Background: Pyruvate transport is vital for growth and survival of T. brucei bloodstream form.
Results: TbPT0 is a unique monocarboxylate transporter with preference for pyruvate and essential for bloodstream form viability.
Conclusion: TbPT0 represents the first molecular identification of essential pyruvate transporters in T. brucei.
Significance: Unique T. brucei pyruvate transporters provide a potential drug target against the bloodstream form parasites.
Keywords: Cell Death, Parasite Metabolism, Pyruvate, Transport, Trypanosoma brucei, Drug Target
Abstract
Pyruvate export is an essential physiological process for the bloodstream form of Trypanosoma brucei as the parasite would otherwise accumulate this end product of glucose metabolism to toxic levels. In the studies reported here, genetic complementation in Saccharomyces cerevisiae has been employed to identify a gene (TbPT0) that encodes this vital pyruvate transporter from T. brucei. Expression of TbPT0 in S. cerevisiae reveals that TbPT0 is a high affinity pyruvate transporter. TbPT0 belongs to a clustered multigene family consisting of five members, whose expression is up-regulated in the bloodstream form. Interestingly, TbPT family permeases are related to polytopic proteins from plants but not to characterized monocarboxylate transporters from mammals. Remarkably, inhibition of the TbPT gene family expression in bloodstream parasites by RNAi is lethal, confirming the physiological relevance of these transporters. The discovery of TbPT0 reveals for the first time the identity of the essential pyruvate transporter and provides a potential drug target against the mammalian life cycle stage of T. brucei.
Introduction
Trypanosoma brucei causes African trypanosomiasis in Saharan and sub-Saharan Africa. African trypanosomiasis is transmitted by the bite of the tsetse fly and is fatal if untreated, causing an economical and medical burden for African countries. Moreover, anti-trypanosomal therapy is far from ideal; it is expensive and toxic, and there is increasing incidence of drug resistance (1). Vaccination against these parasites is unlikely to be effective due to extensive antigenic variation of the variant surface glycoproteins (2). Therefore, there is an urgent need for new drug targets to develop more suitable anti-trypanosomal drugs.
In the past two decades, molecular approaches have helped to define targets for drugs in trypanosomes (3, 4). Glycolysis has been studied as a potential target for drug development, because glycolysis is the only source of ATP for the bloodstream form of T. brucei (5). Under normal aerobic conditions, glucose is almost completely converted into pyruvate (>98%) rather than lactate due to the absence of lactate dehydrogenase. Moreover, pyruvate cannot be further metabolized, as BF2 trypanosomes lack functional mitochondria and the Krebs cycle. The pyruvate is excreted into the bloodstream of the host by specific permeases and would otherwise accumulate to toxic levels inside of the parasite, ultimately causing cell death. This assertion is supported by the observation that inhibition of trypanosome pyruvate transporters by α-cyano-β-(1-phenylindol-3-yl)acrylate (UK5099) resulted in retention and concomitant accumulation of pyruvate within the trypanosomes, causing acidification, osmotic destabilization, and trypanosome death (6, 7). Consequently, plasma membrane pyruvate transporters are important for detoxification and survival of the parasite in the mammalian host.
Simple monocarboxylates are substrates for bidirectional facilitated diffusion via monocarboxylate transporters (MCTs) and are co-transported with protons in an equimolar manner by a symport mechanism (8, 9). The physiological role of MCTs is primarily to transport the endogenous monocarboxylates such as lactate, pyruvate, acetoacetate, and β-hydroxybutytrate. Currently, 14 members of the MCT family have been identified by sequence homology, seven have been functionally characterized in mammals (10, 11), and orthologues have been identified in a variety of species such as S. cerevisiae (12).
The presence of pyruvate transporters in the plasma membrane of T. brucei has been established by biochemical studies demonstrating their physiological importance (6, 7, 13, 14). However, the molecular identification of these important transporters has eluded discovery over an extended period of time. In the present study, I report the cloning and functional analysis of a T. brucei gene that encodes a novel pyruvate transporter TbPT0, which belongs to a multigene family of five members arranged in tandem and encoding distinct but very similar proteins. Notably, inhibition of TbPTs expression by RNAi is lethal, underscoring their critical function for survival and their potential as drug targets.
EXPERIMENTAL PROCEDURES
Chemicals
[1-14C]Pyruvic acid, sodium salt (17 mCi mmol−1) was purchased from PerkinElmer Health Sciences (Boston, MA); [1,2-14C]acetic acid, sodium salt (98 mCi mmol−1) purchased from Moravek Biochemicals (Brea, CA), and [1-14C]lactic acid, sodium salt (55 mCi mmol−1) purchased from American Radiolabeled Chemicals, Inc. (St Louis, MO). The compound UK5099 was kindly provided by Pfizer, Inc. (Groton, CT). All other chemicals were of the highest commercial quality available.
Growth and Transfection of T. brucei Cell Lines
BF and PF T. brucei cell lines were grown as described previously (15). For T7 RNA polymerase-independent driven expression (16), BF T. brucei 427 parasites transfected with pHD1313 (plasmid kindly provided by Dr. Christine Clayton) (17) expressing the tetracycline repressor were generated and grown in 2.5 μg ml−1 bleomycin. 5–10 μg of linearized plasmid DNA was used to transfect mid-log phase BF parasites as described (18). Inducible expression experiments employing the plasmid p82M3HA-driven expression by the T7 promoter were performed using the transgenic BF single marker (SM) clone as described previously (18).
RNA Isolation and Northern Blot
Total RNA from T. brucei parasites was isolated using RNeasy Mini kit (Qiagen) according to the manufacturer's instructions. Northern blot analysis was performed as described elsewhere (19). For autoradiographic detection the Storm 825 scanner (GE Healthcare) was employed and NIH ImageJ software for densitometry analysis. Adobe Photoshop CS3 and Adobe Illustrator CS3 were used to create figure composition.
Yeast Strain and Growth Conditions
S. cerevisiae strain JMY75 (MATa leu3 112ura3–52 his3-Δ1 MAL2–8c SUC2 pyk1Δ::LEU2 mae1Δ::kanMX jen1Δ::loxP), kindly provided by Dr. Eckhard Boles (20), was grown aerobically at 30 °C on a rotary shaker or on yeast nitrogen base medium (YNB) agar plates (+Trp, +His, +ura, 2% ethanol, 10 mm alanine). Strain 5067 (MATa his3Δ1 leu3Δ0 met3Δ0 Δjen1::kanMX) (Invitrogen) was grown aerobically at 30 °C on a rotary shaker or on Dropout Base media, CM-ura (BIO 101 Systems), 200 μg ml−1 G418 agar plates.
Cloning by Genetic Complementation and Isolation of Plasmids
Chemically competent JMY75 cells were transformed with a cDNA library from T. brucei Lister 427 BF parasites cloned in the yeast expression vector p416Met25 (21) kindly provided by Dr. Hosam Shams-Eldin. Transformed yeast were grown at 30 °C on YNB-ura agar plates containing 2% ethanol, 10 mm pyruvate and supplemented for auxotrophic requirements. Plasmids from positive yeast clones were isolated using PureLink Quick Plasmid Miniprep kit (Invitrogen) following the manufacturer's instructions. Then, plasmids were transformed into Escherichia coli DH5α competent cells (Invitrogen) for amplification and were isolated and sequenced by the Core Facility of the Department of Molecular Microbiology & Immunology at the Oregon Health & Sciences University using an Applied Biosystems 16-capillary 3130xI Automated Sequence Analysis System.
Deduced Amino Acid Sequence Analysis
For DNA sequence analysis of TbPT0 through TbPT5 and amino acid sequence alignments, MacVector software (Intelligenetics) was used. Transmembrane segments were predicted by the TMHMM server (version 2.0). Sequence similarity analysis was performed using the BLAST program (version 2.0).
Uptake Assays in Yeast
JMY75 yeast harboring the TbPT0 cDNA clone was employed for kinetic analysis. Also, the pyruvate transport-deficient yeast 5067 (Δjen1) was transformed with the TbPT0 ORF subcloned into the constitutive expression vector pYADH (22) for functional experiments. Uptake of [14C]pyruvate was assayed by incubation of 5 × 107 cells with radiolabeled substrate followed by centrifugation through a cushion of dibutyl phthalate as described (22). For kinetic analysis, uptake was measured for a range of substrate concentrations over a time course ranging from 0 to 60 s. Initial uptake rates at each concentration were determined by linear regression analysis over the linear portion of the time course (Prisam, version 4, GraphPad Software, Inc.). These data were fitted to the Michaelis-Menten equation by non-linear regression using the Prism 4 (GraphPad Software, Inc.), and this fit was used to determine Km values. Assays for inhibition utilized a 1-min incubation with 26 μm [14C]pyruvate in the presence of the indicated competitors. Ki values were estimated by fitting the data to one site competition equation (23) employing Prism (version 4, GraphPad Software, Inc.).
Inhibition of Gene Expression by RNAi
To determine the role of pyruvate transporters in the survival of the BF parasite, a 428-bp gene fragment corresponding to position 632 to 1059 bp of TbPT0 determined by RNAit software was employed. The TbPT-RNAi construct was linearized and transfected into BF/pHD1313 cell line as described to generate BF/TbPT-RNAi clone. RNAi clones were selected by resistance to 2.5 μg ml−1 of bleomycin and 5 μg ml−1 of blasticidin and hairpin loop RNAi induced by addition of 1 μg ml−1 doxycycline.
Inducible Expression of TbPT0-3HA-COOH SM Cell Line and Immunofluorescence Microscopy
To generate TbPT0-3HA-COOH tagged transporter the TbPT0 ORF was subcloned into the plasmid p82M3HA (18) for inducible expression driven by the T7 promoter upon addition of doxycycline. SM parasites were transfected with 5 μg of linear NotI-digested construct as described (18) selected on 2.5 μg ml−1 bleomycin and 2.5 ng ml−1 doxycycline, and cloned by limiting dilution. To induce TbPT0-3HA-COOH, 1 μg ml−1 doxycycline was added to the cell culture and incubated for 48 h. Induced parasites (5 × 106) were centrifuged at 700 × g for 4 min and washed twice at room temperature with PBS, pH 7.2 containing 10 mm glucose. The cell pellet was resuspended in 4% paraformaldehyde in PBS, pH 7.2, and incubated for 15 min at room temperature, cells were centrifuged as described above and washed once with 10 mm glycine in PBS and once with PBS, resuspended in 100 μl of PBS, spotted onto poly-l-lysine-coated coverslips, and blocked with 2% goat serum, 0.01% sodium azide, 0.01% saponin in PBS (blocking solution) for 1 h at room temperature, rinsed three times with PBS and incubated with a 1:500 dilution of the primary mouse anti-HA monoclonal antibodies (Covance, Inc.) for 1 h at room temperature. Then, cells were rinsed as before and incubated with a 1:1000 dilution of goat anti-mouse IgG Alexa Fluor 495TM (Molecular Probes, Invitrogen) in blocking solution for 1 h at room temperature in the dark. For detection of α-tubulin as subpellicular plasma membrane marker, a second round of immunodetection was performed as mentioned above using 1:1000 dilution of the primary mouse anti α-tubulin monoclonal antibodies (Sigma-Aldrich) and 1:1000 dilution of goat anti-mouse IgG Alexa Fluor 580TM (Molecular Probes, Invitrogen). Coverslips were rinsed three times with PBS and mounted onto slides using ProLong® Gold antifade reagent with DAPI (Invitrogen). Fluorescence images were obtained using a wide field deconvolution system (Applied Precision Instruments, Inc.) consisting of an inverted Nikon TE 200 Eclipse microscope, a Kodak CH350 CCD camera, and the Deltavision operating system. Images were acquired using a 60× objective in a 1024 × 1024 format and deconvolved with nine iterations using SoftWoRx software. Adobe Photoshop CS3 and Adobe Illustrator CS3 (Adobe Systems, Inc.) were used to create image compositions.
RESULTS
Cloning of a Pyruvate Transporter Gene by Genetic Complementation of Pyruvate-auxotrophic Yeast
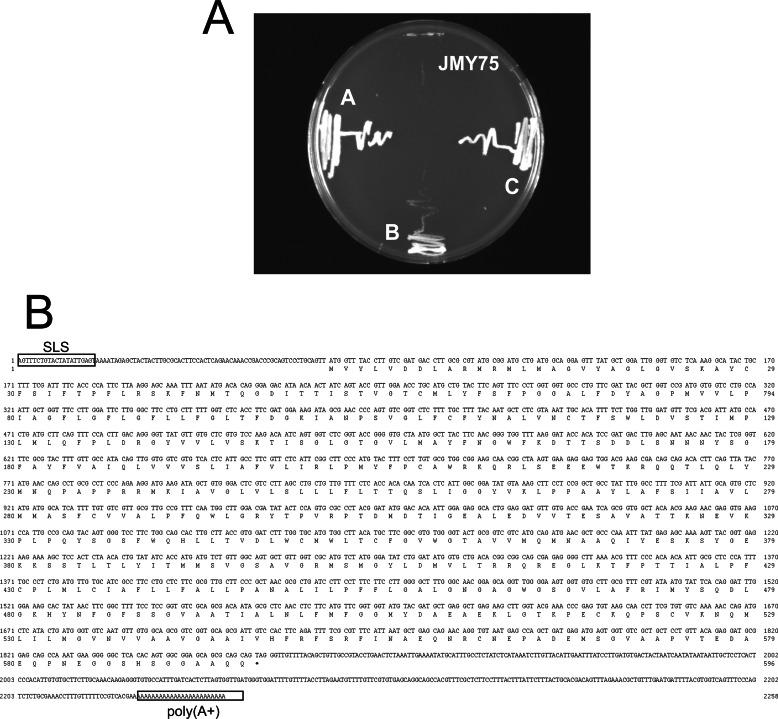
Pyruvate transport has been characterized in T. brucei BF parasites by biochemical means employing whole cells (6, 13). Those studies also highlighted the importance of pyruvate efflux to maintain optimal physiological conditions for parasite survival. However, the molecular identity of such transport proteins has been evasive, precluding molecular studies on this essential cellular function. To identify a bona fide pyruvate transporter, a T. brucei bloodstream form cDNA library, previously used to clone other trypanosomal genes (21, 24, 25), was transformed into the pyruvate auxotrophic mutant of S. cerevisiae JMY75 (20). The JMY75 yeast mutant, deficient in pyruvate kinase, malic enzyme, and pyruvate transporter, can use pyruvate as an efficient carbon source only if the pyruvate transport deficiency conferred by the jen1Δ mutation is complemented by expression of a functional pyruvate permease. Thus, transformants were selected on YNB agar plates (+Trp, +His, -ura, 2% ethanol) containing 10 mm pyruvate as the sole energy source. After 4 days, three yeast colonies, called A, B, and C, grew on the selective plates. To confirm the phenotype, these clones and the JMY75 host strain were streaked on selective medium. The three cDNA clones were able to rescue the growth of the JMY75 mutant, whereas negative control non-transformed JMY75 mutant did not (Fig. 1A), confirming the ability of these three clones to rescue the pyruvate auxotrophy. Plasmid DNA from each clone was isolated and sequenced. Clones A and C contained the same cDNA insert including the spliced leader sequence, a 105-bp 5′-UTR, an ORF encoding a 595-amino acid protein, a 364-bp 3′-UTR, and a poly(A) tail (Fig. 1B). The deduced amino acid sequence obtained from the cDNA ORF is named T. brucei pyruvate transporter 0 (TbPT0) and shown in Fig. 1B. Clone B contained a cDNA insert different from A and C.
FIGURE 1.
Genetic complementation of pyruvate auxotrophic yeast by T. brucei cDNA clones. A, clones A, B, C, and JMY75 pyruvate auxotrophic yeast (top quadrant of plate) were grown on solid YNB medium supplemented with Trp, His, 2% ethanol, and 10 mm pyruvate and incubated at 30 °C for 4 days. B, TbPT0 cDNA and predicted amino acid sequence. The cDNA sequence, including the splice leader sequence (SLS), 5′-UTR, ORF, 3′-UTR, poly(A+) tail, and predicted amino acid sequence, are depicted.
A BLAST search of the T. brucei genome database using the TbPT0 amino acid sequence as query revealed significant identity (Table 1) to hypothetical proteins with systematic names of Tb927.3.4070, Tb927.3.4080, Tb927.3.4090, Tb927.3.4100, and Tb927.3.4110. I have named the ORFs TbPT1, TbPT2, TbPT3, TbPT4, and TbPT5, respectively. Moreover, a homologous TbPT gene family is present in the T. brucei Lister 427 and other T. brucei species database. All five TbPT genes are clustered together in chromosome 3, and their expression is up-regulated ∼13-fold in the BF compared with PF as previous transcriptome analysis reported, albeit some members of the family are preferentially up-regulated (26–29). However, clone B revealed significant identity to pyruvate kinase, PYK1 (high score of 1034 and P (N) 4.9 × 10−106 (the probability of a chance alignment occurring with a better score in a database search); Tb927.10.14140), which genetically complemented the deficiency of the pyruvate kinase gene in JMY75 yeast.
TABLE 1.
TbPT0 homologs in T. brucei and orthologs in T. cruzi and L. major
The alignment (score) and probability (P(N)) scores are indicated for a BLASTP search employing TbPT0 as query.
| Systematic name | Score | P(N) | Name |
|---|---|---|---|
| Tb927.3.4070 | 2849 | 7.1 × 10−298 | TbPT1 |
| Tb927.3.4080 | 2748 | 3.6 ×10−287 | TbPT2 |
| Tb927.3.4090 | 2802 | 6.8 × 10−293 | TbPT3 |
| Tb927.3.4100 | 2895 | 9.4 × 10−303 | TbPT4 |
| Tb927.3.4110 | 2666 | 1.7 × 10−278 | TbPT5 |
| Tc00.1047053503837.10 | 1753 | 6.8 × 10−182 | |
| Tc00.1047053509669.170 | 1704 | 1.1 × 10−176 | |
| Tc00.1047053506175.110 | 1696 | 7.4 × 10−176 | |
| Tc00.1047053506505.30 | 1670 | 4.2 × 10−173 | |
| Tc00.1047053507677.160 | 1650 | 5.6 × 10−171 | |
| LmjF29.1500 | 751 | 2.2 × 10−149 | |
| LmjF29.1510 | 751 | 2.8 × 10−149 |
Amino Acid Sequence Analysis of TbPT0-TbPT5
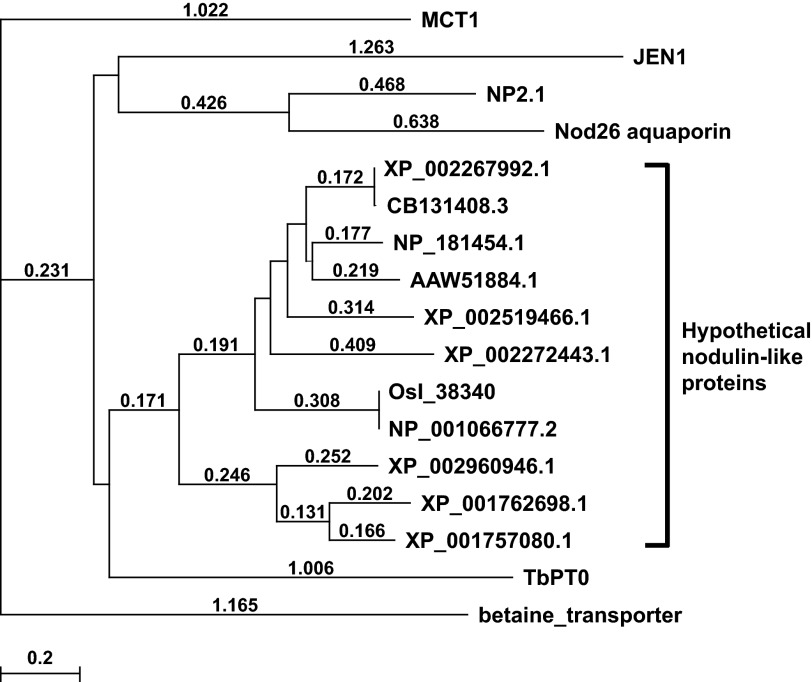
In silico analysis also revealed that all of the TbPT family members encode highly conserved proteins with at least 84% identity and different lengths. Analysis of the genomic 5′-flanking sequences revealed another ATG upstream of the annotated (TriTrypDB database) starting ATG for TbPT1, TbPT2, TbPT3, and TbPT4 genes, similar to the ORF of TbPT0 cDNA and TbPT5. Multi-alignment of these predicted proteins and TbPT0 demonstrates the high identity among them, beginning from the upstream ATG (Fig. 2). Of note, Siegel et al. (30), in their studies to determine the transcriptional landscape of T. brucei BF and PF employing RNA-seq technology, predicted that this first ATG in TbPT1 through TbPT4 is preferentially used to encode the larger ORF. Moreover, the algorithm TMHMM (version 2.0) predicts that all five transporters encompass 13–14 transmembrane domains (TMs). Most of the amino acid differences between the TbPT family reside in the first transmembrane domain and the first putative extracellular loop or in the distinctive COOH termini for TbPT2 and TbPT4. Interestingly, the TbPT0 gene cloned by functional complementation (from T. brucei Lister 427 cDNA library) is not 100% identical to any of the TbPTs annotated in the T. brucei genome database (including T. brucei Lister 427 and strain 927) consistent with genetic polymorphism among these T. brucei strains, including the Lister 427 isolate employed for the cDNA library versus that used for the genomic sequence. A BLAST search of GeneBank employing TbPT0 as query sequence revealed significant similarity to hypothetical nodulin-like proteins from plants, which also predict to be members of the major facilitator superfamily of transporters with the highest scoring for the protein with accession no. EAY83124.1 (85.2 and P (N) 3 × 10−14 (the probability of a chance alignment occurring with a better score in a database search)) from Oryza sativa. However, no similarity was found to any known mammalian monocarboxylate transporter. A multi-alignment of 17 protein sequences, including 11 among the highest scores based on the BLAST results plus the rat monocarboxylate transporter (MCT1), yeast lactate transporter (JEN1), two nodulin proteins from Arabidopsis thaliana (NP2.1 lactate transporter and Nod26 aquaporin), betaine transporter from rabbit, and TbPT0 was performed using ClustalW software, and an evolutionary tree was generated (Fig. 3), suggesting a common ancestor for nodulin-like proteins from plants and trypanosome pyruvate transporters. Interestingly, NP2.1, a lactate transporter, which is up-regulated by oxygen deprivation in A. thaliana (31), also belongs to the generic nodulin-like proteins albeit with only 6 TMs, whereas all of the putative proteins in the TbPT0 clade predict 12–14 TMs. Of interest, a signature sequence localized in the TM13–TM14 loop composed of six amino acids, MYDAEA, is present in all the TbPTs (Fig. 2, double underlining) and also is highly conserved in all the proteins of the TbPT0 clade (Fig. 3). However, this sequence signature is not present in MCT1, JEN1, betaine transporter, Nod26, and NP2.1 lactate transporter. Moreover, BLAST searches employing TbPT0 sequence revealed the presence of highly similar predicted proteins in Trypanosoma cruzi and Leishmania major but not in Plasmodium falciparum or Toxoplasma gondii (Table 1), suggesting that those orthologues might exert similar biochemical functions in those kinetoplastids.
FIGURE 2.
Multi-alignment of the deduced amino acid sequence of TbPT0, TbPT1, TbPT2, TbPT3, TbPT4, and TbPT5. Alignment was performed using CLUSTALW software (MacVector, Intelligenetics). Labeled solid lines over the TbPT0 sequence indicate the predicted TM by TMHMM software. Identities are showed by the black background and similarities by gray background. The double line under the TbPT5 sequence indicates the MYDAEA sequence signature. The numbers at the left and right indicate amino acid positions.
FIGURE 3.
Phylogenetic tree for the bona fide and putative carboxylate transporters. Branch lengths indicate evolutionary distances (values over the branches). As an approximate guide, a value of 0.1 corresponds to a difference of ∼10% between sequences. The tree was generated using MacVector software employing the Neighbor Joining Best Tree method. TbPT0, T. brucei pyruvate transporter 0; NP2.1, lactate transporter, Nod26, aquaporin, NP_181454.1, nodulin-like protein (Arabidopsis thaliana); MCT1, monocarboxylate transporter 1 (Rattus norvegicus); Jen1, lactate transporter (Saccharomyces cerevisiae); XP_002267992, CB131408.3, XP_002519466.1, XP_002272443.1, nodulin-like proteins (Vitis vinifera); Osl_38340, NP_001066777.2, nodulin-like proteins (Oryza sativa); XP_001762698.1, XP_001757080.1, nodulin-like proteins (Physcomitrella patens); XP_002960946.1, nodulin-like protein (Selaginella moellendorffii); AAW51884.1, nodulin-like protein (soybean); betaine transporter (Oryctolagus cuniculus). All of the sequences are members of the MFS superfamily of transporters, with the exemption of NP2.1 and Nod26.
Expression of TbPT0 in Yeast
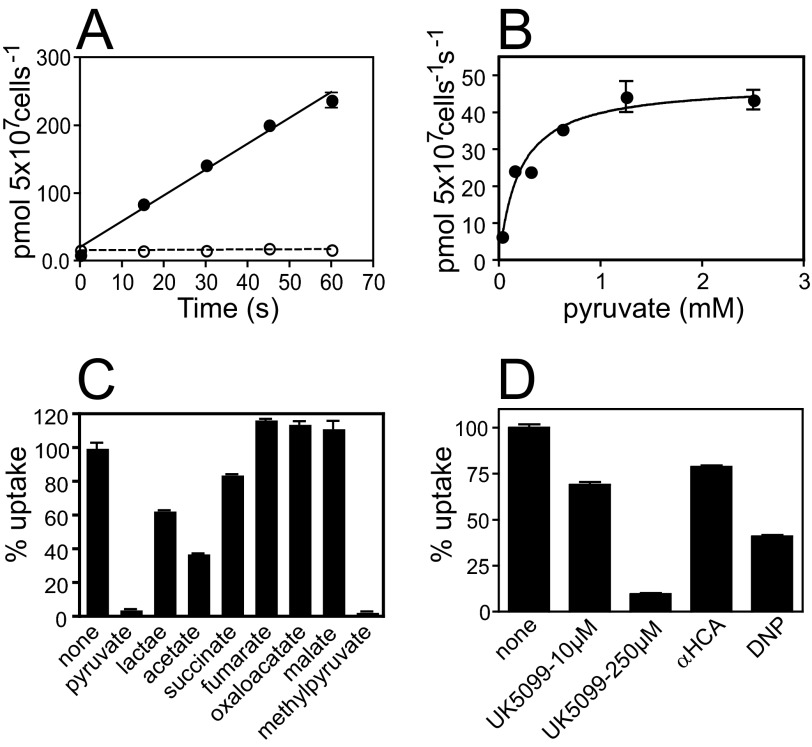
Facilitated transporters have the ability of performing bidirectional transport, and it has been demonstrated in T. brucei that pyruvate transport functions in both directions (6, 13). To determine the functionality of TbPT0, the ORF was subcloned into the constitutive expression vector pYADH (22) and transformed into the yeast Δjen1 pyruvate transport-deficient mutant (Invitrogen). As a negative control for functional experiments, a putative T. brucei transporter (Tb927.5.470), which does not transport pyruvate and localizes to the plasma membrane of the parasite (data not shown), was employed. Uptake of 26 μm [14C]pyruvate from 0–60 s by Δjen1 yeast expressing TbPT0 (Fig. 4A) shows that pyruvate uptake is linear over time and 123-fold higher than the negative control expressing Tb927.5.470. Yeast expressing TbPT0 ORF showed identical transport features compared with the original TbPT0 cDNA clone; therefore, the functional analysis of TbPT0 was carried out employing the original cDNA clone and JMY75 as negative control. Substrate saturation curves for pyruvate (Fig. 4B) in two independent experiments revealed a Km value of 0.21 ± 0.053 mm for pyruvate. Thus, TbPT0 is a bona fide pyruvate transporter.
FIGURE 4.
Functional analysis of TbPT0 in S. cerevisiae. A, time course for uptake of 26 μm [14C]pyruvate by yeast expressing TbPT0 ORF (closed circles) or by yeast expressing Tb927.5.470 (open circles) as control. Error bars represent the S.D. for uptake assays performed in duplicate. B, substrate saturation curve for [14C]pyruvate by yeast expressing TbPT0 cDNA. Uptake for pyruvate was measured for a range of substrate concentrations over a time course ranging from 0 to 60 s. Initial uptake rates at each concentration were determined by linear regression analysis over the linear portion of the time course (Prism4, GraphPad software, Inc.). These data were fitted to the Michaelis-Menten equation by non-linear regression using the Prism4 (GraphPad Software, Inc.), and this fit was used to determine Km values. C, inhibition of 26 μm [14C]pyruvate uptake by 10 mm unlabeled pyruvate, lactate, acetate, succinate, fumarate, oxaloacetate, malate, and methyl pyruvate in yeast expressing TbPT0 cDNA. Uptake assays (1 min) were performed in duplicate, and results are reported as percentage of uptake compared with assays performed in the absence of potential inhibitor. Each data point plotted was adjusted by subtracting the uptake value mediated by the control yeast JMY75 under any given condition. D, inhibition of 26 μm [14C]pyruvate uptake by 10 or 250 μm UK5099, 1 mm α-cyano-4-hydroxy-cinnamic acid (HCA), and 1 mm protonophore 2,4-dinitrophenol (DNP) in yeast expressing TbPT0 cDNA. Uptake assays and analysis was performed as mentioned above.
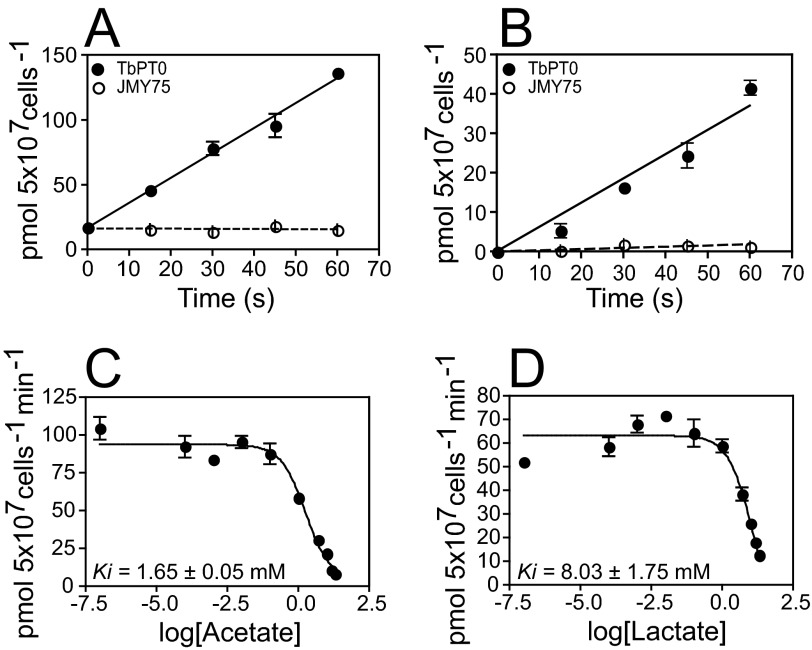
To further dissect the substrate specificity of TbPT0, inhibition of 26 μm [14C]pyruvate uptake was assayed employing different unlabeled compounds as competitors, including carboxylates, two universal MCT inhibitors, and a protonophore. 10 mm of unlabeled pyruvate or methylpyruvate inhibited almost 100% of the [14C]pyruvate transport by TbPT0, whereas 10 mm lactate, acetate, and succinate inhibited partially, and no inhibition was observed by fumarate, malate, or oxaloacetate (Fig. 4C). Hence, the ability of TbPT0 to transport 25.5 μm [14C]acetate and 18 μm [14C]lactate over time was tested. Yeast expressing TbPT0 showed 23-fold increase of acetate uptake (Fig. 5A) and 20-fold increase of lactate uptake (Fig. 5B) compared with yeast negative control. To examine more fully the kinetics of TbPT0 for acetate and lactate, inhibition of 26 μm [14C]pyruvate uptake was measured as a function of acetate or lactate concentration using yeast expressing TbPT0. Uptake of [14C]pyruvate was efficiently inhibited by acetate (Ki = 1.17 ± 0.05 mm) and lactate (Ki = 8.03 ± 1.75 mm) (Fig. 5, C and D), demonstrating that TbPT0 is a monocarboxylate transporter with higher affinity for pyruvate (Km = 0.21 ± 0.053 mm). Also, no significant uptake of [14C]succinate was observed, revealing that succinate is not a real substrate (data not shown), even though it acts as an inhibitor (Fig. 4C). Altogether, these results determine that TbPT0 is a pyruvate/acetate/lactate transporter. The universal MCT inhibitor α-cyano-4-hydroxy-cinnamic acid at 1 mm concentration inhibited ∼20% the [14C]pyruvate transport, whereas 10 μm and 250 μm UK5099 exerted 40 and 90% inhibition, respectively (Fig. 4D), consistent with the observations of Wiemer et al. (7) that pyruvate transport in intact BF parasites is strongly inhibited by UK5099 but poorly by α-cyano-4-hydroxy-cinnamic acid. Finally, the ability of 1 mm protonophore 2,4-dinitrophenol to partially inhibit uptake of pyruvate by TbPT0 (Fig. 4D) suggests that this permease is a proton symporter, in agreement with previous reports by Wiemer et al. (7, 13), Vanderheyden et al. (14), and Nolan and Voorheis (32), which suggested pyruvate transport in intact BF parasites is accompanied by proton flux and also suggested that pyruvate might play an important role in maintaining the intracellular pH in addition to H+-ATPases.
FIGURE 5.
[14C]acetate and [14C]lactate transport by TbPT0. Time course for uptake of 28 μm [14C]acetate (A) and 18 μm [14C]lactate (B) by yeast expressing TbPT0 cDNA (closed circles) or JMY75 (open circles) as control. C, 26 μm [14C]pyruvate uptake inhibition by increasing concentrations of acetate. D, 26 μm [14C]pyruvate uptake inhibition by increasing concentration of lactate. Error bars represent the standard deviation for uptake assays performed in duplicate.
TbPT Gene Family Is Essential for Survival of BF Parasites
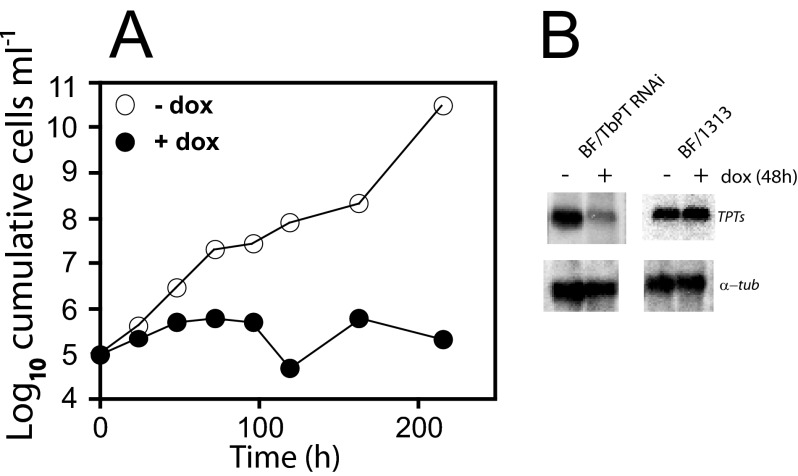
To determine the physiological relevance of TbPTs, the expression of the TbPT gene family was inhibited using a hairpin RNAi construct. The 428-bp DNA fragment used for RNAi experiments exhibits >90% identity between each of the TbPT family members, allowing simultaneous knock down of all five genes. Initially, a transgenic BF clone expressing the tetracycline repressor was generated by transfection of BF parasites with a linear pHD1313 that harbors two copies of this gene separated by the bleomycin-resistant marker (17) and integrated in the tubulin locus under expression of the endogenous promoter. This transgenic BF/1313 clone was used as host cell for subsequent experiments. Then, BF/TbPT0-RNAi cells were generated by transfection of BF/pHD1313 clone with the TbPT0-RNAi construct employing the p3666 vector (16) that transcribes the hairpin loop RNAi unit driven by the EP procyclin promoter fused to four tetracycline operators. Doxycycline was added to induce synthesis of hairpin loop RNA, and the cell number was quantified over time (Fig. 6A, filled circles). Upon induction of RNAi, parasite growth was severely compromised, causing growth arrest and eventually cell death after 2 weeks. In contrast, parasites growing in the absence of doxycycline grew robustly (Fig. 6A, open circles). Northern blot analysis demonstrates that TbPT gene family mRNA level is reduced ∼7-fold after 48 h of RNAi induction (Fig. B, upper left panel), whereas tubulin RNA level remains unchanged (Fig. 6B, lower left panel). Similar analysis employing the transgenic BF/1313 clone as control were performed revealing that both TbPT family mRNA and tubulin mRNA levels did not change (Fig. 6B, upper right and lower right panels). Also, the BF/1313 host did not show any phenotypic change growing in the presence or absence of doxycycline (not shown). These results demonstrate that knocking down TbPT gene family expression caused cell death, presumably because these parasites were unable to export pyruvate. Moreover, RNA interference target sequencing analysis by Alsford et al. (33) revealed that at least two members of the TbPT gene family are essential in BF, and at least one of them is crucial in PF trypanosomes. All of these results emphasize the essential physiological role of these transporters in BF parasites.
FIGURE 6.
Inhibition of the TbPT gene family expression by RNAi. T. brucei BF/1313 clone transfected with the hairpin loop TbPT-RNAi construct was incubated in culture medium containing 1 μg ml−1 doxycycline (+dox; closed circles) or without doxycycline (−dox; open circles). A, growth curve of BF/TbPT-RNAi clone starting with 105 parasites ml−1. Y axis represents the log10 of cumulative cell ml−1, and the x axis represents time (h). B, Northern blot of 5 μg of total RNA isolated from BF/TbPT-RNAi or BF/1313 grown for 48 h in the presence (+) or absence (−) of 1 μg ml−1 doxycycline, hybridized with TbPT0 (upper panels) and α-tubulin (lower panels).
Subcellular Localization of TbPT0
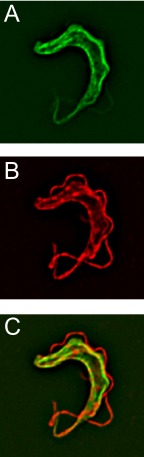
All of the members of this gene family encode proteins with 13–14 predicted TMs, strongly suggesting membrane localization. To determine subcellular localization of TbPT0, a triple hemagglutunin epitope fused to its COOH terminus was constructed, and the tagged protein was expressed in BF parasites from the inducible expression vector. TbPT0-3HA fluorescence signal was detected primarily in the plasma membrane (Fig. 7A), as indicated by the overlap with α-tubulin signal from the subpellicular microtubules (Fig. 7, B and C).
FIGURE 7.
Localization of epitope tagged TbPT0 in BF trypanosomes. Trypanosomes expressing TbPT0 fused to a triple hemagglutinin tag at its COOH terminus (TbPT0-3HA) were immunostained with anti-HA tag antibody (A) and anti-α-tubulin antibody (B); the merged images are shown in C.
DISCUSSION
For more than two decades, it has been known that in the mammalian infective form of T. brucei, BF slender form, pyruvate is not reduced to lactate or oxidized to CO2 because of the lack of lactate dehydrogenase and a functional mitochondrion (6, 7, 13, 34, 35). Pyruvate is the final product of glycolysis and has to be extruded to the extracellular milieu by specific transporters localized in the plasma membrane of the parasite to maintain the integrity and viability of the cell. In 1995, Weiner et al. (7) demonstrated that inhibition of pyruvate transport by UK5099, a universal pyruvate transport inhibitor, causes severe physiological alterations and ultimately leads to cell death, whereas Barnard et al. (6) showed that the subversive pyruvate analogue, bromopyruvate, enters the trypanosomes via pyruvate permeases targeting GAPDH, consequently affecting cell motility and pyruvate efflux and causing death of the parasites. Together, those previous findings highlighted the physiological importance and the pharmacological relevance of pyruvate transporters. However, the molecular identity of pyruvate transporters has been elusive, hampering their thorough molecular and biological analysis that would define these permeases as promising drug targets. In the present study, I report the cloning, functional expression, and characterization of TbPT0 that encodes a unique pyruvate transporter of T. brucei.
Due to its bidirectional transport ability, TbPT0 is able to rescue the mutant phenotype of the pyruvate auxotrophic JMY75 yeast clone (Fig. 1). Thus, TbPT0 is a bona fide pyruvate transporter. Genetic complementation in yeast is a powerful technique that has been applied to the cloning of other trypanosomal proteins (21, 24, 25). For instance, Mäser et al. (24) cloned the gene coding for the TbAT1 adenosine/adenine/pentamidine transporter using a yeast mutant deficient in adenine synthesis. Functional expression of TbPT0 in S. cerevisiae revealed that TbPT0 is a reasonably high affinity pyruvate transporter (Fig. 4A, 4B), supporting its important physiological function in efflux of pyruvate from the BF cytosol. Also, specific inhibition of pyruvate transport mediated by TbPT0 upon addition of UK5099, a proven trypanosomal pyruvate transport inhibitor (Fig. 4D) (7), supports the identity of TbPT0 as a critical pyruvate transporter. In addition, the ability of TbPT0 to transport acetate (Fig. 5A) suggests another potentially important physiological function mediated by the TbPT family. Recently, Tielens et al. (36) reported that short stumpy BF parasites (as opposed to long slender BF) generate acetate and express the mitochondrial enzyme acetate:succinate CoA transferase, demonstrating that this life cycle stage also uses in addition to glycolysis a mitochondrial pathway for catabolism of glucose and ATP generation, yielding acetate and pyruvate as end products of glucose metabolism. Possibly, TbPT family members are involved in the efflux of pyruvate and acetate generated by the short stumpy BF parasites. Moreover, the relatively low affinity exhibited by TbPT0 for lactate, as well as the absence of a trypanosome LDH that could generate lactate, suggests that this lactate transport activity is not physiologically relevant.
TbPT0 and all of the members of the TbPT family are phylogenetically more related to nodulin-like proteins from plants then to mammalian monocarboxylate transporters (Fig. 3). Analysis of the alignments between TbPTs and the protein sequences with the highest similarity (data not shown) identifies a shared signature sequence, MYDAEA, in the last predicted extracellular loop, whereas neither MCT1 (37) nor NP2.1, an A. thaliana lactate transporter (31) encompassed this motif. Such a conserved signature sequence might play a significant role in the transport specificity or kinetics and perhaps be relevant for structure function studies, which will be addressed in the future. Members of the same family of transporters sometimes have distinct biochemical functions despite their high identity (15). A complete biochemical analysis of all of the members of the TbPT family will determine whether all of these have the same substrate specificities or whether some permeases have additional or alternative substrates.
Similarities between trypanosomal and plant proteins have been documented previously by Hannaert et al. (38, 39), suggesting that horizontal genetic transfer occurred during evolution impacting mostly proteins involved in trypanosome glucose metabolism. Recently, Matthews and co-workers (40) identified a plasma membrane protein from T. brucei involved in sensing cis-acconitate, which triggers differentiation from BF to PF parasites, and also showing high similarity to plant permeases. Therefore, TbPTs are trypanosomal proteins with a possible plant origin that significantly differ from their mammalian counterparts (41) and perform an essential physiological function. The uniqueness of TbPTs makes them attractive for pharmacological intervention because the absence of human orthologues to TbPTs may facilitate the discovery of selective inhibitors. In addition, the yeast expression system described here could be employed to screen for potent inhibitors of TbPTs.
The expression of the TbPT gene family is up-regulated at the RNA level about 13-fold in the BF parasites as demonstrated by previous published studies on transcriptomes of T. brucei strains comparing BF with PF parasites (26–28, 30). In addition recent publications by Urbaniak et al. (42) and Butter et al. (43) have shown using genome-wide proteomic analysis that members of the TbPT family of transporters are significantly up-regulated in BF parasites. Moreover, 3× HA-tagged TbPT0 targets to the plasma membrane of the BF parasites (Fig. 7, A and C), consistent with its essential pyruvate efflux function. Also experiments by Barnard et al. (6, 34) determined that pyruvate efflux in PF parasites is ∼100-fold lower compared with the BF parasites mainly due to a down-regulation of glycolytic enzymes, particularly PK which is decreased 25-fold, in addition to further metabolism of pyruvate yielding low levels of cytosolic pyruvate.
Down-regulation of the expression of the TbPT gene family by RNAi (Fig. 6, A and B) resulted in a lethal phenotype in BF trypanosomes. The high sequence identity between members of this family makes it difficult to assess, by RNAi, the relative contribution of each family member to this lethal phenotype. However, the most conservative interpretation at present would be that all TbPT isoforms likely contribute to pyruvate efflux and that coordinate knockdown of all five transcripts probably contributes to cell death. Cell growth arrest was initially observed followed by parasite death after 2 weeks, highlighting an essential role of the TbPT gene family. This result implies that either the absence or a reduced level of TbPTs in the BF plasma membrane impairs pyruvate efflux, causing a cascade of negative physiological effects and ultimately cell death. The definition of these pyruvate transporters presents a potential target for pharmacological intervention in human African trypanosomiasis.
Acknowledgment
I thank Dr. Scott Landfear for unconditional support and critical reading of this manuscript.
This work was supported by Beginning Grant-in-aid 0560056Z (to M. A. S.) from the American Heart Association.
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession number(s) JQ723476.
- BF
- bloodstream form
- MCT
- monocarboxylate transporter
- PF
- procyclic form
- YNB
- yeast nitrogen base medium
- TM
- transmembrane domain
- SM
- single marker.
REFERENCES
- 1. Barrett M. P., Gilbert I. H. (2006) Targeting of toxic compounds to the trypanosome's interior. Adv. Parasitol. 63, 125–183 [DOI] [PubMed] [Google Scholar]
- 2. Paulnock D. M., Freeman B. E., Mansfield J. M. (2010) Modulation of innate immunity by African trypanosomes. Parasitology 137, 2051–2063 [DOI] [PubMed] [Google Scholar]
- 3. Bakker B. M., Westerhoff H. V., Opperdoes F. R., Michels P. A. (2000) Metabolic control analysis of glycolysis in trypanosomes as an approach to improve selectivity and effectiveness of drugs. Mol. Biochem. Parasitol. 106, 1–10 [DOI] [PubMed] [Google Scholar]
- 4. Opperdoes F. R., Michels P. A. (2001) Enzymes of carbohydrate metabolism as potential drug targets. Int. J. Parasitol. 31, 482–490 [DOI] [PubMed] [Google Scholar]
- 5. Aronov A. M., Suresh S., Buckner F. S., Van Voorhis W. C., Verlinde C. L., Opperdoes F. R., Hol W. G., Gelb M. H. (1999) Structure-based design of submicromolar, biologically active inhibitors of trypanosomatid glyceraldehyde-3-phosphate dehydrogenase. Proc. Natl. Acad. Sci. U.S.A. 96, 4273–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barnard J. P., Reynafarje B., Pedersen P. L. (1993) Glucose catabolism in African trypanosomes. Evidence that the terminal step is catalyzed by a pyruvate transporter capable of facilitating uptake of toxic analogs. J. Biol. Chem. 268, 3654–3661 [PubMed] [Google Scholar]
- 7. Wiemer E. A., Michels P. A., Opperdoes F. R. (1995) The inhibition of pyruvate transport across the plasma membrane of the bloodstream form of Trypanosoma brucei and its metabolic implications. Biochem. J. 312, 479–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Halestrap A. P., Price N. T. (1999) The proton-linked monocarboxylate transporter (MCT) family: structure, function and regulation. Biochem. J. 343, 281–299 [PMC free article] [PubMed] [Google Scholar]
- 9. Halestrap A. P. (2012) The monocarboxylate transporter family–Structure and functional characterization. IUBMB Life 64, 1–9 [DOI] [PubMed] [Google Scholar]
- 10. Morris M. E., Felmlee M. A. (2008) Overview of the proton-coupled MCT (SLC16A) family of transporters: characterization, function and role in the transport of the drug of abuse γ-hydroxybutyric acid. AAPS J 10, 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Halestrap A. P., Wilson M. C. (2012) The monocarboxylate transporter family–role and regulation. IUBMB Life 64, 109–119 [DOI] [PubMed] [Google Scholar]
- 12. Akita O., Nishimori C., Shimamoto T., Fujii T., Iefuji H. (2000) Transport of pyruvate in Saccharomyces cerevisiae and cloning of the gene encoded pyruvate permease. Biosci. Biotechnol. Biochem. 64, 980–984 [DOI] [PubMed] [Google Scholar]
- 13. Wiemer E. A., Ter Kuile B. H., Michels P. A., Opperdoes F. R. (1992) Pyruvate transport across the plasma membrane of the bloodstream form of Trypanosoma brucei is mediated by a facilitated diffusion carrier. Biochem. Biophys. Res. Commun. 184, 1028–1034 [DOI] [PubMed] [Google Scholar]
- 14. Vanderheyden N., Wong J., Docampo R. (2000) A pyruvate-proton symport and an H+-ATPase regulate the intracellular pH of Trypanosoma brucei at different stages of its life cycle. Biochem. J. 346, 53–62 [PMC free article] [PubMed] [Google Scholar]
- 15. Sanchez M. A., Tryon R., Green J., Boor I., Landfear S. M. (2002) Six related nucleoside/nucleobase transporters from Trypanosoma brucei exhibit distinct biochemical functions. J. Biol. Chem. 277, 21499–21504 [DOI] [PubMed] [Google Scholar]
- 16. Sunter J., Wickstead B., Gull K., Carrington M. (2012) A new generation of T7 RNA polymerase-independent inducible expression plasmids for Trypanosoma brucei. PLoS One 7, e35167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Deursen F. J., Shahi S. K., Turner C. M., Hartmann C., Guerra-Giraldez C., Matthews K. R., Clayton C. E. (2001) Characterisation of the growth and differentiation in vivo and in vitro-of bloodstream-form Trypanosoma brucei strain TREU 927. Mol. Biochem. Parasitol. 112, 163–171 [DOI] [PubMed] [Google Scholar]
- 18. Ortiz D., Sanchez M. A., Quecke P., Landfear S. M. (2009) Two novel nucleobase/pentamidine transporters from Trypanosoma brucei. Mol. Biochem. Parasitol. 163, 67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, pp. 7.37–7.52, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 20. Makuc J., Cappellaro C., Boles E. (2004) Co-expression of a mammalian accessory trafficking protein enables functional expression of the rat MCT1 monocarboxylate transporter in Saccharomyces cerevisiae. FEMS Yeast Res. 4, 795–801 [DOI] [PubMed] [Google Scholar]
- 21. Mazhari-Tabrizi R., Eckert V., Blank M., Müller R., Mumberg D., Funk M., Schwarz R. T. (1996) Cloning and functional expression of glycosyltransferases from parasitic protozoans by heterologous complementation in yeast: the dolichol phosphate mannose synthase from Trypanosoma brucei brucei. Biochem. J. 316, 853–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sanchez M. A., Drutman S., van Ampting M., Matthews K., Landfear S. M. (2004) A novel purine nucleoside transporter whose expression is up-regulated in the short stumpy form of the Trypanosoma brucei life cycle. Mol. Biochem. Parasitol. 136, 265–272 [DOI] [PubMed] [Google Scholar]
- 23. Cheng Y., Prusoff W. H. (1973) Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108 [DOI] [PubMed] [Google Scholar]
- 24. Mäser P., Sütterlin C., Kralli A., Kaminsky R. (1999) A nucleoside transporter from Trypanosoma brucei involved in drug resistance. Science 285, 242–244 [DOI] [PubMed] [Google Scholar]
- 25. Van Hellemond J. J., Neuville P., Schwarz R. T., Matthews K. R., Mottram J. C. (2000) Isolation of Trypanosoma brucei CYC2 and CYC3 cyclin genes by rescue of a yeast G(1) cyclin mutant. Functional characterization of CYC2. J. Biol. Chem. 275, 8315–8323 [DOI] [PubMed] [Google Scholar]
- 26. Brems S., Guilbride D. L., Gundlesdodjir-Planck D., Busold C., Luu V. D., Schanne M., Hoheisel J., Clayton C. (2005) The transcriptomes of Trypanosoma brucei Lister 427 and TREU927 bloodstream and procyclic trypomastigotes. Mol. Biochem. Parasitol. 139, 163–172 [DOI] [PubMed] [Google Scholar]
- 27. Queiroz R., Benz C., Fellenberg K., Hoheisel J. D., Clayton C. (2009) Transcriptome analysis of differentiating trypanosomes reveals the existence of multiple post-transcriptional regulons. BMC Genomics 10, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jensen B. C., Sivam D., Kifer C. T., Myler P. J., Parsons M. (2009) Widespread variation in transcript abundance within and across developmental stages of Trypanosoma brucei. BMC Genomics 10, 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Veitch N. J., Johnson P. C., Trivedi U., Terry S., Wildridge D., MacLeod A. (2010) Digital gene expression analysis of two life cycle stages of the human-infective parasite, Trypanosoma brucei gambiense reveals differentially expressed clusters of co-regulated genes. BMC Genomics 11, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Siegel T. N., Hekstra D. R., Wang X., Dewell S., Cross G. A. (2010) Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 38, 4946–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Choi W. G., Roberts D. M. (2007) Arabidopsis NIP2;1, a major intrinsic protein transporter of lactic acid induced by anoxic stress. J. Biol. Chem. 282, 24209–24218 [DOI] [PubMed] [Google Scholar]
- 32. Nolan D. P., Voorheis H. P. (2000) Factors that determine the plasma-membrane potential in bloodstream forms of Trypanosoma brucei. Eur. J. Biochem. 267, 4615–4623 [DOI] [PubMed] [Google Scholar]
- 33. Alsford S., Turner D. J., Obado S. O., Sanchez-Flores A., Glover L., Berriman M., Hertz-Fowler C., Horn D. (2011) High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 21, 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barnard J. P., Pedersen P. L. (1994) Alteration of pyruvate metabolism in African trypanosomes during differentiation from bloodstream into insect forms. Arch Biochem. Biophys. 313, 77–82 [DOI] [PubMed] [Google Scholar]
- 35. Haanstra J. R., Kerkhoven E. J., van Tuijl A., Blits M., Wurst M., van Nuland R., Albert M. A., Michels P. A., Bouwman J., Clayton C., Westerhoff H. V., Bakker B. M. (2011) A domino effect in drug action: from metabolic assault towards parasite differentiation. Mol. Microbiol. 79, 94–108 [DOI] [PubMed] [Google Scholar]
- 36. Tielens A. G., van Hellemond J. J. (2009) Surprising variety in energy metabolism within Trypanosomatidae. Trends Parasitol. 25, 482–490 [DOI] [PubMed] [Google Scholar]
- 37. Poole R. C., Sansom C. E., Halestrap A. P. (1996) Studies of the membrane topology of the rat erythrocyte H+/lactate cotransporter (MCT1). Biochemical Journal 320, 817–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hannaert V., Bringaud F., Opperdoes F. R., Michels P. A. (2003) Evolution of energy metabolism and its compartmentation in Kinetoplastida. Kinetoplastid. Biol. Dis. 2, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hannaert V., Saavedra E., Duffieux F., Szikora J. P., Rigden D. J., Michels P. A., Opperdoes F. R. (2003) Plant-like traits associated with metabolism of Trypanosoma parasites. Proc. Natl. Acad. Sci. U.S.A. 100, 1067–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dean S., Marchetti R., Kirk K., Matthews K. R. (2009) A surface transporter family conveys the trypanosome differentiation signal. Nature 459, 213–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Merezhinskaya N., Fishbein W. N. (2009) Monocarboxylate transporters: past, present, and future. Histol. Histopathol. 24, 243–264 [DOI] [PubMed] [Google Scholar]
- 42. Urbaniak M. D., Guther M. L., Ferguson M. A. (2012) Comparative SILAC proteomic analysis of Trypanosoma brucei bloodstream and procyclic lifecycle stages. PLoS One 7, e36619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Butter F., Bucerius F., Michel M., Cicova Z., Mann M., Janzen C. J. (2013) Comparative proteomics of two life cycle stages of stable isotope-labeled Trypanosoma brucei reveals novel components of the parasite's host adaptation machinery. Mol. Cell Proteomics 12, 172–179 [DOI] [PMC free article] [PubMed] [Google Scholar]