Background: Plekhg4 is putative guanine nucleotide exchange factor associated with autosomal dominant spinocerebellar ataxia.
Results: Plekhg4 is regulated by the heat shock proteins and functions as a bona fide guanine nucleotide exchange factor.
Conclusion: Plekhg4 is the first RhoGEF implicated in spinocerebellar ataxia.
Significance: Aberrant GTPase signaling is a novel possible mechanism underlying autosomal dominant spinocerebellar ataxia.
Keywords: Aggregation, Ataxia, Cerebellum, GTPase, Guanine Nucleotide Exchange Factor (GEF), Heat Shock Protein, Ubiquitination
Abstract
Mutations in the PLEKHG4 (puratrophin-1) gene are associated with the heritable neurological disorder autosomal dominant spinocerebellar ataxia. However, the biochemical functions of this gene product have not been described. We report here that expression of Plekhg4 in the murine brain is developmentally regulated, with pronounced expression in the newborn midbrain and brainstem that wanes with age and maximal expression in the cerebellar Purkinje neurons in adulthood. We show that Plekhg4 is subject to ubiquitination and proteasomal degradation, and its steady-state expression levels are regulated by the chaperones Hsc70 and Hsp90 and by the ubiquitin ligase CHIP. On the functional level, we demonstrate that Plekhg4 functions as a bona fide guanine nucleotide exchange factor (GEF) that facilitates activation of the small GTPases Rac1, Cdc42, and RhoA. Overexpression of Plekhg4 in NIH3T3 cells induces rearrangements of the actin cytoskeleton, specifically enhanced formation of lamellopodia and fillopodia. These findings indicate that Plekhg4 is an aggregation-prone member of the Dbl family GEFs and that regulation of GTPase signaling is critical for proper cerebellar function.
Introduction
Guanine nucleotide exchange factors (GEFs)3 comprise a large family of regulatory proteins that control diverse intracellular processes such as gene expression, cytoskeletal rearrangements, intracellular trafficking, and metabolism (1). Accordingly, dysregulation of GEF levels or activity have profound consequences on normal cell behavior and lead to proliferative and developmental pathologies. For example, activating mutations in VAV1 (2), P-Rex1 (3), and Dbl (4) contribute to cell transformation and tumorigenesis, and mutations in FGD1 are associated with facio-gential dysplesia and mental retardation (5).
GEFs mediate their biological effects by facilitating the exchange of bound GDP for GTP in the nucleotide binding pocket of small GTP-binding proteins. The activated GTPases then stimulate specific downstream effectors that control cytoskeletal architecture, vesicular genesis and trafficking, cell polarity, and cell cycle progression (6, 7). The Dbl oncogene (8) is the prototypic member of a large family of structurally and functionally related GEFs, which activate GTPases from the Rho family and are characterized by a tandem arrangement of a Dbl homology (DH) domain and a pleckstrin homology (PH) domain (9). Whereas the DH domain is the minimal functional unit required for nucleotide exchange (10), the PH domain is essential for proper intracellular localization and cell transformation (11). N-terminal to the DH/PH module are spectrin repeats that mediate association of Dbl with the molecular chaperones Hsc70 and Hsp90, and the ubiquitin E3 ligase CHIP (12). These interactions determine the steady-state expression levels of Dbl by modulating the rate of ubiquitination and proteasomal degradation (13). It is generally believed that oncogenic mutations activate Dbl by disrupting intramolecular (14) and intermolecular (13) interactions that alter GEF activity and levels.
Multiple lines of evidence demonstrate that Dbl-like GEFs and their substrate GTPases play important roles in development, morphogenesis, and function of the central nervous system (15) and that they transduce signals from neuronal surface receptors such as EphB, TrkB, NMDA, and the AMPA receptor. Spinocerebellar ataxias (SCAs) are debilitating heritable neurodegenerative disorders characterized by progressive loss of motor coordination and balance that stem from cerebellar dysfunction (16). Of the multiple SCA forms, the 16q22.1-linked autosomal dominant cerebellar ataxia is of special interest. Originally, a single C-to-T substitution in the 5′-untranslated region (5′-UTR) of the PLEKHG4 gene (−16C>T) was shown to associate with the disease (17–22). Moreover, brains of affected patients exhibited selective atrophy of cerebellar Purkinje neurons, accompanied by cytoplasmic aggregation of the Plekhg4 protein (17). Later studies extended this ataxia-linked genomic site to a 900-kb region of the PLEKHG4 promoter (22). Interestingly, ataxia-linked pentanucleotide repeat insertions of various sizes were also observed in that locus (23). The genetic findings that link PLEKHG4 to 16q22.1 SCA are underscored by histopathological and biochemical evidence. Specifically, cerebella samples from 16q22.1-linked SCA patients showed a significant reduction in Plekhg4 mRNA and enhanced formation of cytoplasmic aggregates that contain Plekhg4, G58K, and spectrin (17).
The Plekhg4 primary sequence indicates the presence of a Sec14 domain that often mediates lipid binding, a spectrin domain that typically mediates protein-protein interactions, and the canonical DH/PH module which catalyzes nucleotide exchange on substrate GTPases (10). The conservation of this signature domain architecture raises the intriguing possibility that Plekhg4 functions in the cerebellum by mediating the activation of small GTPases from the Rho family. If true, this is the first case where aberrant GEF-GTPase signaling is a likely molecular culprit underlying SCA pathology. Toward this end, we report here our initial biochemical characterization of Plekhg4 as an activator of Cdc42, Rac1, and RhoA and the post-translational mechanisms that control its expression levels.
MATERIALS AND METHODS
Cell Culture
NIH3T3 cells and COS7 cells were cultured in DMEM supplemented with 10% calf serum or 10% fetal bovine serum (Hyclone), respectively, in 5% CO2 at 37 °C.
Molecular Constructs
A hemagglutinin (HA)-fused Plekhg4 construct was generated by PCR amplification of the PLEKHG4 reading frame from a commercial image clone in the pOBT vector (I.M.A.G.E. clone 6291175; ATCC 10539799) using primers that add an in-frame N-terminal influenza HA tag. The resulting amplicon was ligated into a pCDNA3.1/HA/Hygro+ vector (Invitrogen). Constructs encoding Dbl (residues 1–925) and small GTPases were described previously (12, 24, 25). The pEBG-GST-Plekhg4 construct was generated by cloning the cDNA into a pEBG-GST vector (generous gifts of Yi Zheng, Cincinnati Children's Hospital Medical Center, and Silvio Gutkind, NIDCR, National Institutes of Health). CHIP cDNA (a generous gift from Cam Patterson, University of North Carolina at Chapel Hill) was amplified by PCR and cloned into a pCDNA4.1-HisMax vector (Invitrogen) to generate the Xpress-tagged CHIP construct. Myc-ubiquitin (in the pCW7 plasmid) was a generous gift from Ron Kopito (Stanford University). All constructs were verified by restriction mapping and sequencing.
Reagents
MG132 (Calbiochem) was dissolved in dimethyl sulfoxide and stored at −20 °C. 17-N-allylamino-17-demethoxygeldanamycin (17-AAG; NSC 330507) was obtained from A. G. Scientific and from the Division of Cancer Treatment and Diagnosis, NCI, National Institutions of Health.
Antibodies
Anti-HA (clone HA.11; both ascites and purified), anti-HA-FITC conjugates, and anti-GST antibodies were purchased from Covance Inc. Anti-Dbl antibodies (sc-89 and sc-28582) were from Santa Cruz Biotechnology. Anti-Hsc70 (SPA-815 and SPA-820) and anti-Hsp90 (SPA-835) antibodies were obtained from StressGen. The anti-ubiquitin antibody (clone FK2) was from obtained from Millipore.
RT-PCR
Total RNA was isolated from indicated mouse brain tissues with the RNAqueous-4PCR kit (Ambion). RNA (500 ng) from each sample was reverse-transcribed using High Capacity cDNA Reverse Transcription Kits (Applied Biosystems), and cDNA was amplified using primers based on the mouse brain Plekhg4 sequence. Plekhg4 primers (forward, 5′-TCC CTA AGC CTG CTG ACT G-3′; reverse, 5′-ATG TGG GTC AGC AGG CTC-3′) were chosen to amplify a 412-bp fragment, from nucleotide 494 to 905 in the sequence (GenBank accession number NM_001081333). β-Actin was used as a housekeeping gene for normalizations.
Immunohistochemistry
Formalin-fixed paraffin-embedded brains were sagittally sectioned (20 μm), deparaffinized, and subjected to antigen retrieval step with 10 mm citrate. Sections were incubated overnight at 4 °C with anti-Plekhg4 antibody (Abcam). Plekhg4 expression was visualized using avidin-biotinylated peroxidase and developed with 3,3′-diaminobenzidine.
Ubiquitination and Chaperone Association
Forty-eight hours after transfection, the indicated proteins were isolated by glutathione-agarose precipitations or immunoprecipitations, and associated proteins were analyzed by Western blotting with the indicated antibodies.
GEF-GTPase Association
GST fusion proteins of Rac1, Cdc42, and RhoA were expressed in Escherichia coli and purified as described previously (26). cDNAs were transfected into COS7 cells using FuGENE6 according to the manufacturer's protocol, and cell lysates were prepared as described previously (12). Recombinant GTPases were immobilized on glutathione-agarose and washed extensively with radioimmuneprecipitation assay buffer supplemented with 1% Nonidet P-40 prior to nucleotide loading. To generate nucleotide-free GTPases, immobilized GTPases were washed three times in 20 mm HEPES, pH 7.4, 200 mm NaCl, 10 mm EDTA, 2 mm PMSF and used immediately. To generate constitutively active GTPases, immobilized proteins were washed extensively with 20 mm HEPES, pH 7.4, 200 mm NaCl, 10 mm EDTA, 2 mm PMSF, 100 mm GTPγS, and then with the same buffer supplemented with 10 mm MgCl2. Immobilized GTPases prepared were incubated with GEF-containing lysates at 4 °C for 1.5 h, washed three times in the respective buffer by centrifugation, and associated GEFs were analyzed by Western blotting.
GTPase Activation Assays
To evaluate GEF activity, the fraction of GTPase that is in the active (GTP-bound) state was determined using an established selective precipitation approach (27, 28). Briefly, COS7 cells were co-transfected with Rac1, Cdc42, or RhoA (HA-tagged in the pKH3 vector) together with the GEF (Plekhg4 or Dbl in the pCDNA3.1 vector). Cells were lysed in 20 mm HEPES, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 10% glycerol, 10 mm MgCl2, 1 mm EDTA, 0.2 mm phenylmethylsulfonyl fluoride, and 10 μg/ml each leupeptin and aprotinin and incubated at 4 °C for 2 h with ∼20 μg of immobilized GST-fused GTPase binding domain of mPAK3 (27) or of Rhotekin (28). The amount of activated G protein was visualized with anti-HA Western blotting and compared with the total GTPase levels as determined by anti-HA immunoblotting of whole cell lysates.
Fluorescence Microscopy
NIH3T3 cells were seeded on collagen-coated coverslips and transfected using PolyFect Transfection Reagent (Qiagen) according to the manufacturer's instructions. Thirty-one hours after transfection, cells were serum-starved for 17 h, fixed with 3.7% paraformaldehyde, permeabilized with 0.2% Triton X-100, and stained with the indicated reagents. Stained slides were imaged on a Zeiss LSM 510 META confocal microscope.
Hsp90 Inhibition Studies
HA-tagged Plekhg4 protein was visualized using an anti-HA FITC-conjugated antibody (Covance) and imaged on a Leica DM4100B inverted fluorescence microscope.
RESULTS
Expression of Plekhg4 Is Developmentally Regulated
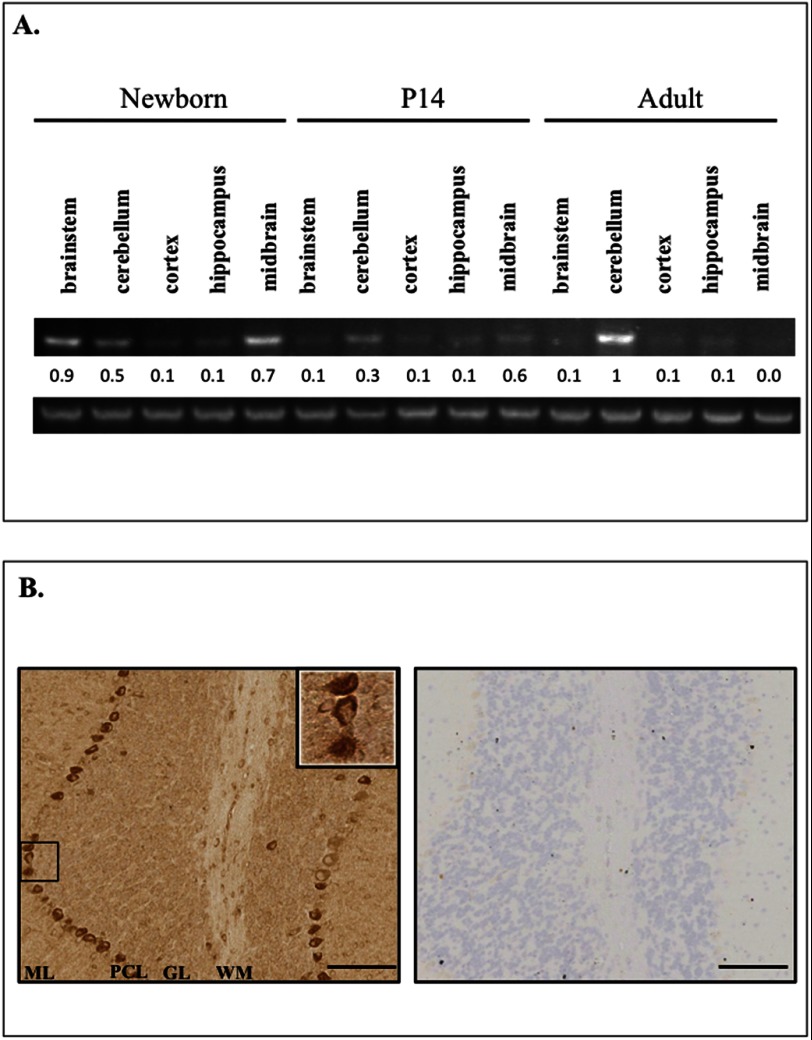
Association between the Plekhg4 gene and spinocerebellar ataxia led us to take a deeper look into Plekhg4 expression in the brain. Previously published data show that Plekhg4 mRNA expression is high in the testis and pancreas; has mild expression in the spleen, thymus, prostate gland, heart, placenta, lung, liver, kidney; and has low expression in the ovary, small intestine, colon, peripheral blood leukocytes, whole brain, and skeletal muscle (17). Examination of the temporal and spatial expression of Plekhg4 mRNA in the mouse brain revealed that the transcript is expressed selectively in the cerebellum, brainstem, and midbrain in an age-dependent manner (Fig. 1A). mRNA expression of Plekhg4 in the newborn brainstem and midbrain decreased with advancing age, leaving the cerebellum as the primary site of expression in the mature animal. To augment these results we employed immunoreactivity-based approaches aimed at detecting expression patterns of the Plekhg4 protein. In agreement with previous reports of Plekhg4 mRNA (17), we detected high level expression of the Plekhg4 protein in the testis (data not shown). Additionally, we observed high expression of Plekhg4 in the cerebellum, specifically in Purkinje neurons, as shown in Fig. 1B. Interestingly, Ishikawa et al. reported the existence of Plekhg4 protein aggregates in Purkinje neurons of SCA-afflicted patients (17). Taken together, these findings strongly suggest that Plekhg4 plays an important role in cerebellar function.
FIGURE 1.
Spatiotemporal expression patterns of Plekhg4 in the murine brain. A, top, Plekhg4 mRNA levels compared at birth (P0) and at the ages of 14 (P14) and 56 days (adult) in the indicated brain regions using real-time RT-PCR. A, bottom, expression of actin mRNA. The image is representative of three independent experiments. Quantification of mRNA levels was achieved using ImageJ. B, left, anti-Plekhg4 immunohistochemical staining of cerebellum section from a 2-week-old mouse. Note dark diaminobenzidine staining seen in Purkinje neurons. ML, molecular layer; PCL, Purkinje cell layer; GL, granular layer; WM, white matter. B, right, negative control (secondary only) for diaminobenzidine staining. Negative control was stained only with secondary antibody. Scale bars, 100 μm.
Domain Structure of Plekhg4
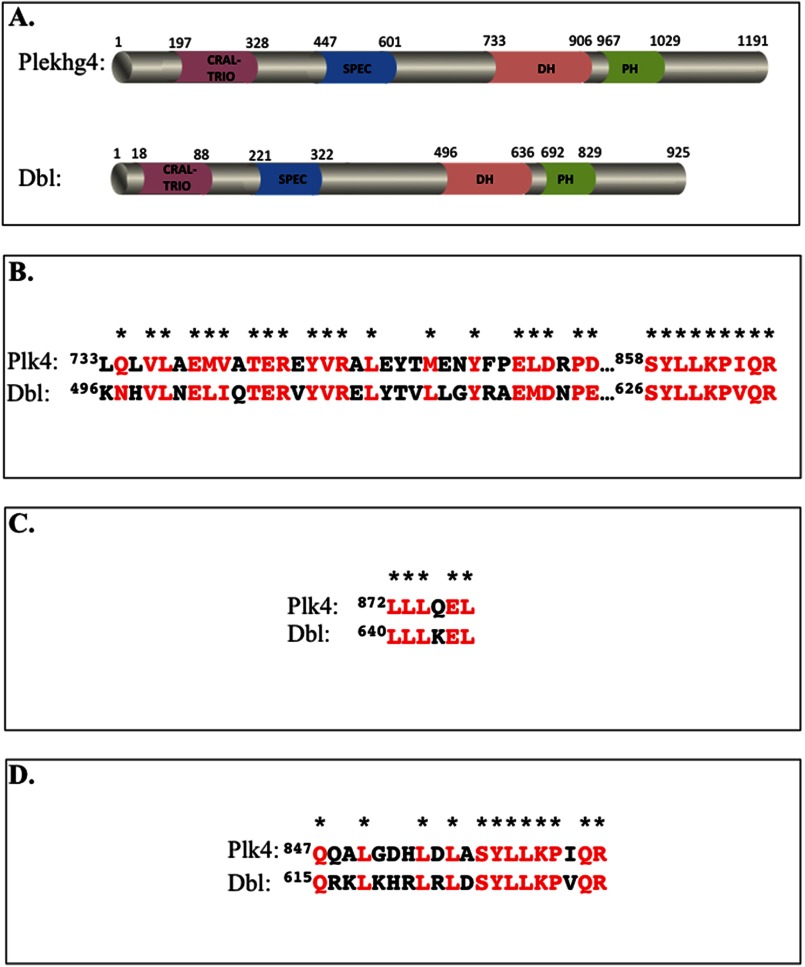
Fig. 2A depicts the predicted domain structures of Plekhg4 compared with the prototypical GEF of the Dbl family, the proto-oncogene product Dbl. Alignment of the Plekhg4 putative GEF (DH) domain with that of Dbl revealed 58% sequence similarity and 42% sequence identity. Importantly, high similarity is evident in residues thought to be critical for catalyzing nucleotide exchange (Fig. 2B), for binding the substrate GTPase (Fig. 2C), and for dictating GTPase specificity (Fig. 2D) (29, 30). Taken together, the high degree of structural similarity between Plekhg4 and Dbl suggests that, like Dbl, the primary function of Plekhg4 is in activating the substrate GTPases RhoA, Rac1, and Cdc42.
FIGURE 2.
Sequence signatures of Plekhg4 and Dbl. A, domain structure of the two proteins inferred using sequence prediction algorithm (Expasy). Domain size and limits were manually drawn to scale. B–D, sequence homology between Plekhg4 and Dbl. Shown are sequence alignments between the DH domains of the proteins (B), residues that mediate GTPase binding (C), and the residues that dictate GTPase selectivity (D). Identical and highly similar residues are shown in red and denoted by asterisks.
GTPase Binding and Activation by Plekhg4
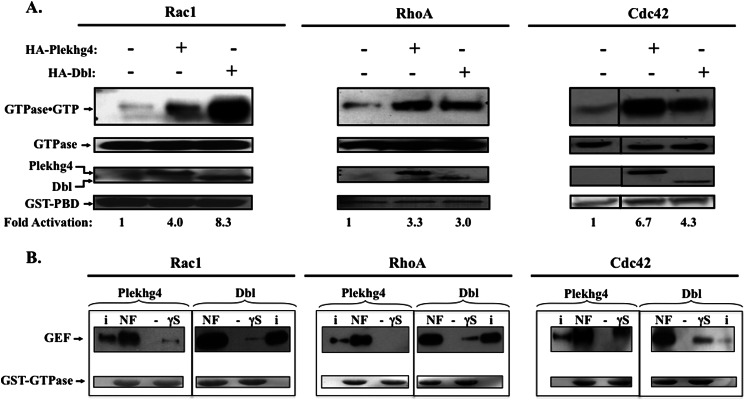
Despite the striking similarity to Rho-GEFs from the Dbl family, the ability of Plekhg4 to stimulate nucleotide exchange on substrate GTPases has not been reported. We therefore assessed the ability of Plekhg4 to activate the different GTPases in cultured cells, using the p21 binding domain (PBD) assay that was originally developed for Ras (31), and adapted for Cdc42, Rac, and RhoA (27, 28). Specifically, we used an immobilized peptide sequence from the GTPase downstream target (mPAK3 for Rac/Cdc42 and Rhotekin for RhoA) to selectively precipitate the GTP-bound form of each GTPases. As shown in Fig. 3A, expression of Plekhg4 in serum-starved cells activated Cdc42, Rac1, and RhoA, evident by the increased fraction of the GTPase that precipitates with the immobilized target PBD. To gain insight into the mechanisms of Plekhg4-stimulated nucleotide exchange, we examined the binding of Plekhg4 to its substrate GTPases. We employed an established approach in which the GEF is precipitated with immobilized GTPases in either the nucleotide-free or the GTPγS-bound forms (10). The data (Fig. 3B) demonstrate that, like Dbl, Plekhg4 associated with Cdc42, RhoA, and Rac1 with similar affinity. Moreover, binding was highly selective for the nucleotide-depleted form of the GTPase (i.e. affinity for the nucleotide-depleted GTPase was >100-fold tighter compared with the GTPγS-bound form), suggesting that like other GEFs, Plekhg4 stimulates substrate GTPases by catalyzing dissociation of the bound GDP (32). Taken together, our observations show that Plekhg4 is a functional guanine nucleotide exchange factor for Cdc42, Rac1, and RhoA.
FIGURE 3.
Plekhg4 is a functional GEF. A, GTPase activation by Plekhg4. COS7 cells were transiently co-transfected with Rac1, RhoA, or Cdc42 (HA-tagged in the pKH3 vector) and the indicated GEF in the pcDNA3.1 construct. After serum deprivation for 24 h, cell lysates were subjected to the PBD assay as described under “Results.” Top panel, activated (GTP-bound) GTPase precipitated with GST-PBD, visualized after anti-HA immunoblotting. Lower panels, expression levels of the indicated proteins in cell lysates. Bait GST-PBD was visualized by Ponceau Red staining of the blotted membranes, and -fold activation was determined after computer densitometry after normalization to GEF expression. Data shown are representative of three independent experiments in which activation varied by ±15%. B, Plekhg4-GTPase association. GST fusion proteins of Rac1, Cdc42, and RhoA were overexpressed in E. coli, purified on glutathione affinity chromatography, and their nucleotide content manipulated as described under “Results.” Immobilized GTPase were incubated with lysates from HA-Plekhg4- or Dbl-expressing COS7 cells and washed, and associated GEFs were visualized by immunoblotting. GST-GTPases baits were visualized by Ponceau Red staining of the blotted membranes. Shown is a representative of three independent experiments.
Plekhg4 Regulates the Actin Cytoskeleton
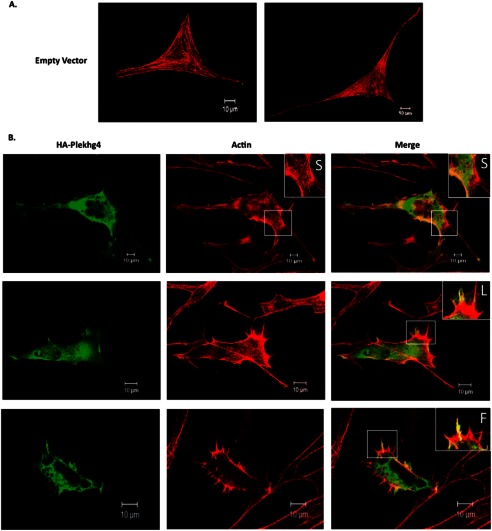
Activation of small GTPases from the Rho family stimulates specific rearrangements of the actin cytoskeleton. Specifically, GTP loading by Cdc42 enhances filopodia formation, whereas activation of Rac1 and RhoA induces formation of lamellipodia and stress fibers, respectively (33–35). Ectopic expression of GEFs in cultured fibroblasts elicits the cytoskeletal architecture pattern typical of their substrate GTPase (24), thereby providing a convenient read-out for their in vivo signaling state. We therefore examined the effect of Plekhg4 expression on the actin cytoskeleton in cultured NIH3T3 fibroblasts. Fig. 4A shows typical fluorescence micrographs of cells expressing Plekhg4 (or control vector). The data indicate that expression of Plekhg4 led to pronounced reorganization of the actin cytoskeleton. Specifically, we observed enhanced formation of lamellipodia, fillopodia, and the beginnings of disorganized stress fiber formation (see insets in Fig. 4). The subcellular expression pattern of Plekhg4 paralleled that of the actin cytoskeleton, with pronounced co-localization in areas of actin rearrangement (Fig. 4). These results affirm our observation above that Plekhg4 acts as a functional GEF for Cdc42, Rac, and Rho GTPases, and further demonstrate that at least one signaling axis of each of these GTPases, regulating cell shape and motility, is activated by Plekhg4.
FIGURE 4.
Plekhg4 regulates the actin cytoskeleton. Plekhg4 regulates the actin cytoskeleton. Cells expressing Plekhg4 (or control vector) were immunostained with HA-antibody (Plekhg4 expression) or Texas Red-conjugated phalloidin and visualized by fluorescence microscopy. A, NIH3T3 fibroblasts transiently transfected with empty vector control. B, NIH3T3 fibroblasts transiently transfected with HA-tagged Plekhg4. Insets show cytoskeletal features exhibited by Plekhg4 expression. Arrows indicate stress fibers (S), lamellipodia (L), or fillopodia (F).
Plekhg4 Associates with the Chaperones Hsc70 and Hsp90 and the E3 Ubiquitin Ligase CHIP
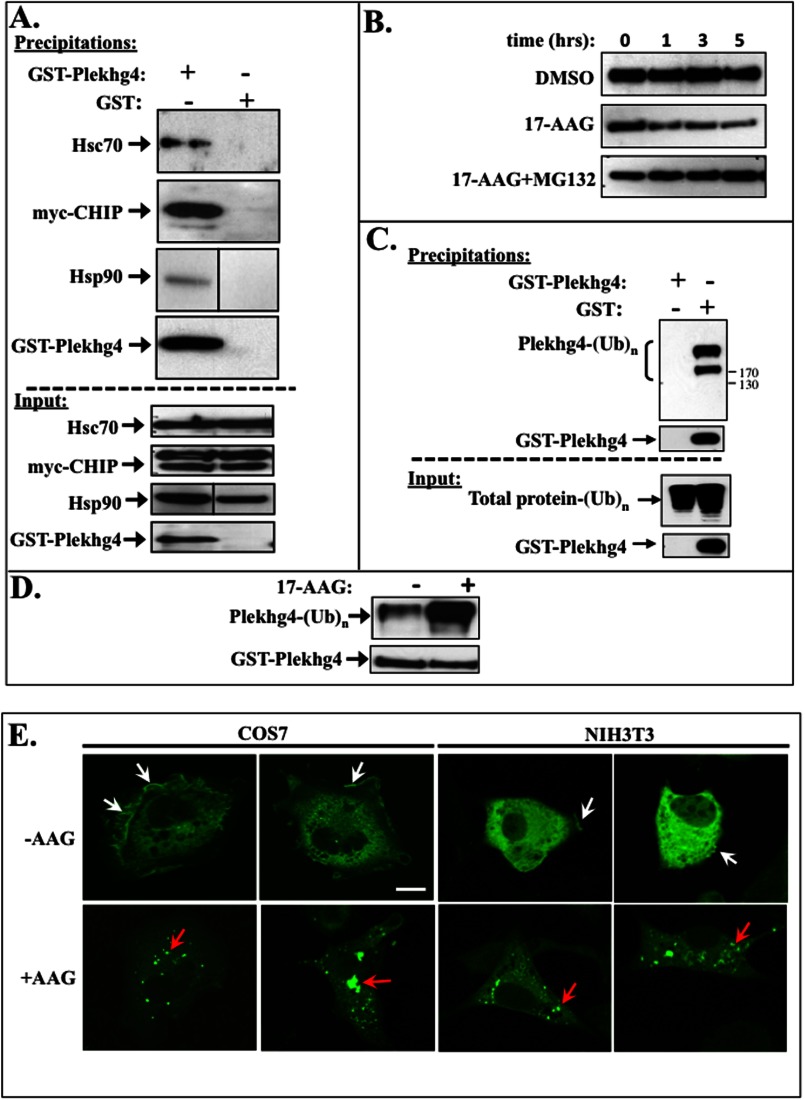
We have previously demonstrated the existence of a novel multiprotein complex that regulates Dbl signaling at the level of protein degradation. Specifically, we have shown that the spectrin domain in the Dbl N terminus mediates interaction between the GEF, the chaperones Hsc70 and Hsp90, and the E3 ubiquitin ligase CHIP (12, 13). This multiprotein complex mediates continuous ubiquitination of Dbl and its subsequent proteasomal degradation, such that steady-state expression levels of the protein are maintained at very low levels (12, 13). The similar domain architecture of Plekhg4 and Dbl (Fig. 2), and especially the sequence homology between the two proteins' spectrin homology domains (28% identical, 46% similar), raise the possibility that Plekhg4 turnover is also subject to regulation by unbiquitination and proteasomal degradation. To examine this possibility, we first examined whether Plekhg4 is associated with the molecular “coordinators” of protein folding and degradation, the chaperones Hsc70 and Hsp90, and the ubiquitin ligase CHIP (36). We found that in growing cells Hsc70, CHIP, and Hsp90 co-precipitated with GST-Plekhg4. These interactions were specific, as the complex did not form in lysates expressing the control GST vector (Fig. 5A). These findings indicate the presence of a constitutive complex composed of the GEF, the molecular chaperones Hsc70 and Hsp90, and the ubiquitin ligase CHIP.
FIGURE 5.
Intracellular fate of Plekhg4 is dictated by Hsc70, Hsp90, and CHIP. A, Plekhg4 associates with Hsc70, Hsp90, and CHIP. GST-tagged Plekhg4 (or control vector) cDNA constructs in pEBG vector were co-transfected into COS7 cells together with cDNA encoding myc-tagged CHIP in pcDNA3.1 vector. GST-tagged proteins were then affinity-precipitated with glutathione-Sepharose, resolved on SDS-PAGE, and probed for interacting endogenous Hsc70, Hsp90, and ectopic myc-CHIP by immunoblotting with the corresponding antibodies. B, COS7 cells were transiently transfected with cDNAs encoding GST-tagged Plekhg4 in pEBG vector. Dimethyl sulfoxide (DMSO) vehicle control (top panel), 3 μm 17-AAG (middle panel), or 3 μm 17-AAG supplemented with 25 mm proteasomal inhibitor MG132 (bottom panel) were added to cells for the indicated time prior to lysis. Cleared cell lysates were resolved on SDS-PAGE and probed for Plekhg4 with anti-GST antibody. C, COS7 were transfected with cDNAs encoding GST-tagged Plekhg4 or GST alone, and glutathione affinity precipitations were performed. The precipitated proteins were resolved on SDS-PAGE and probed for polyubiquitination by immunoblotting with the anti-ubiquitin antibodies. D, COS7 cells transfected with cDNAs encoding GST-tagged Plekhg4 or GST alone were treated with and without 3 μm 17-AAG. Glutathione affinity precipitations were performed, and precipitated proteins were resolved on SDS-PAGE and probed for polyubiquitination by immunoblotting with the anti-ubiquitin antibodies. E, HA-tagged Plekhg4 construct was transiently transfected into both COS7 and NIH3T3 cell lines. Cells were treated for 2 h with 3 mm 17-AAG 46 h after transfection, fixed, stained with anti-HA-FITC conjugates, and visualized by immunofluorescence confocal microscopy.
Chaperone Complex Dictates the Intracellular Fate of Plekhg4
A delicate interplay between the chaperones Hsp90 and Hsc70 directs the fate of client proteins toward stabilization and refolding, or ubiquitination and proteasomal degradation, respectively (36). To evaluate the Hsp90 role in determining the fate of Plekhg4, we examined how the selective Hsp90 inhibitor 17-AAG (37) affects Plekhg4 stability. As shown in Fig. 5B, when protein synthesis was inhibited by cycloheximide, expression levels of Plekhg4 were stable over a 5-h period (Fig. 5B, top panel). Treatment with 17-AAG caused a dramatic decline in Plekhg4 levels over time (Fig. 5B, middle panel), indicating that the protein is rapidly degraded. Co-treatment with the proteasomal inhibitor MG132 stabilized Plekhg4 levels, thereby reversing the effect of 17-AAG (Fig. 5B, bottom panel). These data indicate that the chaperone Hsp90 protects Plekhg4 from proteasomal degradation. In light of the established involvement of these proteins in protein ubiquitination and degradation (38), we examined the ubiquitination status of Plekhg4. When GST-Plekhg4 was precipitated from continuously growing cells, the protein was heavily modified by the endogenous ubiquitination system (Fig. 5C). Similar results were obtained when we employed ectopically expressed ubiquitin (data not shown). Moreover, the Plekhg4 ubiquitination level was markedly enhanced upon treatment with 17-AAG (Fig. 5D). Finally, treatment with 17-AAG caused a dramatic change in the intracellular localization of Plekhg4: in the absence of any treatment, the protein displayed a diffuse cytosolic expression with some localization to plasma membrane structures reminiscent of lamellopodia and actin microspikes (Fig. 5E, upper panels). In the presence of 17-AAG, Plekhg4 localization changed to a distinct pattern characterized by dense irregular punctae (Fig. 5E, red arrows). In some cells, the Plekhg4 punctae were larger, resided near the nucleus (Fig. 5E, yellow arrows) and displayed the characteristic appearance of aggresomes (39, 40). Further immunofluorescence studies in 17-AAG-treated cells revealed pronounced overlap between the intracellular localization of Plekhg4 and that of ubiquitin (data not shown). Taken together, our data indicate that Plekhg4 is a bona fide client protein of Hsc70, Hsp90, and CHIP. The data further show that Plekhg4 is subject to continuous ubiquitination and proteasomal degradation by these mediators of the “triage decision” (41).
DISCUSSION
Cerebellar neurons primarily control and coordinate motor movements (42) although recent data indicate their involvement in some cognitive functions (43). Accordingly, cerebellar pathologies manifest primarily as lack of motor coordination, generically referred to as spinocerebellar ataxias (44). The vast majority of ataxias are familial, stemming from heritable alterations in distinct unrelated genes that affect diverse neurobiological processes such as growth factor signaling, synaptic transmission, lipid trafficking, and ion channels function. On the cellular level, SCAs manifest as anatomic and functional compromise in Purkinje cells, responsible for integrating cerebellar information and coordinating neural output from this region. As in many other neurodegenerative disorders, many SCAs are characterized by the formation of cytoplasmic protein aggregates, especially in Purkinje cells (45). It is generally assumed that such aggregates result from misfolding of aggregation-prone proteins, as seen in mutated versions of ataxin, β-spectrin, atrophin, or PKCγ (46).
Plekhg4 was identified by molecular genetics approaches in the study of 16q22.1-linked autosomal dominant cerebellar ataxia. The protein encoded by the PLEKHG4 gene (sometimes termed puratrophin-1) exhibits a diffuse cytosolic expression pattern in the cell body of cerebellar Purkinje cells, whereas its mutated version resides in dense multiprotein aggregates observed in atrophying Purkinje neurons (17, 47). Data presented here show that Plekhg4 is a functional activator of small GTP-binding proteins from the Rho family, i.e. Cdc42, Rac1, and RhoA. Additionally, our data show that Plekhg4 is an aggregation-prone protein, whose steady-state levels are determined by the so-called triage decision that determines whether the protein will be stabilized and refolded, or ubquitinated and degraded (36). As in other instances, the triage decision is determined by a delicate interplay between chaperones that assist in refolding (specifically Hsp90 (48)), and those that facilitate ubiquitination and subsequently, proteasomal degradation (namely Hsc70/CHIP ((49)).
The mechanisms by which insoluble aggregates of Plekhg4 cause cerebellar dysfunction are completely unknown at present. Two alternative hypotheses can be invoked to explain the cellular pathophysiology of Plekhg4 mutations. (i) Plekhg4 aggregation can lead to reduction in GEF activity in the cytosol or an increase in this activity within the aggregate. Such changes will alter signaling to the substrate Rho family GTPases, which play key roles in controlling neuronal architecture and gene expression. Rho GTPases regulate neuronal differentiation, are involved in controlling neuronal migration during development, and they coordinate neurite outgrowth/collapse that define neuronal plasticity in the adult (see Ref. 50). Moreover, RhoGEFs regulate proper signaling by key neuronal hormones. For example, induction of neurite outgrowth by NGF is mediated by the RacGEF TIAM1 (51) or the Rac/RhoGEF TRIO (52), the Rac-GEF P-Rex is essential for BDNF signaling (53), and activation of Rac and Cdc42 by Vav2 is required for neuron responses to the hippocampus cluster of differentiation 47 (CD47 (54)). Dysregulation of Rho GTPase signaling brought about by their aggregation and is thus expected to markedly affect neuronal function. (ii) Alternatively, protein aggregates can interfere with neurobiological processes directly through toxic misfolded intermediates or through indirect disruption of neurobiological processes (55). Protein aggregates can thus lead to defects in neuronal processes such as axonal transport (56), mitochondrial function (57), and oxidative (58) or endoplasmic reticulum (59) homeostasis. Elucidation of the molecular basis underlying Plekhg4-induced ataxia awaits further experimental work.
This work was supported, in whole or in part, by National Institutes of Health Grant CA82391. This work was also supported by American Cancer Society Award RPG-99-269-01 and Case Comprehensive Cancer Center Pilot Grant 5P30-CA043703 (to D. M.).
- GEF
- guanine nucleotide exchange factor
- 17-AAG
- 17-N-allylamino-17-demethoxygeldanamycin
- DH
- Dbl homology
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- PBD
- p21 binding domain
- PH
- pleckstrin homology
- SCA
- spinocerebellar ataxia.
REFERENCES
- 1. Hall A. (2005) Rho GTPases and the control of cell behaviour. Biochem. Soc. Trans. 33, 891–895 [DOI] [PubMed] [Google Scholar]
- 2. Lazer G., Katzav S. (2011) Guanine nucleotide exchange factors for RhoGTPases: good therapeutic targets for cancer therapy? Cell. Signal. 23, 969–979 [DOI] [PubMed] [Google Scholar]
- 3. Hynes N. E., Gattelli A. (2011) P-Rex1, a guanine exchange factor that is overexpressed in breast cancer, is a convergence node for ErbB and CXCR4 signaling. Mol. Cell 41, 5–7 [DOI] [PubMed] [Google Scholar]
- 4. Eva A., Vecchio G., Rao C. D., Tronick S. R., Aaronson S. A. (1988) The predicted DBL oncogene product defines a distinct class of transforming proteins. Proc. Natl. Acad. Sci. U.S.A. 85, 2061–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Orrico A., Galli L., Falciani M., Bracci M., Cavaliere M. L., Rinaldi M. M., Musacchio A., Sorrentino V. (2000) A mutation in the pleckstrin homology (PH) domain of the FGD1 gene in an Italian family with faciogenital dysplasia (Aarskog-Scott syndrome). FEBS Lett. 478, 216–220 [DOI] [PubMed] [Google Scholar]
- 6. Rossman K. L., Der C. J., Sondek J. (2005) GEF means go: turning on Rho GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6, 167–180 [DOI] [PubMed] [Google Scholar]
- 7. Cerione R. A. (2004) Cdc42: new roads to travel. Trends Cell Biol. 14, 127–132 [DOI] [PubMed] [Google Scholar]
- 8. Srivastava S. K., Wheelock R. H., Aaronson S. A., Eva A. (1986) Identification of the protein encoded by the human diffuse B cell lymphoma (dbl) oncogene. Proc. Natl. Acad. Sci. U.S.A. 83, 8868–8872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Whitehead I. P., Campbell S., Rossman K. L., Der C. J. (1997) Dbl family proteins. Biochim. Biophys. Acta 1332, F1–23 [DOI] [PubMed] [Google Scholar]
- 10. Hart M. J., Eva A., Zangrilli D., Aaronson S. A., Evans T., Cerione R. A., Zheng Y. (1994) Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the dbl oncogene product. J. Biol. Chem. 269, 62–65 [PubMed] [Google Scholar]
- 11. Zheng Y., Zangrilli D., Cerione R. A., Eva A. (1996) The pleckstrin homology domain mediates transformation by oncogenic dbl through specific intracellular targeting. J. Biol. Chem. 271, 19017–19020 [DOI] [PubMed] [Google Scholar]
- 12. Kauppinen K. P., Duan F., Wels J. I., Manor D. (2005) Regulation of the dbl proto-oncogene by heat shock cognate protein 70 (Hsc70). J. Biol. Chem. 280, 21638–21644 [DOI] [PubMed] [Google Scholar]
- 13. Kamynina E., Kauppinen K., Duan F., Muakkassa N., Manor D. (2007) Regulation of proto-oncogenic dbl by chaperone-controlled, ubiquitin-mediated degradation. Mol. Cell. Biol. 27, 1809–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bi F., Debreceni B., Zhu K., Salani B., Eva A., Zheng Y. (2001) Autoinhibition mechanism of proto-dbl. Mol. Cell. Biol. 21, 1463–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hall A., Lalli G. (2010) Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb. Perspect. Biol. 2, a001818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marmolino D., Manto M. (2010) Past, present and future therapeutics for cerebellar ataxias. Curr. Neuropharmacol. 8, 41–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ishikawa K., Toru S., Tsunemi T., Li M., Kobayashi K., Yokota T., Amino T., Owada K., Fujigasaki H., Sakamoto M., Tomimitsu H., Takashima M., Kumagai J., Noguchi Y., Kawashima Y., Ohkoshi N., Ishida G., Gomyoda M., Yoshida M., Hashizume Y., Saito Y., Murayama S., Yamanouchi H., Mizutani T., Kondo I., Toda T., Mizusawa H. (2005) An autosomal dominant cerebellar ataxia linked to chromosome 16q22.1 is associated with a single-nucleotide substitution in the 5′-untranslated region of the gene encoding a protein with spectrin repeat and Rho guanine-nucleotide exchange-factor domains. Am. J. Hum. Genet. 77, 280–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ohata T., Yoshida K., Sakai H., Hamanoue H., Mizuguchi T., Shimizu Y., Okano T., Takada F., Ishikawa K., Mizusawa H., Yoshiura K., Fukushima Y., Ikeda S., Matsumoto N. (2006) A −16C>T substitution in the 5′-UTR of the puratrophin-1 gene is prevalent in autosomal dominant cerebellar ataxia in Nagano. J. Hum. Genet. 51, 461–466 [DOI] [PubMed] [Google Scholar]
- 19. Ouyang Y., Sakoe K., Shimazaki H., Namekawa M., Ogawa T., Ando Y., Kawakami T., Kaneko J., Hasegawa Y., Yoshizawa K., Amino T., Ishikawa K., Mizusawa H., Nakano I., Takiyama Y. (2006) 16q-linked autosomal dominant cerebellar ataxia: a clinical and genetic study. J. Neurol. Sci. 247, 180–186 [DOI] [PubMed] [Google Scholar]
- 20. Onodera Y., Aoki M., Mizuno H., Warita H., Shiga Y., Itoyama Y. (2006) Clinical features of chromosome 16q22.1-linked autosomal dominant cerebellar ataxia in Japanese. Neurology 67, 1300–1302 [DOI] [PubMed] [Google Scholar]
- 21. Nozaki H., Ikeuchi T., Kawakami A., Kimura A., Koide R., Tsuchiya M., Nakmura Y., Mutoh T., Yamamoto H., Nakao N., Sahashi K., Nishizawa M., Onodera O. (2007) Clinical and genetic characterizations of 16q-linked autosomal dominant spinocerebellar ataxia (AD-SCA) and frequency analysis of AD-SCA in the Japanese population. Mov. Disord. 22, 857–862 [DOI] [PubMed] [Google Scholar]
- 22. Amino T., Ishikawa K., Toru S., Ishiguro T., Sato N., Tsunemi T., Murata M., Kobayashi K., Inazawa J., Toda T., Mizusawa H. (2007) Redefining the disease locus of 16q22.1-linked autosomal dominant cerebellar ataxia. J. Hum. Genet. 52, 643–649 [DOI] [PubMed] [Google Scholar]
- 23. Sakai H., Yoshida K., Shimizu Y., Morita H., Ikeda S., Matsumoto N. (2010) Analysis of an insertion mutation in a cohort of 94 patients with spinocerebellar ataxia type 31 from Nagano, Japan. Neurogenetics 11, 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin R., Cerione R. A., Manor D. (1999) Specific contributions of the small GTPases Rho, Rac, and Cdc42 to Dbl transformation. J. Biol. Chem. 274, 23633–23641 [DOI] [PubMed] [Google Scholar]
- 25. Lin R., Bagrodia S., Cerione R., Manor D. (1997) A novel Cdc42Hs mutant induces cellular transformation. Curr. Biol. 7, 794–797 [DOI] [PubMed] [Google Scholar]
- 26. Wu W. J., Leonard D. A., A-Cerione R., Manor D. (1997) Interaction between Cdc42Hs and RhoGDI is mediated through the Rho insert region. J. Biol. Chem. 272, 26153–26158 [DOI] [PubMed] [Google Scholar]
- 27. Bagrodia S., Dérijard B., Davis R. J., Cerione R. A. (1995) Cdc42 and PAK-mediated signaling leads to Jun kinase and p38 mitogen-activated protein kinase activation. J. Biol. Chem. 270, 27995–27998 [DOI] [PubMed] [Google Scholar]
- 28. Ren X. D., Schwartz M. A. (2000) Determination of GTP loading on Rho. Methods Enzymol. 325, 264–272 [DOI] [PubMed] [Google Scholar]
- 29. Aghazadeh B., Zhu K., Kubiseski T. J., Liu G. A., Pawson T., Zheng Y., Rosen M. K. (1998) Structure and mutagenesis of the Dbl homology domain. Nat. Struct. Biol. 5, 1098–1107 [DOI] [PubMed] [Google Scholar]
- 30. Liu X., Wang H., Eberstadt M., Schnuchel A., Olejniczak E. T., Meadows R. P., Schkeryantz J. M., Janowick D. A., Harlan J. E., Harris E. A., Staunton D. E., Fesik S. W. (1998) NMR structure and mutagenesis of the N-terminal Dbl homology domain of the nucleotide exchange factor Trio. Cell 95, 269–277 [DOI] [PubMed] [Google Scholar]
- 31. Taylor S. J., Shalloway D. (1996) Cell cycle-dependent activation of Ras. Curr. Biol. 6, 1621–1627 [DOI] [PubMed] [Google Scholar]
- 32. Bourne H. R., Sanders D. A., McCormick F. (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature 349, 117–127 [DOI] [PubMed] [Google Scholar]
- 33. Hall A. (1992) Ras-related GTPases and the cytoskeleton. Mol. Biol. Cell 3, 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ridley A. J., Hall A. (1992) Distinct patterns of actin organization regulated by the small GTP-binding proteins Rac and Rho. Cold Spring Harb. Symp. Quant. Biol. 57, 661–671 [DOI] [PubMed] [Google Scholar]
- 35. Nobes C. D., Hall A. (1995) Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81, 53–62 [DOI] [PubMed] [Google Scholar]
- 36. Connell P., Ballinger C. A., Jiang J., Wu Y., Thompson L. J., Höhfeld J., Patterson C. (2001) The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3, 93–96 [DOI] [PubMed] [Google Scholar]
- 37. Zhang H., Burrows F. (2004) Targeting multiple signal transduction pathways through inhibition of Hsp90. J. Mol. Med. 82, 488–499 [DOI] [PubMed] [Google Scholar]
- 38. Cyr D. M., Höhfeld J., Patterson C. (2002) Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem. Sci. 27, 368–375 [DOI] [PubMed] [Google Scholar]
- 39. Kopito R. R. (2000) Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524–530 [DOI] [PubMed] [Google Scholar]
- 40. Kopito R. R., Sitia R. (2000) Aggresomes and Russell bodies: symptoms of cellular indigestion? EMBO Rep. 1, 225–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McDonough H., Patterson C. (2003) CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones 8, 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bastian A. J. (2011) Moving, sensing and learning with cerebellar damage. Curr. Opin. Neurobiol. 21, 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Glickstein M. (2006) Thinking about the cerebellum. Brain 129, 288–290 [DOI] [PubMed] [Google Scholar]
- 44. Manto M., Marmolino D. (2009) Cerebellar ataxias. Curr. Opin. Neurol. 22, 419–429 [DOI] [PubMed] [Google Scholar]
- 45. Koeppen A. H. (2005) The pathogenesis of spinocerebellar ataxia. Cerebellum 4, 62–73 [DOI] [PubMed] [Google Scholar]
- 46. Skinner P. J., Vierra-Green C. A., Clark H. B., Zoghbi H. Y., Orr H. T. (2001) Altered trafficking of membrane proteins in Purkinje cells of SCA1 transgenic mice. Am. J. Pathol. 159, 905–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ishikawa K., Mizusawa H. (2006) On autosomal dominant cerebellar ataxia (ADCA) other than polyglutamine diseases, with special reference to chromosome 16q22.1-linked ADCA. Neuropathology 26, 352–360 [DOI] [PubMed] [Google Scholar]
- 48. Young J. C., Agashe V. R., Siegers K., Hartl F. U. (2004) Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5, 781–791 [DOI] [PubMed] [Google Scholar]
- 49. Hatakeyama S., Matsumoto M., Kamura T., Murayama M., Chui D. H., Planel E., Takahashi R., Nakayama K. I., Takashima A. (2004) U-box protein carboxyl terminus of Hsc70-interacting protein (CHIP) mediates poly-ubiquitylation preferentially on four-repeat Tau and is involved in neurodegeneration of tauopathy. J. Neurochem. 91, 299–307 [DOI] [PubMed] [Google Scholar]
- 50. Govek E. E., Hatten M. E., Van Aelst L. (2011) The role of Rho GTPase proteins in CNS neuronal migration. Dev. Neurobiol. 71, 528–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shirazi Fard S., Kele J., Vilar M., Paratcha G., Ledda F. (2010) Tiam1 as a signaling mediator of nerve growth factor-dependent neurite outgrowth. PLoS One 5, e9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Estrach S., Schmidt S., Diriong S., Penna A., Blangy A., Fort P., Debant A. (2002) The human Rho-GEF trio and its target GTPase RhoG are involved in the NGF pathway, leading to neurite outgrowth. Curr. Biol. 12, 307–312 [DOI] [PubMed] [Google Scholar]
- 53. Yoshizawa M., Kawauchi T., Sone M., Nishimura Y. V., Terao M., Chihama K., Nabeshima Y., Hoshino M. (2005) Involvement of a Rac activator, P-Rex1, in neurotrophin-derived signaling and neuronal migration. J. Neurosci. 25, 4406–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Murata T., Ohnishi H., Okazawa H., Murata Y., Kusakari S., Hayashi Y., Miyashita M., Itoh H., Oldenborg P. A., Furuya N., Matozaki T. (2006) CD47 promotes neuronal development through Src- and FRG/Vav2-mediated activation of Rac and Cdc42. J. Neurosci. 26, 12397–12407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ross C. A., Poirier M. A. (2004) Protein aggregation and neurodegenerative disease. Nat. Med. 10, S10–17 [DOI] [PubMed] [Google Scholar]
- 56. Roy S., Zhang B., Lee V. M., Trojanowski J. Q. (2005) Axonal transport defects: a common theme in neurodegenerative diseases. Acta Neuropathol. 109, 5–13 [DOI] [PubMed] [Google Scholar]
- 57. Shi P., Gal J., Kwinter D. M., Liu X., Zhu H. (2010) Mitochondrial dysfunction in amyotrophic lateral sclerosis. Biochim. Biophys. Acta 1802, 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ischiropoulos H., Beckman J. S. (2003) Oxidative stress and nitration in neurodegeneration: cause, effect, or association? J. Clin. Invest. 111, 163–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Salminen A., Kauppinen A., Suuronen T., Kaarniranta K., Ojala J. (2009) ER stress in Alzheimer's disease: a novel neuronal trigger for inflammation and Alzheimer's pathology. J. Neuroinflammation 6, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]