholar]
- Taylor S. M., Jones P. A. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979 Aug;17(4):771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- Theisen M., Stief A., Sippel A. E. The lysozyme enhancer: cell-specific activation of the chicken lysozyme gene by a far-upstream DNA element. EMBO J. 1986 Apr;5(4):719–724. doi: 10.1002/j.1460-2075.1986.tb04273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. H., Elgin S. C. Protein/DNA architecture of the DNase I hypersensitive region of the Drosophila hsp26 promoter. EMBO J. 1988 Jul;7(7):2191–2201. doi: 10.1002/j.1460-2075.1988.tb03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnacliffe A., Sims J. E., Rabbitts T. H. T3 delta pre-mRNA is transcribed from a non-TATA promoter and is alternatively spliced in human T cells. EMBO J. 1986 Jun;5(6):1245–1252. doi: 10.1002/j.1460-2075.1986.tb04353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O., Bohn M. A stretch of "late" SV40 viral DNA about 400 bp long which includes the origin of replication is specifically exposed in SV40 minichromosomes. Cell. 1979 Feb;16(2):453–466. doi: 10.1016/0092-8674(79)90021-7. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Beug H., Groudine M., Graf T. Temperature-sensitive changes in the structure of globin chromatin in lines of red cell precursors transformed by ts-AEV. Cell. 1982 Apr;28(4):931–940. doi: 10.1016/0092-8674(82)90072-1. [DOI] [PubMed] [Google Scholar]
- White M. M., Mayne K. M., Lester H. A., Davidson N. Mouse-Torpedo hybrid acetylcholine receptors: functional homology does not equal sequence homology. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4852–4856. doi: 10.1073/pnas.82.14.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. An exonuclease protection assay reveals heat-shock element and TATA box DNA-binding proteins in crude nuclear extracts. Nature. 1985 Sep 5;317(6032):84–87. doi: 10.1038/317084a0. [DOI] [PubMed] [Google Scholar]
- Wu C., Bingham P. M., Livak K. J., Holmgren R., Elgin S. C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979 Apr;16(4):797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Wu C. Two protein-binding sites in chromatin implicated in the activation of heat-shock genes. Nature. 1984 May 17;309(5965):229–234. doi: 10.1038/309229a0. [DOI] [PubMed] [Google Scholar]
- Wu C., Wong Y. C., Elgin S. C. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979 Apr;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- Yu L., LaPolla R. J., Davidson N. Mouse muscle nicotinic acetylcholine receptor gamma subunit: cDNA sequence and gene expression. Nucleic Acids Res. 1986 Apr 25;14(8):3539–3555. doi: 10.1093/nar/14.8.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Yamamoto K. R. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell. 1984 Aug;38(1):29–38. doi: 10.1016/0092-8674(84)90523-3. [DOI] [PubMed] [Google Scholar]