Background: The phenoloxidase (PO) cascade regulates the melanization of blood in insects and other arthropods.
Results: Hemolymph from Bombyx mori rapidly melanizes after wounding and forms a complex that uses tyrosine as the substrate.
Conclusion: Tyrosine usage requires formation of a phenoloxidase-containing complex.
Significance: Processing of prophenoloxidase is an intermediate step in the phenoloxidase cascade.
Keywords: Amino Acid, Blood, Insect Immunity, Protease, Wound Healing, Cascade, Melanin, Phenoloxidase
Abstract
The phenoloxidase (PO) cascade regulates the melanization of blood (hemolymph) in insects and other arthropods. Most studies indicate that microbial elicitors activate the PO cascade, which results in processing of the zymogen PPO to PO. PO is then thought to oxidize tyrosine and o-diphenols to quinones, which leads to melanin. However, different lines of investigation raise questions as to whether these views are fully correct. Here we report that hemolymph from the silkmoth, Bombyx mori, rapidly melanizes after collection from a wound site. Prior studies indicated that in vitro activated PPO hydroxylates Tyr inefficiently. Measurement of in vivo substrate titers, however, suggested that Tyr was the only PO substrate initially present in B. mori plasma and that it is rapidly metabolized by PO. Fractionation of plasma by gel filtration chromatography followed by bioassays indicated that melanization activity was primarily associated with a high mass complex (∼670 kDa) that contained PO. The prophenoloxidase-activating protease inhibitor Egf1.0 blocked formation of this complex and Tyr metabolism, but the addition of phenylthiourea to plasma before fractionation enhanced complex formation and Tyr metabolism. Mass spectrometry analysis indicated that the complex contained PO plus other proteins. Taken together, our results indicate that wounding alone activates the PO cascade in B. mori. They also suggest that complex formation is required for efficient use of Tyr as a substrate.
Introduction
Melanization of hemolymph is a conserved immune response in insects and other arthropods (1, 2). Melanin can seal off foreign organisms in the hemocoel and starve them of nutrients (3). Melanin synthesis also results in production of reactive oxygen and nitrogen intermediates that are toxic to some microbes and multicellular parasites (1–3). PO3 is thought to be the key enzyme required for hemolymph melanization. PO belongs to the type 3 copper protein family that includes tyrosinases, which oxidize mono- and diphenolic substrates (2, 4, 5). The resulting quinones then undergo further enzymatic and non-enzymatic reactions that ultimately result in the formation of melanin (4).
PO exists in hemolymph as the inactive zymogen PPO, whereas the network of factors that regulate the activation of PPO is referred to as the PO cascade. Prior studies establish that microbes activate the PO cascade when pattern recognition proteins in hemolymph bind polysaccharides on their surface. This event results in activation of several serine proteases, including PAPs (also named PPAEs (prophenoloxidase-activating proteases) or PPAFs (prophenoloxidase-activating factors) (1, 2)), which process PPO to PO by cleavage at one or more conserved Arg-Phe sites. Once present, PO activity usually localizes to the surface of foreign targets or wound sites, which limits global melanization of hemolymph. Endogenous protease inhibitors like serpins act as negative regulators of serine proteases in the PO cascade (6), whereas reducing agents like glutathione (GSH) prevent the formation of melanin (7). Several pathogens of insects, including polydnaviruses, also produce molecules that suppress the melanization response of hosts (1, 8). In the case of Microplitis demolitor bracovirus, the virus expresses the protein Egf1.0 in infected host cells, and it is then secreted into plasma, where it blocks the PO cascade by inhibiting PAPs (9–11).
It is generally assumed that processing of PPO to PO confers the enzymatic activity needed to cause hemolymph to melanize (1, 12–15). Two lines of investigation, however, raise questions as to whether this is fully correct. First, the purification and in vitro activation of PPO from several species indicate that it oxidizes o-diphenols like dihydroxyphenylalanine (DOPA) and dopamine (DA) that derive from Tyr with much greater efficiency than Tyr itself (16–18). However, measures of substrate titers suggest that insect hemolymph contains an abundance of free amino acids including Tyr but little or no DOPA or DA (7, 19–21). Thus, it is unclear how the substrate preferences of in vitro activated PPO relate to the substrates actually available in hemolymph. Second, cleavage of PPO may not be the terminal step of the PO cascade because studies with the moths Bombyx mori and Manduca sexta plus the beetles Tenebrio molitor and Holotrichia diomphalia indicate that POs form chemically cross-linked high molecular mass complexes (16, 22–28). In vitro studies further show that mixing purified components of the PO cascade from T. molitor and H. diomphalia resulted in the formation of complexes that contained PO and a serine protease homolog (SPH). These complexes also bound the surface of bacteria and oxidized a catecholamine, which suggests that complex formation is important in localizing melanin deposition to the surface of pathogens (27, 28).
Here we characterized substrate availability and usage during melanization of hemolymph from B. mori. Our results indicate that Tyr is the endogenous substrate used to rapidly synthesize melanin. Our results further suggest that PO is the enzyme in plasma that uses Tyr to produce melanin, but in order to do so it must form a chemically cross-linked complex, which contains SPH1 and other proteins.
EXPERIMENTAL PROCEDURES
Insects and Plasma Collection
B. mori larvae (p50 strain) were reared as described previously (11). Plasma was collected by first narcotizing final (fifth) instar (day 5) B. mori with CO2 and then surface-rinsing them with ethanol. After rinsing in sterile water, larvae were bled by cutting a proleg and collecting hemolymph into a 1.5-ml Eppendorf tube on ice (29). Hemolymph was immediately centrifuged at 16,000 × g for 30 s to pellet hemocytes, and the resulting plasma supernatant was transferred to a new centrifuge tube for use in experiments.
Melanization and Endotoxin Assays
The first step in melanization of hemolymph is assumed to be the oxidation of Tyr to DOPA in the presence of O2. DOPA may then be converted to DA through the action of DOPA decarboxylase. Each diphenolic substrate is then further oxidized to dopaquinone and dopamine-quinone, respectively. Dopaquinone and dopamine-quinone thereafter proceed through a series of unstable intermediates, such as dopachrome and dopaminechrome, that result in formation of melanin (2). The standard method for monitoring these events in the PO cascade literature is by measuring absorbance in a sample at or near 470 nm, which predominantly detects dopachrome and/or dopaminechrome rather than melanin itself (2, 7). Here, we monitored melanization of plasma by measuring absorbance at 470 nm at 3-min intervals using a BioTek Synergy 4 plate reader. In some assays, melanization was monitored without the addition of any microbial inducer, whereas in others, the β-1,3-glucan polymer curdlan was added before monitoring for melanization. Bacterial contamination on the surface of B. mori larvae was measured using the Limulus amebocyte lysate (LAL) assay using a commercially available kit (Genscript). Larvae were narcotized with CO2 and surface-rinsed with ethanol as described above. We then placed a 20-μl drop of endotoxin-free water on the surface of the larva that covered a 5-mm2 region of the cuticle for 10 min. The water sample was then collected and processed for endotoxin levels according to procedure in the LAL kit.
Substrate Titers
Plasma samples from individual larvae were collected as described above. To rapidly inactivate enzyme activity, 25 μl of freshly collected plasma was immediately diluted with 75 μl of H2O containing 0.05% TFA and boiled for 45 s to precipitate plasma proteins. Control solutions of Tyr, tyramine, DOPA, and DA (0.5 mm) were also boiled as a control. After boiling, protein precipitates were removed from the plasma samples by centrifugation, and supernatants were either tested directly or, in the case of time course experiments, frozen at −80 °C until all samples were collected. Metabolites were separated by HPLC (Hitachi) using a reverse-phase HPLC column (Phenomenex Primesphere C18 HC 5 μm, 250 × 4.6 mm) with a mobile phase gradient of 0–30% B over 30 min (A, H2O with 0.05% TFA; B, acetonitrile with 0.05% TFA) at 0.5 ml/min. 20-μl injections of boiled plasma were sufficient to observe metabolites and provide good resolution. Spectra of eluted peaks with and without co-injected standards were measured directly by photodiode array (L4500A, Hitachi). Substrate titers in boiled plasma were also analyzed with an AminoPac PA10 Analytical column (Dionex Corp.) using a Dionex HPLC and electrochemical detection as directed by the manufacturer (56).
Preparation of Substrate-free Plasma (SFP)
B. mori plasma was prepared as described above except that larvae were bled directly into PBS containing GSH (1 mm final concentration) to inhibit melanization (7). Other samples were bled into PTU (1 mm) or 3-iodotyrosine (1 mm). Plasma samples were diluted 10-fold with PBS and concentrated on a 30-kDa centrifugal filter (Amicon Ultra, Ultracel-30k) three times, after which the sample (SFP >30 kDa) was restored to its original volume using PBS. SFP >300 kDa was prepared from SFP >30 kDa using an identical procedure except for substituting 300-kDa cut-off filters (NanoSep 300k Omega, Pall Corp.). Melanization was measured as described above by adding 30 μl of SFP to 70 μl of PBS containing substrate at a final concentration of 0.5 mm.
Gel Filtration
Plasma was prepared as described above and separated by HPLC (BioLogic, Bio-Rad) using a gel filtration column (Sephacryl 300 HR or Superdex 200, 1 cm × 50 cm). Plasma (80 μl) was injected directly onto the column using 0.7 ml/min (Sephacryl) or 0.4 ml/min (Superdex) and isocratic PBS as a mobile phase. Column calibration was performed under identical conditions by injecting molecular weight standards (Gel Filtration Standard, catalog no. 151-1901; Bio-Rad). Fractions were collected (0.7 ml) and assayed for melanizing activity as described above.
Egf1.0 Assays
Recombinant Egf1.0 was produced in bacteria and purified (9). Freshly collected hemolymph from a single larva was immediately added to two separate 1.5-ml Eppendorf tubes, one containing 60 μl of inhibitor (0.5 mg/ml) and the other containing an equal volume of water. Both also contained 10 μl of 50 mm GSH to block melanization. The final volume was measured to determine the amount of hemolymph added (∼100 μl/tube), and then the samples were centrifuged to remove hemocytes, and SFPs were prepared as described above.
SDS-PAGE and Immunoblotting
Gel filtration fractions were boiled in SDS sample buffer containing 50 mm DTT and loaded onto 7.5% or 4–20% SDS-polyacrylamide gels (Lonza) along with molecular weight standards (Fermentas, PageRuler Plus). Proteins were transferred onto PVDF (Immobilon-P, Millipore) and blocked in 5% nonfat powdered milk. Some blots were probed with a previously generated rabbit polyclonal antibody (50,000-fold dilution) that recognizes B. mori PPO1 and PPO2 (30). Other blots were probed with two new antibodies (25,000-fold dilution) that we generated by immunizing rabbits with either the PO1-specific peptide CVRFTDRTRQRGGGG or PO2-specific peptide CQNVTEPNPRNPPM, followed by affinity purification of antisera (GenScript). After overnight incubation at 4 °C with each antibody, blots were washed three times (PBS with 2% Tween), incubated with a goat anti-rabbit secondary antibody coupled to HRP (50,000-fold dilution) for 4 h at 4 °C, and then visualized using a chemiluminescent substrate (ECL Advance, GE Healthcare) (10).
Mass Spectrometry
Three different plasma samples were collected in the presence of PTU and then run on native PAGE (4–20%) at 40 V for 12 h at 4 °C. Equivalent lanes were stained with Coomassie Blue or immersed in a 0.5 mm Tyr solution in PBS to visualize melanizing activity. The high mass band of interest was then excised from each gel, reduced with DTT, alkylated with iodoacetamide, and subjected to in-gel digestion with Trypsin Gold (Promega) in 20 mm ammonium bicarbonate. Tryptic fragments were extracted with 50% acetonitrile and 0.1% TFA and vacuum-dried. Samples (4 μl) were analyzed by an Orbitrap Elite mass spectrometer, coupled to an Easy-nLC II liquid chromatography (LC) instrument (Thermo Fisher Scientific). Samples were desalted and preconcentrated on a C18 Easy LC precolumn (100-μm internal diameter × 2 cm, 5-μm particle packing). Peptides were eluted from a reverse-phase column (75-μm internal diameter × 10 cm, 3-μm particle packing) with a gradient of 10–35% B for 70 min, 35–95% B for 10 min, 95% B for 5 min (A, 0.1% formic acid in water; B, 0.1% formic acid in acetonitrile) at 300 nl/min. Nanospray ionization was performed with a spray voltage of 2 kV, with a capillary temperature of 200 °C. The Orbitrap mass analyzer was used to provide resolutions of 120,000 and 30,000 for MS and MS/MS analyses, respectively. Briefly, a cycle of one full-scan mass spectrum (300–2000 m/z) was performed, followed by continuous cycles of collision-induced dissociation and high collision dissociation MS/MS spectra acquisitions of the two or five most abundant peptide ions throughout the LC separation until the candidate ions were exhausted. Data were acquired using Xcalibur software (version 2.2, Thermo Fisher Scientific). Proteins were identified using Mascot version 2.3 (Matrix Science), the NCBI (National Institutes of Health) database, and the B. mori genome in November 2012. Searches were performed assuming carbamidomethylation of cysteines but without restriction to protein species, Mr, or pI. Data were visualized with Proteome Discoverer version 1.3 (Thermo). Peptides with scores greater than the identity score (p < 0.05) were considered significant matches. Only products that were matched in each replicate by at least two peptide spectra were considered positive identifications.
RESULTS
B. mori Plasma Rapidly Melanizes after Collection from a Wound Site
As a component of the arthropod immune system, most studies emphasize the role of microbial elicitors in activation of the PO cascade (1, 2). In the case of M. sexta or T. molitor, hemolymph collected from a wound site melanizes weakly or not at all after collection, but the addition of microbial cell wall components alone or together with a diphenolic substrate induces melanization due to binding by pattern recognition receptors like peptidoglycan recognition protein (31, 32). Recognition of β-1,3-glucan by peptidoglycan recognition protein similarly activates the PO cascade in the beetle H. diomphalia, the crayfish Pacifastacus leniusculus, and B. mori (33–35). However, early studies of B. mori suggest that, unlike M. sexta and T. molitor, B. mori hemolymph rapidly melanizes without adding a microbial elicitor or diphenol substrate directly to plasma samples (36).
We assessed this by collecting cell-free plasma from larvae and measuring melanization using a standard spectrophotometric assay at 470 nm. Our results showed a 3-min lag phase before melanization activity rapidly rose and plateaued after 2 h (Fig. 1A). No further acceleration of this response occurred by adding curdlan to hemolymph (data not shown). These data confirmed that B. mori plasma rapidly melanized after collection from a wound site without adding a microbial elicitor directly to the samples we collected. However, these results did not mean that microbial elicitors were absent from our plasma samples, given the likelihood that bacteria endogenously reside on the surface of larvae. To assess levels of microbial contamination on the surface of B. mori, we used the LAL assay to measure endotoxin levels on a 5-mm2 area of B. mori cuticle, which was similar in size to the wound we made when collecting plasma. Our results indicated an average of 1.2 ± 0.11 endotoxin units/sample (n = 6), which is equivalent to ∼120 pg of endotoxin. This result also indicated that B. mori plasma collected from a wound site contacts microbial cell wall components on the surface of larvae.
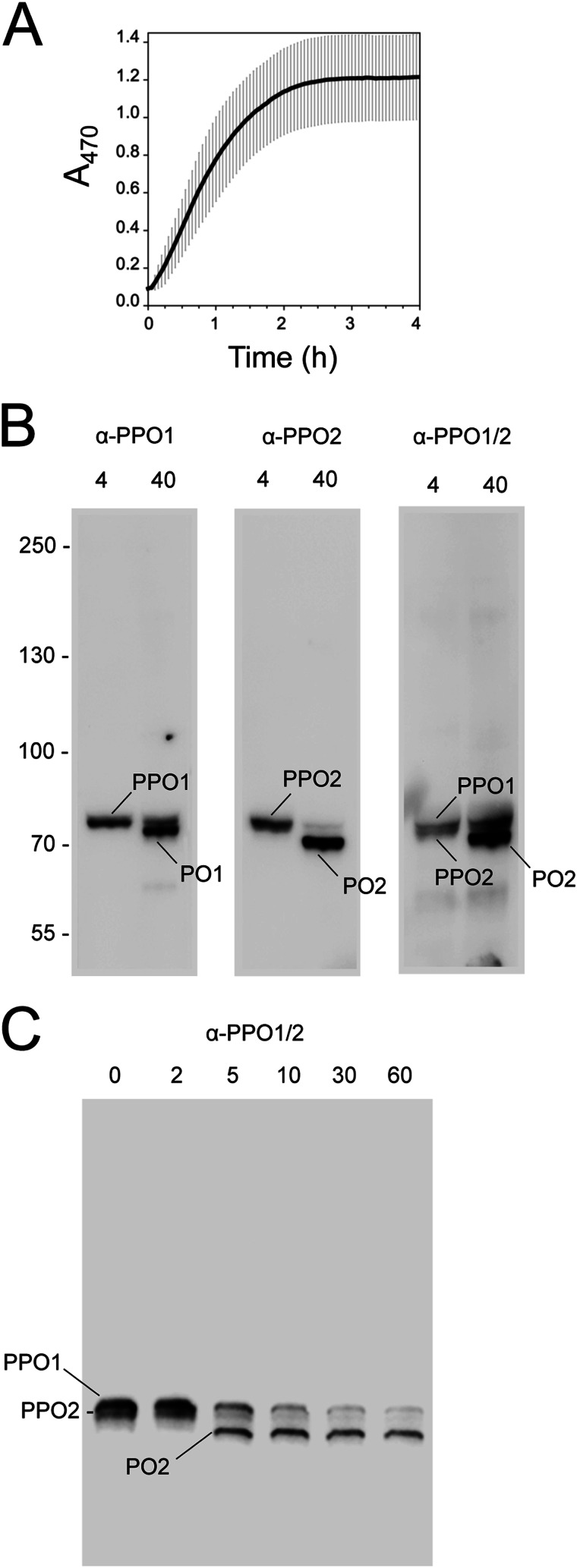
FIGURE 1.
The PO cascade in B. mori is activated by wounding. A, melanization rate of B. mori plasma. Freshly prepared plasma from three fifth instar larvae (20 μl each) was diluted in PBS (80 μl), plated, and monitored for melanization activity 0–4 h ± S.D. post-collection from a wound site. Activity was measured by monitoring formation of dopachrome or dopaminechrome (melanization intermediates) at A470. B, plasma was collected as outlined in A, followed by the addition of sample buffer at 4 or 40 min. After separation on 7.5% continuous SDS-polyacrylamide gels under reducing conditions, samples were immunoblotted using antibodies for PPO1 (α-PPO1), PPO2 (α-PPO2), or a cross-reacting antibody that recognizes both PPOs (α-PPO1/2). Bands corresponding to the predicted sizes of PPO1/PO1 and PPO2/PO2 are indicated. Note that the affinity of α-PPO1/2 for PO2 is stronger than for PO1. C, immunoblot prepared as described in B but with sample buffer added at 0–60 min post-collection and probed using α-PPO1/2. Bands corresponding to PPO1/PO1 and PPO2/PO2 are indicated. Molecular mass markers in B are indicated on the left.
B. mori PPO Is Rapidly Processed after Collection from a Wound Site
Because activation of the PO cascade depends on processing of PPO to PO, we next assessed the timing of PPO processing following collection of plasma from a wound site. B. mori encodes two PPO genes whose predicted products are 80.0 (PPO1) and 78.7 (PPO2) kDa, respectively. PPO1 and PPO2 are thought to persist as a heterodimer, whereas processing by PAPs produces PO1 and PO2, which are 74.0 and 72.8 kDa (37, 38). We thus collected plasma from larvae and prepared samples for electrophoresis at 4 or 40 min post-collection. Samples were then separated on 7.5% SDS-polyacrylamide gels and immunoblotted using PPO1- and PPO2-specific antibodies we generated. At 4 min, each antibody detected single bands whose masses corresponded to the expected sizes of PPO1 and PPO2, but at 40 min, we predominantly detected PO1 and PO2, whose mass differences were clearly distinguishable on blots (Fig. 1B). We also assessed processing using a previously generated antibody that recognizes both PPO1 and PPO2 (30). At 4 min, this antibody recognized a tight doublet whose masses were consistent with PPO1 and PPO2, but at 40 min, the antibody predominantly detected a band with the mobility of PO2 (Fig. 1B). We then used this cross-reacting antibody in finer scale time course assays. Only PPO1 and PPO2 were present at 0 and 2 min post-collection, but at 5 min and later time points, the antibody primarily detected PO2 (Fig. 1C). These results indicated that no PO is present in B. mori plasma before bleeding, but processing of PPO1 and PPO2 occurs within 5 min of collection.
Tyr Is the Likely Physiological Substrate for Melanization of B. mori Plasma
PO has never been purified from B. mori or any other species because it sticks to column matrices and covalently binds other proteins (16, 38). As previously noted, however, PPO has been purified from several species, including B. mori, and activated in vitro (2, 16, 19–21, 37). Bioassays with in vitro activated PPOs show that they efficiently oxidize diphenols (15–18, 39–41). Only three studies from B. mori, M. sexta, and the fly Sarcophaga bullata report Tyr usage, but for each, oxidation rates are much lower than for diphenols (16–18). Thus, despite the long-held view that insect POs oxidize mono- and diphenolic substrates, data supporting this conclusion are somewhat equivocal (2). In addition, only three studies from M. sexta, the moth Pseudoplusia includens, and mosquito Aedes aegypti report the titers of PO substrates in hemolymph. In M. sexta, free Tyr levels near 1 mm were reported, whereas DOPA and other catecholamines were found to not exceed 50 μm (20, 21). In P. includens and A. aegypti, free Tyr was ∼100 μm, whereas DOPA levels were ∼1 μm (A. aegypti) (19) or undetectable (P. includens) (7).
Given the rapid melanization of B. mori plasma, these findings are interesting because they suggest a paradox. On one hand, PO is nearly universally assumed to be the enzyme that catalyzes the melanization of hemolymph. On the other, insect hemolymph seemingly contains an abundance of free Tyr relative to diphenols like DA and DOPA, yet in vitro activated PPO from B. mori and other insects oxidizes Tyr very inefficiently. We therefore assessed substrate usage and availability in B. mori plasma. Our first step was to collect plasma on a 30-kDa filter and then wash it extensively with PBS. This produced a retentate that we named SFP. SFP never melanizes (data not shown), but adding DOPA to SFP stimulated rapid melanization with no lag phase (Fig. 2A). DA also resulted in rapid melanization with no lag phase, but absorbance values plateaued at a lower level than DOPA (Fig. 2A). Tyr resulted in a 12-min lag phase followed by melanization kinetics similar to DOPA (Fig. 2A). These findings indicated that production of SFP removed the endogenous substrate(s) that causes plasma to rapidly melanize but retained the enzymatic machinery that rapidly produces melanin when provided DOPA, DA, or Tyr as a substrate.
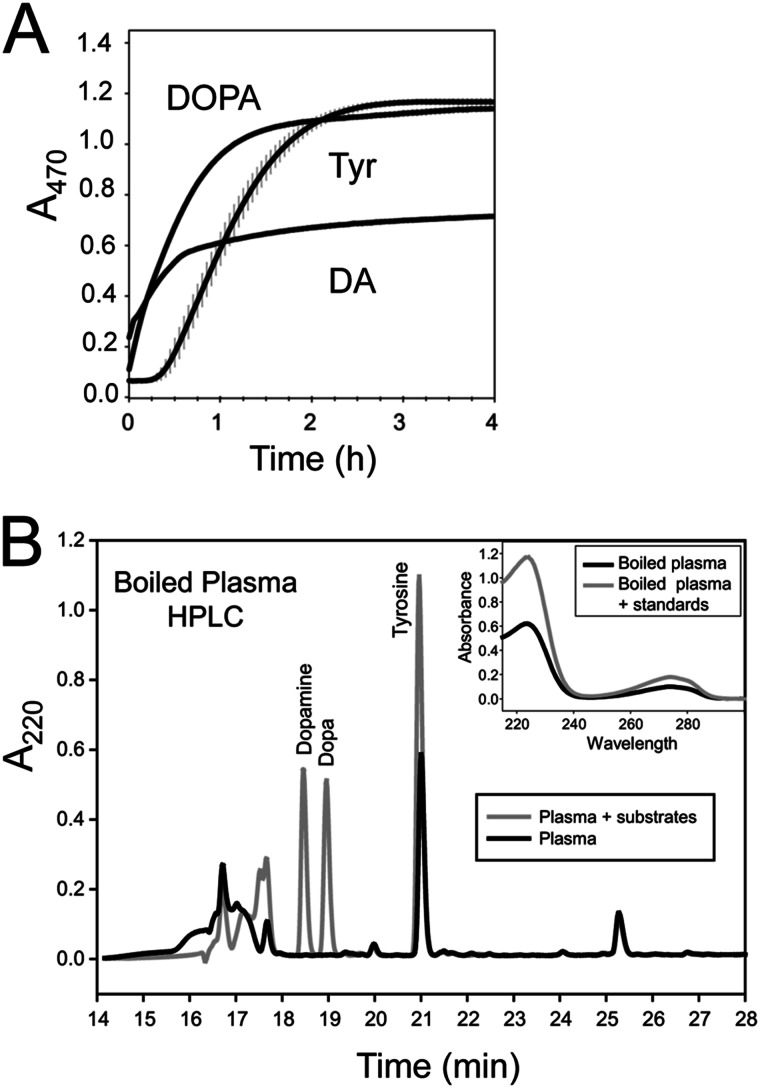
FIGURE 2.
SFP uses multiple substrates but newly collected plasma contains only Tyr. A, melanization rate of SFP. Plasma was prepared in GSH (5 mm final) to block melanization and then washed three times on a 30-kDa filter to prepare SFP. SFP (20 μl) was diluted in PBS (80 μl) along with substrate (0.5 mm final) and monitored for melanin formation at A470. B, fractionation of substrates in plasma by HPLC. Plasma from a fifth instar was boiled for 1 min, cooled on ice, and centrifuged to remove precipitate, and the supernatant was separated by HPLC. The dark line indicates the timing of when peaks from plasma eluted, whereas the light line indicates the timing when peaks corresponding to the tyrosine, DOPA, and dopamine standards eluted. Collected fractions (0.5 ml) were lyophilized, resuspended in PBS, and tested for melanizing activity using B. mori SFP. The active fraction at 21 min co-eluted with a tyrosine standard. The tyramine standard also eluted at 21 min (not shown; see “Results”). Inset, absorbance spectrum of the 21 min peak with and without a co-injected tyrosine standard.
We next assessed the in vivo titers of these substrates plus tyramine in B. mori plasma. Boiling freshly collected plasma and removal of the protein precipitate produced a supernatant that rapidly melanized when added to SFP. Reversed-phase HPLC separation of these supernatant components yielded a single fraction that melanized upon the addition to SFP (Fig. 2B). This fraction co-eluted with a Tyr standard, possessed the same UV spectrum as the standard (see inset), and upon MS analysis contained a predominant molecular mass of 182.0 (predicted M + H for Tyr = 182.2). This fraction also co-eluted with the tyramine standard, but we detected no mass corresponding to tyramine (M + H = 138.2) in the sample. In contrast, neither DOPA nor DA was detected in plasma that was boiled immediately after collection (standards indicated in Fig. 2B). However, we did detect DOPA if we collected plasma and allowed it to sit for 3 min or longer before boiling (data not shown). To control for the possibility that boiling adversely affected tyramine or the diphenolics, we boiled standards of each for 45 s. HPLC analysis showed that all were fully stable (data not shown). We also examined these samples using HPLC and electrochemical detection, which is a potentially more sensitive method for detecting these substrates. This approach also detected free Tyr but did not detect any DA or DOPA in plasma that was boiled immediately after collection. In contrast, both Tyr and DOPA were detected by electrochemical detection if plasma was allowed to sit for 3 min or longer before boiling (data not presented). Thus, although SFP can use Tyr, DOPA, or DA to rapidly produce melanin, the only one of these substrates that is detectable in newly collected plasma is Tyr. We thus concluded that Tyr is the starting substrate that the enzymatic machinery in B. mori plasma uses to rapidly produce melanin.
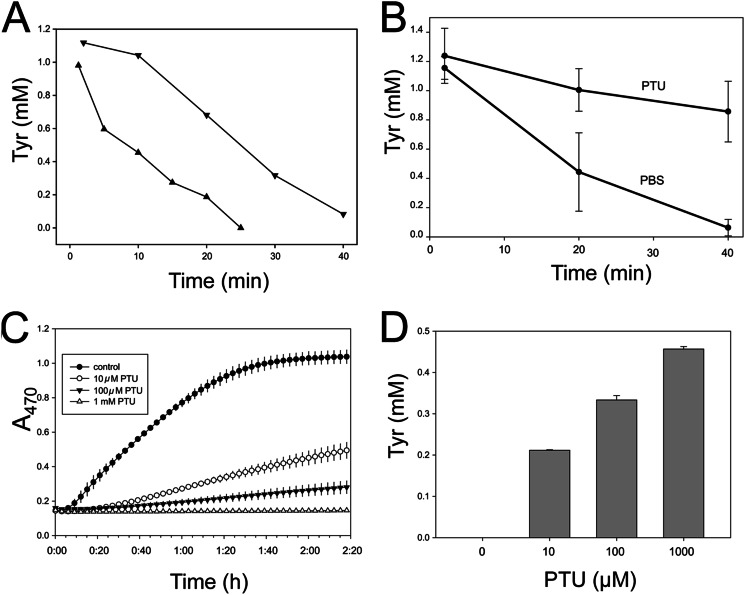
If this interpretation is correct, Tyr should be metabolized at rates that reflect dopachrome formation. Time course experiments using two different B. mori larvae confirmed this by showing that Tyr concentration rapidly dropped from ∼1 mm to 0.083 mm after 40 min in one larva and from 1 mm to 0 by 25 min in the other (Fig. 3A). We also reasoned that if PO is the enzyme in plasma that metabolizes Tyr, then the well-known PO inhibitor PTU should disable Tyr metabolism. In whole plasma, Tyr concentrations declined from a starting concentration of 1.16 mm to 63 μm in 40 min in the absence of PTU, whereas Tyr concentration declined only slightly to 0.86 mm when PTU (100 μm) was present (Fig. 3B). PTU also dose-dependently inhibited Tyr metabolism and melanization when added to SFP (Fig. 3, C and D). These results further indicated that Tyr is the physiological substrate for melanization. The inhibitory effects of PTU, in contrast, suggested that PO is the enzyme that rapidly metabolizes Tyr in vivo although in vitro activated PPO utilizes Tyr very inefficiently (16).
FIGURE 3.
PTU inhibits the rapid metabolism of tyrosine in plasma. A, tyrosine decay in plasma from two different larvae. Aliquots of plasma were removed at the indicated time points, diluted 1:3 with H2O (0.05% TFA), and boiled for 45 s. Protein precipitates were removed by centrifugation, and the supernatants were separated by HPLC as described above. Tyr concentrations were determined by injected standards. B, effect of PTU on Tyr metabolism in plasma. C, effect of PTU on melanization of Tyr by SFP. Tyr was added to SFP at a final concentration of 0.5 mm along with the indicated concentrations of PTU and immediately monitored for melanin formation at A470. D, effect of PTU on Tyr metabolism in SFP. Samples were collected after 2 h, 20 min, filtered (30 kDa) to remove protein and melanin, and then separated by HPLC to measure Tyr as described. Error bars, S.E.
Tyr-dependent Melanization Is Associated with a PO-containing High Molecular Weight Complex
An alternative explanation for the preceding results would be that Tyr is not hydroxylated by PO but by another enzyme, which then feeds DOPA to PO. The only known tyrosinase-like enzymes in B. mori plasma are PPO1/PO1 and PPO2/PO2. However, the B. mori genome does encode a Tyr hydroxylase previously implicated in cuticle pigmentation (42) and whose ortholog in M. sexta is expressed in hemocytes (43). Three lines of evidence, however, argue against Tyr hydroxylase playing a role in the rapid melanization of B. mori plasma. First, Tyr hydroxylases are cytosolic enzymes, but no hemocytes or other cells were present in our plasma samples. Second, as a cytosolic enzyme, Tyr hydroxylase would have to feed DOPA to PO by secreting the diphenol into plasma, but as noted above, no DOPA is initially present in plasma. Third, PTU dose-dependently inhibited melanization and Tyr metabolism (see Fig. 3), but this compound is not a Tyr hydroxylase inhibitor. In contrast, the same experiments using the Tyr hydroxylase inhibitor 3-iodotyrosine had no inhibitory effects on melanization of plasma or Tyr metabolism (data not shown).
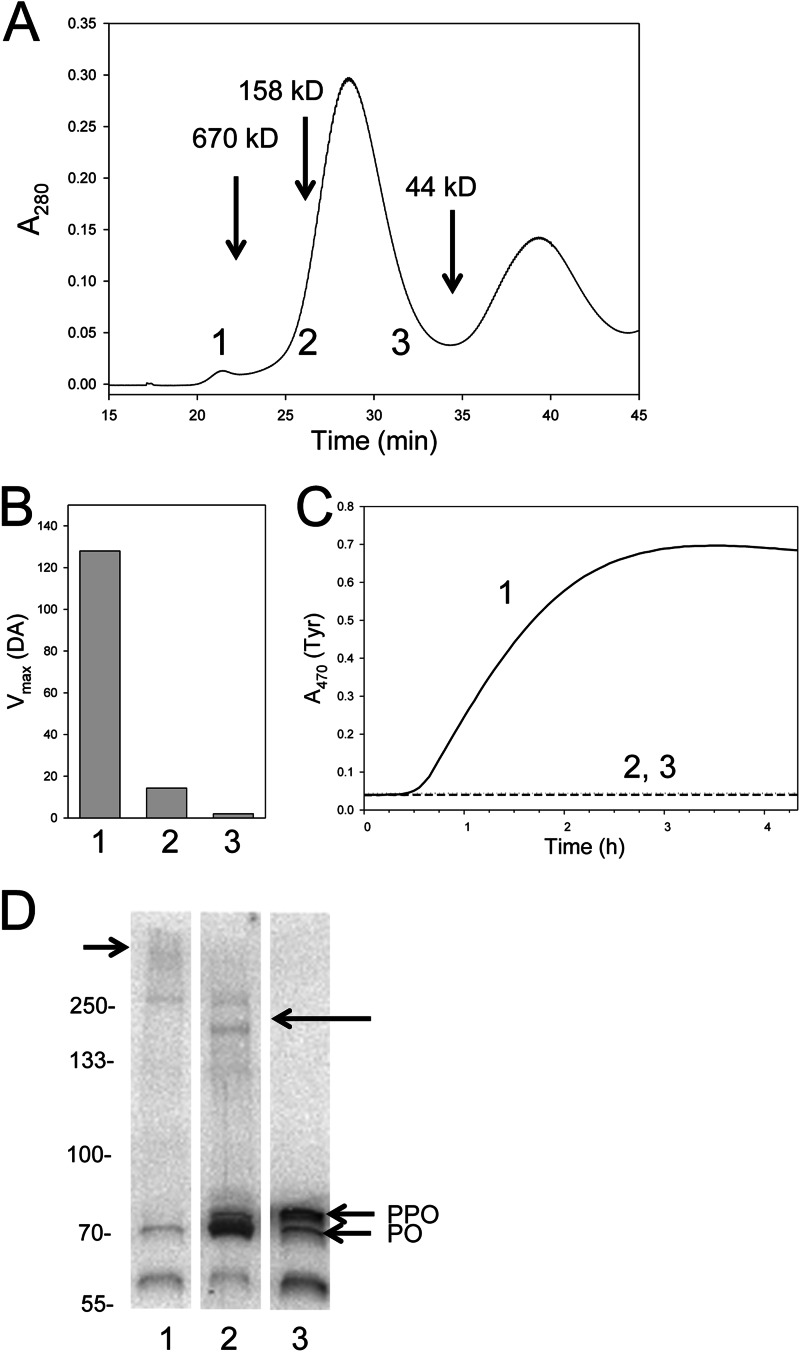
We therefore focused on how substrate usage by PO in plasma might differ from in vitro activated PPO. First, we used size exclusion column chromatography to identify fractions that melanized upon the addition of Tyr or DA. Protein eluent was measured at A280 from the void volume of the column to 45 min (Fig. 4A). Fractions (0.7 ml) were collected from 17 min (void volume of the column) to 45 min and assayed. Three of these fractions (numbered 1, 2, and 3) are indicated at the bottom of the chromatogram shown in Fig. 4A that correspond to where a 670 kDa marker eluted (fraction 1) plus where dimeric (fraction 2) and monomeric (fraction 3) would be expected to elute. Using DA or Tyr as a substrate, fraction 1 exhibited robust activity, whereas fractions 2 and 3 exhibited little or no activity (Fig. 4, B and C). Likewise, all of the other fractions that we bioassayed exhibited little or no melanization activity (data not shown).
FIGURE 4.
Tyr-dependent melanization in B. mori is associated with a high molecular mass fraction. A, plasma was separated by Sephacryl S-300 HR chromatography. Protein eluent was measured at A280 from the void volume of the column to 45 min. The elution times for the 670-, 158-, and 44-kDa protein standards eluted are indicated. Fractions (0.7 ml) were collected from 17 min (void volume of the column) to 45 min and assayed using DA or Tyr as the substrate. Three of these fractions (numbered 1, 2, and 3) are indicated at the bottom of the chromatogram. B, melanization activity for fractions 1, 2, and 3 using DA as the substrate. Substrate was added to the fraction at a final concentration of 0.5 mm, followed by monitoring of dopachrome or dopaminechrome at A470. Vmax values (milli-optical density/min) are presented because kinetics were linear. C, melanization activity for fractions 1–3 using Tyr as the substrate was measured in the same manner as for DA, but results are plotted as a time course. Note that melanization rates are lower than for whole plasma because of sample dilution. D, the proteins in fractions 1–3 were separated on a 4–20% SDS-polyacrylamide gel under reducing conditions, followed by immunoblotting using α-PPO1/2. Bands corresponding to monomeric PPO and PO are indicated. The ∼60 kDa band recognized by α-PPO1/2 corresponds to the predicted mass of PO cleaved at a second Arg-Phe site that exists downstream of the site normally associated with processing of PPO to PO. A short arrow (left) points to a diffuse high mass band recognized by α-PPO1/2 in fraction 1. A long arrow (right) points to two higher molecular mass bands that run between the 250 and 133 kDa markers that was recognized by α-PPO1/2 in fraction 2. Molecular mass markers are indicated on the left.
The proteins in fractions 1–3 were then run on a 7.5% SDS-polyacrylamide gel under reducing conditions, followed by immunoblotting using the cross-reacting anti-PPO/PO antibody. For peak 1, anti-PPO/PO recognized a ∼72 kDa PO band, a smaller, ∼60 kDa band that ran below PO, and a diffuse, high molecular mass band at the top of the gel (Fig. 4D). Peak 2 contained bands corresponding in mass to monomeric PPO and PO that were more intense than observed in peak 1. The same band present in peak 1 that ran below PO plus weak higher mass bands that ran near the 133 kDa marker were also observed (Fig. 4D). Peak 3 contained bands corresponding to monomeric PPO and PO plus the product that runs below PO but no PO-containing high mass bands (Fig. 4D). As noted in other studies (16, 22, 27, 28, 38), these results showed that PPO and PO did not elute from the column at their predicted masses due to binding to other proteins and/or the column matrix. However, these data did show that melanization activity localized primarily to fraction 1, which contained little monomeric PO. Fractions 2 and 3, in contrast, exhibited little or no melanization activity yet contained distinctly more monomeric PO than peak 1. We therefore concluded that melanization activity in these fractions does not correlate with the abundance of monomeric PO present.
Egf1.0 Blocks PPO Processing and Tyr Metabolism
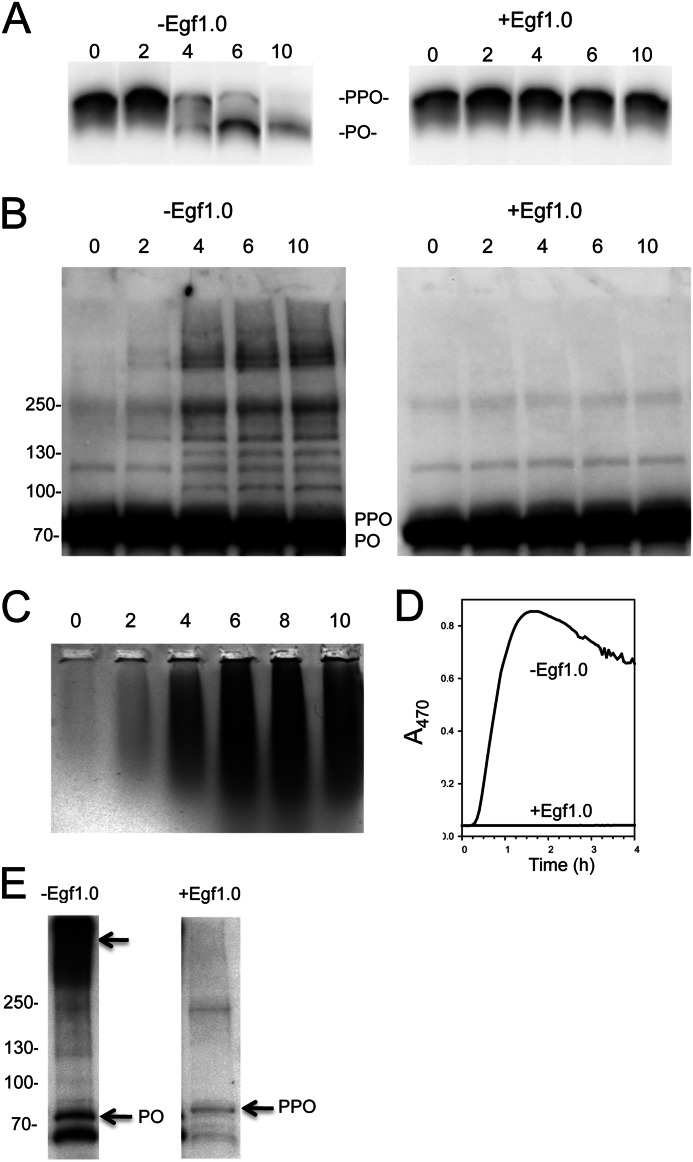
To address whether processing of PPO to PO was required for the melanization activity in fraction 1, we conducted experiments with Egf1.0, which blocks hemolymph melanization in M. sexta and two other moth species by inhibiting PAPs that process PPO (9–11). Plasma samples run on a 7.5% SDS-polyacrylamide gel under reducing conditions followed by immunoblotting confirmed that Egf1.0 also fully inhibits PPO processing in B. mori (Fig. 5A). The same samples run on 4–20% gels further showed that anti-PPO/PO detected the accrual of multiple bands larger than monomeric PO in control samples, but these bands did not appear in samples containing Egf1.0 (Fig. 5B).
FIGURE 5.
Egf1.0 inhibits Tyr-dependent melanization and formation of high mass complexes. A, Egf1.0 blocks PPO cleavage. Aliquots of B. mori plasma were collected and processed 0, 2, 4, 6, or 10 min later. Samples were then separated on a 7.5% continuous SDS-polyacrylamide gel, followed by immunoblotting using α-PPO1/2 as outlined in Fig. 4. B, Egf1.0 blocks formation of most PO-containing complexes that form in plasma. The same samples from A were separated on a 4–20% gradient SDS-polyacrylamide gel using an 8-fold higher sample load. Overloading resulted in no resolution of PPO from PO. The left blot shows the appearance of many PO-containing high mass bands in samples without Egf1.0, whereas the right blot shows that few high mass bands form in samples with Egf1.0. C, Egf1.0 blocks Tyr-dependent melanization. Egf1.0 was added to freshly prepared plasma at 0–10 min post-collection. At 10 min, samples (15 μl) were mixed with 4× native PAGE sample buffer (5 μl; Invitrogen), loaded onto a 0.33% SeaKem agarose gel and run for 2 h at 40 V in Tris-EDTA buffer. Melanization activity was assayed by immersing the gel in 0.5 mm Tyr in PBS for 1 h. D, Egf1.0 blocks melanization activity in fraction 1 using Tyr as the substrate. Freshly prepared B. mori plasma with and without Egf1.0 was separated by Superdex 200 chromatography, and protein eluent was measured at A280. Fraction 1 (0.7 ml), as described in the legend to Fig. 4, was collected and assayed for melanin formation at A470 after the addition of Tyr (0.5 mm). E, the same samples from D were concentrated 10-fold on 30-kDa filters (Amicon Ultra 0.5 ml 30k Ultracel), run on a 4–20% gradient SDS-polyacrylamide gel, and immunoblotted with α-PPO1/2. The PO-containing high mass band was readily visible in fraction 1 from plasma without Egf1.0 (arrow) but was absent in fraction 1 from plasma with Egf1.0. Monomeric PPO and PO are indicated to the right of each lane, whereas molecular mass markers are indicated to the left.
To determine whether Egf1.0 blocks melanin synthesis using Tyr as the substrate, we added Egf1.0 to plasma at 2-min intervals after collection. We then ran each sample on a 0.3% native agarose gel followed by incubation in 0.5 mm Tyr. These assays showed that little or no melanin formed in the lanes where Egf1.0 was added to plasma at 0 and 2 min post-collection, but melanin readily formed in the samples where Egf1.0 was added to plasma at 4, 6, 8, or 10 min post-collection (Fig. 5C). Melanin accumulation in these lanes also formed a smear that was most intense at the well interface. These data were consistent with the interpretation that inhibition of PPO processing by Egf1.0 blocks melanin synthesis, but Egf1.0 cannot inhibit melanin synthesis once processing of PPO to PO has occurred. They also suggested that melanization activity was associated with a protein complex that migrated poorly into agarose.
We thus added Egf1.0 to newly collected plasma followed by gel filtration chromatography and bioassay of the same fractions examined previously. Using Tyr as the substrate, fraction 1 from plasma with no Egf1.0 exhibited robust melanin synthesis activity after a short lag phase, but the same fraction from plasma with Egf1.0 had no activity (Fig. 5D). Although weakly visible in Fig. 4D, 10-fold concentration of fraction 1 on a 100-kDa filter before SDS-PAGE and immunoblotting greatly improved the visibility of the high mass complex recognized by anti-PO (Fig. 5E). However, even after concentration, no high mass complex was visible in the sample with Egf1.0 (Fig. 5E). We therefore concluded that Egf1.0 inhibits formation of the PO-containing complex that elutes in fraction 1 by blocking processing of PPO.
Collection of Plasma in PTU Enhances the Abundance of the High Mass Band in Peak 1 and Its Melanization Activity
Fig. 5B showed that processing of PPO to PO is associated with the rapid formation of many PO-containing bands in plasma that are chemically cross-linked and larger than monomeric PO, whereas our gel filtration data indicated that one of these complexes elutes in fraction 1, which also exhibits most of the melanization activity. Earlier studies with B. mori, T. molitor, and H. diomphalia (16, 22, 27, 28, 38) had already noted that processing of PPO to PO results in formation of PO-containing complexes that are chemically cross-linked. Adding a catecholamine substrate to a complex containing PO and SPH1 from T. molitor further increased its mass (27), whereas adding PTU to B. mori plasma inhibited the formation of most PO-containing bands (22). Together, these data suggested to prior authors that PO plays an enzymatic role in complex formation by oxidizing free catecholomines or Tyr residues in proteins to o-quinones, which then covalently cross-link PO to itself and other proteins.
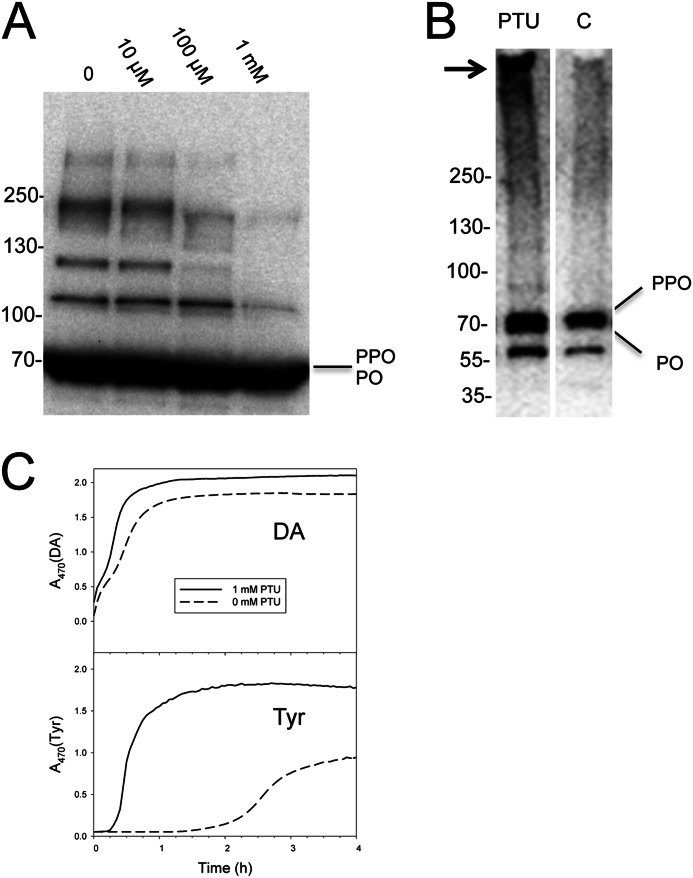
Similar to the effects of Egf1.0, we also observed that adding 100 μm PTU to B. mori plasma prevented most of the high mass bands shown in Fig. 5B from forming (Fig. 6A). However, adding PTU to newly collected plasma followed by size exclusion chromatography produced a result that differed from the effect of Egf1.0. Namely, rather than inhibiting formation of the PO-containing complex that elutes in fraction 1, adding PTU to plasma increased its abundance (Fig. 6B). In contrast, adding PTU to plasma had no effect on the abundance of monomeric PO in fraction 1 (Fig. 6B) or the abundance of PPO and PO that eluted in fractions 2 and 3 (data not shown). Because PTU is a catalytic inhibitor of PO that has no effect on PPO processing, these results suggested that quinone cross-linking is responsible for many of the PO-containing complexes visible in Fig. 5B. However, quinone cross-linking is not required for formation of the PO-containing complex in fraction 1.
FIGURE 6.
Collection of plasma in PTU enhances formation of the high mass complex in fraction 1. A, PTU blocks formation of most PO-containing complexes in plasma. PTU (10 μm to 1 mm) was added to newly collected plasma and then subjected to SDS-PAGE and immunoblot analysis as described in the legend to Fig. 5B. Numerous PO-containing high mass bands are visible in the plasma sample with no PTU, whereas most of these bands are absent in the sample with 1 mm PTU. B, plasma prepared with and without PTU (1 mm) was separated by gel filtration chromatography as outlined in the legend to Fig. 4. Fraction 1 was then added to a 4–20% SDS-polyacrylamide gel under reducing conditions, followed by immunoblotting using α-PPO1/2. The PO-containing high mass complex (arrow) is clearly more visible in fraction 1 from plasma where PTU was added than in fraction 1 from plasma without PTU. In contrast, the abundance of monomeric PPO and PO is similar. Mass markers are indicated to the left. C, melanization activity of fraction 1 from plasma samples with or without PTU (1 mm) using DA or Tyr as the substrate at a final concentration of 0.5 mm. Melanization activity was measured by monitoring dopachrome or dopaminechrome formation at A470.
Although PTU inhibits the catalytic activity of PO, it does not irreversibly bind to its target. We thus removed PTU from the complex in the process of fractionating the sample and washing the fraction on a 30-kDa filter, as done with SFP. We thus could also compare the melanization activity of fraction 1 with the same fraction from control plasma samples where no PTU was added. These results showed that melanization rates were much higher in the sample that initially contained PTU versus the sample without using either DA or Tyr as the substrate (Fig. 6C). Thus, by initially inhibiting PO activity and associated quinone cross-linking in plasma, we suggest that PTU increased the availability of PO for incorporation into the fraction 1 complex, which in turn increased the rate of melanization using either DA or Tyr.
The High Mass Complex in Fraction 1 Contains Other Proteins Besides PO
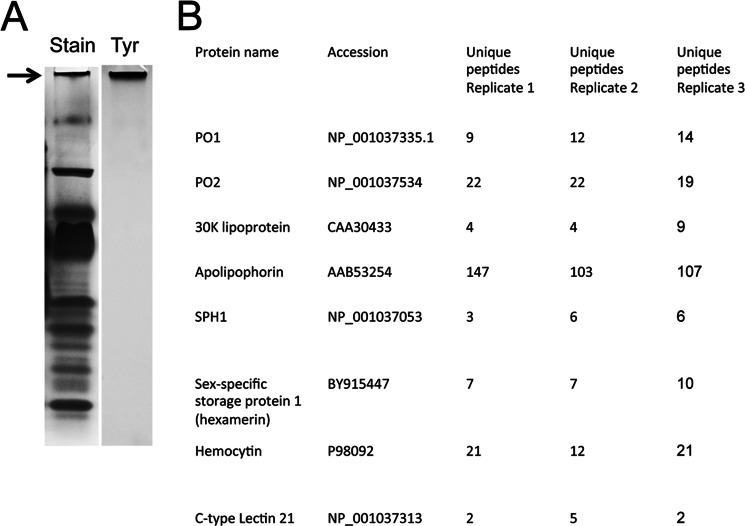
We analyzed the proteins present in the fraction 1 complex by collecting plasma in PTU and then running the sample on a native polyacrylamide gel (Fig. 7A). The PO-containing high mass complex was then excised, in-gel digested, and analyzed by Orbitrap Elite mass spectrometry (Fig. 7B). We detected multiple unique peptides corresponding to PO1 and PO2 but not PPO1 and PPO2. We also detected peptides matching serine protease homolog (SPH1), which is notable because earlier studies showed that SPH1 but not SPH2 homologs were present in PO-containing high mass complexes from T. molitor and H. diomphalia that exhibit melanin synthesis activity using a catecholamine substrate (27, 28). In addition, however, we also identified peptides corresponding to several other proteins, including 30K lipoprotein, apoliophorin, sex-specific storage protein 1 (hexamerin), hemocytin, and a C-type lectin (Fig. 7B). However, we detected no enzymes in the complex other than PO.
FIGURE 7.
B. mori proteins detected in the high mass complex by Orbitrap mass spectrometry. A, aliquots of freshly prepared plasma were collected in PTU (200 μm final concentration). After 1 h, samples (15 μl) were mixed with 4× native PAGE sample buffer (5 μl; Invitrogen) and run on 4–20% native PAGE. The lane to the left was stained with Coomassie Blue, whereas the lane to the right was immersed in 0.5 mm Tyr in PBS for 1 h. The only band that exhibited any melanization was a high mass entity that corresponded to fraction 1 from our gel filtration analysis. B, this band was excised from three independently prepared samples and subjected to Orbitrap MS analysis. The names and GenBankTM accession numbers for the proteins identified in the band are listed to the left. The number of unique peptides corresponding to each of these proteins from each replicate is indicated to the right.
DISCUSSION
Understanding of the PO cascade and its functions has increased considerably in recent years in two areas (1–6). The first has been the identification of several of the serine proteases and serpin inhibitors in the cascade that regulate the activation of PPO (summarized in Refs. 1 and 2). Some of these serine proteases and serpins have also been shown to play a role in activating the Toll pathway (27, 44–46). A second area of emphasis has been in understanding how infection by microorganisms activates the PO cascade. This has led to the identification of PRRs in B. mori and several other species that activate the PO cascade when they bind microbial cell wall components (33–35). Studies in this area have also shed important light on the function of SPHs as PAP cofactors (47, 48) and as proteins that bind to PO and microbial cell wall components to form complexes that localize melanin deposition to the surface of bacteria (27, 28, 38, 40).
This study addresses three areas that have received comparatively little attention in the literature. The first calls attention to the largely unrecognized disparity in how rapidly hemolymph from different species melanizes after collection from a wound site and the implications this variation has for understanding activation of the PO cascade. As previously noted, hemolymph from M. sexta and T. molitor requires several h to melanize or does not melanize at all after collection from a wound site if no microbial elicitor or diphenolic substrate like DA is added directly to the sample (31, 32). Studies with the moth P. includens show that its hemolymph melanizes without the addition of an elicitor, but the time required to do so is 1–2 h (7). Results presented here, however, show that hemolymph from B. mori begins melanizing within 3 min of collection, whereas our observations with another common moth, Helicoverpa zea, show that its hemolymph begins melanizing within 1 min.4 Taken together, these data suggest that wounding is insufficient to trigger melanization in species like M. sexta and T. molitor, which is why adding microbial elicitors directly to plasma samples is required. In contrast, wounding and/or the exposure of hemolymph to the external environment triggers rapid processing of PPO and melanization in B. mori, which is why adding a microbial elicitor directly to plasma samples is unnecessary.
Our detection of pg quantities of endotoxin on B. mori larvae suggests that one reason that plasma rapidly melanizes following collection from a wound site is that bacterial cell wall components endogenously present on the cuticle are sufficiently abundant (50) to trigger activation of PO cascade components in plasma. Another possibility is that damaged cuticle could also release factors that activate the PO cascade, given that 1) the first PAP identified from B. mori was purified from cuticle rather than hemolymph, and 2) studies from other invertebrates have identified molecules from damaged ectoderm that trigger immune defenses (17, 51, 52). In contrast, if microbes on the surface of B. mori larvae or signals released from damaged cuticle activate the rapid melanization of B. mori plasma, it remains unclear why plasma from species like M. sexta or T. molitor does not behave similarly because intuitively we would expect commensal microbes to reside on the surface of all arthropods.
The second area where our study contributes new information is the issue of substrate availability and usage during hemolymph melanization. Our results show that B. mori plasma contains Tyr but no detectable levels of tyramine, DOPA, or DA when first collected from a wound site. This finding is identical to our earlier measures of substrate titers in P. includens (7). Hopkins and co-workers (20) predominantly detected Tyr in hemolymph from M. sexta but also detected lesser amounts of different diphenols. This could indicate that real differences exist between species in the substrates that are available for melanization of hemolymph. However, the profile of substrates reported for M. sexta hemolymph relative to our findings could also reflect small differences in the timing of sample processing, given that we too detected DOPA in B. mori plasma if we waited just a few min before processing a sample for analysis.
The importance of detecting only Tyr in newly collected plasma from B. mori derives from the fact that earlier studies showed that in vitro activated PPO from B. mori hydroxylates Tyr very inefficiently, which results in slow melanization relative to using diphenols (16). Thus, if Tyr is the only substrate initially available in plasma, the very rapid rate of melanization that occurs after collection must be due to either other enzymes rapidly converting Tyr to DOPA or DA (or derivatives like NADA and NBAD) that PO can more efficiently use, or to PO in plasma using Tyr much more efficiently than prior results would suggest. Our detection of DOPA 5 min after collection of plasma indicates that some Tyr is quickly hydroxylated, whereas detection of PPO processing within 3 min of collection together with inhibition of Tyr metabolism by PTU lends circumstantial support that PO is the enzyme that catalyzes this event. In contrast, the absence of any cells or DOPA in our starting samples, together with our finding that 3-indole-tyrosine has no effect on Tyr metabolism, argues against Tyr hydroxylase playing a role in rapidly converting Tyr to DOPA. In addition, we never detected tyramine or DA in B. mori plasma in the first 20 min after collection from a wound site. Thus, although Tyr decarboxylase and DOPA decarboxylase have both been suggested to play roles in converting Tyr to tyramine and DOPA to DA for use by POs (49, 53–55), we see no evidence for this during the rapid melanization of B. mori plasma from a wound site.
The third area in which our study contributes new information is in identifying a critical role for formation of a chemically cross-linked complex that contains PO in substrate usage. Prior studies with B. mori had already shown that PO forms several complexes with other proteins through quinone cross-linking catalyzed by PO itself (22). Reconstitution experiments using a subset of purified components in the PO cascade from H. diomphalia and T. molitor further show that SPH1 binds to PO and microbial carbohydrates, which, together with quinone cross-linking, produces a complex that metabolizes catecholamines (27, 28). Our study corroborates prior results with B. mori by showing that processing of PPO to PO results in the formation of a number of PO-containing complexes of varying molecular mass. Where our data add new insights is where they indicate that Tyr usage is predominantly associated with a PO-containing complex that elutes in a fraction near a 670-kDa standard, and formation of this complex does not depend on quinone cross-linking. Our results further show that this PO-containing complex contains SPH1, as observed for the complexes reported from beetles (27, 28) and very recently melanized nodules from B. mori (49). However, the B. mori complex we analyzed also consistently contains other proteins, including apolipophorins previously implicated as clotting factors in Drosophila (4), a lectin that could function as a PRR or binding protein, and a hexamerin, which is a protein that structurally resembles PO itself (1, 2). Most importantly, our results suggest that complex formation is required for efficient metabolism of Tyr, which is the only PO substrate initially present in B. mori plasma following collection from a wound site.
In summary, our results suggest that processing of PPO to PO is an intermediate rather than terminal step in the PO cascade because formation of a PO-containing complex is required for efficient use of the only PO substrate (Tyr) initially present in plasma to produce melanin. One goal for the future will be to understand how upstream components in the PO cascade function in B. mori to allow melanization to occur so quickly after wounding. Another will be to understand how complex formation enhances the use of Tyr as a substrate.
Acknowledgments
We thank D. Phillips (University of Georgia Proteomics and Mass Spectrometry Core Facility) for assistance with sample analysis, A. Robertson and M. Kallins for assistance with bioassays, and J. A. Johnson for assistance in figure production. We also thank M. Ochiai (Hokkaido University, Japan) for generously providing the cross-reacting PPO antiserum used in the study.
This work was supported by National Science Foundation Grant IOS-1121006, United States Department of Agriculture Grant 2008-35320-18756, and the Georgia Agricultural Experiment Station.
K. D. Clark and M. R. Strand, unpublished observations.
- PO
- phenoloxidase
- DA
- dopamine
- DOPA
- dihydroxyphenylalanine
- PAPs
- phenoloxidase activating proteases
- PPO
- prophenoloxidase
- PTU
- phenylthiourea
- SPH
- serine protease homolog
- SFP
- substrate-free plasma
- LAL
- Limulus amebocyte lysate.
REFERENCES
- 1. Cerenius L., Lee B. L., Söderhäll K. (2008) The proPO-system. Pros and cons for its role in invertebrate immunity. Trends Immunol. 29, 263–271 [DOI] [PubMed] [Google Scholar]
- 2. Kanost M. R., Gorman M. J. (2008) Phenoloxidases in insect immunity. in Insect Immunology (Beckage N. E., ed) pp. 69–96, Academic Press, Inc., San Diego [Google Scholar]
- 3. Nappi A., Poirié M., Carton Y. (2009) The role of melanization and cytotoxic by-products in the cellular immune responses of Drosophila against parasitic wasps. in Advances in Parasitology (Genevieve P., ed) pp. 99–121, Academic Press, Inc., San Diego: [DOI] [PubMed] [Google Scholar]
- 4. Sugumaran M. (2002) Comparative biochemistry of eumelanogenesis and the protective roles of phenoloxidase and melanin in insects. Pigment Cell Res. 15, 2–9 [DOI] [PubMed] [Google Scholar]
- 5. Cerenius L., Söderhäll K. (2004) The prophenoloxidase-activating system in invertebrates. Immunol. Rev. 198, 116–126 [DOI] [PubMed] [Google Scholar]
- 6. Kanost M. R., Jiang H., Yu X. Q. (2004) Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev. 198, 97–105 [DOI] [PubMed] [Google Scholar]
- 7. Clark K. D., Lu Z., Strand M. R. (2010) Regulation of melanization by glutathione in the moth Pseudoplusia includens. Insect Biochem. Mol. Biol. 40, 460–467 [DOI] [PubMed] [Google Scholar]
- 8. Strand M. R. (2010) Polydnaviruses. in Insect Virology (Asgari S., Johnson K., eds) pp. 171–198, Caister Academic Press, Norfolk, UK [Google Scholar]
- 9. Beck M. H., Strand M. R. (2007) A novel polydnavirus protein inhibits the insect prophenoloxidase activation pathway. Proc. Natl. Acad. Sci. U.S.A. 104, 19267–19272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu Z., Beck M. H., Wang Y., Jiang H., Strand M. R. (2008) The viral protein Egf1.0 is a dual activity inhibitor of prophenoloxidase-activating proteinases 1 and 3 from Manduca sexta. J. Biol. Chem. 283, 21325–21333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu Z., Beck M. H., Strand M. R. (2010) Egf1.5 is a second phenoloxidase cascade inhibitor encoded by Microplitis demolitor bracovirus. Insect Biochem. Mol. Biol. 40, 497–505 [DOI] [PubMed] [Google Scholar]
- 12. Nappi A. J., Christensen B. M. (2005) Melanogenesis and associated cytotoxic reactions. Applications to insect innate immunity. Insect Biochem. Mol. Biol. 35, 443–459 [DOI] [PubMed] [Google Scholar]
- 13. Li Y., Wang Y., Jiang H., Deng J. (2009) Crystal structure of Manduca sexta prophenoloxidase provides insights into the mechanism of type 3 copper enzymes. Proc. Natl. Acad. Sci. U.S.A. 106, 17002–17006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vavricka C. J., Christensen B. M., Li J. (2010) Melanization in living organisms: a perspective of species evolution. Protein Cell 1, 830–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hall M., Scott T., Sugumaran M., Söderhäll K., Law J. H. (1995) Proenzyme of Manduca sexta phenol oxidase. Purification, activation, substrate specificity of the active enzyme, and molecular cloning. Proc. Natl. Acad. Sci. U.S.A. 92, 7764–7768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ashida M., Dohke K. (1980) Activation of pro-phenoloxidase by the activating enzyme of the silkworm, Bombyx mori. Insect Biochem. 10, 37–47 [Google Scholar]
- 17. Aso Y., Kramer K. J., Hopkins T. L., Lookhart G. L. (1985) Characterization of haemolymph protyrosinase and a cuticular activator from Manduca sexta (L.). Insect Biochem. 15, 9–17 [Google Scholar]
- 18. Chase M. R., Raina K., Bruno J., Sugumaran M. (2000) Purification, characterization and molecular cloning of prophenoloxidases from Sarcophaga bullata. Insect Biochem. Mol. Biol. 30, 953–967 [DOI] [PubMed] [Google Scholar]
- 19. Munkirs D. D., Christensen B. M., Tracy J. W. (1990) High-pressure liquid chromatographic analysis of hemolymph plasma catecholamines in immune-reactive Aedes aegypti. J. Invertebr. Pathol. 56, 267–279 [DOI] [PubMed] [Google Scholar]
- 20. Hopkins T. L., Morgan T. D., Kramer K. J. (1984) Catecholamines in haemolymph and cuticle during larval, pupal and adult development of Manduca sexta (L.). Insect Biochem. 14, 533–540 [Google Scholar]
- 21. Hopkins T. L., Starkey S. R., Beckage N. E. (1998) Tyrosine and catecholamine levels in the hemolymph of tobacco hornworm larvae, Manduca sexta, parasitized by the braconid wasp, Cotesia congregata, and in the developing parasitoids. Arch. Insect Biochem. Physiol. 38, 193–201 [Google Scholar]
- 22. Ashida M., Yoshida H. (1988) Limited proteolysis of prophenoloxidase during activation by microbial products in insect plasma and effect of phenoloxidase on electrophoretic mobilities of plasma proteins. Insect Biochem. 18, 11–19 [Google Scholar]
- 23. Sugumaran M., Nellaiappan K., Valivittan K. (2000) A new mechanism for the control of phenoloxidase activity. Inhibition and complex formation with quinone isomerase. Arch. Biochem. Biophys. 379, 252–260 [DOI] [PubMed] [Google Scholar]
- 24. Beck G., Cardinale S., Wang L., Reiner M., Sugumaran M. (1996) Characterization of a defense complex consisting of interleukin 1 and phenol oxidase from the hemolymph of the tobacco hornworm, Manduca sexta. J. Biol. Chem. 271, 11035–11038 [DOI] [PubMed] [Google Scholar]
- 25. Yu X.-Q., Kanost M. R. (2000) Immulectin-2, a lipopolysaccharide-specific lectin from an insect, Manduca sexta, is induced in response to gram-negative bacteria. J. Biol. Chem. 275, 37373–37381 [DOI] [PubMed] [Google Scholar]
- 26. Wang Y., Jiang H. (2004) Prophenoloxidase (proPO) activation in Manduca sexta. An analysis of molecular interactions among proPO, proPO-activating proteinase-3, and a cofactor. Insect Biochem. Mol. Biol. 34, 731–742 [DOI] [PubMed] [Google Scholar]
- 27. Kan H., Kim C.-H., Kwon H.-M., Park J.-W., Roh K.-B., Lee H., Park B.-J., Zhang R., Zhang J., Söderhäll K., Ha N.-C., Lee B. L. (2008) Molecular control of phenoloxidase-induced melanin synthesis in an insect. J. Biol. Chem. 283, 25316–25323 [DOI] [PubMed] [Google Scholar]
- 28. Piao S., Song Y.-L., Kim J. H., Park S. Y., Park J. W., Lee B. L., Oh B.-H., Ha N.-C. (2005) Crystal structure of a clip-domain serine protease and functional roles of the clip domains. EMBO J. 24, 4404–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Clark K. D., Pech L. L., Strand M. R. (1997) Isolation and identification of a plasmatocyte-spreading peptide from the hemolymph of the lepidopteran insect Pseudoplusia includens. J. Biol. Chem. 272, 23440–23447 [DOI] [PubMed] [Google Scholar]
- 30. Ashida M., Ochiai M., Niki T. (1988) Immunolocalization of prophenoloxidase among hemocytes of the silkworm, Bombyx mori. Tissue Cell 20, 599–610 [DOI] [PubMed] [Google Scholar]
- 31. Ma C., Kanost M. R. (2000) A β1,3-glucan recognition protein from an insect, Manduca sexta, agglutinates microorganisms and activates the phenoloxidase cascade. J. Biol. Chem. 275, 7505–7514 [DOI] [PubMed] [Google Scholar]
- 32. Zhang R., Cho H. Y., Kim H. S., Ma Y. G., Osaki T., Kawabata S., Söderhäll K., Lee B. L. (2003) Characterization and properties of a 1,3-β-d-glucan pattern recognition protein of Tenebrio molitor larvae that is specifically degraded by serine protease during prophenoloxidase activation. J. Biol. Chem. 278, 42072–42079 [DOI] [PubMed] [Google Scholar]
- 33. Ochiai M., Ashida M. (2000) A pattern-recognition protein for β-1,3-glucan. J. Biol. Chem. 275, 4995–5002 [DOI] [PubMed] [Google Scholar]
- 34. Lee S. Y., Wang R., Söderhäll K. (2000) A lipopolysaccharide- and β-1,3-glucan-binding protein from hemocytes of the freshwater crayfish Pacifastacus leniusculus. J. Biol. Chem. 275, 1337–1343 [DOI] [PubMed] [Google Scholar]
- 35. Lee M. H., Osaki T., Lee J. Y., Baek M. J., Zhang R., Park J. W., Kawabata S., Söderhäll K., Lee B. L. (2004) Peptidoglycan recognition proteins involved in 1,3-d-glucan-dependent prophenoloxidase activation system of insect. J. Biol. Chem. 279, 3218–3227 [DOI] [PubMed] [Google Scholar]
- 36. Ashida M. (1981) A cane sugar factor suppressing activation of prophenoloxidase in haemolymph of the silkworm, Bombyx mori. Insect Biochem. 11, 57–65 [Google Scholar]
- 37. Kawabata T., Yasuhara Y., Ochiai M., Matsuura S., Ashida M. (1995) Molecular cloning of insect pro-phenol oxidase. A copper-containing protein homologous to arthropod hemocyanin. Proc. Natl. Acad. Sci. U.S.A. 92, 7774–7778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yasuhara Y., Koizumi Y., Katagiri C., Ashida M. (1995) Reexamination of properties of prophenoloxidase isolated from larval hemolymph of the silkworm, Bombyx mori. Arch. Biochem. Biophys. 320, 14–23 [DOI] [PubMed] [Google Scholar]
- 39. Asada N., Sezaki H. (1999) Properties of phenoloxidases generated from prophenoloxidase with 2-propanol and the natural activator in Drosophila melanogaster. Biochem. Genet. 37, 149–158 [DOI] [PubMed] [Google Scholar]
- 40. Yu X.-Q., Jiang H., Wang Y., Kanost M. R. (2003) Nonproteolytic serine proteinase homologs are involved in prophenoloxidase activation in the tobacco hornworm, Manduca sexta. Insect Biochem. Mol. Biol. 33, 197–208 [DOI] [PubMed] [Google Scholar]
- 41. Jaenicke E., Decker H. (2003) Tyrosinases from crustaceans form hexamers. Biochem. J. 371, 515–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu C., Yamamoto K., Cheng T.-C., Kadono-Okuda K., Narukawa J., Liu S.-P., Han Y., Futahashi R., Kidokoro K., Noda H., Kobayashi I., Tamura T., Ohnuma A., Banno Y., Dai F.-Y., Xiang Z.-H., Goldsmith M. R., Mita K., Xia Q.-Y. (2010) Repression of tyrosine hydroxylase is responsible for the sex linked chocolate mutation of the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. U.S.A. 107, 12980–12985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gorman M. J., An C., Kanost M. R. (2007) Characterization of tyrosine hydroxylase from Manduca sexta. Insect Biochem. Mol. Biol. 37, 1327–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ligoxygakis P., Pelte N., Ji C., Leclerc V., Duvic B., Belvin M., Jiang H., Hoffmann J. A., Reichhart J. M. (2002) A serpin mutant links Toll activation to melanization in the host defence of Drosophila. EMBO J. 21, 6330–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Christen J. M., Hiromasa Y., An C., Kanost M. R. (2012) Identification of plasma porteinase complexes with serpin-3 in Manduca sexta. Insect Biochem. Mol. Biol. 42, 946–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim C. H., Kim S.-J., Kan H., Kwon H.-M., Roh K.-B., Jiang R., Yang Y., Park J.-W., Lee H.-H., Ha N.-C., Kang H. J., Nonaka M., Söderhäll K., Lee B. L. (2008) A three-step proteolytic cascade mediates the activation of the peptidoglycan-induced Toll pathway in an insect. J. Biol. Chem. 283, 7599–7607 [DOI] [PubMed] [Google Scholar]
- 47. Jiang H., Wang Y., Kanost M. R. (1998) Pro-phenol oxidase activating proteinase from an insect, Manduca sexta. A bacteria-inducible protein similar to Drosophila easter. Proc. Natl. Acad. Sci. U.S.A. 95, 12220–12225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kwon T. H., Kim M. S., Choi H. W., Joo C. H., Cho M. Y., Lee B. L. (2000) A masquerade-like serine proteinease homologue is necessary for phenoloxidase activity in the coleopteran insect, Holotrichia diomphalia larvae. Eur. J. Biochem. 267, 6188–6196 [DOI] [PubMed] [Google Scholar]
- 49. Sakamoto M., Ohta M., Suzuki A., Takase H., Yoshizawa Y., Kitami M., Sato R. (2011) Localization of the serine protease homology BmSPH-1 in nodules of E. coli-injected Bombyx mori larvae and functional analysis of its role in nodule formation. Dev. Comp. Immunol. 35, 611–619 [DOI] [PubMed] [Google Scholar]
- 50. Tsuchiya M., Asahi N., Suzuoki F., Ashida M., Matsuura S. (1996) Detection of peptidoglycan and β-glucan with silkworm larvae plasma test. FEMS Immunol. Med. Microbiol. 15, 129–134 [DOI] [PubMed] [Google Scholar]
- 51. Nyholm S. V., Graf J. (2012) Knowing your friends. Invertebrate innate immunity fosters beneficial bacterial symbioses. Nat. Rev. Microbiol. 10, 815–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dohke K. (1973) Studies on prephenoloxidase-activating enzyme from cuticle of the silworm Bombyx mori. I. Activation reaction by the enzyme. Arch. Biochem. Biophys. 157, 203–209 [DOI] [PubMed] [Google Scholar]
- 53. Christensen B. M., Li J., Chen C. C., Nappi A. J. (2005) Melanization immune responses in mosquito vectors. Trends Parasitol. 21, 192–199 [DOI] [PubMed] [Google Scholar]
- 54. Kim M. H., Joo C. H., Cho M. Y., Kwon T. H., Lee K. M., Natori S., Lee T. H., Lee B. L. (2000) Bacterial-injection induced synthesis of N-β-alanyldopamine and dopa decarboxylase in the hemolymph of coleopteran insect, Tenebrio molitor larvae. Eur. J. Biochem. 267, 2599–2608 [DOI] [PubMed] [Google Scholar]
- 55. Zhu Y., Johnson T. J., Myers A. A., Kanost M. R. (2003) Identification by substractive suppression hybridization of bacteria-induced genes expressed in Manduca sexta fat body. Insect Biochem. Mol. Biol. 33, 541–559 [DOI] [PubMed] [Google Scholar]
- 56. Dionex Corp (2003) Determination of Amino Acids in Cell Cultures and Fermentation Broths, Dionex Application Note 150, p. 4, Dionex Corp., Sunnyvale, CA [Google Scholar]