Background: The ApbE protein with unknown function is widespread in bacteria.
Results: ApbE catalyzes Mg2+-dependent FMN transfer from FAD to Thr residues of flavoproteins in vitro and in vivo.
Conclusion: ApbE is a novel modifying enzyme involved in the maturation of flavoproteins.
Significance: Broad distribution of ApbE suggests a wide utilization of flavoproteins containing FMN attached via a phospho- ester bond.
Keywords: Bacterial Metabolism, Flavoproteins, FMN, Post-translational Modification, Respiratory Chain, Na+-translocating NADH:Quinone Oxidoreductase
Abstract
Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) contains two flavin residues as redox-active prosthetic groups attached by a phosphoester bond to threonine residues in subunits NqrB and NqrC. We demonstrate here that flavinylation of truncated Vibrio harveyi NqrC at Thr-229 in Escherichia coli cells requires the presence of a co-expressed Vibrio apbE gene. The apbE genes cluster with genes for Na+-NQR and other FMN-binding flavoproteins in bacterial genomes and encode proteins with previously unknown function. Experiments with isolated NqrC and ApbE proteins confirmed that ApbE is the only protein factor required for NqrC flavinylation and also indicated that the reaction is Mg2+-dependent and proceeds with FAD but not FMN. Inactivation of the apbE gene in Klebsiella pneumoniae, wherein the nqr operon and apbE are well separated in the chromosome, resulted in a complete loss of the quinone reductase activity of Na+-NQR, consistent with its dependence on covalently bound flavin. Our data thus identify ApbE as a novel modifying enzyme, flavin transferase.
Introduction
Na+-translocating NADH:quinone oxidoreductase (Na+-NQR)2 is a redox-driven sodium pump that generates a transmembrane difference in electrochemical Na+ potential (1). This enzyme has been shown to operate in the respiratory chain of various bacteria, including several pathogenic microorganisms (2, 3). Na+-NQR consists of six subunits (NqrA–F) (4) encoded by six genes of the nqr operon (5, 6) and has a unique amino acid sequence and set of prosthetic groups, some of which are covalently bound. The enzyme contains one [2Fe-2S] cluster, one noncovalently bound FAD and riboflavin, and two covalently bound FMN residues (1, 7). The [2Fe-2S] cluster and FAD are found in subunit NqrF (8, 9), riboflavin is present in subunit NqrB (10), and covalently bound FMN residues are located in subunits NqrB and NqrC (2, 11).
Bacteria may additionally contain a Na+-NQR homolog, the so-called RNF complex. It has been proposed that RNF catalyzes electron transfer from ferredoxin to NAD+ (12, 13) or methanophenazine (14) and, depending on the direction of the catalyzed redox reaction, can act as a Δ̄μNa+ generator (forward reaction) (13) or consumer (reverse reaction) (12). The RNF complex is also formed by six subunits, five of which are homologous to the corresponding NqrA–E subunits. The RNF subunits RnfD and RnfG (paralogs of NqrB and NqrC, respectively) have been shown to contain covalently bound FMN residues as redox-active prosthetic groups (15).
Several types of bonds between flavins and proteins are known. In Na+-NQR (11), RNF (15), and closely related proteins, such as regulator of NO reductase transcription (NosR) (16) and urocanate reductase (UrdA) (17), FMN is bound by a phosphoester bond through a Thr residue. Other acceptors of flavins in flavoproteins include His, Tyr, and Cys residues that form C-N, C-O, and C-S bonds, respectively, mostly through the 8αC atom of the flavin (e.g., in succinate dehydrogenase, fumarate reductase, sarcosine oxidase, trimethylamine dehydrogenase, and p-cresol methylhydroxylase) (18).
It has generally been thought that these types of covalent bonds between flavin and proteins form in autocatalytic reactions (18, 19). However, flavinylation of succinate dehydrogenase (at His) was recently reported to require a specific protein (SdhE) as an FAD chaperone (20, 21). Heterologous expression of the Vibrio cholerae nqrC gene in Escherichia coli results in production of a flavin-deficient apo-form of NqrC (22), also suggesting that some unknown protein factor, absent in E. coli, is required for NqrC flavinylation. Similar results have been recently obtained for Shewanella oneidensis urdA expressed in E. coli (17). Although Na+-NQR subunits NqrB and NqrC and related FMN-binding proteins have completely different overall primary and tertiary structures (23), they contain a common pattern around the FMN-carrying Thr residue (Fig. 1) reminiscent of a motif for a post-translational modification. In the present work, we identified the missing protein factor that recognizes this pattern and present evidence that it is a novel specific enzyme, flavin transferase.
FIGURE 1.

Sequence alignment of FMN-binding motifs in several proteins. Vh_NqrB and Vh_NqrC, V. harveyi Na+-NQR subunits NqrB and NqrC (accession numbers Q9RFW0 and Q9RFV9, respectively); Vc_RnfD and Vc_RnfG, V. cholerae RNF subunits RnfD and RnfG (ACP09041 and ACP09040, respectively); Pd_NosR, P. denitrificans regulator of NO reductase transcription NosR (Pden_4220); So_UrdA, S. oneidensis MR-1 urocanate reductase (NP_720136). The position of the experimentally identified FMN acceptor Thr residue (11, 15) is marked by an asterisk.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Growth, and Media Composition
The bacterial strains used in this study are listed in Table 1. E. coli and Klebsiella pneumoniae cells were routinely grown in LB medium at 37 °C. K. pneumoniae cells were grown in the presence of the antibiotics ampicillin at 150 μg/ml, tetracycline at 3.3 μg/ml, kanamycin at 100 μg/ml, and chloramphenicol at 40 μg/ml. The concentrations of these antibiotics used for E. coli were 100, 10, 50, and 20 μg/ml, respectively. All genetic manipulations in E. coli were carried out using the XL1-Blue and SM10λpir strains.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
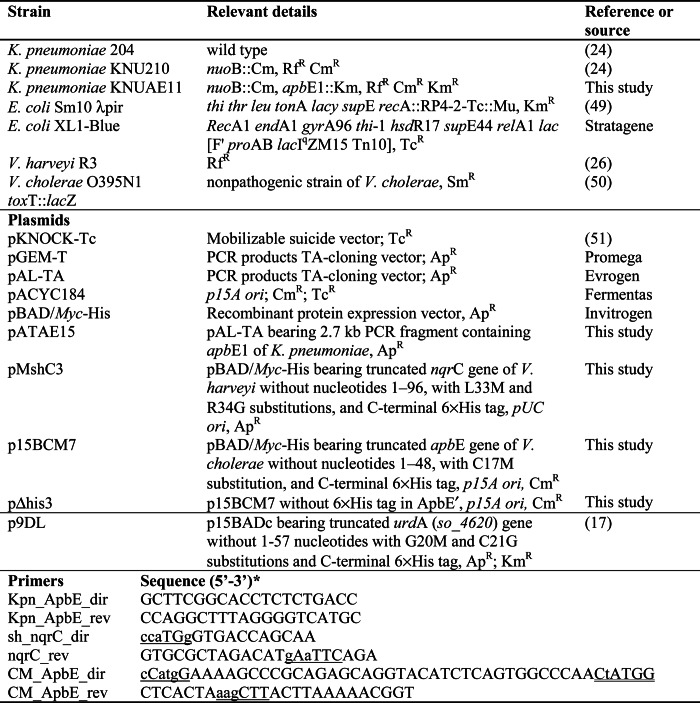
* The nucleotides substituted in mutagenesis experiments are designated by lowercase letters, artificially introduced restriction sites are underlined, and a corrupted intrinsic NcoI site is double underlined.
Construction of Expression Vectors
The gene sequence encoding the truncated NqrC subunit, NqrC′ (a soluble variant of NqrC without its N-terminal transmembrane α-helix), was amplified by PCR with Taq polymerase and primers sh_nqrC_dir and nqrC_rev (see Table 1) using the genomic DNA of Vibrio harveyi as a template. The amplified 706-bp fragment was cloned into the pGEM-T vector (Promega), resulting in the pSHC221 plasmid. The NcoI-EcoRI fragment of this plasmid was subcloned into the pBAD/Myc-His A vector (Invitrogen) treated with the NcoI and EcoRI restriction enzymes, resulting in plasmid pMshC3. This plasmid was used to produce truncated NqrC subunit with a C-terminal His6 tag.
An expression vector for C-terminally His6-tagged ApbE protein devoid of its leader sequence (ApbE′) was constructed by amplifying the DNA fragment of apbE without its leader sequence by PCR with a Tersus PCR kit and the primers CM_ApbE_dir and CM_ApbE_rev (see Table 1) using the genomic DNA of V. cholerae as a template. The resulting 966-bp fragment was cloned into the pAL-TA vector (Evrogen), yielding the pAL_CM14 plasmid. The NcoI-HindIII fragment of this plasmid was subcloned into the pBAD/Myc-His A vector digested by the same restriction nucleases, resulting in plasmid pB_CM1. In addition, the part of this plasmid containing pUC ori and ApR was replaced by the EcoRV-BsaBI fragment of the pACYC184 vector containing p15A ori and CmR, resulting in plasmid p15BCM7.
The plasmid pΔhis3, bearing the gene encoding ApbE′ protein without the His6 tag, was produced by treating the p15BCM7 plasmid with HindIII and self-ligating the resulting 4805-bp fragment. This procedure resulted in removal of the 753-bp fragment containing the His6 tag sequence and creation of a stop codon in the proper place in the apbE′ gene.
Construction of an ApbE1-deficient K. pneumoniae Strain
The K. pneumoniae genome contains two genes encoding ApbE-like proteins, ApbE1 (YP_002237370) and ApbE2 (YP_002238733) (Fig. 2). Because ApbE1 is more similar to ApbE-like proteins from other Gammaproteobacteria (see below), its gene was used as the target for mutagenesis. The DNA fragment containing the apbE1 gene was amplified by PCR with Taq polymerase and primers Kpn_ApbE_dir and Kpn_ApbE_rev (see Table 1) using the genomic DNA of K. pneumoniae 204 as a template. The amplified 2.7-kb fragment was cloned into the pAL-TA vector, resulting in the pAL_A8 plasmid. A kanamycin resistance cassette was inserted into the EcoRV site of the apbE1 gene in pAL_A8, and a Km-containing plasmid (pALAK7) bearing the apbE1 gene together with the unidirectionally transcribing kanamycin resistance cassette was selected. The apbE1::Km fragment from pALAK7 was subcloned into the suicide vector pKNOCK-Tc, resulting in pKn_ApbE1::Km3. This plasmid was transferred into the K. pneumoniae KNU210 (nuoB::Cm) (24) strain via conjugation using E. coli SM10λpir as the donor, and a TcS KmR CmR phenotype clone (KNUAE11 nuoB::Cm, apbE1::Km) characteristic of a double-crossover-introduced mutation was selected. Proper localization of the mutation in the K. pneumoniae chromosome was verified by PCR analysis. The disruption of apbE1 did not result in a thiamine auxotrophic phenotype in the K. pneumoniae KNUAE11 strain when tested in M9 medium supplemented with glucose, as described previously for Salmonella enterica (25).
FIGURE 2.
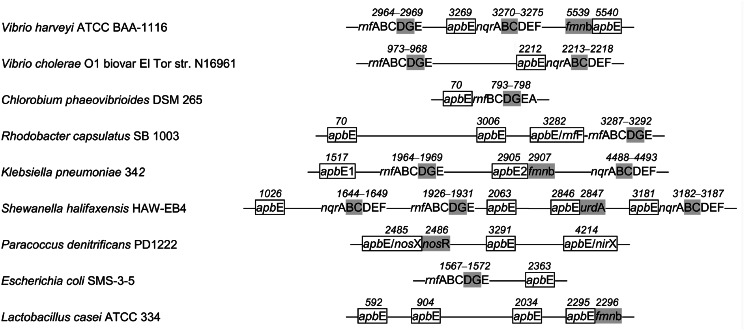
Typical arrangements of genes for ApbE, Na+-NQR, RNF, and other FMN-binding proteins in bacterial chromosomes. Genes for Na+-NQR and RNF subunits are indicated by the last letters in their standard designations. apbE genes together with their accepted alternative designations are shown as rectangles, genes for FMN-binding Na+-NQR and RNF subunits, UrdA, NosR, and other FMN-binding proteins (fmnb) are indicated by gray shading. The connecting lines refer to intercalating sequences. The numbers above the genes indicate their numbers in genomes according to the Kyoto Encyclopedia of Genes and Genomes. The genes were detected as hits in a BLAST search of complete prokaryotic genomes using the V. harveyi proteins as the queries. Genes for FMN-binding proteins were identified using search for Pfam motifs PF04205 and PF03116.
Complementation of K. pneumoniae KNUAE11 Strain with Plasmid-coded ApbE1 Protein
The apbE1-containing DNA fragment was amplified by PCR with the high fidelity Tersus PCR kit (Evrogen) and primers Kpn_ApbE_dir and Kpn_ApbE_rev (Table 1) using the genomic DNA of K. pneumoniae 204 as a template. The amplified 2.7-kb fragment was cloned into the pAL-TA vector, resulting in the pATAE15 plasmid. K. pneumoniae KNUAE11 cells were transformed with the pATAE15 plasmid using electroporation.
Preparation of Membrane Vesicles from K. pneumoniae Cells
Na+-NQR content was increased by growing K. pneumoniae cells in M9 medium supplemented with sodium succinate as the sole energy and carbon source (26). The cells were harvested by centrifugation (10,000 × g, 10 min) and washed twice with medium 1 (100 mm KCl, 10 mm Tris-HCl, 5 mm MgSO4, pH 8.0). The cell pellet was suspended in medium 2 (10 mm HEPES-Tris, 5 mm MgSO4, 50 mm KCl, pH 8.0), and the suspension was passed through a French press (16,000 p.s.i.). Undamaged cells and cell debris were removed by centrifugation at 22,500 × g (10 min), and the supernatant was further centrifuged at 180,000 × g (60 min). The membrane pellet was suspended in medium 2 (at 20–30 mg of protein ml−1) and immediately used for activity measurements.
Activity Assays
NADH and dNADH oxidation by membrane vesicles isolated from K. pneumoniae strains were measured at 30 °C using a Hitachi 557 spectrophotometer at 340 nm. The reaction medium contained 20 mm HEPES-Tris, 5 mm MgSO4, and 100 mm KCl or NaCl (pH 8.0). For measurement of dNADH:menadione oxidoreductase activity, the reaction medium was supplemented with 50 μm menadione. The extinction coefficient, ϵ340, of 6.22 mm−1 cm−1 was used for NADH and dNADH quantitation.
Isolation of Recombinant His6-tagged NqrC′, ApbE′, and UrdA Proteins
For nqrC′, apbE′, or urdA induction, E. coli cells bearing the appropriate plasmid(s) were grown at 32 °C to mid-exponential phase (A600 = 0.3–0.4), after which the growth medium was supplemented with 0.2% (w/v) l-arabinose, and cells were grown for additional 3 h. The cells were harvested by centrifugation (10,000 × g, 10 min) and washed twice with medium containing 300 mm NaCl, 10 mm Tris-HCl, and 5 mm MgSO4 (pH 8.0). The cell pellet was suspended in medium containing 300 mm NaCl, 20 mm Tris-HCl, 5 mm MgSO4, 1 mm phenylmethylsulfonyl fluoride, and 5 mm imidazole HCl (pH 8.0), and the suspension was passed twice through a French press (16,000 p.s.i.). Cell debris and membrane vesicles were removed by centrifugation at 180,000 × g (60 min). His6-tagged proteins were purified from the supernatant using affinity chromatography. This was accomplished by loading the supernatant onto a nickel-nitrilotriacetic acid column equilibrated with solution A containing 300 mm NaCl, 10 mm Tris-HCl, and 5 mm imidazole HCl (pH 8.0); washing the column twice, first with solution A containing 10 mm imidazole HCl and then with solution A containing 20 mm imidazole HCl; and eluting His6-tagged proteins with solution A containing 100 mm imidazole HCl. The protein obtained was concentrated and kept frozen at −80 °C until use. Protein concentration was determined by the bicinchoninic acid method (27) using bovine serum albumin as a standard.
Extraction of Flavins from holoNqrC′
Covalently and noncovalently bound flavins in the holoNqrC′ protein were assayed as described by Casutt et al. (28). The protein (3 mg) was precipitated with 7.5% (v/v) TFA, sedimented by centrifugation, and washed with H2O (0 °C). The supernatants were combined, neutralized by adding 0.8 m K2HPO4, and passed through a Microcon YM-10 centrifugal filter (Millipore). The filtrate was used for determination of flavins noncovalently bound to holoNqrC′.
Covalently bound flavins were released from the protein pellet by alkaline hydrolysis. To this end, an ice-cold 0.5 m LiOH solution (0.2 ml) was added to the pellet, and the mixture was vortexed and incubated on ice for 24 h. After the incubation, the suspension was neutralized by adding 1 m MES and passed through a Microcon YM-10 centrifugal filter. The filtrate was subjected to reverse phase HPLC using a Prontosil 120–5 C18 column (75 × 2 mm). A linear gradient of methanol (total sweep from 10 to 70% (v/v)s, 25 min) in 5 mm MES-Tris (pH 6.0) with the flow rate of 0.1 ml/min was used. Flavins were detected at 360 nm.
Mass Spectrometry
Mass spectra were recorded on an Ultraflex Extreme MALDI-TOF/TOF mass spectrometer (Bruker Daltonics) equipped with a neodymium laser. Aliquots of the sample were mixed on a steel target with a solution of 2,5-dihydroxybenzoic acid (20 mg/ml) in 30% (v/v) acetonitrile (Aldrich) with 0.5% (v/v) TFA. The [MH]+ molecular ions were analyzed in linear (proteins) or reflector (peptides) mode; the values of m/z were accurate to 30 ppm. Fragment ion spectra were obtained in Lift mode with a mass accuracy of better than 1 Da.
In Vitro Flavinylation of apoNqrC′ Protein
The apoNqrC′ protein (4 mg/ml) was incubated for 45 min at 30 °C in medium containing 100 mm NaCl, 0.1 mm EDTA, 1 mm FAD or FMN, 10 mm Tris-HCl (pH 8.0) in the presence or absence of 0.1 mg/ml ApbE′ and/or 5 mm MgSO4. After the incubation, the reaction mixtures were separated by SDS-PAGE.
Electrophoresis
SDS-PAGE was performed using 12.5% (w/v) polyacrylamide gels (29). Covalently bound flavins were detected by photographing gels under UV illumination with a GelDoc-It M-26XV gel documentation system using a SYBR Green emission filter.
RESULTS
Genomic Context Analysis of the nqr and rnf Operons
Bacterial genes are known to often cluster according to their functions (30), a feature that facilitates identification of functionally coupled proteins (31). Genome context analysis of the nqr operons in marine bacteria of the genus Vibrio, a popular source of Na+-NQR, revealed the gene apbE, encoding the putative “alternative pyrimidine biosynthesis” protein ApbE, immediately downstream of the nqr operon in all Vibrio species with known genome sequences (Fig. 2). In contrast to this conservation at downstream positions, upstream positions are occupied by a variety of genes. This same regularity holds throughout the entire set of ∼1600 sequenced bacterial genomes, most of which, primarily proteobacteria, contain the apbE gene adjacent to the nqr operon. In the genomes of Na+-NQR-containing bacteria of the Chlamydiae/Verrucomicrobia group, Deferribacteres, Fusobacteria, Spirochaetes, Thermotogae, and some enterobacteria (e.g., K. pneumoniae), the apbE gene is also invariably present, but at more distant positions from the nqr operon (Fig. 2). Thus, all nqr-containing bacterial genomes contain the apbE gene, which is most commonly adjacent to the nqr operon.
Similarly, the genomes of bacteria containing a homologous RNF complex with covalently bound FMN residues as prosthetic groups (15) always carry an apbE gene (Fig. 2), which is frequently located adjacent to the rnf operon; sometimes proteins containing an ApbE-like domain are even designated RnfF (12). We found only one exception to this rule: some species of the genus Buchnera (obligate endosymbionts of aphids) possess the full rnf operon, but their apbE genes contain an authentic frameshift. The nqr and rnf operons are thus genetically coupled to apbE, suggesting a functional link between the Na+-NQR (RNF) and ApbE proteins. Even when an apbE-like gene is clustered in bacterial genomes with genes for proteins other than NqrB, NqrC, RnfD, and RnfG, their encoded proteins are also often shown or predicted to contain a covalently bound FMN residue. Such examples include the FMN-containing NosR protein (Pden_4220) and ApbE-like protein NosX (Pden_4214) of Paracoccus denitrificans; the presumptive FMN-containing UrdA-like urocanate reductases (Shal_2847, Ssed_0694, and ZP_02156482) and ApbE-like proteins (Shal_2846, Ssed_0695, and ZP_02156483) of Shewanella halifaxensis, Shewanella sediminis, and Shewanella benthica KT99, respectively; and the FAD/FMN-binding/flavocytochrome c (KPK_2907) and ApbE-like protein (KPK_2905) of K. pneumoniae (Fig. 2). In addition, there are several examples in genomic databases where the genes of an ApbE-like protein and a putative FMN-binding protein are fused, resulting in one elongated gene (CAJ73234, ZP_03630453, and ZP_10101304).
ApbE Catalyzes Flavinylation of NqrC When Both Are Co-produced in E. coli Cells
The above analysis suggested that the ApbE protein is the sought-after “flavin-attaching” enzyme. To test this hypothesis, we measured the effect of co-expressing V. harveyi nqrC and V. cholerae apbE in E. coli cells on flavin incorporation into NqrC.
As originally reported by Barquera et al. (22), heterologous expression of the V. cholerae nqrC gene in E. coli cells results in the production of flavin-deficient NqrC. A similar result was obtained in the present work using a truncated V. harveyi NqrC (NqrC′) protein in which the N-terminal transmembrane α-helix was eliminated by genetic manipulations to make NrqC water-soluble. The NqrC′ protein isolated from E. coli/pMshC3 cells showed no bands characteristic of bound flavin in absorption spectra (Fig. 3A, red line), and no fluorescent band was detected in SDS-PAGE analyses (Fig. 4A, lane 1).
FIGURE 3.
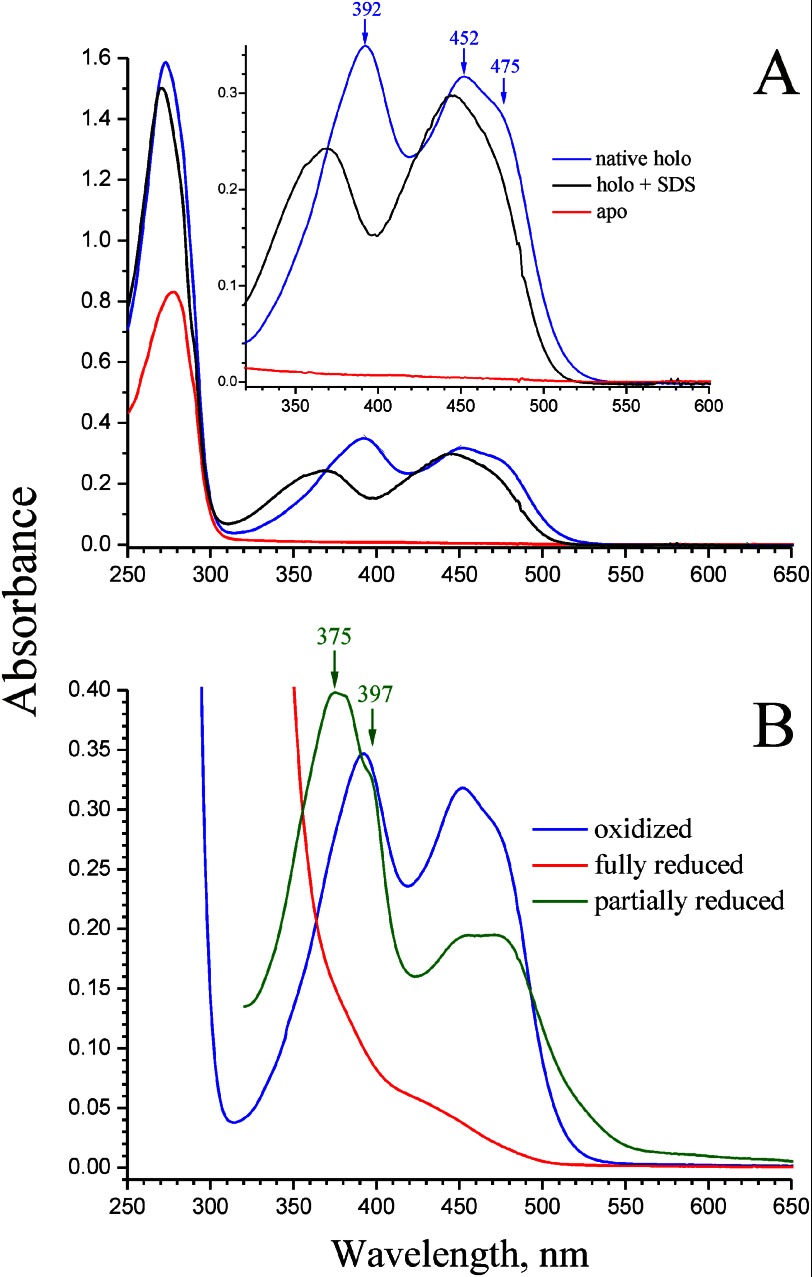
Absorption spectra of recombinant NqrC′ protein of V. harveyi. A, red line, NqrC′ isolated from E. coli/pMshC3 strain (nqrC′ only); blue line, NqrC′ isolated from E. coli/pMshC3 + pΔhis3 strain (nqrC′ + apbE′); black line, NqrC′ isolated from E. coli/pMshC3 + pΔhis3 strain and denaturated with 1% (w/v) SDS. Inset, magnified spectra in the 325–600 nm region. B, HoloNqrC′ purified from E. coli/pMshC3 + pΔhis3 strain. Blue line, air oxidized holoNqrC′ (as isolated); red line, holoNqrC′ fully reduced by excess dithionite; green line, partially reduced holoNqrC′ obtained by slow oxidation of the fully reduced protein by air. Peak labels refer to curves of the same color. The assay medium contained 0.81 mg/ml protein, 20 mm Tris-HCl (pH 8.0), and 50 mm NaCl.
FIGURE 4.
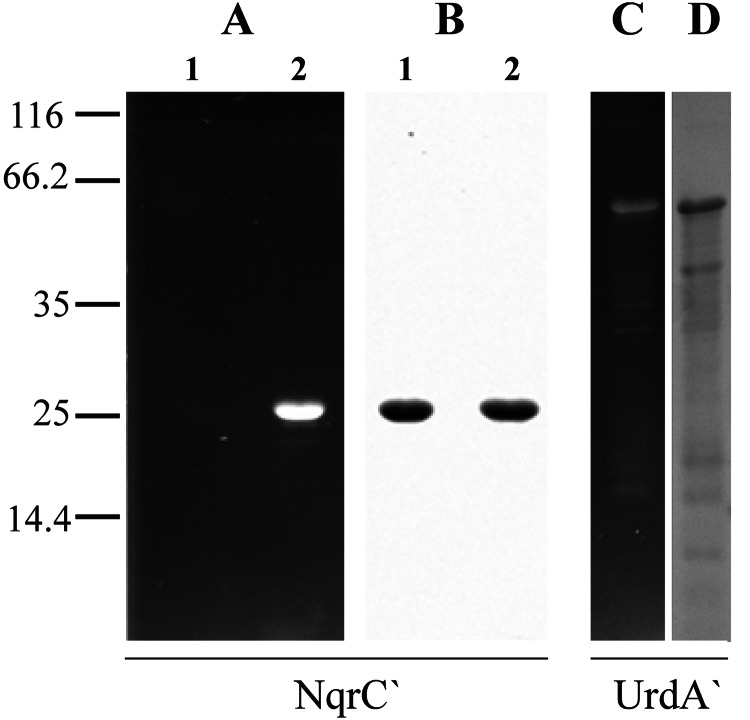
SDS-PAGE analysis of recombinant NqrC′ and UrdA′ proteins purified from different E. coli strains. A and C, unstained gels under UV illumination; B and D, Coomassie Blue-stained gels. Lanes 1 and 2 are NqrC′ isolated from E. coli/pMshC3 strain (nqrC′ only) and E. coli/pMshC3 + pΔhis3 strain (nqrC′ + apbE′), respectively. The UrdA′ protein was purified from E. coli/p9DL + pΔhis3 strain (urdA′ + apbE′). Protein load was 10 μg/lane for NqrC′ and 5 μg/lane for UrdA′. The fluorescent bands seen in A and C correspond to flavin-bound NqrC′ and UrdA′, respectively. The bars with numbers on the left side denote the positions and molecular masses of marker proteins.
To construct an E. coli cell line co-expressing V. harveyi nqrC′ and V. cholerae apbE, we introduced a second plasmid (pΔhis3) containing a gene encoding a cytoplasmic variant of V. cholerae ApbE lacking its leader sequence, ApbE′, into E. coli/pMshC3. Upon induction of simultaneous synthesis of NqrC′ and ApbE′, the cells became yellow-colored. Purification of NqrC′ from these cells yielded a bright yellow protein, which was detected as a ∼27-kDa fluorescent band (calculated mass of the His6-tagged NqrC′ protein, 27.5 kDa) in SDS-polyacrylamide gels (Fig. 4A, lane 2), confirming formation of holoNqrC′. Similar results were obtained for another FMN-containing protein, urocanate reductase UrdA, from S. oneidensis MR-1, which was produced as a flavinylated protein when co-expressed with the ApbE-encoding plasmid pΔhis3 (Fig. 4C) but was detected as an apoprotein when expressed alone in E. coli (17).
The absorption spectrum of the presumed holoNqrC′ (Fig. 3A, blue line) exhibited three peaks with maxima at 273, 392, and 452 nm, and a shoulder ∼475 nm. All three peaks were significantly red-shifted compared with the spectra of free flavin in water solutions (32), indicating strong interaction of the flavin isoalloxazine moiety with NqrC′. Interestingly, the short wavelength band of the spectrum (392 nm) was more intense than the longer wavelength band (452 nm), which is not typical for flavoproteins. A nearly identical spectrum was reported for RnfG, an NqrC paralog from V. cholerae (15), suggesting that such a perturbation in the spectrum may be an intrinsic property of NqrC-like flavoproteins. Assuming that the extinction coefficient, ϵ445, of flavin in holoNqrC′ denaturated by SDS (1% (w/v), 5-min incubation at 80 °C; Fig. 3A, black line) is equal to that of FMN in solution (12.5 mm−1 cm−1 (32)), the value of ϵ452–600 = 13.4 mm−1 cm−1 for native holoNqrC′ can also be obtained from the data in Fig. 3A. The spectrum of fully reduced holoNqrC′ showed no contribution from cytochromes or FeS proteins, indicating the spectral purity of the preparation (Fig. 3B, red line). The spectrum of partially reduced holoNqrC′ contained some additional components, namely an intense peak at 375 nm with a shoulder ∼397 nm and a broad band ∼500 nm (Fig. 3B, green line). These findings suggested that an anionic form of flavosemiquinone (33) is stabilized in holoNqrC′, in accordance with data for the authentic NqrC subunit within the full Na+-NQR complex (34–36).
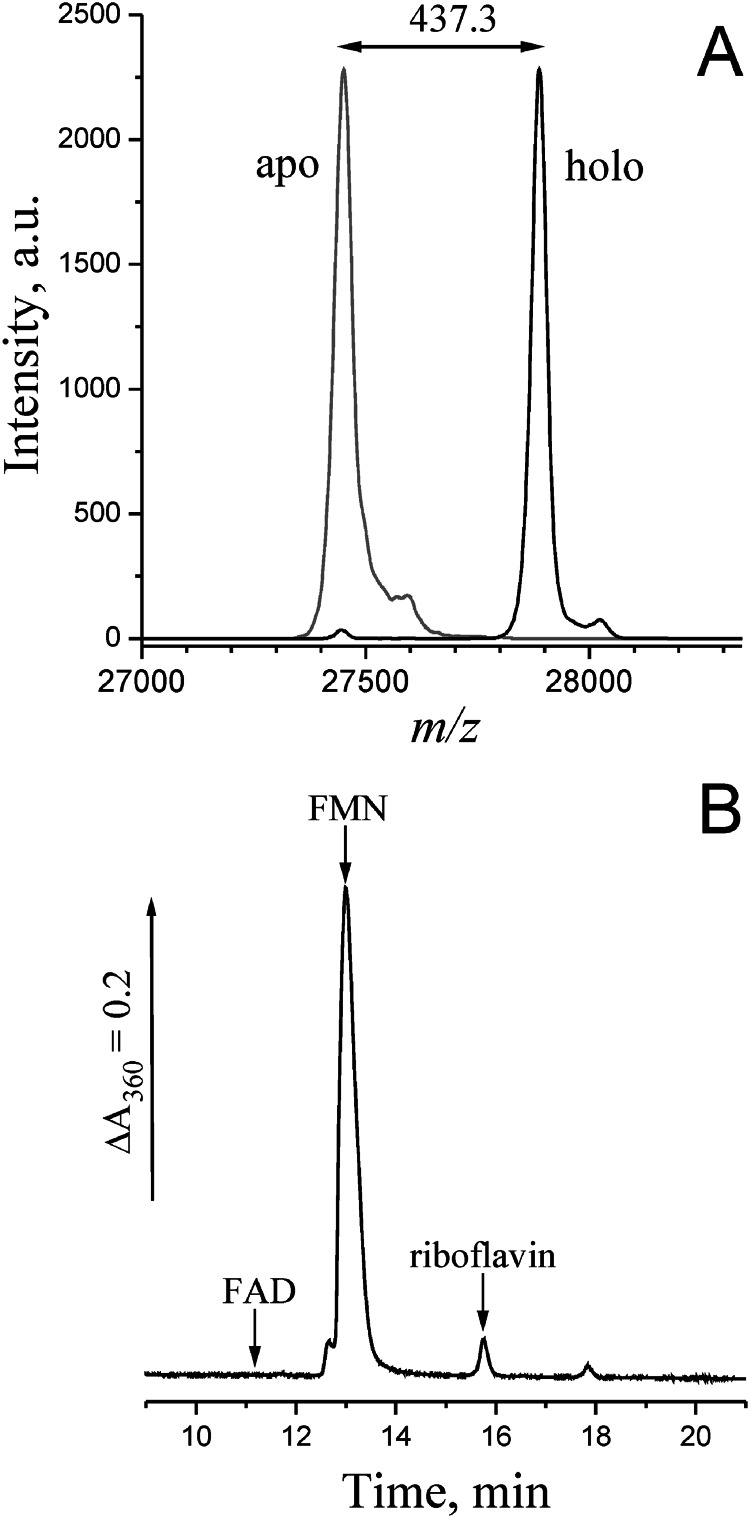
The flavin content of holoNqrC′ was determined in two different ways. First, the masses of the major protein peaks seen in mass spectra (Fig. 5A) differed by 437 Da between the apo- and holoNqrC′, which compares well with the mass of an FMN residue (438.3 Da). Thus, the spectra indicated that a single FMN residue was attached per NqrC′ molecule with a nearly quantitative yield. Second, the amounts of covalently and noncovalently bound flavins were semiqualitatively estimated using a classical acid/alkali treatment procedure (28). The NqrC′ protein obtained in co-expression experiment was precipitated with 7.5% (v/v) TFA and sedimented by centrifugation. Essentially no flavins were detected in the supernatant, indicating that this protein does not contain flavins bound noncovalently or by acid-labile bonds. Subsequent treatment of the protein pellet with LiOH caused a complete release of bound flavin into solution, consistent with its being bound to NqrC′ by a phosphoester bond (28). HPLC analysis of the alkaline extract revealed FMN (97%), traces of riboflavin (3%), and no FAD (Fig. 5B).
FIGURE 5.
Mass spectral and chromatographic identification of the flavin prosthetic group in NqrC′. A, MALDI mass spectra of apoNqrC′ (gray line) and holoNqrC′ (black line). The difference in the molecular masses corresponds to the mass of an FMN residue (438.3 Da). B, HPLC separation of flavins extracted from holoNqrC′ by LiOH treatment. The arrows indicate the retention times for FAD, FMN, and riboflavin standards.
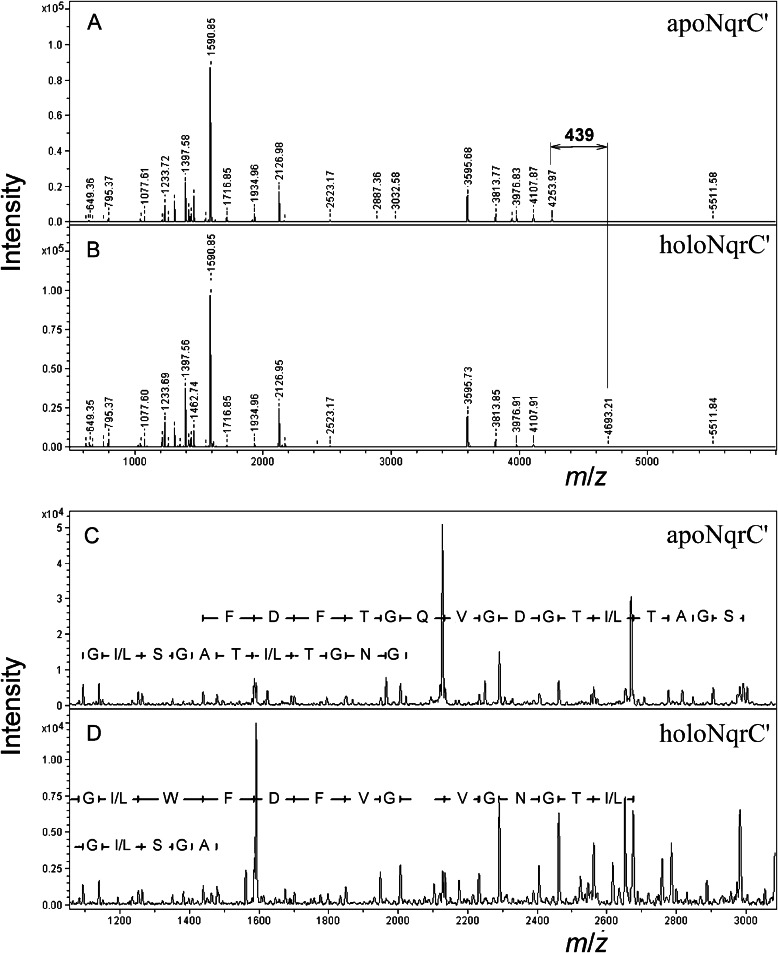
The site of the modification in NqrC′ was identified by mass spectral analysis of the tryptic digests of both NqrC′ proteins. There was only one signal, with an m/z of 4254 in apoNqrC′, that was shifted to an m/z of 4693 in holoNqrC′ (Fig. 6, A and B). This signal corresponded to the peptide occupying positions 212–254 (GGAPEGSEHGVDGLSGATLTGNGVQGTFDFWLGDMGFGPFLAK). The position of the FMN residue within this peptide was determined by analysis of the fragmentation spectra of modified and unmodified peptide variants (Fig. 6, C and D). With the unmodified peptide, a complete series of expected N-terminal (b-ions) and C-terminal (y-ions) fragments was observed, whereas the series ended at Thr-229 in the spectrum of the flavinylated peptide, identifying Thr-229 as the site of the modification.
FIGURE 6.
MALDI mass spectral identification of the modification site in NqrC′. A and B, tryptic digests of apoNqrC′ and holoNqrC′, respectively. C and D, MS/MS spectra of the selected tryptic peptide of apoNqrC′ (m/z = 4254) and holoNqrC′ (m/z = 4693), respectively. The deduced protein sequences are shown.
These data show that ApbE is indeed required for flavin incorporation in NqrC and that the binding mode of the incorporated flavin is identical to that in authentic NqrC isolated from Vibrio alginolyticus or V. cholerae cells (11, 28). Interestingly, the E. coli genome contains its own apbE-like gene (Fig. 2) and, presumably, the corresponding ApbE protein. The inability of E. coli proteins to flavinylate V. cholerae (22), S. oneidensis MR-1 (17), and V. harveyi NrqC proteins suggested that ApbE proteins are species-specific.
Isolated ApbE Can Flavinylate apoNqrC′ in vitro
To test whether other cellular factors are required for NqrC flavinylation, we carried out the reaction with isolated V. cholerae ApbE′ and flavin-free V. harveyi NqrC′. The genes for both proteins were separately expressed in E. coli, and the produced proteins were isolated by affinity chromatography.
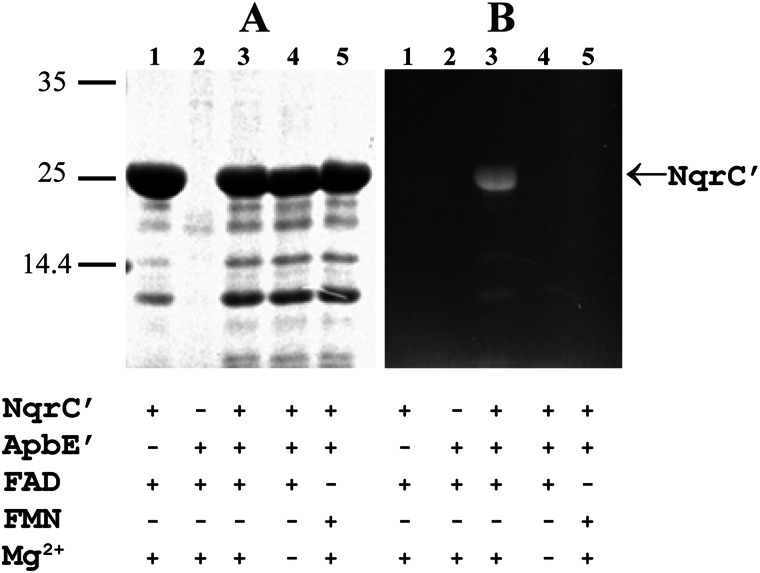
Remarkably, incubation of apoNqrC′ with ApbE′ (at a molar ratio of 55:1) for 45 min in the presence of FAD and Mg2+ resulted in the appearance of a ∼27-kDa fluorescent band on gels after SDS-PAGE of the reaction mixture (Fig. 7B, lane 3), indicative of covalent bond formation between NqrC′ and flavin (2, 21). Using the holoNqrC′ protein purified from the E. coli/pMshC3 + pΔhis3 strain as a fluorescent band standard, we were able to estimate that the yield of flavinylated NqrC′ upon incubation with ApbE′ was ∼20%. Each molecule of ApbE′ thus produced ∼10 molecules of holoNqrC′. In contrast, incubation of apoNqrC′ in the same medium but without ApbE did not produce holoNqrC′ (Fig. 7B, lane 1). Hence, the flavinylation reaction requires the presence of ApbE and is therefore not an autocatalytic process. The data in Fig. 7B (lanes 4 and 5) additionally indicate that flavinylation did not occur without Mg2+ or when FAD was replaced by FMN. An obvious corollary is that ApbE acts as a Mg2+-dependent FAD:protein FMN transferase.
FIGURE 7.
In vitro flavinylation of apoNqrC′. The apoNqrC′ protein (4 mg/ml) was incubated in medium containing ApbE′ (0.1 mg/ml), MgSO4 (5 mm), and flavin (1 mm), as indicated under the gel. The reaction products (5 μl) were separated by SDS-PAGE. A, Coomassie-stained gel. B, unstained gel under UV illumination. The bars with numbers on the left side refer to the positions and molecular masses of marker proteins.
Activities of the Na+-NQR Complex in an ApbE1-deficient Strain of K. pneumoniae
The above data clearly demonstrated that ApbE can flavinylate apoNqrC′ in vitro and in recombinant E. coli cells. To verify the physiological significance of this activity with a full-length apoNqrC that allows formation of a complete Na+-NQR complex in authentic cells, we constructed a mutant strain of K. pneumoniae with a disrupted apbE1 gene (YP_002237370) and measured the activities of Na+-NQR produced by this strain. Na+-NQR exhibits two main enzymatic activities in vitro: quinone reductase and NADH dehydrogenase. Quinone reductase activity is coupled with the transfer of Na+ across the membrane, is specifically activated by sodium ions, is inhibited by low HQNO concentrations, and is observed only with a complete form of the Na+-NQR complex (37). NADH dehydrogenase activity results in a single-electron reduction of soluble quinones (menadione, for example) and other artificial electron acceptors (38), does not depend on the concentration of sodium ions, is resistant to low HQNO concentrations, and is not energy-coupled. Only the FAD-binding domain of the NqrF subunit is apparently required for this activity (8, 9).
There are several reasons that K. pneumoniae KNU210 (24) is the strain of choice for mutagenesis of apbE. First, the nqr operon and apbE1 are separated by 2,500,000 bp in the K. pneumoniae chromosome, making a polar effect of an apbE1 lesion on the nqr operon unlikely (Fig. 2). Second, Na+-NQR is the sole dNADH (reduced nicotinamide hypoxanthine dinucleotide)-oxidizing complex of the respiratory chain in the K. pneumoniae KNU210 strain because of disruption of the nuoB gene encoding a subunit of H+-translocating NADH:quinone oxidoreductase (NDH-1) (24). The other NADH:quinone oxidoreductase (NDH-2) of K. pneumoniae oxidizes only NADH, and not dNADH (39), whereas Na+-NQR acts on both substrates (40). This allowed us to estimate quinone reductase and NADH dehydrogenase activities of Na+-NQR in K. pneumoniae KNU210 membrane vesicles from measurements of dNADH oxidase and dNADH:menadione oxidoreductase activities, respectively (24, 40).
As Table 2 makes clear, the inactivation of apbE1 in K. pneumoniae strain KNUAE11 only slightly decreased the dNADH:menadione oxidoreductase activity of membrane vesicles, whereas the Na+-stimulated and HQNO-sensitive dNADH oxidase activity was completely lost. NDH-2-linked respiration (the difference between the NADH and dNADH oxidase activities) was not affected by apbE1 deletion (Table 2), indicating that complexes of the respiratory chain other than Na+-NQR were not affected in the ΔapbE1 K. pneumoniae strain. Importantly, introduction of a plasmid bearing the K. pneumoniae apbE1 gene (pATAE15) into the KNUAE11 strain resulted in a complete recovery of dNADH oxidase activity (Table 2). This finding ruled out the possibility of any polar effects of the apbE1 lesion. These experiments thus demonstrated that ApbE1 is necessary for Na+-NQR to exhibit quinone reductase activity, which requires covalently bound flavins, but not NADH dehydrogenase activity, which has no such requirement.
TABLE 2.
Rates of NADH and dNADH oxidation by membrane vesicles isolated from different K. pneumoniae strains
| Strain | Activity |
||
|---|---|---|---|
| dNADH oxidase | dNADH:menadione oxidoreductase | NADH oxidase | |
| nmol dNADH oxidized min−1 mg−1 protein | |||
| K. pneumoniae KNU210 (nuoB::Cm) | 125a | 255 | 235 |
| K. pneumoniae KNUAE11 (nuoB::Cm apbE1::Km) | 1.6b | 170 | 145 |
| K. pneumoniae KNUAE11 | 100a | 270 | 250 |
| (nuoB::Cm apbE1::Km)/pATAE15 (apbE1) | |||
a Activity was stimulated by sodium ions and inhibited by HQNO (4 μm).
b Activity was not stimulated by sodium ions or inhibited by HQNO (4 μm).
DISCUSSION
Covalently bound flavin is a common redox-active prosthetic group that can be attached to proteins by different bond types, yet the mechanisms of the post-translational modification remain largely unknown. In many flavoproteins, flavin is attached via a bond involving the 8αC of the flavin residue and the N1 or N3 of a protein His residue (18). The widely accepted view has been that this bond forms by an autocatalytic reaction (i.e., the modification does not require additional protein factors). However, it has been recently demonstrated that this type of flavin attachment to succinate dehydrogenase requires the presence of an SdhE chaperone protein (20, 21).
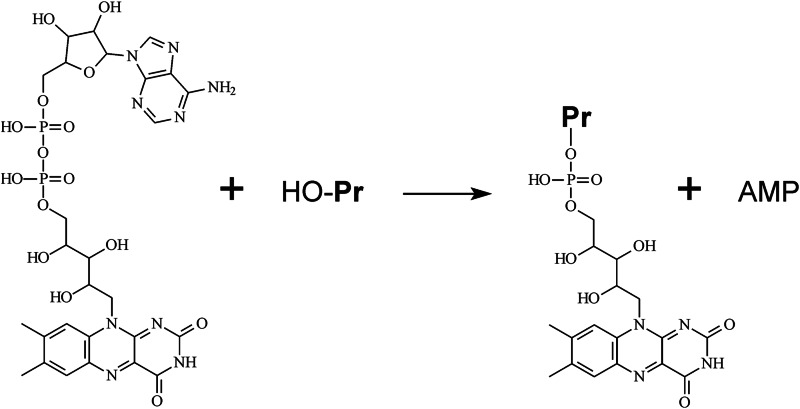
Here we report that formation of a different type of flavin-protein bond, a phosphoester bond involving a protein Thr residue, requires the presence of a specific enzyme, ApbE. The apbE gene was originally identified in a study of the genes involved in thiamine synthesis in S. enterica. Lesions in apbE resulted in a conditional thiamine auxotrophy in this bacterium (41). The product of the apbE gene, ApbE, was found to be a lipoprotein anchored to the periplasmic side of the inner membrane in S. enterica (25). The principal difference between ApbE and SdhE is that the latter delivers a complete FAD molecule to the target protein, whereas ApbE catalyzes transfer of only the FMN moiety and thus acts as an Mg2+-dependent flavin transferase (Fig. 8). ApbE is not homologous to SdhE, emphasizing the different flavinylation mechanisms utilized by these proteins.
FIGURE 8.
The reaction catalyzed by ApbE. HO-Pr, the protein to be modified.
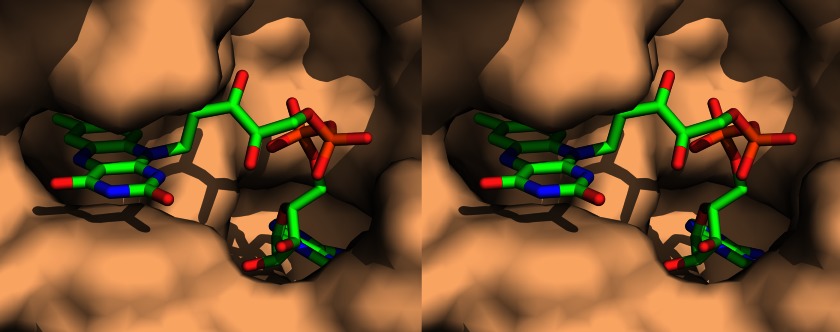
Quite recently, the three-dimensional structures of four ApbE-like proteins have been solved (Protein Data Bank codes 1VRM, 2O18, 2O34, and 3PND) (42, 43). Remarkably, one of the structures (S. enterica ApbE) contains a FAD molecule bound to a unique flavin-binding motif that was not previously observed in flavoproteins (43). The ability of ApbE to bind FAD is fully consistent with the proposed function of this protein as a flavin transferase. The adenosine and flavin moieties of FAD are embedded in the protein globule in the ApbE-FAD complex, whereas the pyrophosphate group of FAD is accessible to attack by an acceptor protein Thr (Fig. 9). Notably, FAD does not bind to S. enterica ApbE very tightly, and the major fraction of the complex dissociates during its purification (43). This is expected behavior for FAD as a substrate, rather than a prosthetic group, of ApbE.
FIGURE 9.
A stereo view of the FAD-binding site in S. enterica ApbE (Protein Data Bank code 3PND) (43). ApbE is shown as a surface model (probe radius, 1.4 Å), and FAD is depicted as a stick model. The pyrophosphate group of FAD is shown in orange. The figure was created with PyMOL (PyMOL Molecular Graphics System, version 1.5.0.4; Schrödinger, LLC).
The catalytic activity of ApbE described in the present work is in good agreement with previously determined phenotypes of mutant strains of different bacteria with lesions in apbE-like genes. For example, impaired nitrogen fixation in a ΔrnfF strain of Rhodobacter capsulatus (12) can be explained by an inability to flavinylate subunits of the RNF complex, given that this complex was proposed to be involved in electron transport to nitrogenase (12). Defective iron-sulfur cluster metabolism in a ΔapbE strain of S. enterica (44), leading to thiamine auxotrophy (41), can also be accounted for by impaired activity of the RNF complex that participates in maturation of iron-sulfur clusters (45, 46). Simultaneous disruption of the genes encoding two ApbE orthologs (NosX and NirX; Fig. 2) in P. denitrificans cells eliminates their NO reductase activity (47), which can now be explained by defective maturation of the regulator of NO reductase transcription NosR, which contains a covalently bound FMN residue (16).
apbE-like genes are found in the majority of known bacterial genomes, including many pathogens, such as Yersinia pestis, V. cholerae, Shigella dysenteriae, S. enterica, Neisseria meningitidis, and Treponema pallidum. These genes are completely absent only in Cyanobacteria. In Archaea, apbE-like genes have been found in only a few species of Methanosarcinaceae and Halobacteriaceae. Among Eukaryota, apbE-like genes are present in some Kinetoplastida (Trypanosoma and Leishmania species), wherein they form the N-terminal part of NADH-dependent fumarate reductase (48). The number of the apbE-like genes may vary from one (e.g., in Chlamydia spp.) to five (in some Shewanella spp.). This fact may mean that the corresponding bacteria require a spectrum of flavin transferase specificities to flavinylate different proteins. This inference is further supported by the observation that different ApbE-like proteins of a bacterium are sometimes only distantly related to each other. Thus, ApbE1 from K. pneumoniae (YP_002237370) clusters with ApbE-like proteins from other Gammaproteobacteria in a phylogenetic tree, whereas ApbE2 (YP_002238733) is close to the ApbE proteins from Firmicutes.
Although apbE-like genes are present in all nqr(rnf)-containing bacterial genomes, many apbE-containing microorganisms, including Bacilli, Actinobacteria (except for Coriobacteridae), Aquificae, Chloroflexi, Deinococcus-Thermus, Fibrobacteres-Acidobacteria, Kinetoplastida, and Halobacteriaceae, contain no nqr or rnf genes in their genomes. This may mean that the corresponding organisms have as yet unrecognized ApbE-dependent proteins that also use a covalently bound FMN residue as a prosthetic group.
In summary, we have shown that covalent binding of an FMN residue to proteins via a phosphoester bond is not an autocatalytic process and instead requires a specific flavin transferase, ApbE. ApbE-like proteins are widespread in the bacterial world, including among many pathogens, but are relatively rare in Archaea and Eukaryota. Because these proteins appear to be species-specific, they may represent good targets for the design of antibacterials.
When our manuscript was under revision, a paper by Deka et al. (52) appeared online on an ApbE-like protein from T. pallidum, reporting that this protein catalyzes a “single-turnover” hydrolysis of FAD to yield FMN and AMP. Although such an unusual activity is feasible as a side reaction of flavin transfer, we could not detect it with V. cholerae ApbE. The reported structure of T. pallidum ApbE and its ability to interact with other proteins are consistent with the flavin transfer function demonstrated herein for the V. cholerae protein.
Acknowledgments
We thank Dr. C. C. Häse for providing us with the V. cholerae O395N1 toxT::lacZ strain. The MALDI MS facility was available to us in the framework of the Moscow State University Development Program PNG 5.13.
This work was supported by Russian Foundation for Basic Research Project Numbers 13-04-00332 and 12-04-01002.
- Na+-NQR
- Na+-translocating NADH:quinone oxidoreductase
- dNADH
- reduced nicotinamide hypoxanthine dinucleotide
- HQNO
- 2-heptyl-4-hydroxyquinoline N-oxide
- NDH-2
- noncoupled NADH:quinone oxidoreductase
- RNF
- putative Na+-motive NADH:ferredoxin oxidoreductase.
REFERENCES
- 1. Verkhovsky M. I., Bogachev A. V. (2010) Sodium-translocating NADH:quinone oxidoreductase as a redox-driven ion pump. Biochim. Biophys. Acta 1797, 738–746 [DOI] [PubMed] [Google Scholar]
- 2. Zhou W., Bertsova Y. V., Feng B., Tsatsos P., Verkhovskaya M. L., Gennis R. B., Bogachev A. V., Barquera B. (1999) Sequencing and preliminary characterization of the Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio harveyi. Biochemistry 38, 16246–16252 [DOI] [PubMed] [Google Scholar]
- 3. Häse C. C., Fedorova N. D., Galperin M. Y., Dibrov P. A. (2001) Sodium ion cycle in bacterial pathogens. Evidence from cross-genome comparisons. Microbiol. Mol. Biol. Rev. 65, 353–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nakayama Y., Hayashi M., Unemoto T. (1998) Identification of six subunits constituting Na+-translocating NADH-quinone reductase from the marine Vibrio alginolyticus. FEBS Lett. 422, 240–242 [DOI] [PubMed] [Google Scholar]
- 5. Rich P. R., Meunier B., Ward F. B. (1995) Predicted structure and possible ion-motive mechanism of the sodium-linked NADH-ubiquinone oxidoreductase of Vibrio alginolyticus. FEBS Lett. 375, 5–10 [DOI] [PubMed] [Google Scholar]
- 6. Hayashi M., Hirai K., Unemoto T. (1995) Sequencing and the alignment of structural genes in the nqr operon encoding the Na+-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett. 363, 75–77 [DOI] [PubMed] [Google Scholar]
- 7. Juárez O., Barquera B. (2012) Insights into the mechanism of electron transfer and sodium translocation of the Na+-pumping NADH:quinone oxidoreductase. Biochim. Biophys. Acta 1817, 1823–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barquera B., Nilges M. J., Morgan J. E., Ramirez-Silva L., Zhou W., Gennis R. B. (2004) Mutagenesis study of the 2Fe-2S center and the FAD binding site of the Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio cholerae. Biochemistry 43, 12322–12330 [DOI] [PubMed] [Google Scholar]
- 9. Türk K., Puhar A., Neese F., Bill E., Fritz G., Steuber J. (2004) NADH oxidation by the Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae. Functional role of the NqrF subunit. J. Biol. Chem. 279, 21349–21355 [DOI] [PubMed] [Google Scholar]
- 10. Casutt M. S., Huber T., Brunisholz R., Tao M., Fritz G., Steuber J. (2010) Localization and function of the membrane-bound riboflavin in the Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae. J. Biol. Chem. 285, 27088–27099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hayashi M., Nakayama Y., Yasui M., Maeda M., Furuishi K., Unemoto T. (2001) FMN is covalently attached to a threonine residue in the NqrB and NqrC subunits of Na+-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett. 488, 5–8 [DOI] [PubMed] [Google Scholar]
- 12. Schmehl M., Jahn A., Meyer zu Vilsendorf A., Hennecke S., Masepohl B., Schuppler M., Marxer M., Oelze J., Klipp W. (1993) Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus. A putative membrane complex involved in electron transport to nitrogenase. Mol. Gen. Genet. 241, 602–615 [DOI] [PubMed] [Google Scholar]
- 13. Müller V., Imkamp F., Biegel E., Schmidt S., Dilling S. (2008) Discovery of a ferredoxin:NAD+-oxidoreductase (Rnf) in Acetobacterium woodii. A novel potential coupling site in acetogens. Ann. N.Y. Acad. Sci. 1125, 137–146 [DOI] [PubMed] [Google Scholar]
- 14. Li Q., Li L., Rejtar T., Karger B. L., Ferry J. G. (2005) Proteome of Methanosarcina acetivorans. Part I. An expanded view of the biology of the cell. J Proteome Res. 4, 112–128 [DOI] [PubMed] [Google Scholar]
- 15. Backiel J., Juárez O., Zagorevski D. V., Wang Z., Nilges M. J., Barquera B. (2008) Covalent binding of flavins to RnfG and RnfD in the Rnf complex from Vibrio cholerae. Biochemistry 47, 11273–11284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wunsch P., Zumft W. G. (2005) Functional domains of NosR, a novel transmembrane iron-sulfur flavoprotein necessary for nitrous oxide respiration. J. Bacteriol. 187, 1992–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bogachev A. V., Bertsova Y. V., Bloch D. A., Verkhovsky M. I. (2012) Urocanate reductase. Identification of a novel anaerobic respiratory pathway in Shewanella oneidensis MR-1. Mol. Microbiol. 86, 1452–1463 [DOI] [PubMed] [Google Scholar]
- 18. Heuts D. P., Scrutton N. S., McIntire W. S., Fraaije M. W. (2009) What's in a covalent bond? On the role and formation of covalently bound flavin cofactors. FEBS J. 276, 3405–3427 [DOI] [PubMed] [Google Scholar]
- 19. Kim J., Fuller J. H., Kuusk V., Cunane L., Chen Z. W., Mathews F. S., McIntire W. S. (1995) The cytochrome subunit is necessary for covalent FAD attachment to the flavoprotein subunit of p-cresol methylhydroxylase. J. Biol. Chem. 270, 31202–31209 [DOI] [PubMed] [Google Scholar]
- 20. Hao H. X., Khalimonchuk O., Schraders M., Dephoure N., Bayley J. P., Kunst H., Devilee P., Cremers C. W., Schiffman J. D., Bentz B. G., Gygi S. P., Winge D. R., Kremer H., Rutter J. (2009) SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science 325, 1139–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McNeil M. B., Clulow J. S., Wilf N. M., Salmond G. P., Fineran P. C. (2012) SdhE is a conserved protein required for flavinylation of succinate dehydrogenase in bacteria. J. Biol. Chem. 287, 18418–18428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barquera B., Häse C. C., Gennis R. B. (2001) Expression and mutagenesis of the NqrC subunit of the NQR respiratory Na+ pump from Vibrio cholerae with covalently attached FMN. FEBS Lett. 492, 45–49 [DOI] [PubMed] [Google Scholar]
- 23. Yeats C., Bentley S., Bateman A. (2003) New knowledge from old. In silico discovery of novel protein domains in Streptomyces coelicolor. BMC Microbiol. 3, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bertsova Y. V., Bogachev A. V. (2004) The origin of the sodium-dependent NADH oxidation by the respiratory chain of Klebsiella pneumoniae. FEBS Lett. 563, 207–212 [DOI] [PubMed] [Google Scholar]
- 25. Beck B. J., Downs D. M. (1999) A periplasmic location is essential for the role of the ApbE lipoprotein in thiamine synthesis in Salmonella typhimurium. J. Bacteriol. 181, 7285–7290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fadeeva M. S., Yakovtseva E. A., Belevich G. A., Bertsova Y. V., Bogachev A. V. (2007) Regulation of expression of Na+-translocating NADH:quinone oxidoreductase genes in Vibrio harveyi and Klebsiella pneumoniae. Arch. Microbiol. 188, 341–348 [DOI] [PubMed] [Google Scholar]
- 27. Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 28. Casutt M. S., Schlosser A., Buckel W., Steuber J. (2012) The single NqrB and NqrC subunits in the Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae each carry one covalently attached FMN. Biochim. Biophys. Acta 1817, 1817–1822 [DOI] [PubMed] [Google Scholar]
- 29. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 30. Lawrence J. (1999) Selfish operons. The evolutionary impact of gene clustering in prokaryotes and eukaryotes. Curr. Opin. Genet. Dev. 9, 642–648 [DOI] [PubMed] [Google Scholar]
- 31. Rentzsch R., Orengo C. A. (2009) Protein function prediction. The power of multiplicity. Trends Biotechnol. 27, 210–219 [DOI] [PubMed] [Google Scholar]
- 32. Munro A. W., Noble M. A. (1999) Fluorescence analysis of flavoproteins. Methods Mol. Biol. 131, 25–48 [DOI] [PubMed] [Google Scholar]
- 33. Massey V., Palmer G. (1966) On the existence of spectrally distinct classes of flavoprotein semiquinones. A new method for the quantitative production of flavoprotein semiquinones. Biochemistry 5, 3181–3189 [DOI] [PubMed] [Google Scholar]
- 34. Barquera B., Ramirez-Silva L., Morgan J. E., Nilges M. J. (2006) A new flavin radical signal in the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. J. Biol. Chem. 281, 36482–36491 [DOI] [PubMed] [Google Scholar]
- 35. Bogachev A. V., Kulik L. V., Bloch D. A., Bertsova Y. V., Fadeeva M. S., Verkhovsky M. I. (2009) Redox properties of the prosthetic groups of Na+-translocating NADH:quinone oxidoreductase. 1. EPR study of the enzyme. Biochemistry 48, 6291–6298 [DOI] [PubMed] [Google Scholar]
- 36. Bogachev A. V., Bloch D. A., Bertsova Y. V., Verkhovsky M. I. (2009) Redox properties of the prosthetic groups of Na+-translocating NADH:quinone oxidoreductase. 2. Study of the enzyme by optical spectroscopy. Biochemistry 48, 6299–6304 [DOI] [PubMed] [Google Scholar]
- 37. Bogachev A. V., Verkhovsky M. I. (2005) Na+-Translocating NADH:quinone oxidoreductase. Progress achieved and prospects of investigations. Biochemistry 70, 143–149 [DOI] [PubMed] [Google Scholar]
- 38. Bourne R. M., Rich P. R. (1992) Characterization of a sodium-motive NADH:ubiquinone oxidoreductase. Biochem. Soc. Trans. 20, 577–582 [DOI] [PubMed] [Google Scholar]
- 39. Matsushita K., Ohnishi T., Kaback H. R. (1987) NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry 26, 7732–7737 [DOI] [PubMed] [Google Scholar]
- 40. Fadeeva M. S., Núñez C., Bertsova Y. V., Espín G., Bogachev A. V. (2008) Catalytic properties of Na+-translocating NADH:quinone oxidoreductases from Vibrio harveyi, Klebsiella pneumoniae, and Azotobacter vinelandii. FEMS Microbiol. Lett. 279, 116–123 [DOI] [PubMed] [Google Scholar]
- 41. Beck B. J., Downs D. M. (1998) The apbE gene encodes a lipoprotein involved in thiamine synthesis in Salmonella typhimurium. J. Bacteriol. 180, 885–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Han G. W., Sri Krishna S., Schwarzenbacher R., McMullan D., Ginalski K., Elsliger M. A., Brittain S. M., Abdubek P., Agarwalla S., Ambing E., Astakhova T., Axelrod H., Canaves J. M., Chiu H. J., DiDonato M., Grzechnik S. K., Hale J., Hampton E., Haugen J., Jaroszewski L., Jin K. K., Klock H. E., Knuth M. W., Koesema E., Kreusch A., Kuhn P., Miller M. D., Morse A. T., Moy K., Nigoghossian E., Oommachen S., Ouyang J., Paulsen J., Quijano K., Reyes R., Rife C., Spraggon G., Stevens R. C., van den Bedem H., Velasquez J., Wang X., West B., White A., Wolf G., Xu Q., Hodgson K. O., Wooley J., Deacon A. M., Godzik A., Lesley S. A., Wilson I. A. (2006) Crystal structure of the ApbE protein (TM1553) from Thermotoga maritima at 1.58 Å resolution. Proteins 64, 1083–1090 [DOI] [PubMed] [Google Scholar]
- 43. Boyd J. M., Endrizzi J. A., Hamilton T. L., Christopherson M. R., Mulder D. W., Downs D. M., Peters J. W. (2011) FAD binding by ApbE protein from Salmonella enterica. A new class of FAD-binding proteins. J. Bacteriol. 193, 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Skovran E., Downs D. M. (2003) Lack of the ApbC or ApbE protein results in a defect in Fe-S cluster metabolism in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185, 98–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martinez-Gomez N. C., Downs D. M. (2008) ThiC is an [Fe-S] cluster protein that requires AdoMet to generate the 4-amino-5-hydroxymethyl-2-methylpyrimidine moiety in thiamin synthesis. Biochemistry 47, 9054–9056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Curatti L., Brown C. S., Ludden P. W., Rubio L. M. (2005) Genes required for rapid expression of nitrogenase activity in Azotobacter vinelandii. Proc. Natl. Acad. Sci. U.S.A. 102, 6291–6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Saunders N. F., Hornberg J. J., Reijnders W. N., Westerhoff H. V., de Vries S., van Spanning R. J. (2000) The NosX and NirX proteins of Paracoccus denitrificans are functional homologues. Their role in maturation of nitrous oxide reductase. J. Bacteriol. 182, 5211–5217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Besteiro S., Biran M., Biteau N., Coustou V., Baltz T., Canioni P., Bringaud F. (2002) Succinate secreted by Trypanosoma brucei is produced by a novel and unique glycosomal enzyme, NADH-dependent fumarate reductase. J. Biol. Chem. 277, 38001–38012 [DOI] [PubMed] [Google Scholar]
- 49. Miller V. L., Mekalanos J. J. (1988) A novel suicide vector and its use in construction of insertion mutations. Osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170, 2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Barquera B., Hellwig P., Zhou W., Morgan J. E., Häse C. C., Gosink K. K., Nilges M., Bruesehoff P. J., Roth A., Lancaster C. R., Gennis R. B. (2002) Purification and characterization of the recombinant Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry 41, 3781–3789 [DOI] [PubMed] [Google Scholar]
- 51. Alexeyev M. F. (1999) The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques 26, 824–828 [DOI] [PubMed] [Google Scholar]
- 52. Deka R. K., Brautigam C. A., Liu W. Z., Tomchick D. R., Norgard M. V. (February 27, 2013) The TP0796 lipoprotein of T. pallidum is a bimetal-dependent FAD pyrophosphatase with a potential role in flavin homeostasis. J. Biol. Chem. 10.1074/jbc.M113.449975 [DOI] [PMC free article] [PubMed] [Google Scholar]