Background: HDACis activate HIV transcription.
Results: P-TEFb release from 7SK snRNP correlates better than histone H3 or tubulin acetylation with HIV reactivation by HDACis in cell lines.
Conclusion: Levels of P-TEFb must be increased before HDACis can reactivate HIV in resting primary CD4+ T cells.
Significance: Levels and activity of P-TEFb are critical for HIV reactivation in all cells.
Keywords: Histone Deacetylase Inhibitors, Histone Modification, HIV, Transcription Elongation Factors, Tubulin, 7SK snRNP, HEXIM1, HIV Latency, P-TEFb
Abstract
Numerous studies have looked at the effects of histone deacetylase inhibitors (HDACis) on HIV reactivation in established transformed cell lines and primary CD4+ T cells. However, their findings remain confusing, and differences between effects of class I- and class II-specific HDACis persist. Because no clear picture emerged, we decided to determine how HDACis reactivate HIV in transformed cell lines and primary cells. We found that neither histone H3 nor tubulin acetylation correlated with HIV reactivation in Jurkat and HeLa cells. Rather, HDACis that could reactivate HIV in chromatin or on episomal plasmids also released free positive transcription elongation factor b (P-TEFb) from its inhibitory 7SK snRNP. In resting primary CD4+ T cells, where levels of P-TEFb are vanishingly low, the most potent HDACi, suberoylanilide hydroxyamic acid (SAHA), had minimal effects. In contrast, when these cells were treated with a PKC agonist, bryostatin 1, which increased levels of P-TEFb, then SAHA once again reactivated HIV. We conclude that HDACis, which can reactivate HIV, work via the release of free P-TEFb from the 7SK snRNP.
Introduction
HIV infection has become a chronic disease for patients who have ready access to highly active antiretroviral therapy (HAART).4 HAART not only suppresses ongoing viral replication and decreases rates of transmission, but it slows the progression to AIDS indefinitely (1–3). However, the cessation of therapy leads to a rapid rebound of viral replication, which can progress to AIDS even after decades of HAART.
In recent years, the nature of this viral reservoir has been under intense study. It is composed, to a large part, of latently infected quiescent CD4+ T cells (for review see Ref. 2). In these cells, the HIV provirus is integrated and remains transcriptionally silent in the host genome. Because no HIV proteins are expressed, no cytopathic effects are observed, and the virus cannot be cleared by the immune system (for review see Refs. 1, 3–5). Thus, efforts to deplete or eradicate the viral reservoir have focused on the reactivation of transcriptionally silent proviruses in latently infected cells (1, 5). Once reactivated, viral gene expression should then kill infected cells directly or lead to their clearance by the immune system (6). Concomitant intensified HAART would block the infection of new cells and thus prevent the spread of the virus.
Transcription of HIV depends critically on the positive transcription elongation factor b (P-TEFb), which is composed of the cyclin-dependent kinase 9 (CDK9) and cyclin T1 or 2 (CycT1 or CycT2) (7, 8). P-TEFb phosphorylates negative elongation factors assembled on the RNA polymerase II (RNAPII) and the transactivation response (TAR) RNA stem loop (DRB sensitivity-induced factor (DSIF) and negative elongation factor (NELF)), which stall transcription complexes on the 5′ HIV long terminal repeat (LTR). Upon the phosphorylation of NELF-E or RD and Spt5, NELF dissociates from TAR, and DSIF is converted to an elongation factor (9). Next, P-TEFb phosphorylates serines at position 2 (Ser-2) in the heptapeptide repeats (52× (YSPTSPS)) of the C-terminal domain of RNAPII, which leads to the elongation and co-transcriptional processing of viral transcripts. Besides NF-κB, bromodomain protein 4 (BRD4) and the super-elongation complex, which bring P-TEFb to the RNAPII on the HIV LTR, the viral transcriptional transactivator Tat also recruits P-TEFb to TAR (7, 8, 10). This recruitment increases greatly HIV transcription and replication. Because TAR is so efficient at recruiting negative elongation factors, which stall RNAPII on the viral promoter, HIV transcription is extremely sensitive to levels and activity of P-TEFb.
In resting and terminally differentiated cells, levels of P-TEFb are vanishingly low (11–13). In growing cells, P-TEFb exists in at least two different functional complexes. When active, small amounts of free P-TEFb are found on active chromatin in association with BRD4, super-elongation complex, or specific transcription factors such as NF-κB, steroid hormone receptors, etc. (7, 8, 14–20). In these cells, most P-TEFb is inactivated in a larger complex with hexamethylene bisacetamide (HMBA)-induced proteins 1 and 2 (HEXIM1/2), noncoding 7SK snRNA, La-related protein 7, and methyl phosphate-capping enzyme (for review, see Ref 18). The equilibrium between these forms of P-TEFb (P-TEFb equilibrium) determines and reflects the state of activation, growth, and proliferation of cells. Free P-TEFb is also required for the initial rounds of HIV transcription in the absence of Tat. Importantly, once Tat is made, which competes with HEXIM1 for binding to P-TEFb, both forms of P-TEFb can be utilized to sustain and maintain HIV replication (21).
Compounds from diverse structural classes can reactivate HIV transcription from latency in cell lines and primary cells. They include differentiation agents such as HMBA, histone deacetylase (HDACis) and bromodomain and extraterminal (BET) bromodomain inhibitors as well as PKC agonists (for a nonexhaustive list see Refs. 22–26). Some of these compounds were also analyzed for their effects on P-TEFb. In this study, we found that although structurally and functionally divergent, HDACis, which can reactivate HIV also affect the P-TEFb equilibrium, i.e. release transiently free P-TEFb from the 7SK snRNP. This release could not be correlated with their effects on histone H3 or tubulin acetylation. Furthermore, we confirmed that levels of P-TEFb are vanishingly low in resting primary CD4 T cells. These cells must be activated to synthesize P-TEFb before effects of these other agonists become apparent. Our findings suggest that combinatorial approaches targeting P-TEFb will be required to reactivate HIV in infected individuals on HAART.
EXPERIMENTAL PROCEDURES
Cell Lines, Antibodies, and Plasmids
JΔK cells and Jurkat cells were grown in RPMI 1640 medium containing penicillin (100 IU/ml), streptomycin (100 μg/ml), and 10% FBS at 37 °C with 5% CO2. NH1 and HeLa cells were grown in DMEM containing penicillin (100 IU/ml), streptomycin (100 μg/ml), and 10% FBS at 37 °C with 5% CO2. HIV release in the supernatant was quantified by p24 capsid ELISA (PerkinElmer Life Sciences). Antibodies used in this study for immunoprecipitations and Western blotting were: anti-HEXIM1 (Hex; Abcam, ab25388), anti-CDK9 (Santa Cruz, sc-484), anti-CycT1 (Santa Cruz, sc-10750), anti-tubulin (Abcam, ab6046), anti-histone H3 (Active Motif, 39163), anti-panAc-histone H3 (Active Motif, 39139), anti-Ac-tubulin (Abcam, ab24610), and anti-actin (Abcam, ab8227). SAHA (S1047), tubastatin A (S2627) and MS-275 (S1053) was purchased from Selleck Chem and HMBA (H4663) from Sigma. Stock solutions were prepared in DMSO and water for HMBA. ST-80 was a kind gift from Prof. Manfred Jung (University of Freiburg, Germany).
RNA Immunoprecipitations
Jurkat or HeLa cells (2.5 × 106) were untreated or treated with compounds for 2 and 4 h. The cells were lysed in buffer A (20 mm HEPES-KOH, pH 7.8, 0.1% Nonidet P-40, 0.2 mm EDTA) containing low salt (10 mm KCl) on ice for 10 min. Cell lysates were centrifuged at 5000 × g for 5 min at 4 °C, and supernatants were collected. Supernatants were then precleared with protein A-Sepharose beads and divided into three aliquots. Each aliquot was incubated with 1 μg of normal rabbit IgG, α-HEXIM1, or anti-CDK9 antibodies overnight at 4 °C and with 20 μl of protein A-Sepharose beads precoated with BSA and yeast tRNA for an additional 2 h at 4 °C. Beads were washed five times with buffer A-containing medium salt (100 mm KCl, MS). RNA was then extracted by TRIzol (Invitrogen) and analyzed by RT-quantitative PCR (RT-qPCR) to quantify 7SK snRNA associated with P-TEFb. Data were normalized to input amounts of 7SK snRNA and calculated as values relative to the amount obtained with untreated cells (set to 1).
Differential Salt Extraction
Differential salt extraction was carried out to determine fractions of free P-TEFb or 7SK snRNP according to Biglione et al. (27), with some modifications. Jurkat cells (5 × 105) were collected and washed twice with cold PBS. Cells were lysed in 80 μl of low salt buffer (10 mm KCl, low salt, 10 mm MgCl2, 10 mm HEPES-KOH, pH 7.5, 1 mm EDTA, 1 mm DTT, 0.5% Nonidet P-40, proteinase inhibitor mixture) and incubated on ice for 10 min. Lysates were then centrifuged at 5000 ×g for 5 min, and supernatants were collected and designated as 7SK snRNP fractions. Pellets were washed once with 200 μl of low salt buffer and resuspended in 80 ml of high salt buffer (450 mm NaCl, high salt, 1.5 mm MgCl2, 20 mm HEPES, pH 7.5, 0.5 mm EDTA, 1 mm DTT, 0.5% Nonidet P-40, proteinase inhibitor mixture). Suspensions were mixed by vortexing briefly and incubated on ice for 10 min. Lysates were then centrifuged at 10,000 × g for 5 min, and supernatants were collected and designated as free P-TEFb fractions.
Preparation, Activation, Maintenance, Infection, and Resting of Primary CD4+ T Cells
Leukopheresis residues were obtained from the Blood Center of the Pacific (San Francisco, CA), and white blood cells were purified further using an OptiPrep (Sigma, 1556) density gradient. Naïve CD4+ T cells were purified from the buffy coat using the Dynabeads® UntouchedTM Human CD4 T Cells kit (Invitrogen, 11346D) according to the manufacturer's protocol. CD4+ T cells were maintained for 24 h in RPMI 1640 medium, 10% FBS, 30 units/ml IL-2 (Roche Applied Science, 10799068001) at 37 °C with 5% CO2 before activation and expansion using anti-CD3/anti-CD28 Dynabeads (Invitrogen, 111.32D) for 14 days in RPMI 1640 medium, 10% FBS, 30 units/ml IL-2. Purity and activation level were determined by immunofluorescence for CD4, CD69 (Santa Cruz Biotechnology, sc-3608) and HLA-DR (Abcam, ab95830) by FACS. Activated CD4+ T cells were infected by spin inoculation using a reporter virus with a deleted Env gene and a luciferase reporter gene in the Nef ORF (pNL4.3ΔenvΔNef-Luc) produced in the presence of HIV-1 Env (pEnvHIV, plasmid kindly provided by Dr. Kotaro Shirakawa). Cells were reverted to a resting state by gradually decreasing the amount of IL-2 in the medium to 2 units/ml over the period of 21 days. Luciferase activity was measured over this period and reached a background plateau.
Transient Transfection and Luciferase Assays
HeLa cells, seeded in 24-well plates, were transfected with 1 μg of plasmid DNA using Xtreme-HP transfection reagent (Roche Applied Science) according to the manufacturer's protocol. Similarly, Jurkat cells (2 × 106) were transfected with 2 μg of HIV.Luc reporter plasmid. Cells were grown in DMEM with 10% FBS and kept in 5% CO2, 37 °C for 48 h. Transfected cells were treated with compounds, and luciferase activity in cell lysate was determined using the Luciferase Assay System (Promega) according to the manufacturer's instruction after 24 h.
Reverse Transcription-qPCR
Total cellular RNA was extracted by TRIzol reagent according to the manufacturer's instruction and reverse transcribed with random hexamers using SuperScript III Reverse Transcriptase (Invitrogen). The cDNA was quantified using a Stratagene Mx3004P Real Time PCR system and SensiFast SYBR Green reagents (Bioline) with specific primers. The primer sequences used in this study were as follows. For GAPDH normalization, primers are described in Ref. 28. For HEXIM1 mRNA quantification: HEXIM1 forward, GACCTGGGAAGAGAAGAAAAAG and HEXIM1 reverse, GAGGAACTGCGTGGTGTTATAG; 7SK snRNA forward, GAGGGCGATCTGGCTGCGACAT and 7SK snRNA reverse, ACATGGAGCGGTGAGGGAGGAA.
RESULTS
Only Some HDACis Reactivate the Replication of an Integrated HIV Provirus in Jurkat Cells
Several HDACis can reactivate transcription of HIV in cell culture models of viral latency (22–25, 29–35). Among these, SAHA inhibits potently class I HDAC1, 2, and 3 and the class II HDAC6 (36). To gain insight into which HDACs might be involved in HIV reactivation, we examined three additional HDACis with different HDAC specificities. Of these, MS-275 inhibits HDAC1, 2, and 3 (36), ST-80 has a strong preference for HDAC6 (37), and TBSA is a highly specific HDAC6 inhibitor (38). We determined the HIV reactivation potential of these different HDACis in JΔK cells. They are Jurkat cells, which contain a transcriptionally silent HIV provirus that lacks NF-κB sites (39), which facilitates the study of HIV reactivation by processes that are independent of immune activation. Indeed, as described before (40), treatment of JΔK cells with increasing concentrations of SAHA leads to a dose-dependent reactivation of latent HIV (up to 25-fold) as measured by newly released viral particles in the supernatant (Fig. 1A, bars 2–4). Like SAHA, ST-80 reactivated HIV in a dose-dependent fashion in these cells (Fig. 1A, bars 8–10). In contrast, the inhibition of HDAC1, 2, and 3 by MS-275 activated HIV transcription significantly less potently, and inhibition of HDAC6 by TBSA had no effect in JΔK cells (Fig. 1A, bars 5–7 and 11–13). Furthermore, combinations of MS-275 and ST-80 did not have additive effects in JΔK cells (Fig. 1B). By measuring levels of histone H3 (class I HDACs) and tubulin (class II HDAC) acetylation, Western blotting confirmed the specificity of different HDACis (Fig. 1A, lower panels). Indeed, whereas SAHA induced the acetylation of histone H3 and tubulin, effects of MS-275 were mainly on histone H3 (Fig. 1A, top and middle panels, lanes 2–7). As expected, ST-80 and TBSA led to increased tubulin acetylation (Fig. 1A, top and middle panels, lanes 8–13). Together, these data indicate that the reactivation of HIV transcription by HDACis does not correlate with histone H3 or tubulin acetylation, i.e. inhibition of either class I or class II HDACs in Jurkat cells.
FIGURE 1.
Only some HDACis reactivate HIV in Jurkat cells. A. acetylation of histone H3 and tubulin does not correlate with HIV reactivation in JΔK cells. Cells were incubated with increasing concentrations of different HDACis or DMSO as indicated for 24 h (upper panel: SAHA from 0.5, 2 to 10 μm; MS-275 from 2, 5 to 25 μm; ST-80 from 2.5, 10 to 40 μm; TBSA from 1, 5 to 20 μm). The production of new viral particles was determined by measuring levels of p24 capsid ELISA in supernatants. Values are presented as -fold activation over the DMSO control, which was set to 1 (bar 1). Error bars represent S.E. of mean of experiments performed in triplicate. Western blotting for acetylated tubulin (lower panel, bar 1) and histone H3 (lower panel, bar 3) confirmed the specificity of HDAC inhibition. Amounts of total tubulin and histone H3 as loading controls are presented in lower panel, bars 2 and 4. B, JΔK cells were incubated with different HDACis (SAHA, 5 μm; MS-275, 12.5 μm; ST-80, 10 μm) or combinations thereof for 24 h. DMSO was included as the control. The production of new viral particles was determined by measuring p24 capsid levels in supernatants. Values are presented as -fold activation over the DMSO control. Error bars are as in A. C, the episomal HIV.Luc plasmid target recapitulates effects of HDACis on the integrated HIV genome in Jurkat cells. Jurkat cells were transfected with an HIV LTR plasmid target (HIV.Luc). 24 h later, cells were incubated with the indicated concentrations of HDACis (SAHA, 5 μm; MS-275, 25 μm; ST-80, 30 μm; TBSA, 20 μm) for an additional 24 h prior to luciferase enzymatic assays. The luciferase activity was determined in cell lysates and normalized to total protein. Values are presented as -fold activation over the DMSO control. Error bars are as in A.
Similar Effects Are Observed with the HIV.Luc Plasmid Target in Jurkat Cells
Episomal plasmids, which do not replicate, collect insignificant amounts of histones on their DNA (41). Thus, we wanted to determine whether the same HDACis would also activate transcription from the transiently expressed HIV.Luc plasmid target, which contains the HIV LTR linked to the luciferase reporter gene, in Jurkat cells. Indeed, as presented in Fig. 1C, SAHA and ST-80, but not MS-275 and TBSA, increased its luciferase activity (4-fold) in Jurkat cells (bars 2–5). Importantly, this increase was independent of Tat, which amplifies effects of these HDACis (Fig. 1A). These findings suggest that a mechanism different from histone H3 or tubulin acetylation is responsible for HIV reactivation by informative HDACis in Jurkat cells.
The Release of Free P-TEFb from the 7SK snRNP Is Observed with HDACis That Reactivate HIV in Jurkat Cells
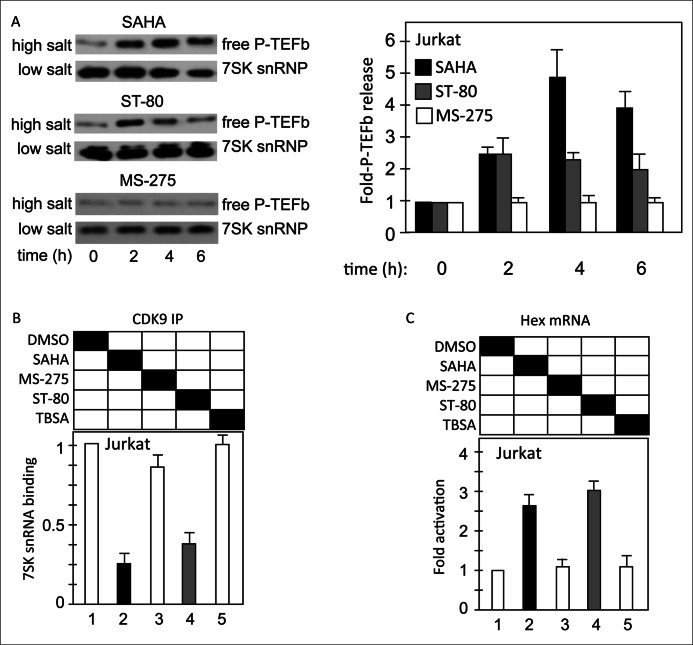
Other small molecules that reactivate HIV include HMBA and JQ1 (23, 24, 29, 30, 33, 42). Previously, we observed that HMBA, SAHA, and JQ1 also lead to the rapid release of free P-TEFb from the 7SK snRNP, which is required for the first rounds of HIV transcription. The release of free P-TEFb also increases the synthesis of its inhibitor, HEXIM1 (23, 24, 43). Thus, we examined levels of free P-TEFb and HEXIM1 transcription and correlated them with HIV reactivation by different HDACis. First, we found that SAHA and ST-80 but not MS-275 increased transiently levels of free P-TEFb, which is associated with chromatin and can be released by high salt (Fig. 2A, left upper panels, high salt, also quantified in the right panel, bars corresponding to 0, 2, 4, and 6 h after the addition of these compounds to Jurkat cells). This finding was confirmed using RNA-immunoprecipitation, where complexes isolated with anti-CDK9 antibodies were subjected to RT-qPCR to quantify levels of associated 7SK snRNA. As presented in Fig. 2B, 4 h after the addition of these compounds, we found that SAHA and ST-80, but not MS-275 or TBSA, led to the release of CDK9 (free P-TEFb) from the 7SK RNA (Fig. 2B, bars 2–5). We obtained further proof for this shift in the P-TEFb equilibrium by examining the transcription of the HEXIM1 gene. Indeed, SAHA and ST-80 but not MS-275 or TBSA increased levels of steady-state HEXIM1 transcripts, which were measured 24 h after the addition of these compounds (Fig. 2C, bars 2–5). These data reveal that HIV reactivation (in chromatin and on episomal plasmids) by HDACis can be correlated with the transient release of free P-TEFb from the 7SK snRNA and not histone H3 or tubulin acetylation in Jurkat cells.
FIGURE 2.
The release of free P-TEFb from the 7SK snRNP is observed with HDACis that reactivate HIV in Jurkat cells. A, HDACis that reactivate HIV in Jurkat cells also release free P-TEFb from the 7SK snRNP as determined by differential salt extraction. Jurkat cells were incubated with HDACis (SAHA, 5 μm; ST-80, 30 μm; MS-275, 25 μm) for the indicated periods of time, and levels of chromatin-bound (free P-TEFb) and 7SK snRNP-associated (7SK snRNP) CDK9 protein were determined by Western blotting. CDK9 levels were quantified for each time point using ImageJ software, and relative levels of active CDK9 were calculated (high salt/(high salt + low salt)). Quantification is presented in the right panel, and error bars are as in Fig. 1. B, the release of free P-TEFb from the 7SK snRNP was confirmed by RNA immunoprecipitation. Jurkat cells were treated with HDACis (SAHA, 5 μm; MS-275, 25 μm; ST-80, 30 μm; TBSA, 20 μm) for 4 h, and CDK9 was immunoprecipitated from cell lysates. CDK9-associated 7SK snRNA in immunoprecipitations was determined by RT-qPCR. Values are presented as relative enrichment with the DMSO control set to 1. Error bars represent S.E. of mean of experiments performed in triplicate. C, levels of HEXIM1 transcripts rise with the release of free P-TEFb. Jurkat cells were treated with HDACis (SAHA, 5 μm; MS-275, 25 μm; ST-80, 30 μm; TBSA, 20 μm) for 6 h and HEXIM1 mRNA was determined by RT-qPCR. Levels of HEXIM1 transcripts were normalized to those of GAPDH and are presented as -fold activation over the DMSO control. Error bars represent S.E. of mean of experiments performed in triplicate.
Different HDACis Reactivate the Transcription of an Integrated HIV LTR in HeLa Cells
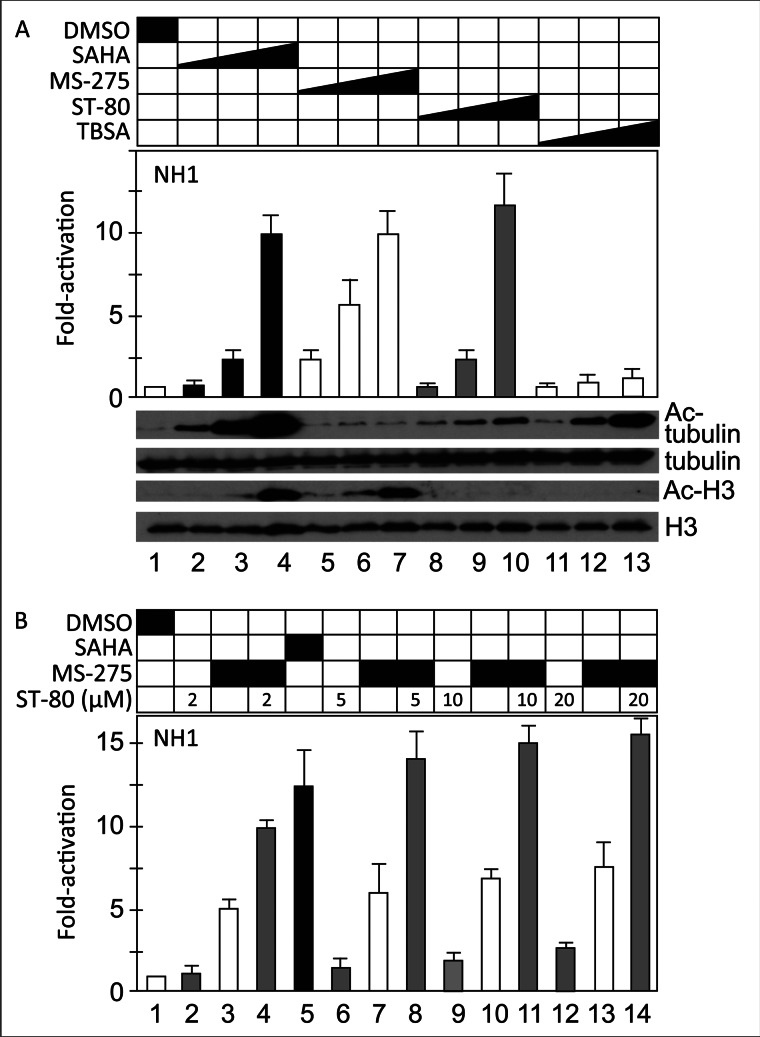
To extend our observation in Jurkat cells to other commonly used cells, we analyzed HIV reactivation by the same HDACis in HeLa cells. NH1 cells carry an integrated HIV LTR linked to the luciferase reporter gene (44). As presented in Fig. 3A, SAHA, MS-275, and ST-80, but not TBSA, increased HIV transcription in these cells (bars 2–10 and 11–13). Western blotting revealed that these HDACis acetylated appropriate proteins also in NH1 cells (Fig. 3A, lower panels, acetylated and total tubulin and histone H3). We conclude that in contrast to Jurkat cells, class I HDAC inhibition has greater effects on the HIV LTR in HeLa than in Jurkat cells. In contrast to JΔK cells, MS-275 and ST-80 also had synergistic effects in NH1 cells (Fig. 3B). Again, the effects of class II HDAC inhibition were divergent.
FIGURE 3.
Only some and different HDACis reactivate HIV in HeLa cells. A, NH1 cells were incubated with increasing concentrations of different HDACis or DMSO for 24 h (upper panel: SAHA from 0.5, 2 to 10 μm; MS-275 from 2, 5 to 25 μm; ST-80 from 2.5, 10 to 40 μm; TBSA from 1, 5 to 20 μm). HIV reactivation was determined by measuring the luciferase activity in cell lysates. Values are presented as -fold activation over the DMSO control (left bar). Error bars represent S.E. of mean of experiments performed in triplicate. Western blotting for acetylated tubulin (lower panels, bar 1) and histone H3 (lower panels, bar 3) confirmed the specificity of HDAC inhibition. Amounts of total tubulin and histone H3 as loading controls are presented in lower panel, bars 2 and 4. B, NH1 cells were incubated with fixed concentrations of MS-275 (12.5 μm, bars 3, 4, 7, 8, 11–14) alone or in combination with increasing concentrations of ST-80 as indicated (μm) for 24 h. SAHA (bar 5; 5 μm) and DMSO (bar 1) were included as controls. HIV LTR reactivation was determined by measuring relative luciferase activities in cell lysates. Values are presented as -fold activation over the DMSO control. Error bars represent S.E. of mean of experiments performed in triplicate.
The Release of Free P-TEFb from the 7SK snRNP Is Also Observed with HDACis That Reactivate HIV in HeLa Cells
To determine whether increased HIV transcription also correlates with the release of free P-TEFb from the 7SK snRNP in HeLa cells, we performed additional studies. As presented in Fig. 4A, SAHA, ST-80, and MS-275 but not TBSA released CDK9 (free P-TEFb) from the 7SK snRNA (Fig. 4A, RNA immunoprecipitation, bars 2–5). This finding confirms that changes in the P-TEFb equilibrium also affect HIV transcription in HeLa cells. Previously, we observed that the activation of Hex.Luc by JQ1 parallels the release of free P-TEFb from the 7SK snRNP (23). Indeed, the transiently expressed Hex.Luc was also activated by SAHA, ST-80, and MS-275 but not by TBSA (Fig. 4B, bars 2–5, luciferase activity 24 h after the addition of these compounds). Together with increased HIV transcription, these findings reveal that HDACis that affect the P-TEFb equilibrium also reactivate HIV in transformed cell lines.
FIGURE 4.
The release of free P-TEFb from the 7SK snRNP is also observed with HDACis that reactivate HIV in HeLa cells. A, HDACis that reactivate HIV also disrupt the 7SK snRNP as determined by RNA immunoprecipitation in HeLa cells. HeLa cells were treated with HDACis (SAHA, 5 μm; MS-275, 25 μm; ST-80, 30 μm; TBSA, 20 μm) for 4 h, and CDK9 was immunoprecipitated from cell lysates. CDK9-associated 7SK snRNA in immunoprecipitations was determined by RT-qPCR. Values are presented as relative enrichment with the DMSO control set to 1. Error bars represent S.E. of mean of experiments performed in triplicate. B, these HDACis also activate a Hex.Luc plasmid target in HeLa cells. Hex.Luc was transfected into HeLa cells. After 24 h, cells were treated with HDACis (SAHA, 5 μm; MS-275, 5 μm; ST-80, 30 μm; TBSA, 5 μm) for 24 h, and luciferase activity was measures in cell lysates. Values are presented as -fold activation over the DMSO control set to 1 (bar 1). Error bars represent S.E. of mean of experiments performed in triplicate.
Levels of P-TEFb Must Be Increased before HDACis Can Reactivate HIV in Primary Resting CD4+ T Cells
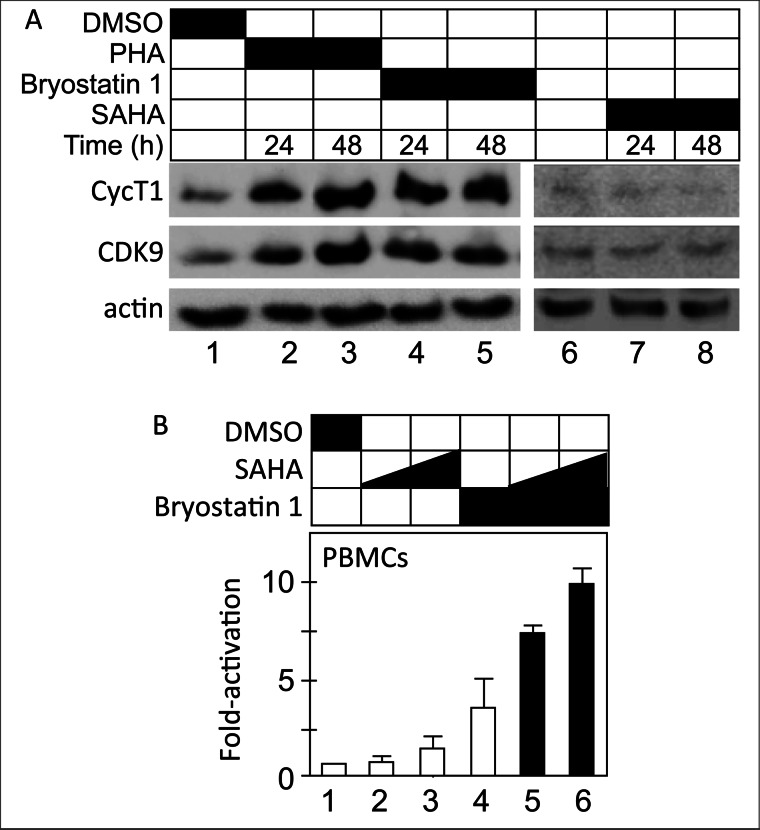
Primary resting CD4+ T cells have vanishingly low levels of P-TEFb (11–13). Because we correlated the ability of HDACis to activate HIV transcription with changes in the P-TEFb equilibrium, we reasoned that the absence of P-TEFb could block effects of our strongest HDACi, SAHA. Indeed, this lack of P-TEFb could explain the poor reactivation potential of HDACis in primary cell models of HIV latency (45, 46). Importantly, PKC agonists can increase levels of P-TEFb in resting cells without mitogenic effects (47). Thus, we wanted to determine whether combinations of PKC agonists and HDACis could reactivate HIV transcription in resting CD4+ T cells. First we examined whether bryostatin 1, a known PKC agonist (48, 49), can increase levels of P-TEFb in resting CD4+ T cells, which were isolated from anonymous uninfected donors, activated, and allowed to return to a resting state in limiting amounts of IL-2 over 2 weeks. The resting state of the cells was verified by detection of low levels of CD69 and HLA-DR by FACS (data not presented). While resting, cells were incubated with bryostatin 1 or phytohemagglutinin as a positive control for 24 and 48 h (Fig. 5A). Western blotting for levels of CDK9 and CycT1 indicated that bryostatin 1 increased levels of P-TEFb equivalently to PHA in these cells (Fig. 5A, top two panels, lanes 2–5). When treated only with SAHA, levels of P-TEFb did not increase (Fig. 5A, top two panels, lanes 7 and 8). Next, we examined HIV reactivation in the presence of bryostatin 1 and SAHA in latently infected resting CD4+ T cells. To this purpose, CD4+ T cells were isolated, activated, and infected with a nonreplicating HIV (pNL4–3Luc). After infection, cells were allowed to return to a resting state in limiting amounts of IL-2 over the course of 2 weeks. At this point they were activated with bryostatin 1 or SAHA alone or in combination. As presented in Fig. 5B, SAHA and bryostatin 1 had small effects on HIV transcription (Fig. 5B, bars 2–4, 2–3-fold), respectively. However, when cells were treated with a combination of bryostatin 1 and SAHA, HIV was reactivated potently (Fig. 5B, bars 5 and 6, 7–10-fold). We conclude that increased synthesis of P-TEFb is a prerequisite for HIV reactivation by HDACis in primary resting infected cells.
FIGURE 5.
Levels of P-TEFb must be increased before HDACis can reactivate HIV in primary resting CD4+ T cells. A, levels of P-TEFb are low in primary CD4+ T cells and increase following PKC activation. CD4+ T cells were isolated and expanded from anonymous healthy donors (see “Experimental Procedures”). They were incubated for the indicated periods of time with bryostatin 1, PHA, or SAHA. Levels of CycT1 and CDK9 were determined by Western blotting. Levels of actin were measured as loading controls. B, increased levels of P-TEFb are required before HDACis can reactivate HIV in primary CD4+ T cells. Isolated CD4+ T cells were treated as in A. Resting cells were treated with bryostatin 1, SAHA, or combinations thereof for 24 h at the indicated concentrations. Luciferase activity was determined in cell lysates, normalized to total protein content, and calculated as -fold activation over the DMSO control (bar 1, set to 1). Experiments with two different lots of peripheral blood mononuclear cells are presented. Error bars represent S.E. of mean of experiments performed in triplicate.
DISCUSSION
In this report, we could not correlate acetylation of histone H3 or tubulin, which reflect the inhibition of class I- and class II-specific HDACis, respectively, with HIV reactivation in Jurkat, HeLa, or primary CD4+ T cells. All of these cells have been used as models of HIV latency. There were even differences in their responses to a potent class I-specific HDACi, MS-275, which had little to no effect in Jurkat cells, but potently activated HIV in HeLa cells. Because similar effects were observed on targets in chromatin and on episomal plasmids, to which little to no histones bind and on which nucleosomes do not form efficiently, we conclude that these HDACis affect another pathway for HIV reactivation. Indeed, we correlated their ability to release free P-TEFb from the 7SK snRNP with HIV reactivation in our transformed cell lines. Because primary resting cells contain insignificant levels of P-TEFb, they had to be increased first with PKC agonists before effects of our most potent HDACi, SAHA, could be observed. We conclude that for HIV reactivation, HDACis must be able to release free P-TEFb from the 7SK snRNP in cells.
In this study, we used transformed cells, where the entire HIV genome was present (JΔK cells) or just the HIV LTR was linked to a reporter gene (NH1), both in chromatin. Importantly, because in JΔK cells, the HIV LTR lacks NF-κB sites, it is not activated via this transcription factor in Jurkat cells. Consequently, the recruitment of HDAC1 by p50 homodimers is not involved in its transcriptional repression (50). Importantly, these experiments were repeated with the HIV LTR linked to a reporter gene in transient expression assays, confirming that HDACis stimulate HIV transcription independently of chromatin. Rather, HIV reactivation correlated with the release of free P-TEFb from the 7SK snRNP, which also increased the synthesis of its inhibitor, HEXIM1. Indeed, similar to the situation with NF-κB, which increases the transcription of its inhibitor, I-κB, free P-TEFb increases the synthesis of HEXIM1 (23, 43). Thus, P-TEFb reassembles into the 7SK snRNP, and the P-TEFb equilibrium is restored rapidly, thus causing cell cycle arrest, which can lead to apoptosis. Indeed, this increased synthesis of HEXIM1 might be the mechanism of action of these HDACis for their effects in leukemias, lymphomas, and other human cancers.
Of interest, we and others observed little effect of HDAC inhibition in primary resting CD4+ T cells (46, 52–54). In these cells, levels of P-TEFb are vanishingly low, due to actions of specific miRNAs and NF-90 (11–13, 55), which block the translation of CycT1 mRNA. This lack of P-TEFb is thought to be critical for the establishment and maintenance of HIV latency (for review see Refs. 1, 3). However, upon increased synthesis of P-TEFb, the ability our most potent HDACi, SAHA, to reactivate HIV was restored in these cells. T cell activation, PKC and TLR agonists can all increase the synthesis of CycT1 and CDK9 in primary resting CD4+ T cells. This finding also confirms the central role that P-TEFb plays in HIV reactivation in all cells.
Several previous studies suggested that class I HDACis and specifically those that inhibit HDAC3, rather than class II HDACis, might be optimal for HIV reactivation (56–58). Indeed, our specific class II HDACi, TBSA, had no effect in our cells. However, MS-275 also had little to no effect in Jurkat cells and other studies found conflicting results using yet other class I-specific HDACis in different cell lines (56, 57). Thus, it is possible that the relevant targeted HDAC does not increase histone H3 or tubulin acetylation. Nevertheless, this HDAC could deacetylate CycT1, whose release from 7SK snRNP and subsequent association with BRD4 depend on three acetylated lysines in the middle of the protein (19, 59). Alternatively, this HDAC could affect the acetylation of histone H4, which is required for the binding of BRD4 and possibly other members of this family to chromatin (60, 61). Thus, because BET bromodomain inhibition by JQ1, iBET, and related compounds, which release BRD4 from chromatin, also leads to HIV reactivation (23, 29, 30, 33, 42), the inhibition of effective HDACs might impact overall chromatin structure rather than specific epigenetic modifications at the HIV LTR. These changes in chromatin structure (compaction/decompaction) are expected to occur rapidly, as has been observed with JQ1 and HMBA (60, 61). They would also change transcriptional profiles of the cell. In this scenario, HDACi-induced chromatin stresses, like those from DNA damage (UV light), arrested transcription (DRB, flavopiridol), apoptosis, and BET bromodomain inhibition would then lead to the release free P-TEFb from the 7SK snRNP.
This correlation between P-TEFb and HIV reactivation contains important lessons for any future strategy to eradicate the viral reservoir from the infected host. Obviously, optimal HAART will have to be maintained during this treatment so as to minimize the spread of the virus to new cells. Minimally, the HIV antilatency therapy will contain agents that will increase levels of P-TEFb and others that will affect the P-TEFb equilibrium. For example, PKC agonists, such as bryostatin 1, will be combined with the optimal HDACi, which at the moment is SAHA or vorinostat, a drug that is approved for human use against cutaneous T cell lymphoma (51). However, because cells do not tolerate high levels of free P-TEFb, after its disruption, the 7SK snRNP reassembles quickly due to the increased synthesis of HEXIM1. For HIV, it is important to note that once Tat is made, it competes with HEXIM1 for the binding to P-TEFb. Thus, it can utilize P-TEFb from any source to sustain HIV transcription and replication even after cells have returned to their resting state. With this knowledge, frequent but sustained cycles of increased synthesis, release, and reassembly of P-TEFb might indeed impact the reservoir of HIV in the infected host.
Acknowledgments
We thank Drs. Manfred Jung and Kotaro Shirakawa for reagents.
This work was supported in part by Collaboratory of AIDS Researchers for Eradication (CARE) Center Grant U19 AI96113.
- HAART
- highly active antiretroviral therapy
- BET
- bromodomain and extraterminal
- BRD4
- bromodomain protein 4
- CDK9
- cyclin-dependent kinase 9
- CycT1
- cyclin T1
- DMSO
- dimethyl sulfoxide
- HDACi
- histone deacetylase inhibitor
- HEXIM1/2
- HMBA-induced proteins 1 and 2
- HMBA
- hexamethylene bisacetamide
- Luc
- luciferase
- MS
- medium salt
- NELF
- negative elongation factor
- P-TEFb
- positive transcription elongation factor b
- qPCR
- quantitative PCR
- RNAPII
- RNA polymerase II
- SAHA
- suberoylanilide hydroxamic acid
- 7SK snRNP
- 7SK small nuclear ribonucleoprotein
- TAR
- transactivation response
- TBSA
- tubastatin A.
REFERENCES
- 1. Contreras X., Lenasi T., Peterlin B. M. (2006) HIV latency: present knowledge, future directions. Future Virol. 1, 733–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eisele E., Siliciano R. F. (2012) Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity 37, 377–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Richman D. D., Margolis D. M., Delaney M., Greene W. C., Hazuda D., Pomerantz R. J. (2009) The challenge of finding a cure for HIV infection. Science 323, 1304–1307 [DOI] [PubMed] [Google Scholar]
- 4. Mbonye U., Karn J. (2011) Control of HIV latency by epigenetic and nonepigenetic mechanisms. Curr. HIV Res. 9, 554–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xing S., Siliciano R. F. (2012) Targeting HIV latency: pharmacologic strategies toward eradication. Drug Discov. Today pii, S1359–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shan L., Deng K., Shroff N. S., Durand C. M., Rabi S. A., Yang H. C., Zhang H., Margolick J. B., Blankson J. N., Siliciano R. F. (2012) Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity 36, 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ott M., Geyer M., Zhou Q. (2011) The control of HIV transcription: keeping RNA polymerase II on track. Cell Host Microbe 10, 426–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peterlin B. M., Price D. H. (2006) Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23, 297–305 [DOI] [PubMed] [Google Scholar]
- 9. Fujinaga K., Irwin D., Huang Y., Taube R., Kurosu T., Peterlin B. M. (2004) Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 24, 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barboric M., Lenasi T. (2010) Kick-sTARting HIV-1 transcription elongation by 7SK snRNP deporTATion. Nat. Struct. Mol. Biol. 17, 928–930 [DOI] [PubMed] [Google Scholar]
- 11. Budhiraja S., Famiglietti M., Bosque A., Planelles V., Rice A. P. (2013) Cyclin T1 and CDK9 T-loop phosphorylation are down-regulated during establishment of HIV-1 latency in primary resting memory CD4+ T cells. J. Virol. 87, 1211–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chiang K., Rice A. P. (2012) MicroRNA-mediated restriction of HIV-1 in resting CD4+ T cells and monocytes. Viruses 4, 1390–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chiang K., Sung T. L., Rice A. P. (2012) Regulation of cyclin T1 and HIV-1 replication by microRNAs in resting CD4+ T lymphocytes. J. Virol. 86, 3244–3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He N., Liu M., Hsu J., Xue Y., Chou S., Burlingame A., Krogan N. J., Alber T., Zhou Q. (2010) HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol. Cell 38, 428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He N., Zhou Q. (2011) New insights into the control of HIV-1 transcription: when Tat meets the 7SK snRNP and super-elongation complex (SEC). J. Neuroimmune Pharmacol. 6, 260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luo Z., Lin C., Shilatifard A. (2012) The super-elongation complex (SEC) family in transcriptional control. Nat. Rev. Mol. Cell Biol. 13, 543–547 [DOI] [PubMed] [Google Scholar]
- 17. Marshall R. M., Grana X. (2006) Mechanisms controlling CDK9 activity. Front. Biosci. 11, 2598–2613 [DOI] [PubMed] [Google Scholar]
- 18. Peterlin B. M., Brogie J. E., Price D. H. (2012) 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip. Rev. RNA 3, 92–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schröder S., Cho S., Zeng L., Zhang Q., Kaehlcke K., Mak L., Lau J., Bisgrove D., Schnölzer M., Verdin E., Zhou M. M., Ott M. (2012) Two-pronged binding with bromodomain-containing protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. J. Biol. Chem. 287, 1090–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takahashi H., Parmely T. J., Sato S., Tomomori-Sato C., Banks C. A., Kong S. E., Szutorisz H., Swanson S. K., Martin-Brown S., Washburn M. P., Florens L., Seidel C. W., Lin C., Smith E. R., Shilatifard A., Conaway R. C., Conaway J. W. (2011) Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146, 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barboric M., Yik J. H., Czudnochowski N., Yang Z., Chen R., Contreras X., Geyer M., Matija Peterlin B., Zhou Q. (2007) Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 35, 2003–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Archin N. M., Espeseth A., Parker D., Cheema M., Hazuda D., Margolis D. M. (2009) Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res. Hum. Retroviruses 25, 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bartholomeeusen K., Xiang Y., Fujinaga K., Peterlin B. M. (2012) Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein. J. Biol. Chem. 287, 36609–36616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Contreras X., Barboric M., Lenasi T., Peterlin B. M. (2007) HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 3, 1459–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xing S., Bullen C. K., Shroff N. S., Shan L., Yang H. C., Manucci J. L., Bhat S., Zhang H., Margolick J. B., Quinn T. C., Margolis D. M., Siliciano J. D., Siliciano R. F. (2011) Disulfiram reactivates latent HIV-1 in a Bcl-2-transduced primary CD4+ T cell model without inducing global T cell activation. J. Virol. 85, 6060–6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ying H., Zhang Y., Lin S., Han Y., Zhu H. Z. (2010) Histone deacetylase inhibitor Scriptaid reactivates latent HIV-1 promoter by inducing histone modification in in vitro latency cell lines. Int. J. Mol. Med. 26, 265–272 [DOI] [PubMed] [Google Scholar]
- 27. Biglione S., Byers S. A., Price J. P., Nguyen V. T., Bensaude O., Price D. H., Maury W. (2007) Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib, and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology 4, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blazek D., Kohoutek J., Bartholomeeusen K., Johansen E., Hulinkova P., Luo Z., Cimermancic P., Ule J., Peterlin B. M. (2011) The cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 25, 2158–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Banerjee C., Archin N., Michaels D., Belkina A. C., Denis G. V., Bradner J., Sebastiani P., Margolis D. M., Montano M. (2012) BET bromodomain inhibition as a novel strategy for reactivation of HIV-1. J. Leukoc. Biol. 92, 1147–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boehm D., Calvanese V., Dar R. D., Xing S., Schroeder S., Martins L., Aull K., Li P. C., Planelles V., Bradner J. E., Zhou M. M., Siliciano R. F., Weinberger L., Verdin E., Ott M. (2013) BET bromodomain-targeting compounds reactivate HIV from latency via a Tat-independent mechanism. Cell Cycle 12, 452–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Choudhary S. K., Archin N. M., Margolis D. M. (2008) Hexamethylbisacetamide and disruption of human immunodeficiency virus type 1 latency in CD4+ T cells. J. Infect. Dis. 197, 1162–1170 [DOI] [PubMed] [Google Scholar]
- 32. Gallastegui E., Marshall B., Vidal D., Sanchez-Duffhues G., Collado J. A., Alvarez-Fernández C., Luque N., Terme J. M., Gatell J. M., Sánchez-Palomino S., Muñoz E., Mestres J., Verdin E., Jordan A. (2012) Combination of biological screening in a cellular model of viral latency and virtual screening identifies novel compounds that reactivate HIV-1. J. Virol. 86, 3795–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li Z., Guo J., Wu Y., Zhou Q. (2013) The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat-transactivation. Nucleic Acids Res. 41, 277–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shishido T., Wolschendorf F., Duverger A., Wagner F., Kappes J., Jones J., Kutsch O. (2012) Selected drugs with reported secondary cell-differentiating capacity prime latent HIV-1 infection for reactivation. J. Virol. 86, 9055–9069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ying H., Zhang Y., Zhou X., Qu X., Wang P., Liu S., Lu D., Zhu H. (2012) Selective histone deacetylase inhibitor M344 intervenes in HIV-1 latency through increasing histone acetylation and activation of NF-κB. PLoS One 7, e48832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bradner J. E., West N., Grachan M. L., Greenberg E. F., Haggarty S. J., Warnow T., Mazitschek R. (2010) Chemical phylogenetics of histone deacetylases. Nat. Chem. Biol. 6, 238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Heltweg B., Dequiedt F., Marshall B. L., Brauch C., Yoshida M., Nishino N., Verdin E., Jung M. (2004) Subtype selective substrates for histone deacetylases. J. Med. Chem. 47, 5235–5243 [DOI] [PubMed] [Google Scholar]
- 38. Butler K. V., Kalin J., Brochier C., Vistoli G., Langley B., Kozikowski A. P. (2010) Rational design and simple chemistry yield a superior, neuroprotective HDAC6 inhibitor, tubastatin A. J. Am. Chem. Soc. 132, 10842–10846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leonard J., Parrott C., Buckler-White A. J., Turner W., Ross E. K., Martin M. A., Rabson A. B. (1989) The NF-κ B binding sites in the human immunodeficiency virus type 1 long terminal repeat are not required for virus infectivity. J. Virol. 63, 4919–4924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Contreras X., Schweneker M., Chen C. S., McCune J. M., Deeks S. G., Martin J., Peterlin B. M. (2009) Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J. Biol. Chem. 284, 6782–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Žumer K., Low A. K., Jiang H., Saksela K., Peterlin B. M. (2012) Unmodified histone H3K4 and DNA-dependent protein kinase recruit autoimmune regulator to target genes. Mol. Cell. Biol. 32, 1354–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhu J., Gaiha G. D., John S. P., Pertel T., Chin C. R., Gao G., Qu H., Walker B. D., Elledge S. J., Brass A. L. (2012) Reactivation of latent HIV-1 by inhibition of BRD4. Cell Rep. 2, 807–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. He N., Pezda A. C., Zhou Q. (2006) Modulation of a P-TEFb functional equilibrium for the global control of cell growth and differentiation. Mol. Cell. Biol. 26, 7068–7076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Felber B. K., Pavlakis G. N. (1988) A quantitative bioassay for HIV-1 based on trans-activation. Science 239, 184–187 [DOI] [PubMed] [Google Scholar]
- 45. Bosque A., Planelles V. (2009) Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 113, 58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lassen K. G., Hebbeler A. M., Bhattacharyya D., Lobritz M. A., Greene W. C. (2012) A flexible model of HIV-1 latency permitting evaluation of many primary CD4 T-cell reservoirs. PLoS One 7, e30176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sung T. L., Rice A. P. (2006) Effects of prostratin on cyclin T1/P-TEFb function and the gene expression profile in primary resting CD4+ T cells. Retrovirology 3, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mehla R., Bivalkar-Mehla S., Zhang R., Handy I., Albrecht H., Giri S., Nagarkatti P., Nagarkatti M., Chauhan A. (2010) Bryostatin modulates latent HIV-1 infection via PKC and AMPK signaling but inhibits acute infection in a receptor-independent manner. PLoS One 5, e11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wender P. A., Baryza J. L., Brenner S. E., DeChristopher B. A., Loy B. A., Schrier A. J., Verma V. A. (2011) Design, synthesis, and evaluation of potent bryostatin analogs that modulate PKC translocation selectivity. Proc. Natl. Acad. Sci. U.S.A. 108, 6721–6726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Williams S. A., Chen L. F., Kwon H., Ruiz-Jarabo C. M., Verdin E., Greene W. C. (2006) NF-κB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 25, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gryder B. E., Sodji Q. H., Oyelere A. K. (2012) Targeted cancer therapy: giving histone deacetylase inhibitors all they need to succeed. Future Med. Chem. 4, 505–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Friedman J., Cho W. K., Chu C. K., Keedy K. S., Archin N. M., Margolis D. M., Karn J. (2011) Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J. Virol. 85, 9078–9089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sahu G. K., Cloyd M. W. (2011) Latent HIV in primary T lymphocytes is unresponsive to histone deacetylase inhibitors. Virol. J. 8, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tyagi M., Pearson R. J., Karn J. (2010) Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J. Virol. 84, 6425–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hoque M., Shamanna R. A., Guan D., Pe'ery T., Mathews M. B. (2011) HIV-1 replication and latency are regulated by translational control of cyclin T1. J. Mol. Biol. 410, 917–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Archin N. M., Keedy K. S., Espeseth A., Dang H., Hazuda D. J., Margolis D. M. (2009) Expression of latent human immunodeficiency type 1 is induced by novel and selective histone deacetylase inhibitors. AIDS 23, 1799–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huber K., Doyon G., Plaks J., Fyne E., Mellors J. W., Sluis-Cremer N. (2011) Inhibitors of histone deacetylases: correlation between isoform specificity and reactivation of HIV type 1 (HIV-1) from latently infected cells. J. Biol. Chem. 286, 22211–22218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Keedy K. S., Archin N. M., Gates A. T., Espeseth A., Hazuda D. J., Margolis D. M. (2009) A limited group of class I histone deacetylases acts to repress human immunodeficiency virus type 1 expression. J. Virol. 83, 4749–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cho S., Schroeder S., Kaehlcke K., Kwon H. S., Pedal A., Herker E., Schnoelzer M., Ott M. (2009) Acetylation of cyclin T1 regulates the equilibrium between active and inactive P-TEFb in cells. EMBO J. 28, 1407–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ai N., Hu X., Ding F., Yu B., Wang H., Lu X., Zhang K., Li Y., Han A., Lin W., Liu R., Chen R. (2011) Signal-induced Brd4 release from chromatin is essential for its role transition from chromatin targeting to transcriptional regulation. Nucleic Acids Res. 39, 9592–9604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhao R., Nakamura T., Fu Y., Lazar Z., Spector D. L. (2011) Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat. Cell Biol. 13, 1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]