Background: Fungal mannanases contribute to enzymatic degradation of lignocellulose.
Results: New fungal mannanases reveal striking differences in substrate specificities. A rigid linker tightly connects the family 26 glycoside hydrolase to its binding module.
Conclusion: Podospora anserina mannanases display differences in substrate binding modes, transglycosylation activity, and modular organization.
Significance: Information on the structure-function relationships of fungal mannanases is essential to improve the comprehension of biomass deconstruction.
Keywords: Carbohydrate, Enzyme Structure, Fungi, Glycoside Hydrolases, Plant Cell Wall, Polysaccharide, CAZymes, Mannan
Abstract
The microbial deconstruction of the plant cell wall is a key biological process that is of increasing importance with the development of a sustainable biofuel industry. The glycoside hydrolase families GH5 (PaMan5A) and GH26 (PaMan26A) endo-β-1,4-mannanases from the coprophilic ascomycete Podospora anserina contribute to the enzymatic degradation of lignocellulosic biomass. In this study, P. anserina mannanases were further subjected to detailed comparative analysis of their substrate specificities, active site organization, and transglycosylation capacity. Although PaMan5A displays a classical mode of action, PaMan26A revealed an atypical hydrolysis pattern with the release of mannotetraose and mannose from mannopentaose resulting from a predominant binding mode involving the −4 subsite. The crystal structures of PaMan5A and PaMan26A were solved at 1.4 and 2.85 Å resolution, respectively. Analysis of the PaMan26A structure supported strong interaction with substrate at the −4 subsite mediated by two aromatic residues Trp-244 and Trp-245. The PaMan26A structure appended to its family 35 carbohydrate binding module revealed a short and proline-rich rigid linker that anchored together the catalytic and the binding modules.
Introduction
Endo-β-1,4-mannanases (β-mannanases, E.C. 3.2.1.78) catalyze the random hydrolysis of manno-glycosidic bonds in mannans and heteromannans. These polysaccharides are the main components of hemicellulose in softwoods and are found in smaller amounts in angiosperms (1). Mannans comprise a backbone of β-1,4-linked d-mannose residues, known as mannan, or a heterogeneous combination of β-1,4-d-mannose and β-1,4-d-glucose units, termed glucomannan. Both can be decorated with α-1,6-linked galactose side chains, and these polysaccharides are referred to as galactomannan and galactoglucomannan, respectively.
Several types of glycoside hydrolases (GH)2 are required for complete degradation of mannans, and endo-β-1,4-mannanases are the key enzymes. In the CAZy database (2), β-1,4-mannanase activities are found in families GH5, GH26, and GH113. The three families belong to clan GH-A; they share the same (β/α)8-barrel protein fold, catalytic machinery, and retaining double displacement mechanism (3–5). Because of this retaining double displacement mechanism, some of these enzymes are able to perform transglycosylation in which a carbohydrate hydroxyl group can act as an acceptor molecule rather than water as is the case in hydrolysis. Transglycosylation thus leads to the synthesis of new glycosides or oligosaccharides longer than the original substrate. GH5 and GH113 mannanases have been described as able to catalyze transglycosylation reactions (6–9), whereas to date no evidence of transglycosylation has been reported for GH26 mannanases (10). β-Mannanases are frequently encountered as modular enzymes. Indeed, some harbor carbohydrate binding modules (CBMs) from families CBM1, CBM6, CBM10, CBM31, and CBM35 (11, 12). It is generally observed that the linker regions between catalytic module and CBM display a great deal of structural flexibility to maximize substrate accessibility, as has been confirmed by the few crystal structures of bacterial modular enzymes (13, 14).
GH5 endo-β-1,4-mannanases, which are found in bacteria, fungi, animals, and plants are the most largely characterized family. To date only three eukaryotic endo-mannanase three-dimensional structures from family GH5 are available: one from Trichoderma reesei (PDB code 1QNO; Ref. 15), one from the blue mussel Mytilus edulis (PDB code 2C0H; Ref. 7), and one from the tomato fruit Solanum lycopersicum (PDB code 1RH9; Ref. 16). Although several family GH26 endo-β-1,4-mannanases have also been characterized from different organisms (e.g. Cellulomonas fimi (17), Cellvibrio japonicus (18), Piromyces equi (19)), only sparse studies have focused on GH26 endo-β-1,4-mannanases of fungal origin, and the five three-dimensional structures available (B. subtilis, PDB code 2WHK (20); B. subtilis PDB code 2QHA, (21); C. fimi PDB code 2BVT (17); C. japonicus PDB code 1GVY (22); PDB code 2VX4 (23)) are all from bacteria and represent only catalytic domains (CDs).
The characterization of endo-β-1,4-mannanases biochemical properties and substrate specificities revealed that many release essentially mannobiose and mannotriose as end products (9, 17, 24, 25) and that their active site displays generally 5–6 subsites able to accommodate the substrate (10, 15). Although GH5 and GH26 mannanases share some characteristics, several studies revealed different modes of action. In particular, biochemical studies pointed to divergence in specificity between GH5 and GH26 bacterial mannanases, a suggesting different biological role (20, 26).
The coprophilic fungus Podospora anserina has one of the largest fungal sets of candidate enzymes for cellulose and hemicellulose degradation described to date and one of the highest numbers of CBMs of all the fungal genomes available (27). In a previous study comparative genomics were used that identified two mannanases from families GH5 (PaMan5A) and GH26 (PaMan26A) in the P. anserina genome (28). Investigation of the contribution made by each P. anserina mannanase to the saccharification of spruce demonstrated that they individually supplemented the secretome of the industrial T. reesei CL847 strain. The most striking effect was obtained with PaMan5A that improved the release of total sugars by 28% and of glucose by 18% (28). In the present study P. anserina GH5 and GH26 mannanases were subjected to detailed comparative analysis of their substrate specificities, active site organization, and transglycosylation capacity. The three-dimensional structures of PaMan5A and PaMan26A linked to a CBM35 module were solved in their native form at 1.4 and 2.85 Å resolution, respectively.
EXPERIMENTAL PROCEDURES
Production and Purification of PaMan5A and PaMan26A
PaMan5A and PaMan26A were produced in P. pastoris 2-liter cultures and purified as described previously in (28). Enzyme purification was completed by an additional size exclusion chromatographic step. After the nickel chelate purification step, the eluate containing PaMan5A or PaMan26A was concentrated using a Vivaspin with 10-kDa cut-off polyethersulfone membrane (Sartorius, Palaiseau, France) and dialyzed against the buffer used for the size exclusion chromatography (20 mm Hepes, pH 7.5, 150 mm NaCl). The concentrated fraction was subsequently loaded onto a Superdex S200 HiLoad 16/60 column (Amersham Biosciences). The fractions containing PaMan5A or PaMan26A were pooled and concentrated as described above.
Construction of Site-specific Variants
Site-directed mutagenesis was performed using the QuikChange kit (Stratagene), with primers listed in Table 1, according to the instructions of manufacturer. Using the wild-type PaMan5A and PaMan26A plasmids described in Couturier et al. 28), active-site variants were designed for each enzyme. Two single-site mutants were constructed for each enzyme: E177A and E283A for PaMan5A and E300A and E390A for PaMan26A. Transformation was performed in P. pastoris, and production and purification of enzyme variants were carried out as described above.
TABLE 1.
Mutagenesis primers used in the study
Modified codons are underlined.
| Primer | Nucleotide sequence 5′ to 3′ |
|---|---|
| E177Aforward | GGGAACTTGCCAACGCGCCCAGGTGCAAGGG |
| E177AReverse | CCCTTGCACCTGGGCGCGTTGGCAAGTTCCC |
| E283AForward | CCGTGTTTGTTGGAGGCGTATGGGTATGAGAGTGATAGG |
| E283AReverse | CCTATCACTCTCATACCCATACGCCTCCAACAAACACGG |
| E320AForward | GGAGACCTCTTCACGCCGCGGAGGGTGGTTGG |
| E320AReverse | CCAACCACCCTCCGCGGCGTGAAGAGGTCTCC |
| E410AForward | GATGATTGCTGCTGCAGCCGTTGGCGCTGCC |
| E410AReverse | GGCAGCGCCAACGGCTGCAGCAGCAATCATC |
Deglycosylation Assay
N-Glycosylation sites were predicted using the NetNGlyc 1.0 Server. To remove N-linked glycans, purified enzymes were treated with peptide N-glycosidase F New England Biolabs, Ipswich, MA) under denaturing conditions according to the manufacturer's instructions. Briefly, 10 μg of protein were incubated in 0.5% SDS and 40 mm DTT and boiled for 10 min for complete denaturation. Denaturated samples were subsequently incubated with 1500 units of peptide N-glycosidase F in appropriate buffer for 1 h at 37 °C. Deglycosylated and control samples were analyzed by SDS-PAGE (Bio-Rad).
Analysis of End Products Release from Polysaccharides
The activity of PaMan5A and PaMan26A was assayed toward glucomannan, galactomannan, and linear mannan. Briefly, a 1% w/v solution was prepared in 50 mm sodium acetate buffer, pH 5.2. The assay was performed by incubating 75 μg of enzyme with 90 μl of 1% w/v substrate solution or suspension at 40 °C for 30 min. After hydrolysis, mono- and oligo-saccharides were analyzed using high performance anion exchange chromatography (HPAEC) coupled with pulsed amperometric detection (PAD) (ICS 3000; Dionex, Sunnyvale, CA) equipped with a carbo-PacPA-1 analytical column (250 × 4 mm). 10-μl samples of enzymatic reactions were stopped by the addition of 90 μl of 100 mm NaOH before injection (5 μl) into the HPAEC system. Elution was carried out in 130 mm NaOH using a 25-min linear gradient program from 100% A (130 mm NaOH) to 60% A and 40% B (NaOAc, 500 mm; NaOH, 130 mm). All the assays were carried out in triplicate.
Hydrolysis Product Formation from Oligosaccharides and Determination of Kinetic Parameters
Products generated after hydrolysis of manno-oligosaccharides were analyzed using HPAEC-PAD as described above. 20 μl of suitably diluted enzyme were incubated at 40 °C for various time lengths with 180 μl of 100 μm substrate in 50 mm acetate buffer, pH 5.2. Calibration curves were plotted using β-1,4-manno-oligosaccharides as standards from which response factors were calculated (Chromeleon program, Dionex) and used to determine the amount of products released at different time points. All the assays were carried out in duplicate. The data were fitted to the equation of Matsui (29, 30), k = ln[S0]/[St], where k = (kcat/Km)[enzyme] × time, and [S0] and [St] represent substrate concentration before the start of the reaction and at a specified time during the reaction, respectively.
Hydrolysis of M5 and M6 in H218O
To determine and compare the hydrolytic cleavage patterns of M5 and M6 by PaMan5A and PaMan26A, HPAEC-PAD data on the hydrolysis products (as described above) was combined with the analysis of hydrolysis performed in H218O as described previously (31, 32). Each productive binding of M5 or M6 gives rise to two products (e.g. M6 cleaved to either two molecules of M3 or to M5 and M1 or to M4 and M2). Quantitative HPAEC-PAD analysis of one product per cleavage (M3, M4, and M5, respectively) was used to calculate the relative frequency of the productive binding modes of M5 and M6 that give rise to these products. Each of these products can further be produced by either of two binding modes, and to distinguish between these two modes, the ratio of non-labeled (16O) and labeled (18O) product (M3, M4, and M5) was used. Reactions were performed at 8 °C (low temperature was used to avoid spontaneous incorporation of 18O (31)) in H218O (93%, with a total of 7% H216O contamination (3% in the original H218O and 4% from the enzyme and substrate stock solutions) containing 1 mm sodium acetate buffer, pH 5, 0.8 mm substrate, and 0.1 μm enzyme. Samples (0.5 μl) were withdrawn at different time points (0–60 min) and spotted directly on a stainless steel plate for matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) analysis. Matrix (10 mg·ml−1 2,5-dihydroxybenzoic acid in H2O) was applied immediately to the sample, and it was dried under warm air. Samples from 40 and 60 min had sufficient product build-up, and the determined ratios were in good agreement (2–8% variation between the 40- and the 60-min samples from each incubation). The data from 40 min samples were used.
MALDI-TOF MS Data Acquisition and Analysis
MALDI-TOF MS spectra were recorded in positive reflector mode using a 4700 Proteomics Analyzer (Applied Biosystems, Framingham, MA). The laser intensity was set at 5500, and 50 subspectra with 20 shots on each were accumulated from each sample spot. The program Data Explorer version 4.5 was used for analysis of the data. The relative frequencies of the different productive binding modes resulting in the same products were calculated from the relative areas of the monoisotopic peaks of 16O- and 18O-labeled products as previously described (31). Two corrections were made; one for the [M + 2] natural isotope peak (5.3% of the monoisotopic peak for M3, 8% of the monoisotopic peak for M4, and 11% of the monoisotopic peak for M5) of the light (16O) species that overlaps with the heavy (18O) peak followed by another correction for the 7% H216O contamination in the hydrolysis assays. M1 was not detectable because of matrix suppression of low masses.
Transglycosylation Analysis
Reactions were set up to aim for detection of transglycosylation products with MALDI-TOF-MS, similarly to what was done in Rosengren et al. (32). Five mm M5 was incubated with 0.5 μm PaMan5A at 40 °C in 10 mm sodium acetate buffer pH 5 for 0–15 min. Samples (0.5 μl) were withdrawn at different time points and spotted directly onto a stainless steel MALDI plate. Matrix solution (10 mg·ml−1 2,5-dihydroxybenzoic acid in water) was applied (0.5 μl), and the samples were dried under warm air. Data acquisition and analysis was performed as described above.
Protein Crystallization, Data Collection, and Processing
All crystallization trials were carried out by the vapor diffusion method at 20 °C. PaMan5A was concentrated to 8 mg·ml−1 in 20 mm Hepes, pH 7.5, 150 mm NaCl buffer. Initial crystallization trials were performed using Wizard and MDL screens (Qiagen) on a cartesian robot. For each condition, three drops (100 nl of screen buffer + 100, 200, and 300 nl of protein) were formed. Optimization was then carried out by varying the pH and the concentration of precipitant. The final crystallization conditions were Tris 0.1 m pH 8.5, 0.2 m sodium acetate, 30% PEG 4000. Glycerol was used at a concentration of 25–30% as the cryoprotectant in the subsequent data collection stage. PaMan5A crystals belonged to the P212121 space group with the cell dimensions a = 56.9 Å, b = 58.0 Å, and c = 98.2 Å and diffracted to 1.4 Å resolution. X-ray diffraction data of a PaMan5A crystal were collected at 100K at the European Synchrotron Research Facilities (ESRF, Grenoble, France) beam line ID29.
PaMan26A was concentrated to ∼26 mg·ml−1 in 20 mm Hepes, pH 7.5, 150 mm NaCl buffer. Small PaMan26A crystals were obtained in the conditions (i) 0.1 m Tris pH 7, 0.2 m Li2SO4, 1 m potassium sodium tartrate and (ii) 0.1 m imidazole, pH 8, 0.1 m potassium sodium tartrate, 0.2 m NaCl, both conditions of the Wizard screen. The best crystals were obtained after optimization in a solution containing 0.1 m Tris, pH 7, 0.2 m NaCl, 0.8 m potassium sodium tartrate, 1 mm HgCl2. For cryoprotection, crystals were transferred in a solution containing 25% (v/v) glycerol, 1.5 m Li2SO4, 100 mm Bistris propane, pH 7.4. The crystals belonged to the P6522 space group with the following cell dimensions: a = b = 97.5 Å, c = 268.7 Å. Several x-ray diffraction data sets were collected on beam line Proxima1 at the French synchrotron SOLEIL (Saint-Aubin, France) and on beam lines ID14-4 and ID29 at the European Synchrotron Research Facilities. The best x-ray diffraction data were collected to 2.85 Å resolution at the European Synchrotron Research Facilities beam line ID14-4.
All the data sets were processed with the programs XDS (33) and SCALA (34). The data collection statistics are summarized in Table 2.
TABLE 2.
Data collection and model refinement statistics of PaMan5A and PaMan26A
| PaMan5A | PaMan26A | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.97914 | 1.00648 |
| Space group | P212121 | P6522 |
| a, b, c (Å) | 56.87, 57.90, 97.86 | 97.49, 97.49, 268.72 |
| Resolution (Å)a | 30-1.4 (1.48-1.4) | 50-2.85 (3.0-2.85) |
| Unique reflectionsa | 62,519 (8,848) | 18,575 (2,627) |
| Multiplicitya | 8.4 (8.3) | 11.8 (11.8) |
| Completeness (%)a | 97.3 (95.7) | 99.9 (100.0) |
| I/σa | 18.7 (3.4) | 25.6 (4.6) |
| Rmerge (%)a,b | 8.6 (67.8) | 7.6 (65.2) |
| Refinement and model quality | ||
| Resolution (Å) | 30-1.4 | 30-2.85 |
| Reflections | 59,254 | 18,483 |
| Rcryst/Rfree (%)c | 15.0/17.2 | 20.6/25.7 |
| Number of atoms | 3,193 | 3,676 |
| Protein/carbohydrate moiety | 2,745/0 | 3,493/28 |
| Water/solvent/ion | 434/14/0 | 143/12/2 |
| Average B-factors (Å2) | ||
| Protein/carbohydrate moiety | 10.6 | 68.2/888 |
| Water/solvent/ion | 28.5/10.6 | 63.3/85.1/92.1 |
| r.m.s.d.d | ||
| Bond (Å) | 0.010 | 0.008 |
| Angle (°) | 1.02 | 1.04 |
| Ramachandran plot (%) | ||
| Most favored regions | 98.0 | 95.5 |
| Additionally allowed regions | 2.0 | 4.3 |
| Outlier regions | 0 | 0.2 |
| PDB accession code | 3ZIZ | 3ZM8 |
a Values in parentheses are for the highest resolution shell.
b Rmerge = Σhkl(Σi|Ihkl−〈Ihkl〉|)/Σhkl|〈Ihkl〉|.
c Rcryst = Σhkl‖Fo| − |Fc‖/Σhkl|Fo|; Rfree was calculated for 5% of randomly selected reflections excluded from refinement.
d Root mean square deviation from ideal values.
Structure Determination and Refinement
The structure of PaMan5A was determined with the molecular replacement method using the AMoRe program (35) and the T. reesei GH5 mannanase coordinates (PDB code 1QNO). The rotation function yielded one solution, and the translation function yielded a unique solution, with a correlation coefficient and an Rfactor of 38.1 and 44.5%, respectively, for data between 10 and 4 Å. After rigid body refinement, the correlation coefficient was 59.2% for an Rfactor of 35.9%. After refinement using the programs Refmac (36) and Buster (37), the final crystallographic Rfactor and Rfree were 15.0 and 17.2%.
The structure of PaMan26A was also determined with the molecular replacement method using the AMoRe program (35). The superposition of four structures of GH26 CDs (PDB codes 2QHA, 2BVT, 2VX4, and 2WHK) plus the homology model given by the Phyre server (38) has been used as an ensemble search model for molecular replacement. The rotation function yielded one solution, and the translation function yielded a unique solution, with a correlation coefficient and an Rfactor of 32.4 and 49.2%, respectively, for data between 10 and 4 Å. After rigid body refinement, the correlation coefficient was 42.7% for an Rfactor of 43.7%. A modified structure of the CBM35 from Clostridium thermocellum (PDB code 2W47) with most of the loops deleted was located manually in the difference Fourier electron density map and was used as a starting point to build the CBM domain of PaMan26A. After performing several cycles of refinement using Refmac (36) and Buster (37) programs and manual replacement and building on the graphic display with the Turbo-Frodo program (39), the Rfactor has decreased to 20.7% (Rfree 25.8%). All representations of the structure in the figures were prepared with the program PyMOL. Coordinates for the structure PaMan5A and PaMan26A have been deposited in the Protein Data Bank under the accession number 3ZIZ and 3ZM8, respectively.
RESULTS AND DISCUSSION
Hydrolytic Activity of PaMan5A and PaMan26A toward Polysaccharides
In a recent study we showed that PaMan5A and PaMan26A displayed similar kinetic parameters toward a range of mannan substrates. To further compare the P. anserina mannanases, we measured the release of manno-oligosaccharides after hydrolysis of ivory nut mannan and carob galactomannan. Toward the end of the reaction, PaMan5A yielded mainly M2 and M3 and smaller amount of M1 (data not shown), consistent with other GH5 mannanases such as T. reesei (40). PaMan26A produced mainly M4 and smaller amounts of M1, M2, and M3 (data not shown). In other GH26 mannanases, different profiles have been observed; C. fimi CfMan26A and C. japonicus CjMan26B released M2 and M1 (17, 26); and B. subtilis BCMan released M2 and M4 (21). The nature of oligosaccharide products released upon mannan hydrolysis confirms (i) the endo-mode of action of the two enzymes and (ii) differences between the two enzymes in substrate binding.
Hydrolytic Activity of PaMan5A and PaMan26A toward Oligosaccharides
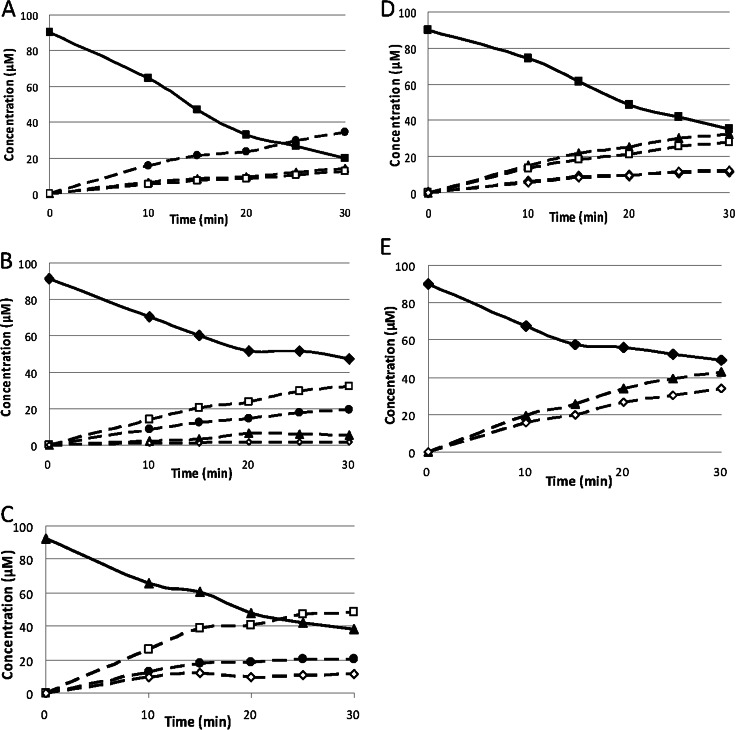
The capacity of PaMan5A and PaMan26A to hydrolyze a range of manno-oligosaccharides was evaluated by ionic chromatography to get further insights into their active site architecture (Fig. 1). PaMan5A had very low activity on M3, higher activity on M4, and cleaved M5 and M6 rapidly (Table 3). A decrease of kcat/Km was observed with decreasing degree of polymerization. The relative kcat/Km values of PaMan5A on M3, M4, M5, and M6 were 1:358:1127:1782. The increase of the degree of polymerization from 4 to 5 (M4 and M5) resulted in a 3.1-fold increase in kcat/Km, suggesting that at least four subsites are required to achieve efficient hydrolysis. In contrast, PaMan26A had no detectable activity on M3, very low activity on M4, and cleaved M5 and M6 rapidly. For PaMan26A the relative kcat/Km values on M4, M5, and M6 were 1:195:365 with an increase of kcat/Km of 1.9-fold between M5 and M6 hydrolysis. These data suggest that PaMan26A requires at least five subsites to achieve maximum manno-oligosaccharide hydrolysis efficiency.
FIGURE 1.
Progress curve of the manno-oligosaccharides generated by PaMan5A and PaMan26A after the hydrolysis of manno-oligosaccharides. The recombinant enzymes were incubated with 100 μm manno-oligosaccharides in acetate buffer, pH 5.2, at 40 °C. The quantity of mannose (open diamonds), M2 (open squares), M3 (full circles), M4 (full triangles), M5 (full diamonds), and M6 (full squares) produced during the course of the reaction was quantified using HPAEC-PAD. The concentrations of enzymes used were PaMan5A, 18.2 nm with M6 (A), 18.2 nm with M5 (B), and 60 nm with M4 (C), and PaMan26A (D), 15 nm with M6 and 30 nm with M5 (E).
TABLE 3.
Catalytic efficiencies of PaMan5A and PaMan26A on manno-oligosaccharides
| Substrate |
Kcat/Km |
|
|---|---|---|
| PaMan5A | PaMan26A | |
| m−1 min−1 | ||
| M6 | 2.9 × 106 | 2.4 × 106 |
| M5 | 1.9 × 106 | 1.3 × 106 |
| M4 | 5.9 × 105 | 6.4 × 103 |
| M3 | 1.6 × 103 | NDa |
a Not determined due to low activity.
The nature of the hydrolysis products yielded from manno-oligosaccharides (summarized in Table 4) also revealed striking differences between the P. anserina mannanases. M6 hydrolysis by PaMan5A produced mainly M3 with smaller amounts of M2 and M4, whereas M6 hydrolysis by PaMan26A produced mainly M4 and M2, with smaller amounts of M5 and M1 and without any M3. M5 hydrolysis by PaMan5A yielded mainly M2 and M3, with small amounts of M4 and M1, whereas hydrolysis by PaMan26A yielded only M4 and M1. M4 hydrolysis by PaMan5A yielded mainly M2 with small amounts of M1 and M3, whereas PaMan26A had low activity on M4 and produced M1, M2, and M3. PaMan5A poorly hydrolyzed M3, yielding M1 and M2, and PaMan26A had no detectable activity on M3. Neither mannanase had detectable activity on M2 and 4-nitrophenyl-mannose even at high enzyme loading (data not shown). Again, PaMan26A showed an atypical hydrolytic profile for a GH26 endo-mannanase compared with CfMan26A and CjMan26A. CjMan26A hydrolyzes M4 rapidly and requires occupation of four subsites to achieve efficient hydrolysis (18), whereas CfMan26A is less efficient toward M4 and requires substrate binding at five subsites to achieve efficient hydrolysis (17). For PaMan26A, the occupation by substrate of at least five subsites to achieve efficient hydrolytic activity is even more pronounced, with a dramatic increase in kcat/Km between M4 and M5.
TABLE 4.
Hydrolysis products released by PaMan5A and PaMan26A from manno-oligosaccharides
Products were quantified using HPAEC-PAD and are expressed as μm. Incubation was carried out for 30 min at 40 °C; ND, not detected.
| Enzyme | Enzyme loading | Substrate | Products |
||||
|---|---|---|---|---|---|---|---|
| M1 | M2 | M3 | M4 | M5 | |||
| nm | μm | ||||||
| PaMan5A | 60 | M4 | 11 | 49 | 21 | ||
| 18.2 | M5 | 2 | 32 | 20 (80a) | 5 (20a) | ||
| 18.2 | M6 | ND | 12 | 34 (55a) | 14 (45a) | ND | |
| PaMan26A | 30 | M5 | 34 | ND | ND | 43 (100a) | |
| 15 | M6 | 13 | 28 | ND | 33 (75a) | 11 (25a) | |
a The values in parentheses represent the relative molar distribution (%) between the products M3, M4, and M5 from each of the M5 and M6 incubations, which were used to estimate the relative frequency of productive binding modes yielding these products (presented in Fig. 2A). One product (M3, M4, or M5) per productive binding was used for calculations; thus, only half of the produced M3 (bold) from M6 incubations was accounted for (two molecules of M3 are produced from each molecule of M6).
Productive Binding Mode of M5 and M6 by PaMan5A and PaMan26A
β-Mannanases usually bind oligomeric substrates in multiple productive binding modes that can generate identical products. The simplest example of this is M3 hydrolysis to M2 and M1, where mannose would be released from either the reducing end or the non-reducing end. In the former case M3 binds productively from the −1 subsite to the +2 subsite and in the latter case from the −2 to the +1 subsite, following the established subsite nomenclature (41). As another example, from M5, each of the products M3 and M4, respectively, can be produced by either of two binding modes (see the scheme in Fig. 2A). Binding of M5 from subsite −2 to +3 or from subsite −3 to +2, both, generates M3, and binding from subsite −4 to +1 and from subsite −1 to +4, both, generates M4. Thus, product analysis using HPAEC-PAD data alone cannot distinguish between binding modes giving the same products. However, this can be achieved when the HPAEC-PAD product analysis (as in previous paragraph) is combined with in situ product isotope labeling using 18O-labeled water followed by mass spectrometric analysis as shown previously (31, 32).
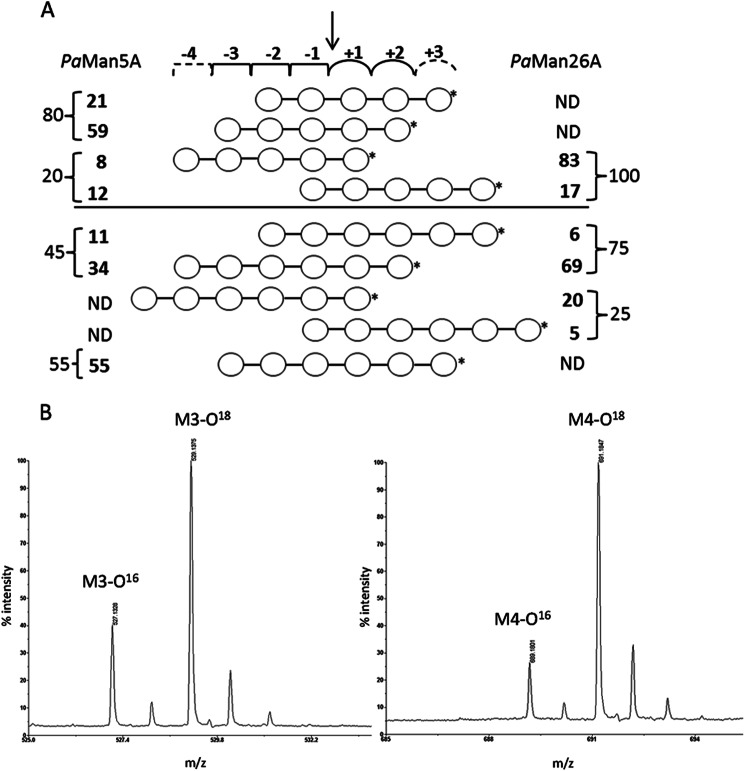
FIGURE 2.
Relative frequency of the productive modes of binding of manno-oligosaccharides to PaMan5A and PaMan26A. A, the numbers represent the percentages of binding in each binding mode. These were calculated from the quantitative product analysis using HPAEC-PAD (numbers to the far left and far right, obtained from Table 4) followed by a detailed analysis of the hydrolytic cleavage patterns of M5 and M6 using MALDI-TOF-MS analysis of 18O-labeled products, allowing to distinguish further between binding modes. The arrow indicates the mannosidic bond to be cleaved. *, reducing end of oligosaccharide. The −4 and +3 dashed subsites are only present in PaMan26A and PaMan5A, respectively. ND, not detected. B, MALDI-TOF-MS spectra of M5 hydrolysis by PaMan5A and PaMan26A show enlarged parts of the spectra with the M3 product formed by PaMan5A at a ratio of 1:2.9 of M3/M3O18 (left) and the M4 product formed by PaMan26A at a ratio of 1:5.0 of M4/M4O18 (right). The peaks in the spectra correspond to the monoisotopic masses of sodium adducts [M + Na]+ of the manno-oligosaccharides.
Relative quantities of the produced M3, M4, and M5 from the HPAEC-PAD data of M5 and M6 hydrolysis (Table 4) were used to calculate the relative molar distribution of these products (Table 4, values in parentheses), and thus the frequencies of productive binding modes that give rise to these products could be estimated (Fig. 2A, far left and far right column). MALDI-TOF-MS analysis was conducted to determine the ratio of non-labeled (16O) and labeled (18O) species of each product (light versus heavy M3, M4, or M5), which was then used to estimate the relative frequency of the productive binding modes of M5 and M6 that give rise to these same products. The combined results of the HPAEC-PAD and MALDI-TOF-MS data are summarized in Fig. 2A, showing the relative frequencies (%) of productive binding modes of M5 and M6. The calculation procedure is explained in supplemental Table S1. To exemplify, determined from HPAEC-PAD data (Table 4), 80% of the productive binding during the hydrolysis of M5 by PaMan5A generated M3. MALDI-TOF analysis then determined the ratio between the two binding modes that give M3. The analysis gave a ratio of M3/M3O18 of 1:2.9 (Fig. 2B), which shows that the enzyme binds M5 preferably from subsite −3 to +2 to produce M3, giving a 59% frequency of this binding mode (Fig. 2A). Small amounts of M1 and M4 were also produced, and the ratio of M4/M4O18 was 1:0.7. Hydrolysis of M6 by PaMan5A produced mainly M3 (55% binding frequency) but also smaller amounts of M2 and M4. The ratio of M4/M4O18 was 1:3.2, which shows that PaMan5A binds M6 preferably from subsite −4 to +2 to produce M4 (34% binding frequency). Hydrolysis of M5 by PaMan26A yielded M1 and M4 with a M4/M4O18 ratio of 1:5.0 (Fig. 2B), which shows that the enzyme binds M5 preferentially from subsite −4 to +1 (83% binding frequency, see Fig. 2A). For hydrolysis of M6, major product ratio analysis showed that the ratio of M4/M4O18 was 1:10.8, which shows that PaMan26A prefers to bind M6 from subsite −4 to +2 (69% binding frequency). The ratio of the minor product M5/M5O18 was 1:4.2, showing that M1 and M5 are mainly produced without binding at the +2 subsite. Thus, these data reveal clear differences in the binding mode of the two P. anserina mannanases; the predominant modes of binding of M5 and M6 were significantly different (Fig. 2A). PaMan5A showed a classical pattern of hydrolysis products (M3 and M2 mainly were released from M5) as described in several studies (B. subtilis, C. japonicus, M. edulis), whereas PaMan26A showed release of M4 and M1 from M5, which is unusual when compared with other GH26 endo-mannanases (B. subtilis BCMan, C. fimi CfMan26A, C. japonicus CjMan26A), suggesting an unusual arrangement of subsites in the catalytic center.
Transglycosylation Ability
To detect potential transglycosylation ability of the two enzymes, they were incubated with M5 as substrate. The resulting short time course study of the product formation clearly showed that PaMan5A, in addition to hydrolysis products, also produces transglycosylation products with higher degree of polymerization than the original substrate (Fig. 3). PaMan5A was able to transglycosylate yielding to oligosaccharide structures of up to a degree of polymerization of 8 (n + 1 to n + 3), in good agreement with GH5 mannanases described before, T. reesei (n + 1 to n + 3) (31), and Aspergillus nidulans ManA (n + 1 to n + 3) and ManC (n + 1 and n + 2) (6). No transglycosylation products could be detected with PaMan26A incubated with M5 in the same experimental conditions, which is consistent with some other family GH26 mannanases that have been described as non transglycosylating enzymes (10).
FIGURE 3.
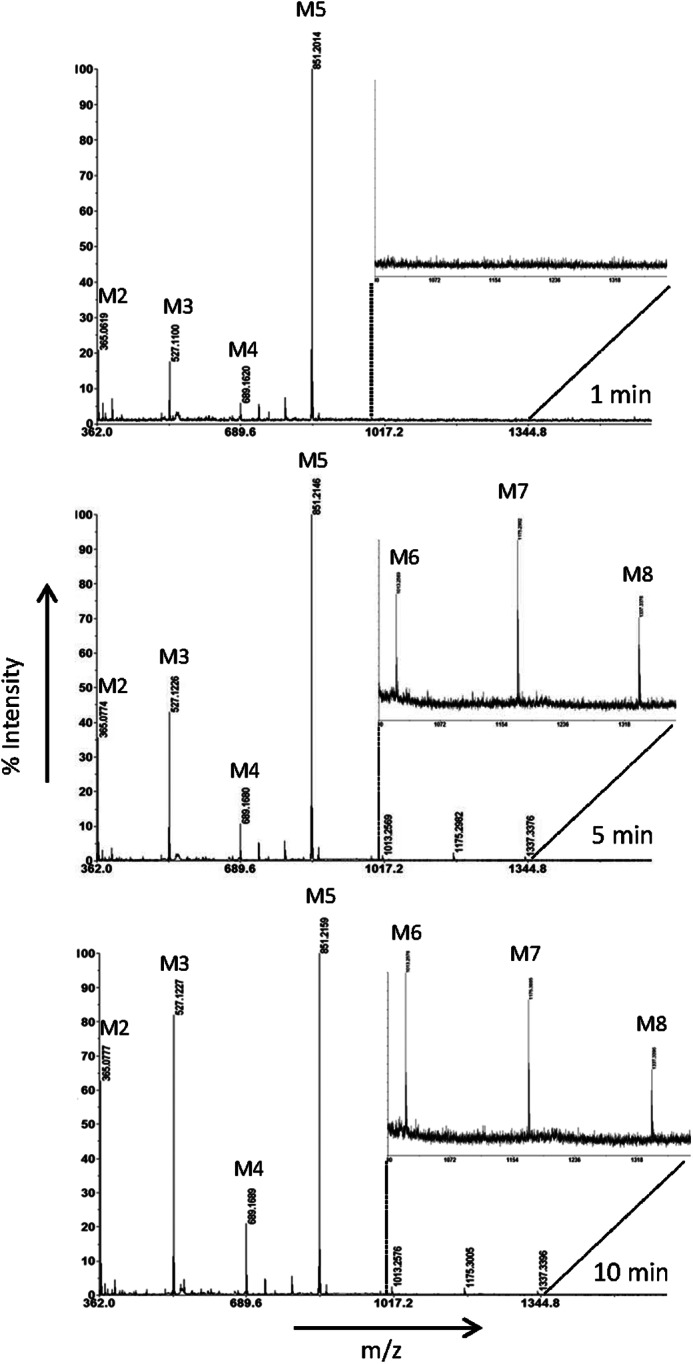
MALDI-TOF-MS analysis of the transglycosylation product formation from M5 during 1–10 min by PaMan5A. Peaks in the spectra correspond to monoisotopic masses of sodium adducts [M + Na]+ of manno-oligosaccharides: M2, m/z 365.1; M3, m/z 527.1; M4, m/z 689.2; M5, m/z 851.2; M6, m/z 1013.3, M7, m/z 1175.3; M8, m/z 1337.3. The enlarged part in each spectrum corresponds to ∼3% of the relative intensity.
Structure of PaMan5A
The crystal structure of PaMan5A was solved in its free form. The crystal contained one monomer in the asymmetric unit, and light-scattering experiments indicated that the protein is a monomer in solution (data not shown). The overall structure of PaMan5A (Fig. 4A) revealed a (β/α)8-barrel fold as expected for enzymes belonging to clan GH-A. When superimposed with TrMan5A (PDB code 1QNR) and Thermomonospora fusca mannanase (PDB code 3MAN) structures (supplemental Fig. S1), the overall fold of PaMan5A is very similar to that of TrMan5A, with structural differences being confined mainly in the loop regions (Fig. 4A) that have been defined as eight loops: loop 1 (residues 35–42), loop 2 (66–95), loop 3 (120–144), loop 4 (177–184), loop 5 (213–232), loop 6 (252–258), loop 7 (287–289), and loop 8 (316–336). Compared with TrMan5A, which contains four disulfide bonds, Cys-26–Cys-29, Cys-172—Cys-175, Cys-265—Cys-272, and Cys-284—Cys-334, PaMan5A contained only three disulfide bonds, i.e. Cys-180—Cys-184, Cys-272—Cys-279, and Cys-291–Cys-342. After N-deglycosylation of PaMan5A using peptide N-glycosidase F, no shift in the apparent molecular mass (46 kDa) was observed on SDS-PAGE compared with untreated sample (data not shown). This observation was in good agreement with NetGlyc predictions from the PaMan5A primary sequence (no predicted N-glycosylation site) and analysis of the PaMan5A crystallographic data that confirmed absence of glycosylation units.
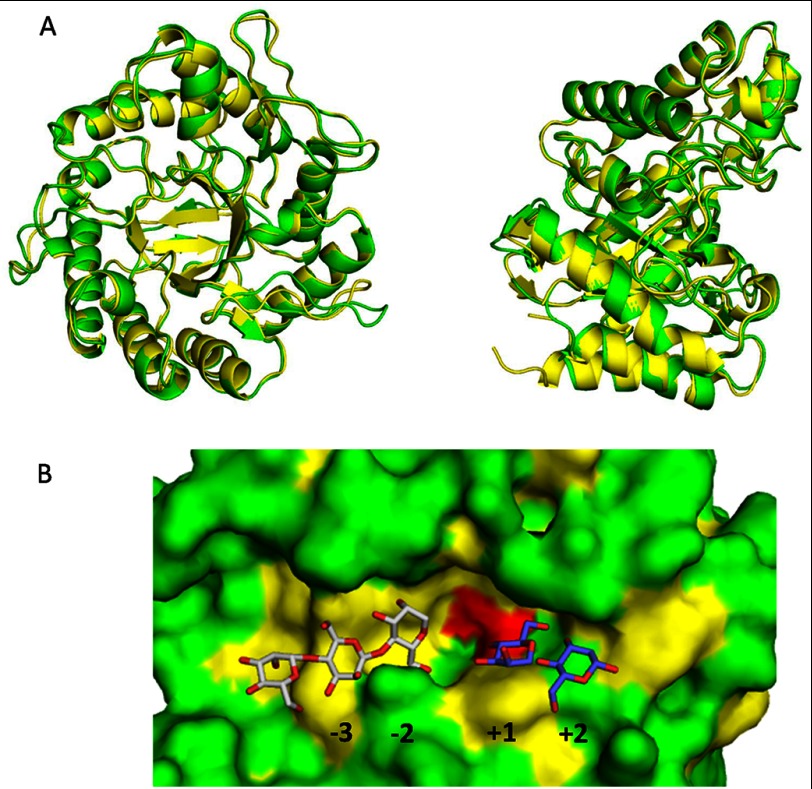
FIGURE 4.
Crystal structure of PaMan5A. A, superposition of PaMan5A (green) and TrMan5A (yellow) structures is shown. The two views are related by a rotation of ∼90° about the vertical axis. B, shown is a surface view of the catalytic cleft of PaMan5A with mannotriose modeled in the −2 and −3 subsites and mannobiose modeled in the +1 and +2 subsites. The structures of GH5 from T. reesei and T. fusca in complex with mannobiose and mannotriose, respectively, were superimposed on the top of the structure of PaMan5A to map the substrate binding subsites.
The active site of PaMan5A was clearly identified in the groove, with the two conserved catalytic glutamate residues (acid-base and nucleophile) positioned near the C-terminal ends of β-strands four and seven of the (β/α)8 barrel (41), Glu-177 and Glu-283, respectively. Mutant E283A showed no catalytic activity for glucomannan, thus indicating that Glu-283 should be the nucleophile. E177A had a specific activity of 0.47 units·mg−1 toward glucomannan, which is roughly 100-fold lower than the wild-type enzyme (45 units·mg−1), thus indicating that Glu-177 should be the acid-base catalytic residue. These results are in agreement with other homologous GH5 enzymes where catalytic residues have been determined (42, 43). Despite several attempts, no structure of PaMan5A inactive mutants alone or in complex with its substrate has been obtained. Consequently, we performed comparative structural analysis of PaMan5A with other GH5 mannanases complexes (T. reesei PDB code 1QNO, Thermotoga petrophila PDB code 3PZ9, and S. lycopersicum PDB code 1RH9) to map the substrate binding subsites (Fig. 4B). In the −1 and +1 subsites where the catalytic cleavage occurs, 7 of 8 residues highly conserved in GH5 mannanases (44) are found in PaMan5A, among which are the catalytic residues Glu-177 and Glu-283 and Arg-62, Asn-176, His-248, Tyr-250, Trp-315 (Fig. 4B). PaMan5A also has an arginine equivalent to Arg-171 in the +2 subsite of TrMan5A (15), which is semi-conserved among GH5 mannanases and which was shown to play a significant role in the transglycosylation ability of TrMan5A (32).
Structure of PaMan26A Catalytic Module
The structure of PaMan26A was successfully solved using molecular replacement. The search model was composed of the superimposition of four structures of bacterial mannanases (PDB codes 2QHA, 2BVT, 2VX4, and 2WHK). The final structure comprising 443 residues was refined at 2.85 Å resolution. The overall structure of PaMan26A CD revealed a (β/α)8-barrel fold (Fig. 5A) as expected for enzymes belonging to clan-GHA. The active site was clearly identified in the groove, with the two conserved catalytic glutamate residues (Glu-300 and Glu-390) positioned at the end of the (β/α)8 barrel and several aromatic residues forming the subsites of catalytic cleft. Mutant E390A showed no catalytic activity for glucomannan, indicating that Glu-390 should be the nucleophile. E300A had a specific activity of 0.33 units·mg−1, which is roughly 200-fold lower than the wild-type enzyme (65 units·mg−1), thus indicating that Glu-300 should be the acid-base catalytic residue. These results are in agreement with other homologous GH26 enzymes where catalytic residues have been determined (45).
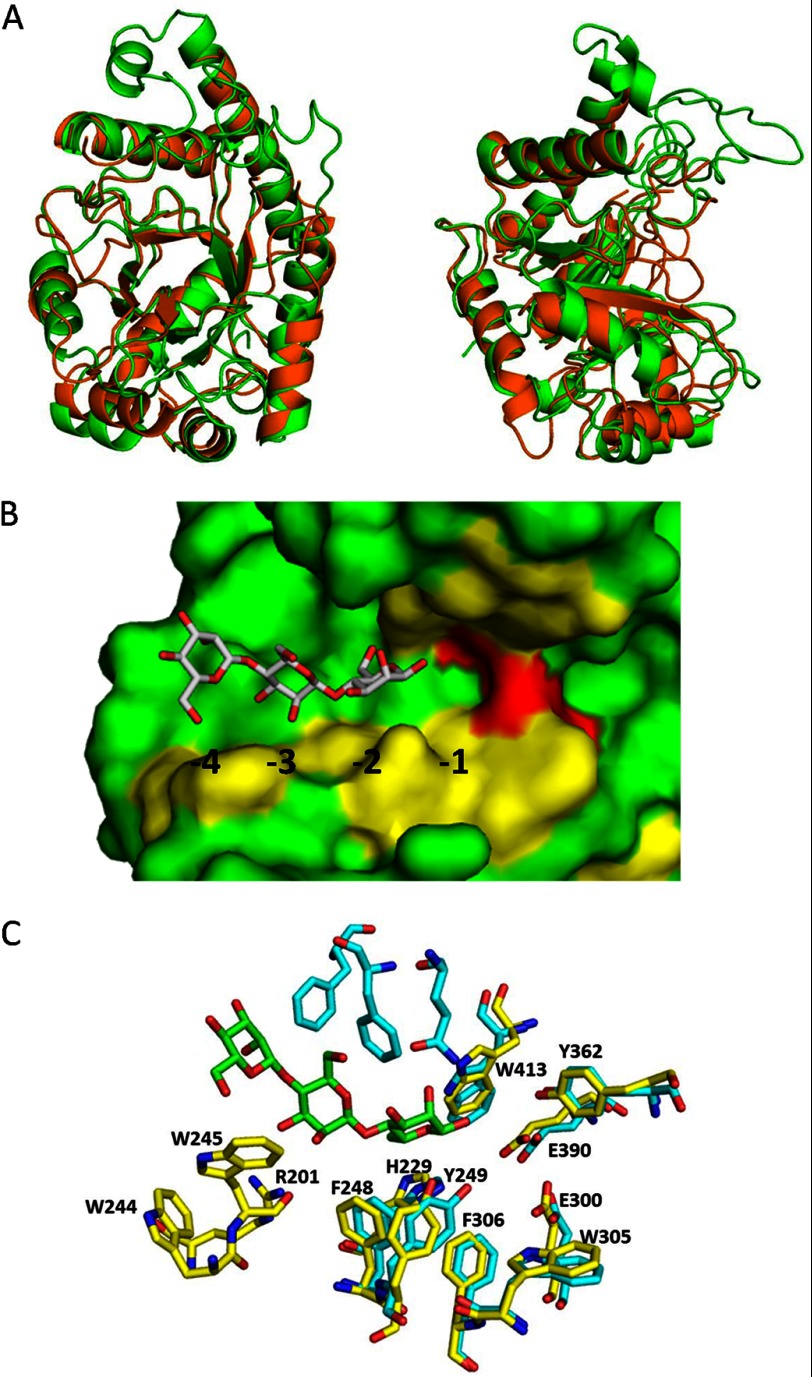
FIGURE 5.
Crystal structure of PaMan26A. A, superposition of PaMan26A catalytic module (green) and BCMan (orange) structures. The two views are related by a rotation of ∼90° about the vertical axis. B, shown is a surface view of the catalytic cleft of PaMan26A with mannotriose modeled in the −2 to −4 subsites. The structure of GH26 from C. fimi in complex with mannotriose was superimposed on the top of the structure of PaMan26A to map the substrate-binding subsites. C, shown is the organization of the glycone binding subsites in PaMan26A (yellow) compared with C. fimi (cyan).
Electron density was observed for two carbohydrate sugar residues at one glycosylation site, Asn-268, which is located in the CD on the external side of the barrel. As modeled from electron density, 2 β-1,4-linked N-acetylglucosamine (GlcNAc) units are attached to this N-glycosylation site. N-Deglycosylation of PaMan26A using peptide N-glycosidase F was associated with a 2–3-kDa shift in the apparent molecular mass on SDS-PAGE compared with untreated sample (data not shown). These results confirm that PaMan26A is N-glycosylated and are in agreement with the NetNGlyc prediction (one predicted N-glycosylation site at position Asn-268).
Several regions are highly conserved between PaMan26A and other GH26 mannanases from B. subtilis z-2 (PDB code 2QHA), B. subtilis subsp. bacillus (PDB code 2WHK), C. japonicus (PDB codes 1GW1 and 2VX6), and C. fimi (PDB code 2BVT; supplemental Fig. S2) as shown in the superimposition of PaMan26A and B. subtilis z-2 (PDB code 2QHA) structures (Fig. 5A). The central β-barrel and most of the surrounding α-helices are superimposable between PaMan26A and B. subtilis z-2, whereas loop regions are dramatically different. Indeed, B. subtilis enzyme exhibits a flat surface with a shallow dish-shaped active center, whereas PaMan26A displays large loops that form a deep cleft. According to the three-dimensional structure of PaMan26A, 8 loops are involved in the binding of the substrate to the active site: loop 1 (171–174), loop 2 (195–208), loop 3 (230–266), loop 4 (301–314), loop 5 (342–346), loop 6 (362–368), loop 7 (392–396), and loop 8 (413–425). The most striking difference stands in loop 2 that contains four aromatic residues (Trp-244, Trp-245, Phe-248, and Tyr-249) and is nine amino acids longer than B. subtilis z-2 (PDB code 2QHA), B. subtilis subsp. bacillus (PDB code 2WHK), C. japonicus (PDB code 2VX6), and C. fimi (PDB code 2BVT) mannanases. A shorter loop 2 does not allow interaction with the substrate at the glycone binding subsites in the case of B. subtilis z-2 (PDB code 2QHA). The −1 and +1 subsites of PaMan26A are quite similar to homologous enzymes with the conserved residues His-299, Trp-305, Phe-306, Tyr-362, Trp-413 (Fig. 5, B and C). As described for CfMan26A and CjMan26A, PaMan26A Tyr-362 is probably involved in a hydrogen bond with the catalytic nucleophile Glu-390, whereas PaMan26A Trp-305 and Trp-413 could play a role as aromatic platforms to stabilize mannopyrannose rings at the +1 and −1 subsites, respectively (Fig. 5C). In the −2 subsite of BCMan, binding is not favorable because of steric hindrance due to the position of Tyr-40 (21). In the case of CfMan26A, the two aromatic Phe-123 and Tyr-124 residues that are superimposed with PaMan26A F248 and Tyr-249 stabilize the interaction with a mannose unit at the −2 subsite (Fig. 5C).
Our experimental data indicate that PaMan26A displays strong interactions at the −4 subsite. Indeed, PaMan26A was poorly active toward M4 probably due to the formation of an unproductive complex between −4 and −1 subsites. We further analyzed the −4 subsite in the PaMan26A structure and identified two aromatic residues, Tro-244 and Trp-245, located in loop 2 that could stabilize mannopyrannose rings in the −4 subsite (Fig. 5, B and C). As PaMan26A, CfMan26A active site also contains four glycone binding subsites, but experimental results provided evidence for the existence of a strong −3 subsite, and residues involved in the −4 subsite were described as making a minor contribution to binding (17). In PaMan26A, there is no equivalent to the Phe-42, Phe-325, and Gln-329 CfMan26A −3 subsite residues. The lack of a strong −3 subsite and the presence of a strong −4 subsite in the structure is in agreement with our experimental results that suggest a predominant substrate binding mode involving the −4 subsite. Lacking a strong −4 subsite, CfMan26A produces M2 and M3 as major products from M5 with only minor amounts of M1 and M4 (31).
PaMan26A Modular Organization
PaMan26A harbors a family 35 CBM at its N-terminal end, and the closest characterized enzyme is Humicola insolens β-mannanase (GenBankTM AAQ31840 (46)) with 78% amino acid identity. After a BlastP search using the PaMan26A amino acid sequence, it is interesting to note that all related bacterial and fungal sequences harbor a CBM35 module at their N terminus. In fungi, in addition to PaMan26A CBM35, only one CBM35 module binding to galactan has been characterized to date in a Phanerochaete chrysosporium exo-β-1,3-galactanase (47). We previously suggested that the N-terminal CBM35 module of PaMan26A displayed dual binding specificity toward xylan and mannan (28), and the phylogenetic analysis was performed by Correia et al. (48) clustered PaCBM35 in the subfamily II that is proposed to target β-1,4 mannan.
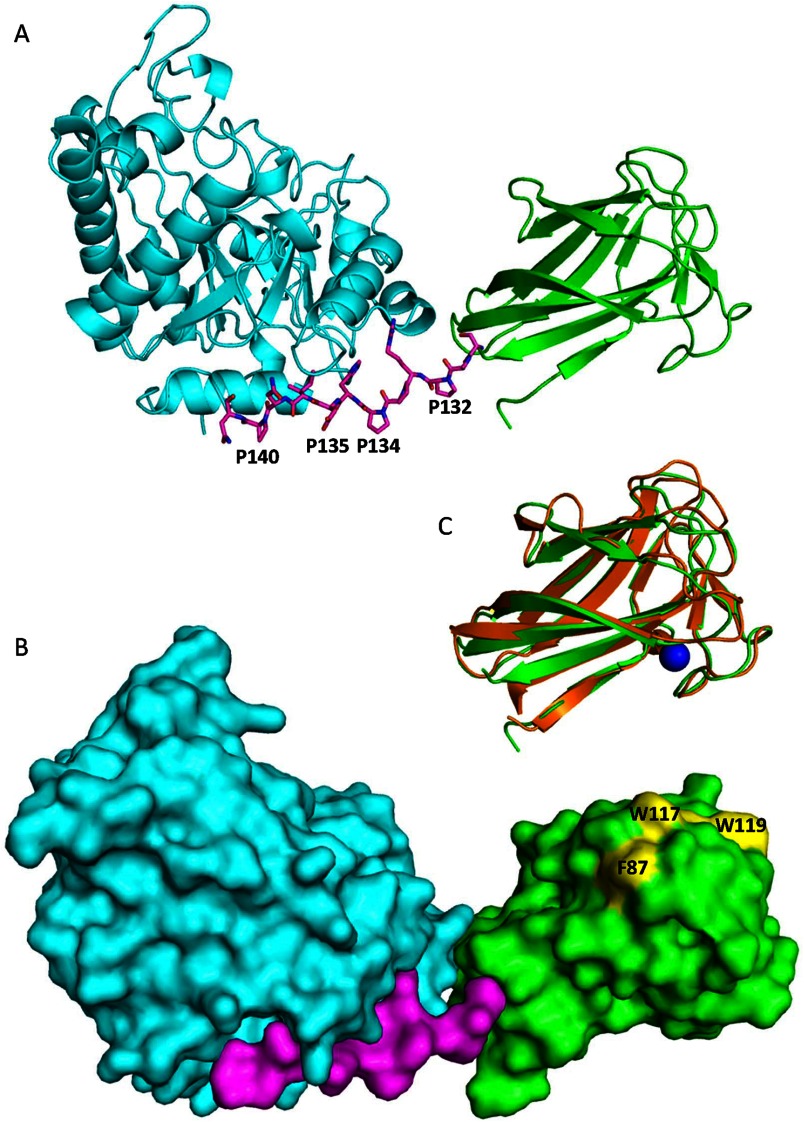
Although the structures of fungal GH bearing a CBM are generally determined separately, this is the first intact structure that allows visualization of the juxtaposition of the CBM35 module relative to the GH26 CD. The linker region of PaMan26A is short without any glycosylation sites, whereas modular fungal GHs usually display long and highly glycosylated linkers. The PaMan26A linker sequence was rich in proline residues, i.e. it contains 4 prolines (Pro-132, -134, -135, and -140) of 12 residues that may confer rigidity to the modular enzyme (Fig. 6A). The linker starts on residue Ser-130 at the end of the last β-strand of the N-terminal CBM domain. Only two residues (Ala-131 and Pro-132) have no interaction with the rest of the molecule. The region from residue R133 to residue N141, which may be considered as the end of the linker, is tightly bound to the CD. Arg-133 and His-136 side chains make hydrogen bond with Asp-382, Asn-374, and Gln-404, whereas the side chain of Ile-138 fits into a hydrophobic cavity made of Arg-159 (aliphatic part of the side chain), Tyr-162, and Mer-385. The CBM and the catalytic module are thus in close association thanks to the embedded linker (Fig. 6B), and it may explain why attempts to express the catalytic module alone were unsuccessful (data not shown). Alignment of PaMan26A-CBM35 with 60 microbial GH26 mannanases sequences bearing a CBM35 module revealed that they all display a short linker region (12–14 residues) rich in proline residues (data not shown).
FIGURE 6.
Views of Modular architecture of PaMan26A. A, shown is a ribbon diagram of PaMan26A catalytic (blue) and CBM (green) domains. The proline-rich linker is shown in stick format. B, shown is a molecular surface representation of PaMan26A structure with the catalytic domain in blue, the PaCBM35 domain in green, and the linker in purple. The three aromatic residues present at the surface of the PaCBM35 domain are shown in yellow. C, shown is superposition of the PaCBM35 domain (green) and C. thermocellum CBM35 (orange). The calcium ion is represented by a blue sphere.
The CBM35 domain comes into contact with the CD through hydrophobic interactions. Indeed a hydrophobic patch comprising Leu-58 and Leu-103 on the surface of the CBM35 domain stands in front of a cluster of hydrophobic residues (Ala-402, Tyr-403, and Leu-399) of the CD. The rationale of the tight modular association of bacterial and fungal GH26-CBM35 mannanases will need further work to gain insights into their function.
PaCBM35 Domain
The CBM35 domain overall structure consists of 2 antiparallel sheets consisting of 4 and 5 antiparallel β-strands, respectively. The two sheets are packed in a β-sandwich conformation enclosing a highly hydrophobic core. The closest structural homologue found using the DALI server (49) is a CBM35 from C. thermocellum (PDB code 2W1W (50)), with a Z-score of 18.6. Its superimposition with PaCBM35 shows that 57 Cα of 125 Cα (45%) have equivalent positions in both molecules, with the distance between the superimposed Cα atoms <1 Å. The main differences occur in the loops connecting the β-strands as shown in Fig. 6C. A metal ion is present that has been modeled as calcium based on its coordination geometry exclusively with oxygen atoms. A calcium ion is also present at a similar location in the structure of the CBM35 domain from C. thermocellum (50). However, the second calcium evidenced in all of the other CBM35s and involved in carbohydrate recognition (50) is not conserved in PaCBM35. A platform of three aromatic residues (Phe-87, Trp-117, and Trp-119) was observed at the surface of the PaCBM35 (Fig. 6B). These residues are aligned with the PaMan26A catalytic cleft, suggesting that they could play a role in substrate binding.
Conclusions
The P. anserina CAZome (the genome-wide inventory of CAZymes) includes three genes encoding β-(1,4)-mannanases: two GH5 mannanases without CBM (including PaMan5A) that both belong to the GH5 subfamily 7 (51) and one GH26 mannanase bearing a CBM35 (i.e. PaMan26A) with affinity for hemicellulosic polysaccharides (28). Based on our kinetic analysis, we can conclude that PaMan5A and PaMan26A are complementary in terms of hydrolysis profile and could act in synergy to deconstruct mannan polysaccharides. Indeed, PaMan26A produces larger manno-oligosaccharides that could be processed by PaMan5A. In C. japonicus, a bacterium also producing both GH5 and GH26 β-mannanases, the catalytic modules of GH5 mannanases were linked to various CBMs, whereas GH26 mannanases were found as single CD (25). Therefore, it has been suggested that GH26 mannanases were involved in degradation of storage tissues, whereas GH5 mannanases harboring cellulose-specific CBMs were involved in degradation of plant cell wall (20, 26). It is interesting to note that the P. anserina mannanase system does not seem to fit with this model, suggesting a difference in the strategies to degrade mannan between these two microbes.
Together with our previous studies on P. anserina CAZymes (28, 52, 53), the present findings give more insights into the P. anserina enzymatic machinery for the deconstruction of plant cell wall polysaccharides. This knowledge is essential to design tailor-made biocatalysts, which can then be used in the biofuel and bioprocessing industries.
Acknowledgments
We thank N. Lopes-Ferreira, P.M. Coutinho and C. Dumon for helpful discussions, A. Blanchard for Dionex analyses, and M. Haon for protein purification. We thank the European Synchrotron Radiation Facility at Grenoble (France) in particular the beamline ID29 staff and French synchrotron (SOLEIL) at Saint-Aubin (France) and the beamline Proxima1 staff for assistance.
This work was funded by the FUTUROL project and OSEO Innovation. This work was also supported by FORMAS and VINNOVA (to H. S.).

This article contains supplemental Table S1 and Figs. S1 and S2.
The atomic coordinates and structure factors (codes 3ZIZ and 3ZM8) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- GH
- glycoside hydrolase
- CBM
- carbohydrate binding module
- CD
- catalytic domain
- HPAEC-PAD
- high performance anion exchange chromatography-pulsed amperometric detection
- M1
- mannose
- M2
- mannobiose
- M3
- mannotriose
- M4
- mannotetraose
- M5
- mannopentaose
- M6
- mannohexaose
- PaCBM35
- P. anserina CBM35
- PaMan5A
- P. anserina GH5 mannanase A
- PaMan26A
- P. anserina GH26 mannanase A
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- BCMan
- B. subtilis z-2 mannanase.
REFERENCES
- 1. Timell T. E. (1967) Recent progress in the chemistry of wood hemicellulose. Wood Sci. Technol. 1, 45–70 [Google Scholar]
- 2. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The carbohydrate-active EnZymes database (CAZy). An expert resource for glycogenomics. Nucleic Acids Res. 37, 233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davies G., Henrissat B. (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3, 853–859 [DOI] [PubMed] [Google Scholar]
- 4. Vocadlo D. J., Davies G. J. (2008) Mechanistic insights into glycosidase chemistry. Curr. Opin. Chem. Biol. 12, 539–555 [DOI] [PubMed] [Google Scholar]
- 5. Gilbert H. J., Stålbrand H., Brumer H. (2008) How the walls come crumbling down. Recent structural biochemistry of plant polysaccharide degradation. Curr. Opin. Plant. Biol. 11, 338–348 [DOI] [PubMed] [Google Scholar]
- 6. Dilokpimol A., Nakai H., Gotfredsen C. H., Baumann M. J., Nakai N., Abou Hachem M., Svensson B. (2011) Recombinant production and characterization of two related GH5 endo-β-1,4-mannanases from Aspergillus nidulans FGSC A4 showing distinctly different transglycosylation capacity. Biochim. Biophys. Acta 1814, 1720–1729 [DOI] [PubMed] [Google Scholar]
- 7. Larsson A. M., Anderson L., Xu B., Muñoz I. G., Usón I., Janson J. C., Stålbrand H., Ståhlberg J. (2006) Three-dimensional crystal structure and enzymic characterization of β-mannanase Man5A from blue mussel Mytilus edulis. J. Mol. Biol. 357, 1500–1510 [DOI] [PubMed] [Google Scholar]
- 8. Harjunpää V., Helin J., Koivula A., Siika-aho M., Drakenberg T. (1999) A comparative study of two retaining enzymes of Trichoderma reesei. Transglycosylation of oligosaccharides catalysed by the cellobiohydrolase I, Cel7A, and the β-mannanase, Man5A. FEBS Lett. 443, 149–153 [DOI] [PubMed] [Google Scholar]
- 9. Zhang Y., Ju J., Peng H., Gao F., Zhou C., Zeng Y., Xue Y., Li Y., Henrissat B., Gao G. F., Ma Y. (2008) Biochemical and structural characterization of the intracellular mannanase AaManA of Alicyclobacillus acidocaldarius reveals a novel glycoside hydrolase family belonging to clan GH-A. J. Biol. Chem. 283, 31551–31558 [DOI] [PubMed] [Google Scholar]
- 10. Anderson L., Hägglund P., Stoll D., Lo Leggio L., Drakenberg T., Stålbrand H. (2008) Kinetics and stereochemistry of the Cellulomonas fimi β-mannanase studied using 1H NMR. Biocatal. Biotransformation 26, 86–95 [Google Scholar]
- 11. Stoll D., Boraston A., Stålbrand H., McLean B. W., Kilburn D. G., Warren R. A. (2000) Mannanase Man26A from Cellulomonas fimi has a mannan binding module. FEMS Microbiol. Lett. 183, 265–269 [DOI] [PubMed] [Google Scholar]
- 12. Hägglund P., Eriksson T., Collén A., Nerinckx W., Claeyssens M., Stålbrand H. (2003) A cellulose binding module of the Trichoderma reesei β-mannanase Man5A increases the mannan-hydrolysis of complex substrates. J. Biotechnol. 101, 37–48 [DOI] [PubMed] [Google Scholar]
- 13. Fujimoto Z., Kuno A., Kaneko S., Kobayashi H., Kusakabe I., Mizuno H. (2002) Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase. Sugar binding structure of the family 13 carbohydrate binding module. J. Mol. Biol. 316, 65–78 [DOI] [PubMed] [Google Scholar]
- 14. Pell G., Szabo L., Charnock S. J., Xie H., Gloster T. M., Davies G. J., Gilbert H. J. (2004) Structural and biochemical analysis of Cellvibrio japonicus xylanase 10C. How variation in substrate binding cleft influences the catalytic profile of family GH-10 xylanases. J. Biol. Chem. 279, 11777–11788 [DOI] [PubMed] [Google Scholar]
- 15. Sabini E., Schubert H., Murshudov G., Wilson K. S., Siika-Aho M., Penttilä M. (2000) The three-dimensional structure of a Trichoderma reesei β-mannanase from glycoside hydrolase family 5. Acta Crystallogr. D. Biol. Crystallogr. 56, 3–13 [DOI] [PubMed] [Google Scholar]
- 16. Bourgault R., Oakley A. J., Bewley J. D., Wilce M. C. (2005) Three-dimensional structure of (1,4)-β-d-mannan mannanohydrolase from tomato fruit. Protein Sci. 14, 1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Le Nours J., Anderson L., Stoll D., Stålbrand H., Lo Leggio L. (2005) The structure and characterization of a modular endo-β-1,4-mannanase from Cellulomonas fimi. Biochemistry 44, 12700–12708 [DOI] [PubMed] [Google Scholar]
- 18. Hogg D., Woo E. J., Bolam D. N., McKie V. A., Gilbert H. J., Pickersgill R. W. (2001) Crystal structure of mannanase 26A from Pseudomonas cellulosa and analysis of residues involved in substrate binding. J. Biol. Chem. 276, 31186–31192 [DOI] [PubMed] [Google Scholar]
- 19. Fanutti C., Ponyi T., Black G. W., Hazlewood G. P., Gilbert H. J. (1995) The conserved noncatalytic 40-residue sequence in cellulases and hemicellulases from anaerobic fungi functions as a protein docking domain. J. Biol. Chem. 270, 29314–29322 [DOI] [PubMed] [Google Scholar]
- 20. Tailford L. E., Ducros V. M., Flint J. E., Roberts S. M., Morland C., Zechel D. L., Smith N., Bjørnvad M. E., Borchert T. V., Wilson K. S., Davies G. J., Gilbert H. J. (2009) Understanding how diverse β-mannanases recognize heterogeneous substrates. Biochemistry 48, 7009–7018 [DOI] [PubMed] [Google Scholar]
- 21. Yan X. X., An X. M., Gui L. L., Liang D. C. (2008) From structure to function. Insights into the catalytic substrate specificity and thermostability displayed by Bacillus subtilis mannanase BCman. J. Mol. Biol. 379, 535–544 [DOI] [PubMed] [Google Scholar]
- 22. Ducros V. M., Zechel D. L., Murshudov G. N., Gilbert H. J., Szabó L., Stoll D., Withers S. G., Davies G. J. (2002) Substrate distortion by a β-mannanase. Snapshots of the Michaelis and covalent-intermediate complexes suggest a B(2,5) conformation for the transition state. Angew. Chem. Int. Ed. Engl. 41, 2824–2827 [DOI] [PubMed] [Google Scholar]
- 23. Cartmell A., Topakas E., Ducros V. M., Suits M. D., Davies G. J., Gilbert H. J. (2008) The Cellvibrio japonicus mannanase CjMan26C displays a unique exo-mode of action that is conferred by subtle changes to the distal region of the active site. J. Biol. Chem. 283, 34403–34413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Setati M. E., Ademark P., van Zyl W. H., Hahn-Hägerdal B., Stålbrand H. (2001) Expression of the Aspergillus aculeatus endo-β-1,4-mannanase-encoding gene (man1) in Saccharomyces cerevisiae and characterization of the recombinant enzyme. Protein Expr. Purif. 21, 105–114 [DOI] [PubMed] [Google Scholar]
- 25. Xu B., Hägglund P., Stålbrand H., Janson J. C. (2002) Endo-β-1,4-Mannanases from blue mussel Mytilus edulis. Purification, characterization, and mode of action. J. Biotechnol. 92, 267–277 [DOI] [PubMed] [Google Scholar]
- 26. Hogg D., Pell G., Dupree P., Goubet F., Martín-Orúe S. M., Armand S., Gilbert H. J. (2003) The modular architecture of Cellvibrio japonicus mannanases in glycoside hydrolase families 5 and 26 points to differences in their role in mannan degradation. Biochem. J. 371, 1027–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Espagne E., Lespinet O., Malagnac F., Da Silva C., Jaillon O., Porcel B. M., Couloux A., Aury J. M., Ségurens B., Poulain J., Anthouard V., Grossetete S., Khalili H., Coppin E., Déquard-Chablat M., Picard M., Contamine V., Arnaise S., Bourdais A., Berteaux-Lecellier V., Gautheret D., de Vries R. P., Battaglia E., Coutinho P. M., Danchin E. G., Henrissat B., Khoury R. E., Sainsard-Chanet A., Boivin A., Pinan-Lucarré B., Sellem C. H., Debuchy R., Wincker P., Weissenbach J., Silar P. (2008) The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 9, R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Couturier M., Haon M., Coutinho P. M., Henrissat B., Lesage-Meessen L., Berrin J.-G. (2011) Podospora anserina hemicellulases potentiate the Trichoderma reesei secretome for saccharification of lignocellulosic biomass. Appl. Environ. Microbiol. 77, 237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matsui I., Ishikawa K., Matsui E., Miyairi S., Fukui S., Honda K. (1991) Subsite structure of Saccharomycopsis α-amylase secreted from Saccharomyces cerevisiae. J. Biochem. 109, 566–569 [DOI] [PubMed] [Google Scholar]
- 30. Berrin J.-G., Ajandouz el H., Georis J., Arnaut F., Juge N. (2007) Substrate and product hydrolysis specificity in family 11 glycoside hydrolase. An analysis of Penicillium funiculosum and Penicillium griseofulvum xylanases. Appl. Microbiol. Biotechnol. 74, 1001–1010 [DOI] [PubMed] [Google Scholar]
- 31. Hekmat O., Lo Leggio L., Rosengren A., Kamarauskaite J., Kolenova K., Stålbrand H. (2010) Rational engineering of mannosyl binding in the distal glycone subsites of Cellulomonas fimi endo-β-1,4-mannanase. Mannosyl binding promoted at subsite −2 and demoted at subsite −3. Biochemistry 49, 4884–4896 [DOI] [PubMed] [Google Scholar]
- 32. Rosengren A., Hägglund P., Anderson L., Pavon-Orozco P., Peterson-Wulff R., Nerinckx W., Stålbrand H. (2012) The role of subsite +2 of the Trichoderma reesei β-mannanase TrMan5A in hydrolysis and transglycosylation. Biocatal. Biotransformation 30, 338–352 [Google Scholar]
- 33. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 35. Navaza J. (2001) Implementation of molecular replacement in AMoRe. Acta Crystallogr. D. Biol. Crystallogr. 57, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 36. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 37. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O. S., Vonrhein C., Womack T. O. (2011) BUSTER Version 2.11.2, Global Phasing Ltd., Cambridge, UK [Google Scholar]
- 38. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the web. A case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 39. Roussel A., Cambillau C. (1991) Turbo-Frodo. in Silicon Graphics Geometry Partners Directory, p. 86, Mountain View Publications, CA [Google Scholar]
- 40. Stålbrand H., Siika-aho M., Tenkanen M., Viikari L. (1993) Purification and characterization of two β-mannanases from Trichoderma reesei. J. Biotechnol. 29, 229–242 [Google Scholar]
- 41. Davies G. J., Wilson K. S., Henrissat B. (1997) Nomenclature for sugar binding subsites in glycosyl hydrolases. Biochem. J. 321, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jenkins J., Lo Leggio L., Harris G., Pickersgill R. (1995) β-Glucosidase, β-galactosidase, family A cellulases, family F xylanases, and two barley glycanases form a superfamily of enzymes with 8-fold β/α architecture and with two conserved glutamates near the carboxyl-terminal ends of β-strands four and seven. FEBS Lett. 362, 281–285 [DOI] [PubMed] [Google Scholar]
- 43. Henrissat B., Callebaut I., Fabrega S., Lehn P., Mornon J. P., Davies G. (1996) Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc. Natl. Acad. Sci. U.S.A. 93, 5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hilge M., Gloor S., Winterhalter K., Zimmermann W., Piontek K. (1996) Crystallization and preliminary crystallographic analysis of two β-mannanase isoforms from Thermomonospora fusca KW3. Acta Crystallogr. D. Biol. Crystallogr. 52, 1224–1225 [DOI] [PubMed] [Google Scholar]
- 45. Bolam D. N., Hughes N., Virden R., Lakey J. H., Hazlewood G. P., Henrissat B., Braithwaite K. L., Gilbert H. J. (1996) Mannanase A from Pseudomonas fluorescens ssp. cellulosa is a retaining glycosyl hydrolase in which E212 and E320 are the putative catalytic residues. Biochemistry 35, 16195–16204 [DOI] [PubMed] [Google Scholar]
- 46. Kauppinen M. S., Schulein M., Schnorr K., Andersen L. N., Mads E. (May 20, 2003) U. S. Patent 6,566,114
- 47. Ichinose H., Yoshida M., Kotake T., Kuno A., Igarashi K., Tsumuraya Y., Samejima M., Hirabayashi J., Kobayashi H., Kaneko S. (2005) An exo-β-1,3-galactanase having a novel β-1,3-galactan-binding module from Phanerochaete chrysosporium. J. Biol. Chem. 280, 25820–25829 [DOI] [PubMed] [Google Scholar]
- 48. Correia M. A., Abbott D. W., Gloster T. M., Fernandes V. O., Prates J. A., Montanier C., Dumon C., Williamson M. P., Tunnicliffe R. B., Liu Z., Flint J. E., Davies G. J., Henrissat B., Coutinho P. M., Fontes C. M., Gilbert H. J. (2010) Signature active site architectures illuminate the molecular basis for ligand specificity in family 35 carbohydrate binding module. Biochemistry 49, 6193–6205 [DOI] [PubMed] [Google Scholar]
- 49. Holm L., Rosenström P. (2010) Dali server. Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Montanier C., van Bueren A. L., Dumon C., Flint J. E., Correia M. A., Prates J. A., Firbank S. J., Lewis R. J., Grondin G. G., Ghinet M. G., Gloster T. M., Herve C., Knox J. P., Talbot B. G., Turkenburg J. P., Kerovuo J., Brzezinski R., Fontes C. M., Davies G. J., Boraston A. B., Gilbert H. J. (2009) Evidence that family 35 carbohydrate binding modules display conserved specificity but divergent function. Proc. Natl. Acad. Sci. U.S.A. 106, 3065–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Aspeborg H., Coutinho P. M., Wang Y., Brumer H., 3rd, Henrissat B. (2012) Evolution, substrate specificity, and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 12, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bey M., Zhou S., Poidevin L., Henrissat B., Coutinho P. M., Berrin J.-G., Sigoillot J.-C. (2013) Cello-oligosaccharide oxidation reveals differences between two lytic polysaccharide monooxygenases (family GH61) from Podospora anserina. Appl. Environ. Microbiol. 79, 488–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lafond M., Navarro D., Haon M., Couturier M., Berrin J.-G. (2012) Characterization of a broad-specificity β-glucanase acting on β-(1,3)-, β-(1,4)-, and β-(1,6)-glucans that defines a new glycoside hydrolase family. Appl. Environ. Microbiol. 78, 8540–8546 [DOI] [PMC free article] [PubMed] [Google Scholar]