Background: Current drugs against sleeping sickness are limited, toxic, and inefficient.
Results: In vivo genetic and chemical studies identified isoleucyl-tRNA synthetase (IleRS) as a drug target and an IleRS inhibitor that cures mice of infection.
Conclusion: Inhibition of IleRS constitutes a potential treatment for sleeping sickness.
Significance: Identification of drug targets and inhibitors aids in development of drugs against T. brucei and related parasites.
Keywords: Aminoacyl-tRNA Synthetase, Chemical Biology, Drug Development, Enzyme Inhibitors, Molecular Biology, Parasitology, Trypanosoma brucei
Abstract
Trypanosoma brucei sp. causes human African trypanosomiasis (HAT; African sleeping sickness). The parasites initially proliferate in the hemolymphatic system and then invade the central nervous system, which is lethal if not treated. New drugs are needed for HAT because the approved drugs are few, toxic, and difficult to administer, and drug resistance is spreading. We showed by RNAi knockdown that T. brucei isoleucyl-tRNA synthetase is essential for the parasites in vitro and in vivo in a mouse model of infection. By structure prediction and experimental analysis, we also identified small molecules that inhibit recombinant isoleucyl-tRNA synthetase and that are lethal to the parasites in vitro and highly selective compared with mammalian cells. One of these molecules acts as a competitive inhibitor of the enzyme and cures mice of the infection. Because members of this class of molecules are known to cross the blood-brain barrier in humans and to be tolerated, they may be attractive as leading candidates for drug development for HAT.
Introduction
Human African trypanosomiasis (HAT),2 also known as African sleeping sickness, is caused by brucei-group trypanosomes and is endemic in 36 sub-Saharan countries (1, 2). Two subspecies with different geographic distributions and transmission dynamics cause somewhat different human diseases. Trypanosoma brucei gambiense occurs in West Africa and causes a chronic disease, whereas Trypanosoma brucei rhodesiense occurs in East Africa and produces a more acute disease (1, 2). During acute infection (stage 1), parasites proliferate in the host bloodstream and lymphatics and undergo antigenic variation, thereby evading elimination by the immune system (1, 2). In chronic infections (stage 2), parasites are present in the central nervous system, resulting in multiple clinical sequellae and death if not treated (1–3). Current drug treatments for stage 1 HAT include pentamidine and suramin (for T. brucei gambiense and T. brucei rhodesiense, respectively), whereas those for stage 2 HAT include melarsoprol and eflornithine (for T. brucei rhodesiense and T. brucei gambiense, respectively). More recently, nifurtimox and eflornithine in combination have also been used as a treatment for chronic disease (4, 5). These treatments are highly toxic, require complicated dosing, and must also contend with increasing parasite drug resistance (5–8). Hence, there is a dire need for new effective drugs for HAT, especially for the second stage.
Several approaches have been taken to develop anti-HAT drugs, ranging from large compound library screening against the organisms in vitro (which is target-agnostic) to target-specific structure-based drug design (9, 10). We have taken a hybrid approach to identify potential drugs for HAT. We began by using genetic methods to selectively assess the essentiality in vitro and in vivo of predicted “druggable” enzymes in T. brucei (11). We then applied target-specific chemistry design to identify compounds that inhibited the enzyme activity and parasite growth and confirmed their specific inhibitions using biochemical- and molecular-based approaches. Finally, we tested whether our compounds cured the infection in vivo using a mouse model and validated their potential use for drug development.
Aminoacyl-tRNA synthetases have been identified as possible drug targets for several infectious diseases (12–14). They are responsible for charging a specific tRNA with its cognate amino acid, which is essential for protein synthesis (15). Drugs targeting isoleucyl-tRNA synthetase (IleRS) have been successfully developed against bacterial infections, e.g. mupirocin (16). In T. brucei, IleRS is encoded by one gene that undergoes alternative mRNA trans-splicing, thereby allowing tRNA isoleucylation to be performed in both the cytoplasm and mitochondrion (17). The amino acid sequence of IleRS is conserved among T. brucei, Trypanosoma cruzi, and Leishmania sp. Because the genomes of these parasites are highly conserved (18), the validation of drug targets and the discovery of inhibitors for T. brucei may also aid in the development of new drugs for leishmaniasis and Chagas disease (19, 20).
Here, we present genetic and chemical validation of T. brucei IleRS as a target for drug development. We knocked down the IleRS gene by RNAi and found it to be essential for growth and infection of mice. We also identified small molecule inhibitors that are highly selective to the parasites, including a molecule that acts as a competitive inhibitor of the IleRS enzyme and cures mice of infection. These results may aid in the development of new drugs for HAT.
EXPERIMENTAL PROCEDURES
Plasmid Construction for RNAi and Transfection
The inducible RNAi plasmid for silencing IleRS gene expression was generated using the pQuadra system (21). Briefly, 400 bp of the gene were selected using RNAit software (22) and amplified by PCR using oligonucleotides with specifically designed BstXI sites (7538-F, ATACCAATGTGATGGTACGTCACAACCCAACTGGA; and 7539-R, ATACCATAGAGTTGGCATTTCCCCCGGATAGTTTT). Ligation with BstXI-digested pQuadra1 and pQuadra3 plasmids generated the pQ041 vector, containing inverted repeats of the PCR product separated by a spacer region. Transfection of NotI-linearized constructs into a bloodstream form “single marker” (SM427) cell line (23) and selection of transgenic cell lines were carried out as described previously (24).
Cell Maintenance and Growth Curve Analysis
The T. brucei bloodstream form was maintained at exponential growth (between 105 and 106 cells/ml) in HMI-9 medium supplemented with 10% fetal bovine serum. RNAi was induced by the addition of 1 μg/ml tetracycline to the medium, and a cumulative growth curve was obtained by counting (and diluting) the cells daily using a Beckman cell counter or Neubauer chamber.
Recombinant Protein Expression and Aminoacylation Assay
The T. brucei gene Tb927.10.9190 (which codes for IleRS) was cloned into the pET-28b(+) vector and expressed in Escherichia coli as described previously (25). Protein activity was tested using a TLC-based aminoacylation assay (26). Inhibition assays and enzymology analysis were performed as described previously (25). The percentage of remaining recombinant IleRS (rIleRS) activity in the presence of compounds was calculated as follows: % remaining rIleRS activity = (rIleRSwc × 100)/rIleRSnc, where rIleRSwc is the average of the rIleRS activity measurements with compounds and rIleRSnc is the average of the rIleRS activity measurements without compounds (vehicle only).
Cell-based Compound Screen
Compounds were obtained from the National Cancer Institute. Compounds were searched by similarity to Ile-AMP using a Tanimoto similarity index of 80%. We used the PubChem search at the National Center for Biotechnology Information and the National Cancer Institute database. Compound stocks were prepared at a 10 mm concentration in dimethyl sulfoxide. T. brucei bloodstream forms (100 μl at 2.0 × 104 parasites/ml) were plated in 96-well plates and mixed with 100 μl of compounds diluted in HMI-9 medium with 10% FBS (Me2SO at <0.2%). Parasites not treated with compounds were also plated as controls. After 48 h of incubation at 37 °C and 5% CO2, 20 μl of alamarBlue (Invitrogen) was added, and the assays were developed for 4 h. Fluorescence measurements were obtained using a SpectraMax M2 microplate reader (Molecular Devices) with excitation at 544 nm and emission at 590 nm (590-nm cutoff). Data were analyzed using GraphPad Prism for Windows.
In Vivo RNAi and Mouse Treatment with Inhibitors
RNAi in Vivo
The T. brucei IleRS RNAi line growing at mid-log phase was used to infect BALB/cAnNHsd mice (6–8-week-old males; Harlan Laboratories). 1.0 × 104 parasites in 200 μl of HMI-9 medium were injected intraperitoneally. Doxycycline at 200 μg/ml and 5% sucrose were added to the drinking water either 18 h before infection or 24 or 48 h after infection to induce RNAi expression (replaced daily). For non-induced conditions, mice received drinking water containing 5% sucrose only. Parasitemia was monitored daily starting at day 2 post-infection by tail prick. Mice with parasitemia of >1.0 × 108 parasites/ml of blood were killed. All procedures were performed in the vivarium of the Seattle Biomedical Research Institute in compliance with the laws and institutional guidelines (Institutional Animal Care and Use Committee KS-01).
Treatment of T. brucei-infected Mice with Ile-AMP Analogs
BALB/cAnNHsd mice (6–8-week-old males) were infected with 1.0 × 104 parasites in 200 μl of HMI-9 medium injected intraperitoneally. At 24 or 48 h post-infection, parasitemia was checked with a blood smear, and administration of compounds (in PBS and 5% Me2SO) was begun by intraperitoneal injections (once or twice per day for 4 days or more according to the compound used; see “Results”). Control mice received vehicle only (in PBS and 5% Me2SO). For these experiments, the compounds NSC70422 (Angene International), NSC7359 (Ryan Scientific), and NSC404241 and NSC30605 (Sigma) were used.
Molecular Modeling and Docking
The T. brucei IleRS structure was modeled by homology to Thermus thermophilus IleRS (Protein Data Bank code 1JZQ) (16) using the SWISS-MODEL workspace (27). Docking was performed with compound NSC70422 and N-[isoleucinyl]-N′-[adenosyl]-diaminosufone (Ile-AMS; the structure previously determined with T. thermophilus IleRS) using AutoDock 4.2 and Vina 1.1.2 (28). Data visualization was performed with PyMOL (Version 1.5.0.4, Schrödinger, LLC).
Data Presentation and Statistical Analysis
All data are shown as the mean ± S.D. Comparisons among groups were made by two-tailed t test for repeated measures using GraphPad Prism Version 5.00 for Windows. The p values <0.05 with a confidence interval of 95% were considered statistically significant unless specified otherwise.
RESULTS
IleRS Is Essential for T. brucei Growth and Infection
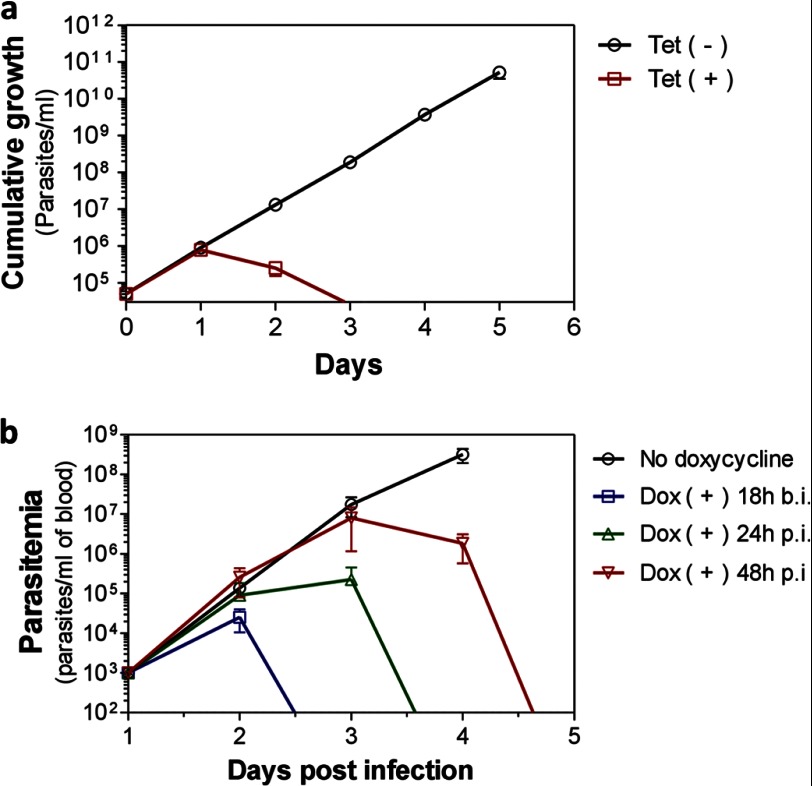
To genetically validate IleRS as a drug target in the T. brucei bloodstream form, we knocked down its expression by RNAi. A T. brucei line was generated in which a tetracycline-regulated hairpin RNAi construct against IleRS had been transfected. Tetracycline induction of RNAi against IleRS resulted in rapid and severe inhibition of parasite growth in vitro (Fig. 1a). Fluorescence-activated cell sorting analysis revealed that ∼65% of the parasites were dead at 24 h (supplemental Fig. S1), and no parasites could be detected at 72 h after RNAi induction (Fig. 1a). Quantitative RT-PCR analysis showed an 80% decrease in IleRS mRNA at 24 h after tetracycline induction, confirming its knockdown (supplemental Fig. S2a). No inhibition of parasite growth upon tetracycline induction was observed with a T. brucei line transfected with a control vector without RNAi insertions (supplemental Fig. S2b). To assess IleRS essentiality in vivo, we infected BALB/c mice with the T. brucei IleRS RNAi line. Doxycycline (a stable tetracycline analog) was added to the mouse drinking water to induce RNAi expression, and parasitemia was monitored daily. Doxycycline addition to the water either before or after infection resulted in rapid and complete parasite clearance (Fig. 1b). Mice were monitored for up to 20 days post-infection without any recurrence of infection. Notably, mice that received doxycycline at 48 h post-infection presented high parasitemia levels at 72 h (≈1.0 × 107 parasites/ml of blood), but they were still cured upon RNAi induction (Fig. 1b), which shows that knockdown of this gene not only impairs parasite infection but also cures highly infected mice. No effect on parasitemia was observed when the parental cell line (SM427, which does not express an RNAi construct) was infected in the presence of doxycycline (supplemental Fig. S2c), which rules out any effect of doxycycline on the parasite growth. These results show that IleRS is essential for parasite growth and infection in vivo. The rapid cure of highly infected mice after RNAi induction implies that IleRS is a potential target for drug development, suggesting that inhibition of this enzyme could lead to a rapid cure of infection.
FIGURE 1.
IleRS is essential for T. brucei growth and infection. a, cumulative growth curve of T. brucei bloodstream forms with tetracycline-inducible RNAi against IleRS. Tet (+), RNAi induced with 1 μg/ml tetracycline; Tet (−), non-induced. b, parasitemia of mice infected with the T. brucei bloodstream form IleRS RNAi line. Mice were infected with 1.0 × 104 parasites. Mice received doxycycline (Dox; 200 μg/ml) added to the drinking water for RNAi induction 18 h before infection (b.i.) or 24 or 48 h post-infection (p.i.) and replaced daily. Non-induced mice received water without doxycycline. Data in a are the mean ± S.D. of triplicate measures, and those in b are the mean ± S.D. of two experiments performed in triplicate.
Identification of Ile-AMP Analogs That Inhibit T. brucei Growth
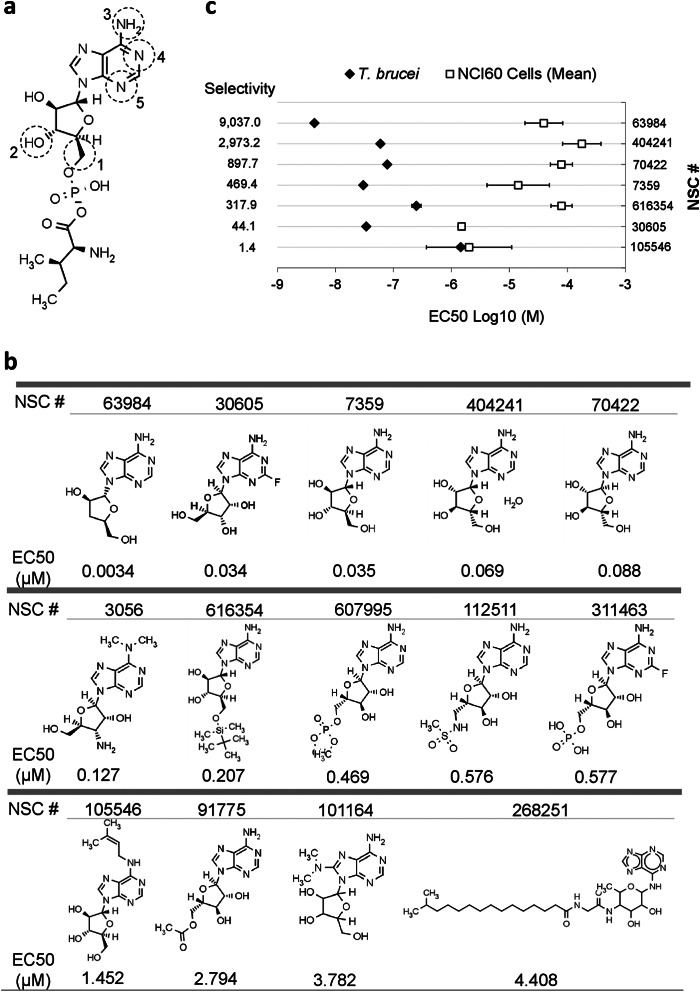
During tRNA aminoacylation, IleRS hydrolyzes ATP and conjugates isoleucine to AMP, thereby forming the Ile-AMP intermediate (15). Because Ile-AMP has been shown to strongly interact with IleRS (Kd ≈ 10−9 m) (16, 29), we reasoned that compounds that mimic its structure could inhibit its enzyme activity. 20 Ile-AMP analogs (>80% similarity to Ile-AMP) were obtained from the National Cancer Institute compound collection and tested against T. brucei bloodstream form growth (Fig. 2, a and b, and supplemental Table S1). We identified 14 Ile-AMP analogs that inhibited T. brucei growth with EC50 (effective concentration that inhibits parasite growth by 50%) values between 3.9 nm and 4.4 μm (Fig. 2b and supplemental Table S1). Analysis of the substructure of the compounds revealed that the changes in the chemical groups at position 1 caused a slight reduction in their potency (Fig. 2b). The addition of chemical groups throughout positions 2–5 in general resulted in less potent or inactive molecules (Fig. 2b and supplemental Table S1). The EC50 values of these compounds have also been determined for 59 different mammalian cell lines (named NCI60 cells) (30). A comparison of the EC50 values for the most effective compounds revealed six with lower EC50 values for T. brucei compared with NCI60 cells (Fig. 2c). Some compounds have been previously tested in vivo for other conditions, and thus, pharmacokinetic and toxicity information is also available (supplemental Table S1), e.g. NSC404241, which has been approved for human use and which crosses the blood-brain barrier (31). The compound NSC63984 has also been shown to cross the blood-brain barrier, although it did not completely cure mice infected with T. brucei (32). Overall, we identified small molecules that inhibit parasite growth and that are selective compared with mammalian cells; thus, they may be feasible leading candidates for drug development for HAT.
FIGURE 2.
Identification of Ile-AMP analogs that inhibit T. brucei growth. a, Ile-AMP structure. Positions 1–5 indicate the locations of chemical groups changed in the analogs analyzed. Comprehensive information on the compounds and their structures is provided in supplemental Table S1. b, structure-activity relationship of the Ile-AMP analogs tested against T. brucei. The EC50 for T. brucei is shown under the compound structure. c, comparison of the EC50 values of selected compounds for T. brucei and NCI60 cells. The EC50 values for NCI60 cells represent the mean of the EC50 data for 59 different cell lines (30). Compound selectivities were obtained by dividing the mean of the NCI60 EC50 by the T. brucei EC50 data for each compound. Data shown in b are the mean of at least three experiments performed in triplicate.
Identification of a Competitive Inhibitor of T. brucei IleRS
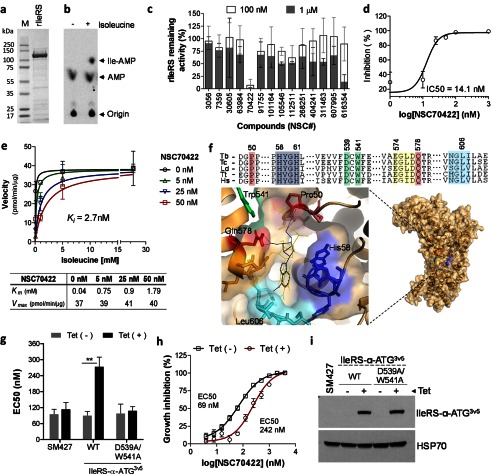
To evaluate whether these Ile-AMP analogs are targeting the IleRS enzyme, we expressed and purified T. brucei rIleRS from E. coli (Fig. 3a). The enzyme was active as detected by tRNAIle aminoacylation in vitro (Fig. 3b). Enzyme inhibition analysis showed that these small molecules reduced rIleRS activity (Fig. 3c), especially NSC70422, which presented an IC50 of 14 nm (Fig. 3d). It is noteworthy that this molecule inhibited T. brucei growth with an EC50 of 88 nm and ≈900-fold parasite selectivity (Fig. 2, b and c). To investigate its mechanism of inhibition of IleRS, we performed enzymology studies. NSC70422 increased the rIleRS Km for isoleucine, but no effect was observed on the enzyme Vmax (Fig. 3e), which is consistent with a mechanism of competitive inhibition. The very low inhibition constant (Ki = 2.7 ± 1.4 nm) resembles the high affinity interaction between IleRS and its reaction intermediate, Ile-AMP (12, 16, 29). Docking analysis of NSC70422 on the IleRS structure predicted its interaction with Gln-578 and Pro-50, two amino acids involved in isoleucine interaction (16), and with the AMP-binding site (Fig. 3f). Therefore, NSC70422 seems to interact with the Ile-AMP-binding site of IleRS and to interfere with isoleucine binding, in accordance with its mode of action as a competitive inhibitor.
FIGURE 3.
Identification of a competitive inhibitor of T. brucei IleRS. a, T. brucei rIleRS (122 kDa) resolved by 4–20% SDS-PAGE and stained with Imperial Coomassie stain (Pierce). b, rIleRS aminoacylation assay performed with 32P-labeled tRNAIle. P1 nuclease digestion of Ile-tRNAIle resulted in free [32P]AMP and [32P]Ile-AMP, which were separated by TLC. c, inhibition of rIleRS activity with 14 Ile-AMP analogs. Assays were performed with compounds at 0.1 and 1 μm as described previously (25). The molecule NSC616354 is a positive control (25). Data are shown as the percentage of remaining IleRS activity (see “Experimental Procedures”). d, dose-response inhibition of rIleRS with NSC70422. The experiment was performed with 1–1000 nm NSC70422. e, enzymology analysis of NSC70422 inhibition of rIleRS. Assays were performed with 5–50 nm NSC70422 and various concentrations of isoleucine (0.1–20 mm). The Michaelis-Menten constant (Km), maximum velocity (Vmax), and inhibition constant (Ki) were calculated by nonlinear regression with GraphPad Prism software. The table shows the apparent Km and Vmax for IleRS (with respect to isoleucine) in the absence or presence of NSC70422. f, docking of NSC70422 (black lines) and Ile-AMS (yellow lines) in the modeled structure of T. brucei IleRS. The alignment shows amino acids of the IleRS catalytic site. Pro-50 and Gln-578 (red boxes) are involved in isoleucine binding. His-58–His-61 (HYGH motif; blue box) are involved in AMP binding. Yellow, green, and cyan boxes show amino acids of the binding site highlighted in the model (16, 33). Green boxes also show Asp-539 and Trp-541 mutated to alanine in g and i. Asp-539 is not visible in the IleRS model due to the orientation of the binding site, which favors showing the NSC70422 inhibitor. Ile-AMS is an Ile-AMP analog known to interact with T. thermophilus IleRS (16) and was used here for comparison. Tb, T. brucei; Tc, T. cruzi; Ld, Leishmania donovani; Tt, T. thermophilus; Hs, Homo sapiens. g, analysis of NSC70422 EC50 values in T. brucei overexpressing cytoplasmic IleRS (IleRS-α-ATG). NSC70422 EC50 values were calculated for T. brucei SM427 or cell lines overexpressing WT IleRS-α-ATG3V5 or the inactive mutant IleRS-α-ATG3V5(D539A/W541A). Tet, 1 μg/ml tetracycline. h, graph showing the growth inhibition curve shift of NSC70422 EC50 values after overexpression of WT IleRS-α-ATG3V5 in T. brucei. i, Western blot analysis of WT IleRS-α-ATG3V5 and mutant IleRS-α-ATG3V5(D539A/W541A) expression in T. brucei. T. brucei lysates in the presence or absence of 1 μg/ml tetracycline were separated by 10% SDS-PAGE, transferred to a nitrocellulose membrane, and blotted with monoclonal antibodies against the three-V5 tag. The membrane was stripped and reblotted with monoclonal antibodies against HSP70 (mAb78). Data in e are the mean ± S.D. of three single experiments. Data in c, d, g, and h are the mean ± S.D. of at least three experiments performed in triplicate. **, p < 0.007.
To assess whether IleRS is the primary target of NSC70422, we overexpressed IleRS minus its mitochondrial targeting signal (IleRS-α-ATG3V5, containing a C-terminal three-V5 tag) in the T. brucei bloodstream form and performed growth inhibition assays to determine the EC50. Overexpression of IleRS-α-ATG3V5 in T. brucei conferred a 3.5-fold increase in the EC50 of NSC70422 (242 nm) compared with non-induced cells (69 nm, which is similar to 88 nm for wild-type cells) (Fig. 3, g-i). Overexpression of a catalytically inactive IleRS mutant (IleRS-α-ATG3V5(D539A/W541A)) (33) by tetracycline induction failed to increase the EC50 (Fig. 3, g and i). Because Asp-539 and Trp-541 are in the IleRS active site, their mutation may interfere with IleRS binding to its substrates as well as to NSC70422. Because the mutant enzyme is catalytic inactive, its overexpression does not rescue the inhibitory effect of NSC70422. In contrast, overexpression of WT IleRS provides extra enzyme active sites, which partially rescue the inhibitory effect of NSC70422, observed by an increase in EC50. These results confirm that NSC70422 targets IleRS and inhibits its activity in vivo, consistent with the enzymology analysis (Fig. 3, c–e). These results show that NSC70422 is a potent and specific intracellular inhibitor of T. brucei IleRS.
IleRS Inhibitor Cures T. brucei Infection in Vivo
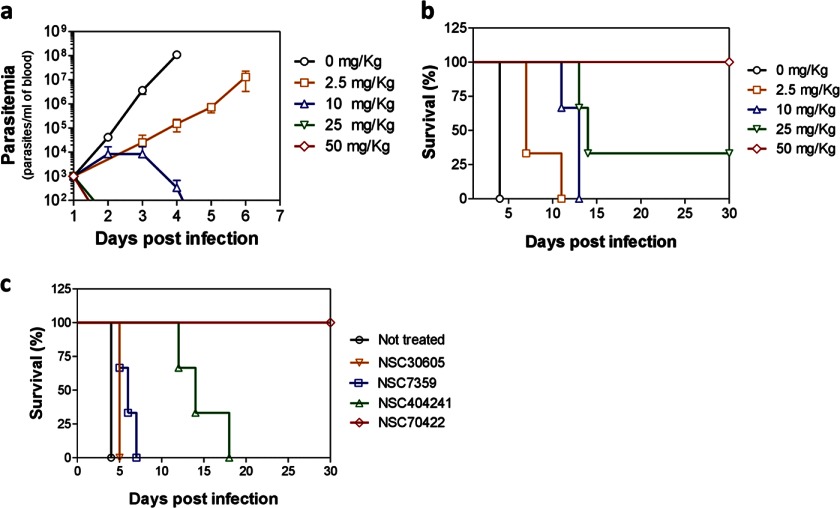
Finally, we sought to evaluate whether these small molecules cure infection in vivo. T. brucei-infected mice were treated with NSC70422, and parasitemia and survival times were monitored daily. A 4-day treatment with dosages as low as 10 mg/kg twice per day was sufficient to suppress parasite growth in vivo (Fig. 4a). Mice presented prolonged life spans upon NSC70422 treatment, and complete cure of infection was observed with 50 mg/kg twice per day (Fig. 4b); cure was determined by the absence of parasitemia in treated mice for >30 days post-infection. Mice were monitored for up to 60 days post-infection without recurrence of infection. No signs of toxicity were observed when mice were treated with 50 mg/kg NSC70422 four times per day for 4 days (data not shown), in agreement with a previous report of no toxicity with daily doses of up to 400 mg/kg for 5 days (30). An increased life span was also observed upon treatment of the mice with compounds NSC404241, NSC7359, and NSC30605 (Fig. 4c and supplemental Fig. S3). NSC404241 crosses the blood-brain barrier in humans (31). Although this has not yet been tested for the other compounds, it implies that this class of compounds may be candidates for drugs against stage 2 HAT. These results show that NSC70422 is effective against T. brucei in vivo and therefore a new lead for the development of drugs for treating HAT.
FIGURE 4.
IleRS inhibitor cures T. brucei infection in vivo. a, parasitemia in mice with or without NSC70422 treatment. Mice were infected with an inoculum of 1.0 × 104 T. brucei SM427 parasites. NSC70422 was administered by intraperitoneal injections twice per day for 4 consecutive days starting at 24 h post-infection. Parasitemia was monitored daily by tail prick (n = 3 per dose group). b, Kaplan-Meier survival plot of the experiment shown in a. Mice were killed when parasitemia was equal to or higher than 1.0 × 108 parasites/ml of blood. c, Kaplan-Meier survival plot comparing the survival of T. brucei-infected mice after treatment with the following compounds: NSC7359, 50 mg/kg administered 48 h post-infection once per day for 5 days; NSC30605, 2.5 mg/kg administered 24 h post-infection twice per day for 4 days (highly toxic at higher concentrations); NSC404241, 50 mg/kg administered 24 h post-infection twice per day for 12 days; and NSC70422, 50 mg/kg administered 24 h post-infection twice per day for 4 days. Data shown in a–c are averages of experiments performed in triplicate.
DISCUSSION
One of the main factors hampering the development of effective drugs for trypanosomatid parasites is a lack of drug target knowledge (20). Although there is a variety of work that identifies essential genes in T. brucei in vitro, there are still limited examples of target validation, including in vivo analysis of gene essentiality and chemical validation with identification of inhibitors as candidates for accelerating the design of new drugs. Here, we have presented the validation of IleRS as a drug target in T. brucei and identified new small molecule inhibitors that are potential candidates for the development of new drugs for treatment of HAT. We have shown that IleRS is essential for parasite growth in vitro and for in vivo infection of mice. RNAi knockdown of IleRS resulted in rapid parasite death, despite incomplete knockdown (80%) of its mRNA at 24 h. The rapid cell death upon partial RNA knockdown suggests that the low levels of target mRNA and hence protein may not be sufficient for sustaining cell growth, as also observed for other essential genes in T. brucei (34, 35). Because IleRS is involved in protein synthesis, the rapid cell death likely resulted from the parasites' inability to synthesize new proteins. Infected mice could also be cured after RNAi induction against T. brucei IleRS in vivo, which suggests that inhibition of this enzyme could render parasites unable to sustain an infection and thus is a target for drug development.
We found that the small molecule NSC70422 is a specific inhibitor of IleRS and that it is safe and effective against T. brucei in a murine model of infection. Although little is known about this molecule's pharmacokinetics and toxicity in humans, other molecules of this class (e.g. NSC404241) are well tolerated in humans and are known to cross the blood-brain barrier (31, 36), an important prerequisite for treating stage 2 HAT. Besides NSC404241, the molecule NSC63984 has also been shown to cross the blood-brain barrier (32), suggesting that this class of molecules may be explored for the development of drugs against stage 2 HAT. Because this small molecule shows promising results in vivo in a mouse model, pharmacokinetic studies will be performed with this molecule and other analogs to develop new drugs against HAT and related parasites.
T. brucei and human IleRS amino acid sequences have 38% identity, with several amino acid differences encompassing the active site. This indicates that the greater susceptibility of trypanosomes compared with human cells to these inhibitors could be due to differences in IleRS protein structure, its association with other proteins or protein complexes, and inhibitor uptake and metabolism.
Several attempts to purify human IleRS from E. coli (using the pET-28 system and E. coli BL21 Rosetta(DE3)pLysS) failed due to its extensive degradation. Similar results were obtained when human IleRS was coexpressed with the GroES-GroEL-tig chaperones (37), which indicates that the human protein is unstable in E. coli. Nonetheless, comparison of the human and T. brucei EC50 values for NSC70422 showed a 900-fold parasite selectivity, and no toxicity in mice was observed upon high dosage administration, suggesting that NSC70422 may preferentially inhibit the parasite protein.
The discoveries of new drug targets in T. brucei through genetic and chemical validation may also help advance development of drugs against the related parasites T. cruzi and Leishmania sp. because of their genomic similarities (18). The T. brucei IleRS sequence is highly conserved in T. cruzi and Leishmania sp. and is also likely to be essential for their survival. Although genetic manipulation of these parasites is less tractable compared with T. brucei, the inhibitors identified here may be useful for their chemical validation and provide a scaffold for development of new drugs against them.
In summary, we have validated the enzyme IleRS as a target for drug development in T. brucei and identified a selective inhibitor that cures the infection in vivo. This small molecule represents a new lead for drug development against HAT.
Acknowledgments
We are very grateful to Dr. Fred Buckner for sharing protocols and reviewing the manuscript, Drs. Lindsay N. Carpp and Suzanne McDermott for revision of the manuscript, Drs. Jason Carnes and Marilyn Parsons for discussions throughout this work, and Christina McCormick for administrative support.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI078962 (to K. S) and National Institutes of Health Fellowship 5T32AI007509-12 administered by the University of Washington (to I. C.).

This article contains supplemental “Methods,” Figs. S1–S3, Table S1, and additional references.
- HAT
- human African trypanosomiasis
- IleRS
- isoleucyl-tRNA synthetase
- rIleRS
- recombinant IleRS
- Ile-AMS
- N-[isoleucinyl]-N′-[adenosyl]-diaminosufone.
REFERENCES
- 1. Brun R., Blum J., Chappuis F., Burri C. (2010) Human African trypanosomiasis. Lancet 375, 148–159 [DOI] [PubMed] [Google Scholar]
- 2. Fèvre E. M., Wissmann B. V., Welburn S. C., Lutumba P. (2008) The burden of human African trypanosomiasis. PLoS Negl. Trop. Dis. 2, e333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Frevert U., Movila A., Nikolskaia O. V., Raper J., Mackey Z. B., Abdulla M., McKerrow J., Grab D. J. (2012) Early invasion of brain parenchyma by African trypanosomes. PLoS ONE 7, e43913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barrett M. P. (2010) Potential new drugs for human African trypanosomiasis: some progress at last. Curr. Opin. Infect. Dis. 23, 603–608 [DOI] [PubMed] [Google Scholar]
- 5. Simarro P. P., Jannin J., Cattand P. (2008) Eliminating human African trypanosomiasis: where do we stand and what comes next? PLoS Med. 5, e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barrett M. P., Vincent I. M., Burchmore R. J., Kazibwe A. J., Matovu E. (2011) Drug resistance in human African trypanosomiasis. Future Microbiol. 6, 1037–1047 [DOI] [PubMed] [Google Scholar]
- 7. Chitanga S., Marcotty T., Namangala B., Van den Bossche P., Van Den Abbeele J., Delespaux V. (2011) High prevalence of drug resistance in animal trypanosomes without a history of drug exposure. PLoS Negl. Trop. Dis. 5, e1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alsford S., Eckert S., Baker N., Glover L., Sanchez-Flores A., Leung K. F., Turner D. J., Field M. C., Berriman M., Horn D. (2012) High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature 482, 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Frearson J. A., Brand S., McElroy S. P., Cleghorn L. A., Smid O., Stojanovski L., Price H. P., Guther M. L., Torrie L. S., Robinson D. A., Hallyburton I., Mpamhanga C. P., Brannigan J. A., Wilkinson A. J., Hodgkinson M., Hui R., Qiu W., Raimi O. G., van Aalten D. M., Brenk R., Gilbert I. H., Read K. D., Fairlamb A. H., Ferguson M. A., Smith D. F., Wyatt P. G. (2010) N-Myristoyltransferase inhibitors as new leads to treat sleeping sickness. Nature 464, 728–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jacobs R. T., Nare B., Wring S. A., Orr M. D., Chen D., Sligar J. M., Jenks M. X., Noe R. A., Bowling T. S., Mercer L. T., Rewerts C., Gaukel E., Owens J., Parham R., Randolph R., Beaudet B., Bacchi C. J., Yarlett N., Plattner J. J., Freund Y., Ding C., Akama T., Zhang Y. K., Brun R., Kaiser M., Scandale I., Don R. (2011) SCYX-7158, an orally-active benzoxaborole for the treatment of stage 2 human African trypanosomiasis. PLoS Negl. Trop. Dis. 5, e1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Agüero F., Al-Lazikani B., Aslett M., Berriman M., Buckner F. S., Campbell R. K., Carmona S., Carruthers I. M., Chan A. W., Chen F., Crowther G. J., Doyle M. A., Hertz-Fowler C., Hopkins A. L., McAllister G., Nwaka S., Overington J. P., Pain A., Paolini G. V., Pieper U., Ralph S. A., Riechers A., Roos D. S., Sali A., Shanmugam D., Suzuki T., Van Voorhis W. C., Verlinde C. L. (2008) Genomic-scale prioritization of drug targets: the TDR Targets database. Nat. Rev. Drug Discov. 7, 900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schimmel P., Tao J., Hill J. (1998) Aminoacyl-tRNA synthetases as targets for new anti-infectives. FASEB J. 12, 1599–1609 [PubMed] [Google Scholar]
- 13. Hoepfner D., McNamara C. W., Lim C. S., Studer C., Riedl R., Aust T., McCormack S. L., Plouffe D. M., Meister S., Schuierer S., Plikat U., Hartmann N., Staedtler F., Cotesta S., Schmitt E. K., Petersen F., Supek F., Glynne R. J., Tallarico J. A., Porter J. A., Fishman M. C., Bodenreider C., Diagana T. T., Movva N. R., Winzeler E. A. (2012) Selective and specific inhibition of the Plasmodium falciparum lysyl-tRNA synthetase by the fungal secondary metabolite cladosporin. Cell Host Microbe 11, 654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koh C. Y., Kim J. E., Shibata S., Ranade R. M., Yu M., Liu J., Gillespie J. R., Buckner F. S., Verlinde C. L., Fan E., Hol W. G. (2012) Distinct states of methionyl-tRNA synthetase indicate inhibitor binding by conformational selection. Structure 20, 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ibba M., Soll D. (2000) Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 [DOI] [PubMed] [Google Scholar]
- 16. Nakama T., Nureki O., Yokoyama S. (2001) Structural basis for the recognition of isoleucyl-adenylate and an antibiotic, mupirocin, by isoleucyl-tRNA synthetase. J. Biol. Chem. 276, 47387–47393 [DOI] [PubMed] [Google Scholar]
- 17. Rettig J., Wang Y., Schneider A., Ochsenreiter T. (2012) Dual targeting of isoleucyl-tRNA synthetase in Trypanosoma brucei is mediated through alternative trans-splicing. Nucleic Acids Res. 40, 1299–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. El-Sayed N. M., Myler P. J., Blandin G., Berriman M., Crabtree J., Aggarwal G., Caler E., Renauld H., Worthey E. A., Hertz-Fowler C., Ghedin E., Peacock C., Bartholomeu D. C., Haas B. J., Tran A. N., Wortman J. R., Alsmark U. C., Angiuoli S., Anupama A., Badger J., Bringaud F., Cadag E., Carlton J. M., Cerqueira G. C., Creasy T., Delcher A. L., Djikeng A., Embley T. M., Hauser C., Ivens A. C., Kummerfeld S. K., Pereira-Leal J. B., Nilsson D., Peterson J., Salzberg S. L., Shallom J., Silva J. C., Sundaram J., Westenberger S., White O., Melville S. E., Donelson J. E., Andersson B., Stuart K. D., Hall N. (2005) Comparative genomics of trypanosomatid parasitic protozoa. Science 309, 404–409 [DOI] [PubMed] [Google Scholar]
- 19. Stuart K., Brun R., Croft S., Fairlamb A., Gürtler R. E., McKerrow J., Reed S., Tarleton R. (2008) Kinetoplastids: related protozoan pathogens, different diseases. J. Clin. Invest. 118, 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Renslo A. R., McKerrow J. H. (2006) Drug discovery and development for neglected parasitic diseases. Nat. Chem. Biol. 2, 701–710 [DOI] [PubMed] [Google Scholar]
- 21. Inoue M., Nakamura Y., Yasuda K., Yasaka N., Hara T., Schnaufer A., Stuart K., Fukuma T. (2005) The 14-3-3 proteins of Trypanosoma brucei function in motility, cytokinesis, and cell cycle. J. Biol. Chem. 280, 14085–14096 [DOI] [PubMed] [Google Scholar]
- 22. Redmond S., Vadivelu J., Field M. C. (2003) RNAit: an automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol. Biochem. Parasitol. 128, 115–118 [DOI] [PubMed] [Google Scholar]
- 23. Wirtz E., Leal S., Ochatt C., Cross G. A. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89–101 [DOI] [PubMed] [Google Scholar]
- 24. Schnaufer A., Panigrahi A. K., Panicucci B., Igo R. P., Jr., Wirtz E., Salavati R., Stuart K. (2001) An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291, 2159–2162 [DOI] [PubMed] [Google Scholar]
- 25. Cestari I., Stuart K. (2013) A spectrophotometric assay for quantitative measurement of aminoacyl-tRNA synthetase activity. J. Biomol. Screen. 18, 490–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ledoux S., Uhlenbeck O. C. (2008) 3′-32P labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods 44, 74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bordoli L., Kiefer F., Arnold K., Benkert P., Battey J., Schwede T. (2009) Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 4, 1–13 [DOI] [PubMed] [Google Scholar]
- 28. Trott O., Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nomanbhoy T. K., Schimmel P. (2001) Active site of an aminoacyl-tRNA synthetase dissected by energy transfer-dependent fluorescence. Bioorg. Med. Chem. Lett. 11, 1485–1491 [DOI] [PubMed] [Google Scholar]
- 30. Shoemaker R. H. (2006) The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 6, 813–823 [DOI] [PubMed] [Google Scholar]
- 31. Landry M. L., Booss J., Hsiung G. D. (1982) Duration of vidarabine therapy in biopsy-negative herpes simplex encephalitis. JAMA 247, 332–334 [PubMed] [Google Scholar]
- 32. Rottenberg M. E., Masocha W., Ferella M., Petitto-Assis F., Goto H., Kristensson K., McCaffrey R., Wigzell H. (2005) Treatment of African trypanosomiasis with cordycepin and adenosine deaminase inhibitors in a mouse model. J. Infect. Dis. 192, 1658–1665 [DOI] [PubMed] [Google Scholar]
- 33. Bishop A. C., Nomanbhoy T. K., Schimmel P. (2002) Blocking site-to-site translocation of a misactivated amino acid by mutation of a class I tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 99, 585–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alsford S., Horn D. (2012) Cell cycle-regulated control of VSG expression site silencing by histones and histone chaperones ASF1A and CAF-1b in Trypanosoma brucei. Nucleic Acids Res. 40, 10150–10160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Badjatia N., Ambrósio D. L., Lee J. H., Günzl A. (2013) Trypanosome Cdc2-related kinase 9 controls spliced leader RNA cap4 methylation and phosphorylation of the RNA polymerase II subunit RPB1. Mol. Cell. Biol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morse G. D., Shelton M. J., O'Donnell A. M. (1993) Comparative pharmacokinetics of antiviral nucleoside analogues. Clin. Pharmacokinet. 24, 101–123 [DOI] [PubMed] [Google Scholar]
- 37. Nishihara K., Kanemori M., Yanagi H., Yura T. (2000) Overexpression of trigger factor prevents aggregation of recombinant proteins in Escherichia coli. Appl. Environ. Microbiol. 66, 884–889 [DOI] [PMC free article] [PubMed] [Google Scholar]