Background: Eukaryotic nitrate reductase maturation is poorly understood.
Results: Binding of molybdenum cofactor to apo-nitrate reductase is independent from other prosthetic groups.
Conclusion: Active site formation of eukaryotic nitrate reductase is an autonomous process intrinsically tied to nitrate reductase dimerization.
Significance: The understanding of molybdenum cofactor-dependent enzyme maturation is of significance as molybdenum enzymes are involved in essential cellular processes.
Keywords: Electron Paramagnetic Resonance (EPR), Heme, Molybdenum, Neurospora, Ultracentrifugation, Molybdenum Cofactor, Nitrate Reductase, Prosthetic Group Insertion
Abstract
Nitrate reductase (NR) is a complex molybdenum cofactor (Moco)-dependent homodimeric metalloenzyme that is vitally important for autotrophic organism as it catalyzes the first and rate-limiting step of nitrate assimilation. Beside Moco, eukaryotic NR also binds FAD and heme as additional redox active cofactors, and these are involved in electron transfer from NAD(P)H to the enzyme molybdenum center where reduction of nitrate to nitrite takes place. We report the first biochemical characterization of a Moco-free eukaryotic NR from the fungus Neurospora crassa, documenting that Moco is necessary and sufficient to induce dimer formation. The molybdenum center of NR reconstituted in vitro from apo-NR and Moco showed an EPR spectrum identical to holo-NR. Analysis of mutants unable to bind heme or FAD revealed that insertion of Moco into NR occurs independent from the insertion of any other NR redox cofactor. Furthermore, we showed that at least in vitro the active site formation of NR is an autonomous process.
Introduction
In autotrophic organisms the major pathway for assimilating inorganic nitrogen is the nitrate assimilation pathway, with nitrate reductase (NR)2 catalyzing the first and rate-limiting step. NR is a molybdenum cofactor (Moco)-dependent enzyme that shares its cofactor with a family of four other plant enzymes that are involved in sulfite detoxification, purine catabolism, and abscisic acid biosynthesis (1). As a unifying characteristic, these enzymes catalyze two-electron transfer (redox) reactions involving a molybdenum atom coordinated in Moco (2). This cofactor is a unique pterin derivative conserved among all kingdoms of life, and with the exception of the bacterial nitrogenase, it is found in all enzymes that hold a molybdenum atom in their active site. In addition to Moco, eukaryotic NRs harbor with heme and FAD two other prosthetic groups. For nitrate reduction, NR uses NAD(P)H as an electron donor that provides two electrons to the FAD cofactor that are subsequently transferred to the heme group and finally reduce the molybdenum center of the enzyme (3). In eukaryotes, NRs form a family of enzymes that share a high degree of sequence homology. Differences exist concerning the use of NADH or NADPH as electron donors for nitrate reduction. NADH-specific NR forms are found in higher plants and algae, in contrast to the less frequently occurring NAD(P)H-bispecific NR forms found in higher plants, algae, and fungi. NR forms specific for NADPH are only found in fungi (4). Bacterial NRs are completely different from their eukaryotic counterparts both in sequence and in structural composition. Eukaryotic NR is only functional as a homodimer. The NR monomer has a size of ∼100 kDa and harbors its three redox cofactors, Moco, heme, and FAD, in three structurally distinct domains (4, 5) (Fig. 1A). Furthermore, there is a dimerization domain that is located C-terminal to the Moco binding domain. The NAD(P)H and FAD binding domains form the so-called cytochrome b reducing fragment (CbR). Combination of the heme binding domain with the CbR fragment builds the so called cytochrome c reducing fragment (CcR). Both CbR and CcR are functional units of the NR protein capable to reduce artificial electron acceptors like ferricyanide or cytochrome c, whereby the former accepts electrons directly from the CbR fragment and the latter is reduced by the CcR fragment (Fig. 1A). Other than the cofactor housing domains, the connecting hinge regions as well as an N-terminal extension preceding the Moco domain are not conserved among eukaryotic NRs (Fig. 1B). In plants the hinge 1 region, which connects the Moco and heme domain, was shown to be important for NR activity regulation (6), and likewise the N-terminal extension is thought to possibly have a function in the post-transcriptional regulation of NR activity (7).
FIGURE 1.
Conserved domains of N. crassa NR. A, shown is a schematic representation of N. crassa NR domain structure. The first and last residues of the NR domains are indicated. Moco and dimerization domain share a common sequence stretch comprising 11 amino acids. B, shown is a sequence comparison of N. crassa NR (NcNR) with NR from A. thaliana (AtNR), Nicotiana tabacum (NtNR), Zea mays (ZmNR), A. nidulans (AnNR), and P. angusta (PaNR). Strictly conserved residues are highlighted in black, and conserved residues are highlighted in gray. The alignment was generated with Clustal W (51).
There are indications for Moco to function in the dimerization process of NR (4, 8–11); however, contrary results indicate that the dimerization domain functions autonomously (12). The insertion of Moco into molybdenum -containing enzymes is an ill-defined process. Also, the insertion of other prosthetic groups during the maturation of this class of metalloenzymes is poorly understood. The reason for this gap in knowledge is that the study of cofactor insertion was hampered by the availability of well defined, stable apo-enzyme proteins in sufficiently large amounts. So far the recombinant expression and purification of a Moco-free eukaryotic NR was not reported, and all attempts to characterize the oligomerization state of Moco-free NR were constrained to whole cell extracts of Moco-deficient mutants from plants and fungi.
Virtually nothing is known about the influence of the two remaining redox active cofactors heme and FAD on the formation of the physiologically active, homodimeric eukaryotic NR. Also the sequence of redox cofactor incorporation into eukaryotic NR and likewise the underlying principles are still an open question.
In this work we characterized the recombinant Moco-free eukaryotic NR from the filamentous fungus Neurospora crassa. Biochemical characterization of the apo-enzyme revealed that Moco is solely sufficient to induce NR dimer formation. Furthermore, the sequence of prosthetic group insertion was found to be independent from Moco bound to NR.
EXPERIMENTAL PROCEDURES
Cloning of the Neurospora NR
The gene encoding Neurospora NR has been identified by Okamoto et al. (13), and the Neurospora gene-locus NCU05298 has been assigned to the NR encoding sequence (14, 15). Therefore, we cloned the gene of Neurospora NR using Phusion® High-Fidelity DNA Polymerase (New England Biolabs) with primers derived against the locus NCU05298. The oligonucleotides designed to amplify the gene were: forward primer (5′-TATTCACGTGATGGAGGCTCCAGCTCTC-3′) and reverse primer (5′-ATTACTAGTTCAAAAAACTAATACATCCTCATCCTTCC-3′). The forward primer included the sequence for a PmlI, and the reverse primer included the sequence for a SpeI restriction site. As PCR template, genomic DNA from N. crassa strain FGSC #988 (16) was used. The single intron of the Neurospora NR gene was removed by overlap extension polymerase chain reaction, thus yielding the coding DNA sequence of Neurospora NR. The CloneJETTM PCR cloning kit has been used for subcloning according to the manufacturer's instructions.
Site-directed Mutagenesis of the Neurospora NR
Based on the coding DNA sequence of N. crassa NR, site-directed mutagenesis was carried out using overlap extension-PCR. For heme-free Neurospora NR variant H654A/H677A, codons 654 (CAT) and 677 (CAC) were changed to GCC, resulting in the conversion of both histidines to alanines. Three Neurospora NR single amino acid variants with altered FAD binding characteristics were constructed; for NR variant R778E, codon 778 CGC was altered to GAA, in NR variant Y780A, codon 780 TAC was changed to GCG, and codon 811 (GGA) was changed to GTG, yielding variant G811V.
Cloning of the Neurospora NR CcR Fragment
NR CcR fragments containing amino acids 618–984 of full-length NR were amplified by PCR using forward primer 5′-TGTCACGTGGTCACTCGACTTATC-3′ and reverse primer 5′-ATTACTAGTTCAAAAAACTAATACATCCTCATCCTTCC-3′. The coding DNA sequence of Neurospora NR and its variants H654A/H677A, R778E, Y780A, and G811V were used as PCR templates to generate wild type CcR and mutated CcR variants, respectively.
Expression and Purification of Recombinant Proteins
For expression of Neurospora NR and CcR fragments in Escherichia coli, the NR and CcR fragment-coding DNA sequence was subcloned into PmlI and SpeI sites of a bacterial expression vector resulting in the C-terminal fusion of a Twin-Strep-tag® (IBA GmbH) to the protein. As a second tag, a His6 tag was encoded on this vector, resulting in the N-terminal fusion of this tag to the protein, thus allowing the expression of a double-tagged protein. The His6 tag/Twin-Strep-tag® encoding vector was constructed based upon the pQE-80L vector (Qiagen GmbH). Different E. coli strains were used for recombinant NR production, thus allowing the expression of Moco-free NR (E. coli strain RK5204 (17)), molybdopterin (MPT)-containing NR (E. coli strain RK5206 (17)), or Moco containing NR (E. coli strain TP1000 (18)). Expression of Neurospora NR was carried out in LB medium containing 50 μg/ml ampicillin at 22 °C. For expression in TP1000 cells, 10 mm sodium molybdate was additionally added. After cell density reached an A600 nm = 0.1, NR expression was induced with 20 μm isopropyl 1-thio-β-d-galactopyranoside, and cells were allowed to grow aerobically for 20 h. For production of NR CcR fragments, E. coli BL21 cells were used. Expression was carried out in LB medium containing 50 μg/ml ampicillin at 30 °C. After cell density reached an A600 nm = 0.2, cells were induced with 50 μm isopropyl 1-thio-β-d-galactopyranoside and allowed to grow aerobically for 20 h. Cell lyses was achieved by two passages through a French pressure cell. Upon this, cells were sonicated for 1.0 min on ice. Cell lysis was carried out at 4 °C. After centrifugation, double-tagged proteins were purified at 4 °C under native conditions. In the first purification step Ni-NTA Superflow resin (Qiagen) was used according to the manufacturer's instructions. Cell lysis buffer contained 100 mm Tris-HCl, 150 mm NaCl, 5 mm imidazole, 2% (v/v) glycerol (pH 8.0). Washing steps were carried out using washing buffer containing 100 mm Tris-HCl, 150 mm NaCl, 10 mm imidazole, 2% (v/v) glycerol (pH 8.0). Proteins were eluted in elution buffer (100 mm Tris-HCl, 150 mm NaCl, 250 mm imidazole, 2% (v/v) glycerol (pH 8.0)). All buffers were degassed before use. Eluted fractions were pooled and subsequently loaded on Strep-Tactin Superflow® high capacity resin (IBA). Washing steps were carried out using washing buffer containing 100 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, and 2% (v/v) glycerol. Proteins were eluted in elution buffer (100 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 2% (v/v) glycerol, 20 mm d-desthiobiotin). Purity of the eluted protein was routinely documented by Coomassie Blue staining after SDS-PAGE. Pure protein fractions were concentrated (Vivaspin 6, Sartorius AG) and stored in 20-μl aliquots in liquid nitrogen.
Size Exclusion Chromatography
Purified recombinant proteins were analyzed by gel filtration chromatography using an analytical Superdex 200 column (GE Healthcare) connected to an Äkta purifier system (Amersham Biosciences). As running buffer, 100 mm Hepes-KOH, 150 mm NaCl, 5 mm EDTA, and 5% (v/v) glycerol (pH 7.5) was chosen. Molecular weight standards (GE Healthcare) were used for calibration according to the manufacturer's instruction.
CD Spectroscopy
CD spectra were recorded on a Jasco Model J-810 spectropolarimeter (Jasco) at 20 °C. CD spectra of purified protein preparations were recorded at protein concentrations of 1 μm in 50 mm sodium phosphate, 100 mm NaF, and 2% (v/v) glycerol (pH 7.2) using quartz glass cuvettes of 1-mm cell path length between 350 and 180 nm at 0.1-nm intervals. A minimum of 10 scans was recorded, and base-line spectra were subtracted from each spectrum. Data analysis was performed using CDPro software (19).
Analytical Ultracentrifugation
Sedimentation velocity experiments were carried out in a Beckman Coulter ProteomeLab XL I analytical ultracentrifuge at 35,000 rpm and 20 °C in a buffer containing 0.15 m NaCl, 1 mm EDTA, and 50 mm Tris-HCl (pH 8.0) using an An-50 Ti rotor. Concentration profiles were measured using the manufacturer's data acquisition software ProteomeLab XL-I Graphical User Interface Version 6.0 (firmware 5.06) and the UV absorption scanning optics at 280 nm. Experiments were performed in 3- or 12-mm double sector centerpieces filled with 100 or 400 μl of sample, respectively. Due to thermal equilibration of the rotor before the start of the run, diluted samples were incubated at least 2 h at 20 °C before centrifugation. Because it was recently found that version 6.0 of the data acquisition software records incorrect elapsed time in the data files, the program SEDFIT was used to determine correction factors and to correct scan times (20). Correction factors were in the range of 1.076–1.078. Data analysis was performed using a model for diffusion-deconvoluted differential sedimentation coefficient distributions (c(s) distributions)) implemented in SEDFIT (21). Partial specific volume, buffer density, and viscosity were calculated by the program SEDNTERP (22) and were used to correct the experimental sedimentation coefficients to s20,w. Contributions of bound cofactors to partial specific volume of the enzyme were not taken into account. Protein concentrations were determined spectrophotometrically using the absorption coefficients at 280 nm as calculated from amino acid composition (23) and are given in monomers throughout the text.
EPR Spectroscopy
For EPR spectroscopy proteins were incubated in 100 mm Hepes-KOH, 150 mm NaCl, 5 mm EDTA, 5% (v/v) glycerol (pH 7.5) containing 50 μm FAD and 20 mm KNO3. Reduction of NR molybdenum was initiated by the addition of 20 mm NADPH. Samples were incubated for 25 s at room temperature and frozen in an ethanol/liquid nitrogen mixture. EPR spectra were recorded at X band frequency (9.4314 GHz) with a Bruker model EMX-6/1 spectrometer equipped with a standard TE102 rectangular cavity. The temperature was maintained at 77 K by immersion of the samples in partially silvered liquid nitrogen finger Dewar. A correction for the deviation of the magnetic field measured by the EMX-032T Hall probe at the position of the sample was calculated from the g value of the strong pitch (g = 2.0028) and the microwave frequency of the ER-041–1161 counter. Spin concentrations were calculated by double integration and comparison with a 1 mm Cu2+ EDTA standard under non-saturating conditions (i.e. a microwave power of <0.2 milliwatt for Mo5+ and <20 microwatts for Cu2+). A correction for the signals of 95Mo and 97Mo nuclear hyperfine split signals as well as a correction for the undetectable signals of the 95Mo and 97Mo hyperfine split signals was made. To obtain a suitable signal-to-noise ratio for comparison of spectral shape and intensity, multiple spectra (up to 32) were recorded at a 2-milliwatt microwave power and 0.65-millitesla modulation amplitude (modulation frequency, 100 kHz).
Quantification of Moco/MPT
The amount of total Moco/MPT was quantified by HPLC FormA analysis as described earlier (24).
Inductively coupled Plasma Mass Spectrometry (ICP-MS)
Molybdenum content of NR was quantified with Agilent 7700 Series ICP-MS (Agilent Technologies) according to a standard calibration curve of inorganic molybdenum (Fluka). The detection limit of ICP-MS for molybdenum ranged between 1 and 20 μg/liter. Protein solutions and standards were mixed automatically with rhodium as an internal standard. All values were corrected for the molybdenum content of control samples consisting of buffer alone. All data were collected and processed using MassHunter work station software.
Quantification of Heme
The amount of heme bound to NRs was quantified using a QuantiChrom heme assay kit purchased from GENTAURTM, and changes in absorbance at 400 nm were measured in a TECAN sunriseTM microplate reader. For calculation of NR, heme content UV-visible absorption measurements were performed with a PerkinElmer Life Sciences Lambda 25 spectrophotometer using ϵ = 120,000 m−1 cm−1 at 412 nm for heme in oxidized redox state (25) and ϵ = 156,760 m−1 cm−1 at 280 nm for NR.
Nitrate Reducing Activity
Nitrate reducing activity was determined according to Evans and Nason (26) with slight modifications. For typical enzyme assays, 50–600 ng of purified NR was incubated in 50 mm sodium phosphate, 200 mm NaCl, 5 mm EDTA (pH 7.2) buffer containing 1 mm NADPH, 10 mm potassium nitrate, 50 μm FAD. The reaction was started with potassium nitrate and incubated at room temperature for 2–30 min. Enzyme assays were terminated by the addition of 0.6 m zinc acetate. For quantification of nitrite converted from nitrate, 2 volumes of 1% (w/v) sulfanilamide in 3 m HCl and 2 volumes 0.02% (w/v) N-(1-naphthyl)-ethylenediamine dihydrochloride were added. After incubation for 10 min at room temperature, the solutions were centrifuged (13,000 × g) for 5 min, and the absorbance was measured at 540 nm using TECAN sunriseTM microplate reader. According to nitrite standard curve NR activity (μmol of nitrite/min × mg of protein) was calculated.
NADPH-dependent Cytochrome c Reducing Activity
NR NADPH-dependent cytochrome c reducing activity was determined according to Garrett and Nason (27). For a typical cytochrome c assay, 100–500 ng of purified NR were incubated in 50 mm sodium phosphate, 200 mm NaCl, 5 mm EDTA (pH 7.2) buffer containing 5 μm FAD, 82 μm cytochrome c, and 0.1 mm NADPH. Cytochrome c reductase activity was measured by observing the increase in absorbance at 550 nm with a PerkinElmer Life Sciences Lambda 25 spectrophotometer. The molar extinction coefficient for cytochrome c of ϵ = 19,600 m−1 cm−1 (reduced-oxidized) was used to calculate the enzyme activity (μmol of reduced cytochrome c/min × mg NR). Conversion of NADPH was followed at 340 nm and quantified by its extinction coefficient, ϵ = 6,220 m−1 cm−1 (Sigma).
NR in Vitro Reconstitution
For reconstitution of Moco-free NR various amounts of purified Moco carrier protein (MCP) and apo-NR were co-incubated in degassed 100 mm Hepes-KOH, 150 mm NaCl, 5 mm EDTA, 5 mm glutathione, 5% (v/v) glycerol (pH 7.5) buffer for 3 h at room temperature or at 4 °C overnight. MCP was expressed and purified as described earlier (28). NR in vitro reconstitution was analyzed using HPLC-based FormA quantification, NR activity measurements, and dimer formation (SEC) as described above.
Isothermal Titration Calorimetry
Isothermal titration Calorimetry (ITC) titrations were carried out at 25 °C using a MicroCal VP-ITC instrument (GE Healthcare). Before the titration, proteins and FAD were dialyzed against 100 mm Tris HCl, 150 mm NaCl, 1 mm EDTA buffer (pH 8.0) at 4 °C for 2 days. After dialysis, the protein concentration was determined using the Bradford protein assay (29). The FAD concentration was determined spectrophotometrically using a molar extinction coefficient of 11,300 m−1 cm−1 at 450 nm. The reference cell of the ITC instrument was filled with dialysis buffer. In a typical experiment the respective CcR fragment (20–35 μm) in the ITC cell was titrated with FAD (0.5 mm) loaded into the ITC syringe. The first injection (2 μl, omitted from analysis) was followed by 19 injections of 10 μl, with 210-s intervals between injections. The cell contents were stirred at 307 rpm to provide immediate mixing. The data were collected automatically and base-line correction, peak integration, and binding parameters were performed using the ORIGIN analysis software. Normalized area data were plotted in kcal/mol of injectant versus the molar ratio of FAD/CcR. Non-linear fitting of data points was performed with the one set of sites binding model, and binding constants were calculated.
RESULTS
Prosthetic Group Content of Recombinant Holo- and Moco-free Neurospora NR
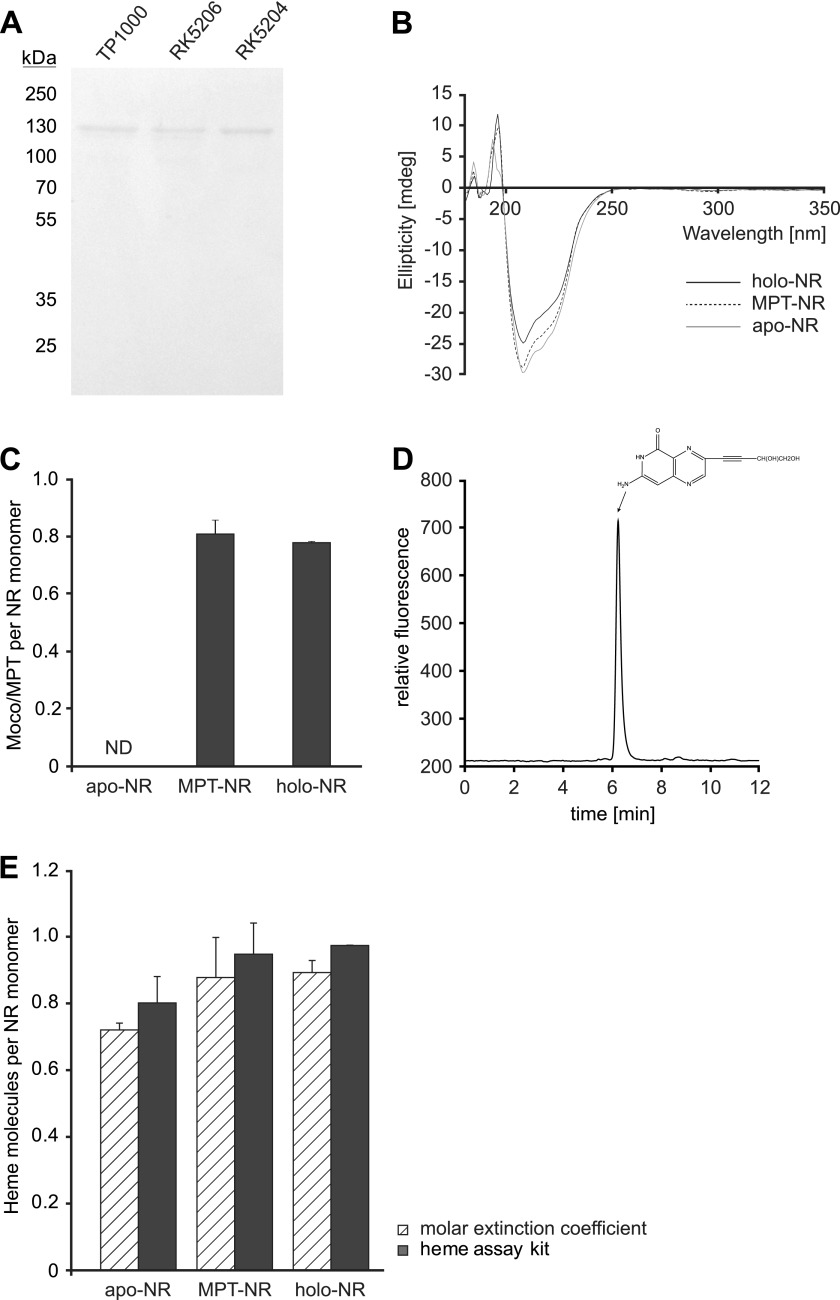
Neurospora NR was purified by two-step affinity chromatography from the Moco accumulating bacterial strain TP1000 (18), the MPT accumulating bacterial strain RK5206 (17), and the Moco/MPT free bacterial strain RK5204 (17). Protein preparations obtained this way were analyzed by SDS-PAGE and CD spectroscopy (Fig. 2, A and B). Comparison of the spectra revealed no significant differences so that one can conclude that all NR proteins purified possess a highly similar secondary structure. Likewise yield and purity of the proteins obtained were comparable, thus allowing carrying out a comparative biochemical characterization of the different NR forms. In a first set of experiments, we quantified the contents of prosthetic groups (i.e. Moco/MPT, heme, and FAD) of two different, holo-, apo-, and MPT-NR preparations, respectively. Moco was quantified in NR preparations derived from TP1000. Therefore, the molybdenum content of these preparations was determined using ICP-MS, revealing on average >80% molybdenum saturation (data not shown). The same protein preparations were also analyzed for Moco content by HPLC (24), thus revealing a Moco/MPT saturation of 0.81 ± 0.04 molecules per monomer (Fig. 2C). Therefore, we conclude that exclusively Moco has been co-purified with NR derived from strain TP1000. No Moco but its immediate precursor MPT was present in NR preparations derived from RK5206 (0.78 ± 0.11 MPT per monomer) (Fig. 2C) as quantified by HPLC-based FormA analysis (Fig. 2D (24)). RK5204-derived NR preparations contained neither Moco nor MPT. In the following we will use the term holo-NR for Moco-containing NR. The term apo-NR will be used for Moco/MPT-free NR, whereas the term MPT-NR will be used for NR co-purified with MPT. Next we asked whether or not the NR heme content is related to the presence of Moco/MPT. To answer this, NR-bound heme was quantified using two different methods. At first protein-bound heme was quantified by using the molar extinction coefficient of cytochrome b5, a method that is commonly used to quantify NR heme content (25). As a second method, a chromogenic assay was taken to quantify NR-bound heme. In this way, heme saturation of holo NR was found to be in the range of 0.89 ± 0.04 molecules heme per monomer (determination was based on the extinction coefficient) or 0.97 ± 0.01 molecules heme per monomer (determination by chromogenic assay) (Fig. 2E). MPT-NR showed essentially the same heme stoichiometries as identified for holo-NR. However, apo-NR displayed a slightly lower heme binding stoichiometry, i.e. 0.72 ± 0.01 molecules heme per monomer (via extinction coefficient) or 0.8 ± 0.03 molecules heme per monomer (via chromogenic assay) (Fig. 2E). Therefore, neither Moco nor MPT has a significant influence on the stoichiometries of NR to bind heme. Subsequently we also quantified the amount of FAD co-purified with holo-, MPT-, and apo-NR using UV-visible spectroscopy (30). Results from these experiments showed that none of the tested NRs was co-purified with FAD (data not shown). It is noteworthy that FAD deficiency was already reported for purified NRs from various other species (31, 32).
FIGURE 2.
Purification and biochemical characterization of N. crassa NR. A, SDS-PAGE analysis of NR was purified from E. coli strains TP1000, RK5206, and RK5204. B, CD spectroscopy of Moco-containing (holo-NR), MPT-containing (MPT-NR), and Moco/MPT-free (apo-NR) NR is shown. C, Moco/MPT content of recombinant NR protein pools is shown. Three replicas of two different holo-, apo-, and MPT-NR protein preparations were analyzed. D, HPLC-based analysis of the Moco/MPT content co-purified with holo-NR is shown. The HPLC elution profile of FormA derived from 3.75 μg of purified recombinant holo-NR is shown. E, analysis of NR heme content is shown. Extinction coefficient-based heme quantification was carried out using 3.37 μm concentrations of the respective recombinant NR proteins. A chromogenic assay for heme quantification was carried out using 90–210 μg of each protein. Three replicas of two different holo-, apo-, and MPT-NR protein preparations were analyzed. ND = not definable.
Nitrate Reducing Activity of Recombinant Holo-NR from Neurospora
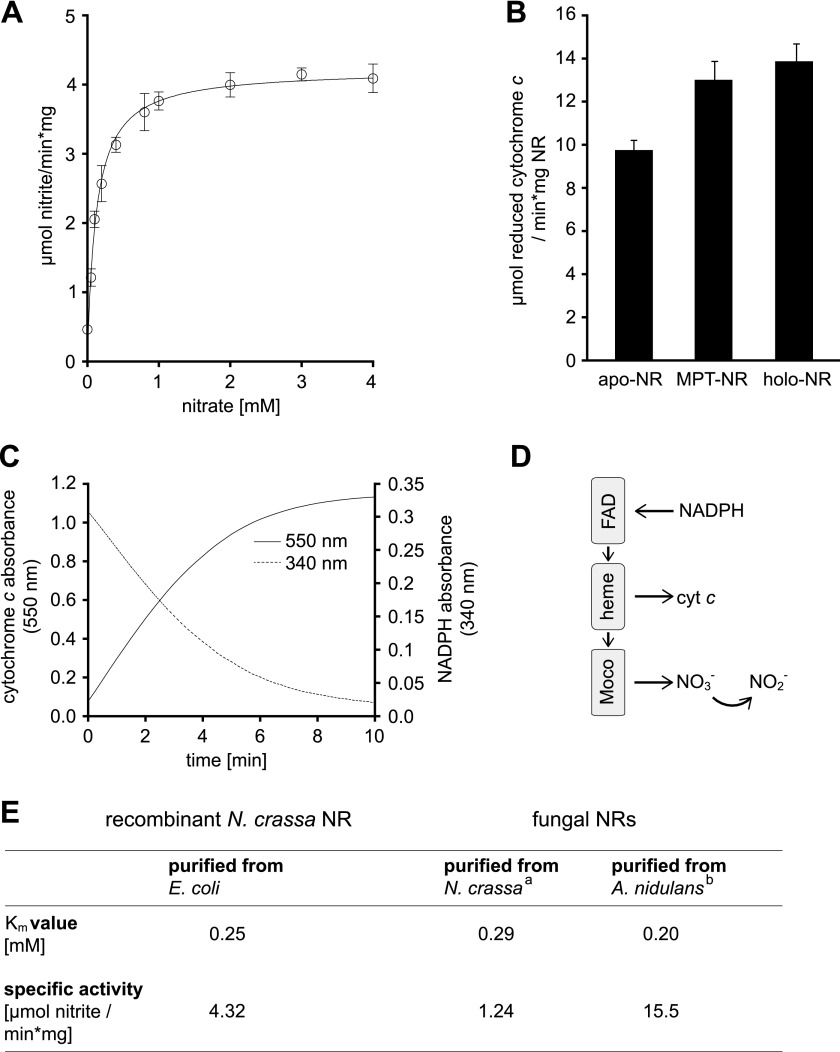
Hitherto expression of eukaryotic holo-NR was preferentially carried out in the methylotrophic yeast Pichia pastoris, yielding active recombinant holo-NR (3, 33). Recombinant N. crassa holo-NR purified from E. coli is likewise functional. For its nitrate reducing activity, the corresponding Km value was found to be 0.25 mm with a Vmax of 4.32 μmol of nitrite/min·mg of protein (Fig. 3E). The Km determined was similar to the Km values documented for the endogenous NR purified from Neurospora and the NR purified from the fungus Aspergillus nidulans. However, differences exist regarding the Vmax values of these NR species (Fig. 3E).
FIGURE 3.
Biochemical characterization of N. crassa NR. A, shown are Michaelis-Menten kinetics of holo-NR using the physiological substrates nitrate and NADPH. S.D. are derived from six independent measurements using two different holo-NR protein preparations. The Michaelis-Menten curve was fitted using GraphPad Prism. B, NADPH-dependent cytochrome c reductase activity of NR containing Moco (holo-NR), molybdopterin containing NR (MPT-NR), or Moco and MPT free NR (apo-NR) is shown. S.D. are derived from six independent measurements using two different protein preparations. C, time-dependent, holo-NR mediated oxidation of NADPH and reduction of cytochrome c is shown. D, shown is a schematic overview of NR electron flow. E, shown is a comparison of Km values and specific activities known for purified fungal NRs with kinetic properties of recombinant holo-NR. A holo-NR Km value for nitrate was calculated from Lineweaver-Burk plots of substrate saturation curves. a, according to Horner (52); b, according to Minagawa and Yoshimoto (32).
NADPH-dependent Cytochrome c Reducing Activity of Recombinant Neurospora NR
Other than holo-NR, MPT- and apo-NR have no nitrate reducing activity. However, all three NR species were purified with approximately equimolar amounts of heme (Fig. 2E). In the following we tested apo-NR and MPT-NR for their NADPH-dependent cytochrome c reducing activity, thus monitoring the electron flow from FAD to heme (Fig. 3D). As a control, the NADPH-dependent cytochrome c reducing activity of holo-NR was recorded. Correlating with its nitrate reducing activity, holo-NR also showed, as expected, NADPH-dependent cytochrome c reducing activity (Fig. 3, B and C). Likewise, apo- and MPT-NR showed cytochrome c reducing activity (Fig. 3B). However, when compared with holo-NR, apo-NR had a marginally lower (∼10% reduced) cytochrome c reducing activity, which correlates to its ∼15% reduced heme content (Fig. 2E). Consequently we reason that heme bound to apo- and MPT-NR is fully capable of transferring NADPH-derived electrons.
Oligomerization State of Recombinant Neurospora NR
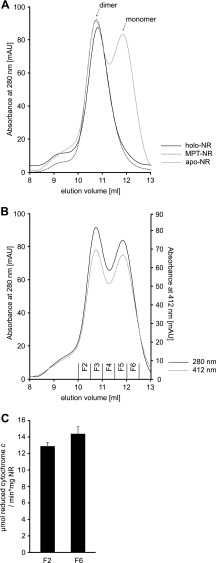
Next we wanted to characterize the oligomerization state of holo-, apo-, and MPT-NR. After a two-step affinity purification, SEC revealed that the NR protein pools purified from strains TP1000 and RK5206 contained dimeric NR exclusively (Fig. 4A). In contrast, protein preparations from RK5204 contained dimeric as well as monomeric NR, with both forms appearing in an ∼1:2 molar ratio (Fig. 4A). However, the ratio of dimeric to monomeric NR varied significantly from preparation to preparation. Next we asked whether or not heme is important for NR dimerization. Therefore, cytochrome c reductase activities of the main peak fractions of monomeric and dimeric apo-NR were determined and found to be invariant, thus showing that each fraction contains NR proteins with identical heme binding stoichiometries (Fig. 4, B and C). Therefore, the lack of Moco/MPT is solely responsible for monomer formation of apo-NR.
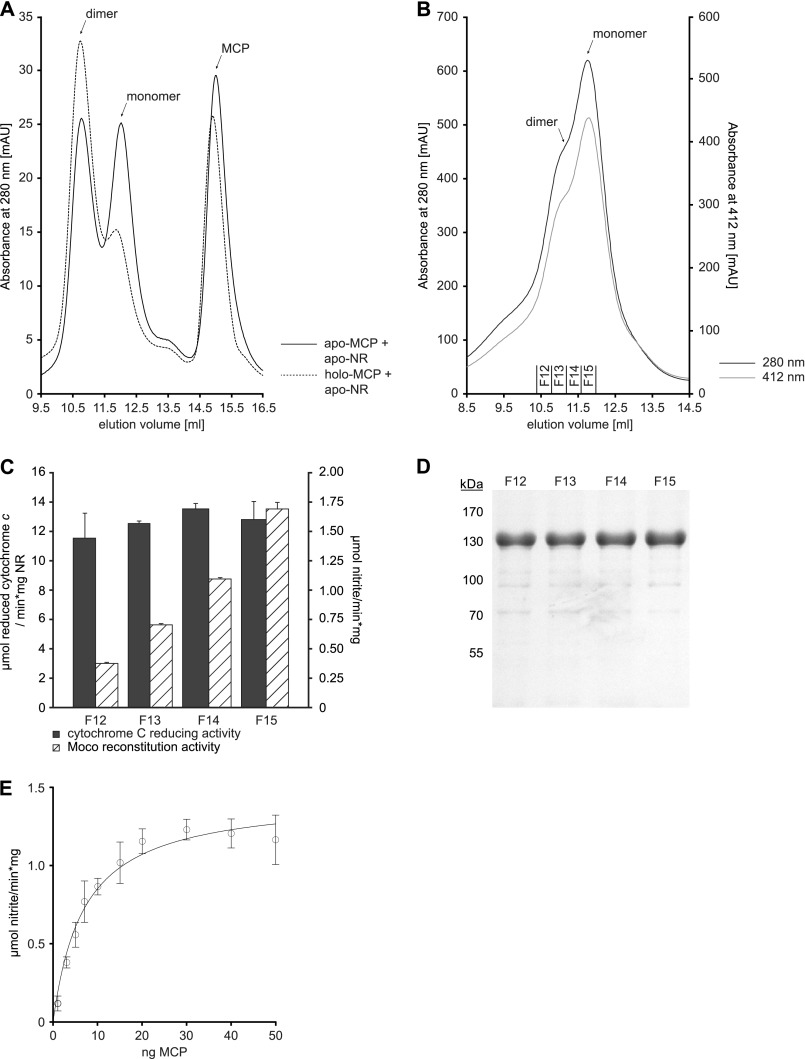
FIGURE 4.
Oligomerization state of recombinant NR. A, 200 μg of recombinant NR containing Moco (holo-NR) or molybdopterin (MPT-NR) or no Moco or MPT (apo-NR) were subjected to SEC. mAU, milliabsorbance units. B, shown is an elution profile of holo-NR as shown in A. In addition to absorption at 280 nm, the optical density at 412 nm is shown. Cytochrome c reducing activity was determined for fractions F2 and F6 as shown in C. Determination of cytochrome c reducing activity was carried out as described under “Experimental Procedures.”
Analysis of NR Protein Variants by Analytical Ultracentrifugation
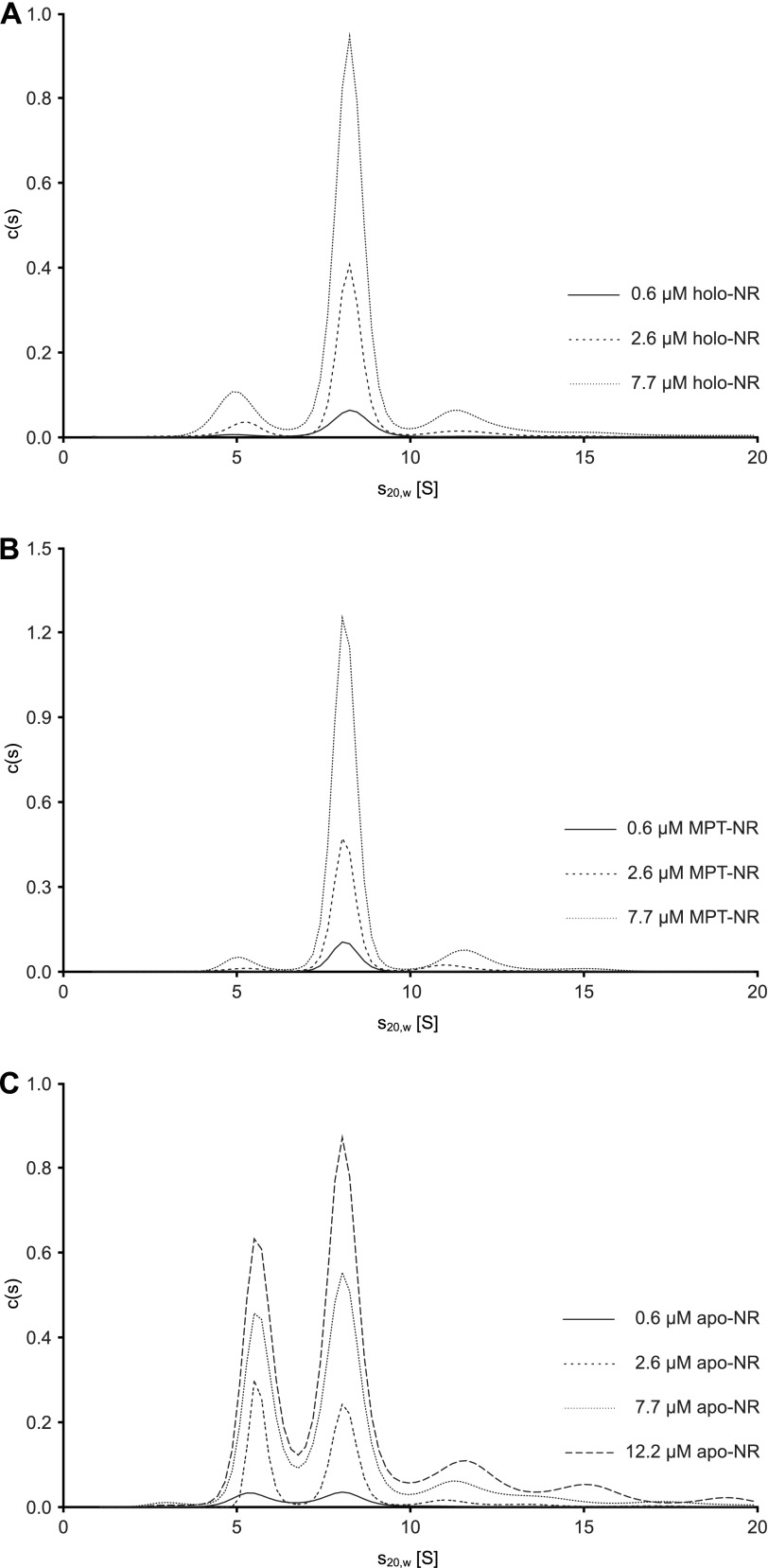
To further characterize the oligomerization state of the different NR variants, we examined the proteins in a concentration range from 0.6 to 7.7 μm by sedimentation velocity experiments in the analytical ultracentrifuge. Holo- and MPT-NR sedimented predominantly as a single species with an s20,w of 8.3 and 8.1 S, respectively, independently of the protein concentration used (Fig. 5, A and B). The continuous c(s) distribution model in the program SEDFIT (21) was used to determine the molar mass of the proteins from the diffusion broadening of the sedimenting boundary. At a concentration of 2.6 μm, molar masses of 220 and 213 kg/mol were obtained for holo- and MPT-NR, respectively. Because a molar mass of 114.5 kg/mol can be calculated from amino acid composition, both proteins exist predominantly as dimers in solution. This is consistent with the results for the native enzyme purified from N. crassa (34). For these dimers frictional ratios of 1.65 and 1.68 can be calculated from the sedimentation coefficients. Because for a globular hydrated protein a frictional ratio of 1.1–1.2 is expected (35), both proteins deviate substantially from the shape of a sphere with the holoenzyme being slightly more compact than the MPT form. All three NR variants show, in addition to the dimeric form, a species sedimenting with about 11–12 S, which might represent a small amount of tetrameric enzyme (Fig. 5). A tendency of NR to form tetramers without significant impact on functionality has been described previously (3). Furthermore, a small fraction of holo- and MPT-NR sedimented with an s20,w value about 5–6 S (Fig. 5, A and B). Because the NR preparations were not completely saturated with Moco/MPT (Fig. 2C), these fractions might correspond to monomeric enzyme devoid of these co-factors. Consistent with the SEC results, c(s) distributions of apo-NR, in contrast to those of holo- and MPT-NR, show two major species (Fig. 5C). A faster one sedimenting with essentially the same sedimentation coefficient as MPT-NR dimers and a slower one with an s20,w of about 5.6 S, corresponding to an asymmetrical monomer with a frictional ratio of 1.5.
FIGURE 5.
Characterization of different NR variants by analytical ultracentrifugation. Different concentrations of molybdenum cofactor (Moco) containing, molybdopterin (MPT) containing and Moco/MPT free (apo) nitrate reductase (NR) were analyzed in sedimentation velocity runs at 35,000 rpm and 20 °C. To achieve better comparability, all sedimentation coefficient distributions have been converted to 12-mm path lengths. Whereas holo- and MPT-NR exist predominantly as dimers in solution, in apoNR both monomers and dimers are clearly populated. Unexpectedly, the monomer/dimer ratio did not change significantly when protein concentration was varied by a factor of 20, implying that both oligomerization states are not in equilibrium.
To investigate at which concentration apo-NR dissociates completely into monomers, 0.6–12.2 μm apo-NR were diluted from a stock solution, incubated for at least 2 h at 20 °C, and analyzed in a sedimentation velocity experiment. Unexpectedly, the ratio between monomeric and dimeric apo-NR did not change significantly upon a 20-fold dilution (Fig. 5C). To rule out a very slow dissociation reaction, a similar experiment was performed after incubating the diluted protein for 24 h at 4 °C (data not shown). Similar to the results shown in Fig. 5C, the monomer/dimer ratio did not change significantly. Therefore, we conclude that the monomeric and dimeric forms of apo-NR are not in equilibrium.
Moco-dependent Dimerization of Apo-NR
Hitherto, all NR oligomerization and reconstitution studies were restricted to whole cell extracts of source organisms containing Moco, solely MPT, or neither of them. Therefore, it remained enigmatic whether transition of monomeric to active dimeric NR is solely dependent on the presence of Moco or is additionally also dependent on other factors (e.g. Moco supply proteins, chaperones, etc.). To show that Moco is the determining factor to promote NR dimerization, a fully defined in vitro system was established. It contained apo-NR and a Moco source. As the Moco source, the MCP from the green algae Chlamydomonas reinhardtii was chosen. Recombinant MCP contains up to 25% co-purified Moco, and moreover, Chlamydomonas MCP was shown to bind exclusively Moco but not the metal-free MPT (28). Co-incubation was carried out using equimolar amounts of apo-NR and Moco bound to MCP. After co-incubation, the reaction mixture was subjected to SEC (Fig. 6A), unveiling the Moco-dependent transition of monomeric to dimeric NR. As a control, Moco-free MCPs were incubated with apo-NR in parallel, revealing no influence of the carrier protein on NR dimerization (Fig. 6A). Therefore, Moco is sufficient to initiate NR dimer formation.
FIGURE 6.
Moco-dependent dimerization of NR. A, 100 μg of recombinant Moco-free NR (apo-NR) were co-incubated with the 2-fold stoichiometric excess of the MCP with bound Moco (holo-MCP) and Moco-free MCP (apo-MCP), respectively. After co-incubation, the reaction mixture was separated using SEC. B, 2 mg of apo-NR were subjected to SEC, and fractions F12-F15 were collected. Moco-dependent NR reconstitution activity and cytochrome c reducing activity were determined for these fractions as shown in C. For Moco-dependent NR reconstitution, 200 ng of holo-MCP were co-incubated with 500 ng of apo-NR, resulting in a 2.5 stoichiometric excess of MCP over NR. NADPH-dependent nitrate reduction and cytochrome c reduction was carried out as described under “Experimental Procedures.” D, shown is SDS-PAGE analysis of SEC fractions F12–F15. E, 400 ng of recombinant apo-NR were co-incubated for 3 h at room temperature with increasing amounts of recombinant holo-MCP in degassed reconstitution buffer, and Moco-dependent NR activity was recorded as described.
In Vitro Reconstitution of Apo-NR
Next we asked whether or not apo-NR gains functional activity upon co-incubation with MCP, thus demonstrating the transfer of physiologically active Moco to apo-NR. Therefore, various amounts of MCP were co-incubated with apo-NR. NR activity was measurable upon co-incubation with MCP and increased with the MCP amount used for co-incubation (Fig. 6E). Based upon these results, the optimal amount of MCP for apo-NR reconstitution was identified to be 2 molecules MCP per NR monomer. Because the in vitro reconstitution system contained both the monomeric and dimeric apo-NR, we also tested both apo-NR forms for their capability to take up Moco. Therefore, we again took advantage of SEC yielding apo-NR fractions containing mainly monomeric (fraction 15, abbreviated as F15) and dimeric (F12) NR (Fig. 6, B and D). Proteins from these and fractions F13 and F14 were subsequently used for co-incubation with MCP. The highest NR activity was measurable upon co-incubation of MCP with fraction F15, containing mainly monomeric apo-NR, whereas co-incubation of MCP with fraction F12 resulted in drastically reduced Moco reconstitution activity (Fig. 6C). However, fractions F13 and F14 likewise gave rise to NR activity. Therefore, we conclude that monomeric apo-NR is competent for uptake of Moco.
Modeling of the Structure of N. crassa NR
To identify the molecular basis for Moco-dependent NR dimerization, we mapped the amino acid sequence of the N-terminal domain of N. crassa NR (Moco and dimerization domain) (Fig. 1B) onto the structure of the Pichia angusta Moco and dimerization domain with bound Moco (PDB ID 2BIH) by using the one-to-one threading option of the Phyre2 modeling server (36) and analyzed the dimer interface of the model with PISA (protein interfaces, surfaces, and assemblies service at European Bioinformatics Institute) (37) (Fig. 7).
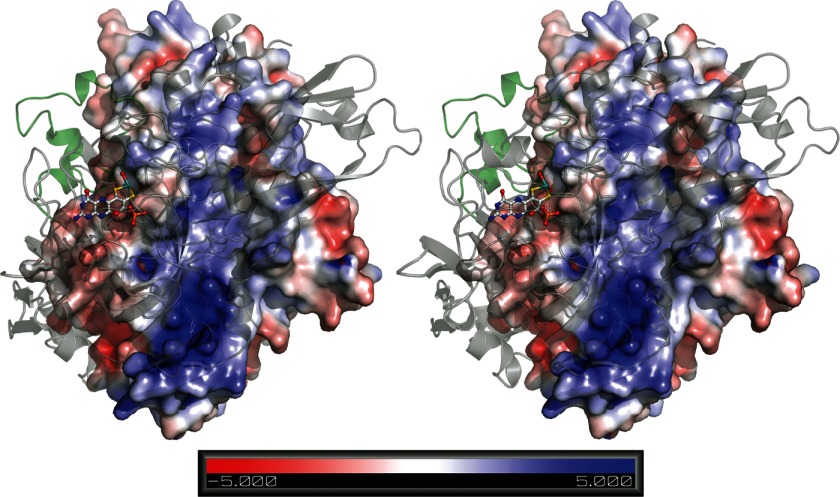
FIGURE 7.
Stereo view of the dimerization interface of a N. crassa NR model based on the structure of Moco-bound NR from P. angusta. The model was computed using the one-to-one threading option of the Phyre2 server. The left monomer is colored by the potential on its solvent-accessible surface as calculated by pdb2pqr (53) (scale ranging from −5 to 5 kTe−1). The right monomer is shown in schematic representation with the small all-helical domain colored in green. Moco is shown in ball-and-stick representation. The figure was prepared with PyMOL (54).
The largely hydrophobic N. crassa NR dimer interface has a buried area of about 2000 Å2 per monomer and is spotted with conserved charged (Arg-294, Lys-314, Glu-497, Arg-523, Glu-535) and polar amino acids (e.g. Val-381, Thr-384, Arg-406, Arg-454, Glu-456, Tyr-472), which form intermolecular salt bridges and hydrogen bonds. The hydrophobicity of the interface leads to a significant free energy gain upon dimerization and creates a strong bias toward the dimer.
Moco is no direct part of the interface but strengthens it by stabilizing a small all-helical domain, which spans amino acids Asn-372 to Ile-409 and contributes about 500 Å2 to the buried area. This domain was identified by a thorough B-factor analysis of 2BIH and the fact that it is the only part of the interface with an independent fold. According to our PISA analysis, it plays an essential role for complex formation, and its elimination renders the dimer unstable.
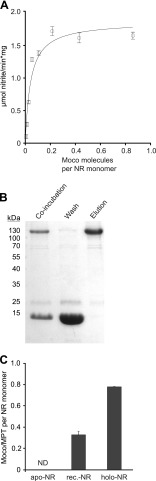
Quantification of Moco Transfer
Results from the in vitro reconstitution experiments demonstrate that monomeric apo-NR dimerizes upon Moco addition, yielding physiologically active NR. As a next step, the amount of Moco transferred to apo-NR had to be quantified. To address this question, we enriched reconstitution-competent, monomeric apo-NR by SEC (data not shown) and co-incubated it with various amounts of MCP. NR activity was measurable upon co-incubation with MCP and increased with the amount of MCP used for co-incubation (Fig. 8A).
FIGURE 8.
Reconstitution of monomeric Moco-free NR. A, NADPH-dependent NR activity of reconstituted monomeric NR after titration with Moco bound to MCP. After incubation of enriched monomeric apo-NR with various amounts of MCP, NADPH dependent NR activity was measured; data points were fit to a single binding site model. B, SDS-PAGE analysis upon separation of reconstituted NR and MCP is shown. A 4-fold excess of MCP over monomeric apo-NR was co-incubated for 14 h at 4 °C in reconstitution buffer. After co-incubation, the sample was diluted 1:1 with buffer containing 100 mm Tris-HCl, 150 mm NaCl, and 10% (v/v) glycerol and subjected to StrepTactin Macroprep resin. C, quantification of Moco bound to reconstituted NR (rec.-NR). Subsequent to purification of reconstituted NR, Moco/MPT content was quantified using HPLC-based FormA analysis and compared with apo- and holo-NR. ND = not definable.
Maximum NR activity was observed upon co-incubation of one monomer apo-NR with ∼0.35 molecules of Moco. In the following, monomeric apo-NR was co-incubated with the 4-fold stoichiometric excess of Moco bound to MCP. After co-incubation, strep-tagged NR was quantitatively separated from MCP by Strep-Tactin affinity chromatography as demonstrated by SDS-PAGE analysis (Fig. 8B). Subsequently the amount of Moco/MPT co-purified with NR was determined HPLC-based, thus revealing an average Moco/MPT binding stoichiometry of 0.34 ± 0.04 molecules per NR monomer (Fig. 8C). Therefore, in vitro reconstituted NR has a significantly lower Moco binding stoichiometry as recombinant holo-NR.
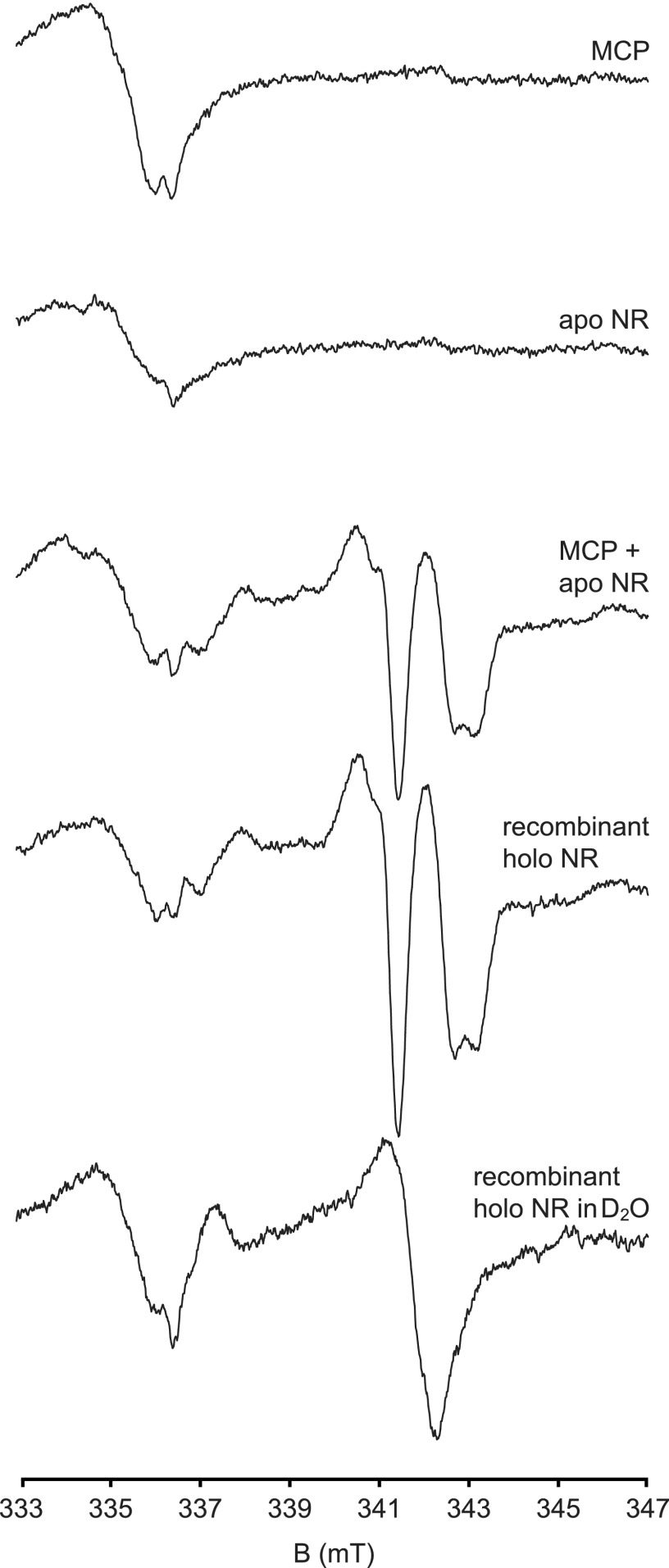
EPR Studies of Reconstituted NR
Because EPR spectroscopy is an extremely sensitive tool to probe the microenvironment of molybdenum, we employed this technique to detect Moco transfer from MCP to apo-NR. First, suitable conditions to reproducibly elicit Mo5+ EPR signals in N. crassa holo-NR had to be established. Early work on spinach NR used incubation with an excess of NADPH and nitrate to generate an axial Mo5+ EPR signal (38).
As judged from g values and proton superhyperfine parameters, this signal is characteristic for NR as EPR signals with almost identical parameters were detected in partially reduced Candida nitratophila (39) and Chlorella vulgaris NR (40). N. crassa holo-NR also showed the characteristic nearly axial Mo5+ EPR signals (Fig. 9, g = 1.998, 1.971, 1.969). Upon exchange of the solvent for D2O, the coupling of the single, solvent exchangeable proton vanished (AH = 1.2, 1.0, and 1.8 millitesla, respectively).
FIGURE 9.
EPR spectroscopy of reconstituted NR. EPR spectroscopy revealed that the Mo5+ microenvironment upon treatment with nitrate and NADPH of N. crassa Moco-free apo-NR reconstituted with holo-MCP is identical to N. crassa holo-NR. All samples were incubated for 25 s with 50 μm FAD, 20 mm nitrate, and 20 mm NADPH and shock-frozen in an ethanol/liquid nitrogen mixture. X band EPR spectra at 77 K (bottom two traces, 12 μm holo-NR) show a characteristic near axial signal (g = 1.998, 1.971, 1.969) that derives from Mo5+ coupled to a single, solvent exchangeable proton (AH = 1.2, 1.0 and 1.8 millitesla, respectively). A broad signal from the background and a FAD semiquinone radical signal are visible in the 333–337 millitesla (mT) range. The Mo5+ signal is lacking in purified holo-MCP (84 μm) or apo-NR (25 μm; top two traces) but appears upon incubation of holo-MCP (84 μm) with apo-NR (25 μm; (middle trace).
Our data confirm the ample information on N. crassa NR (41). Double integration showed that under these conditions 5–10% of the molybdenum could be trapped in the EPR active 5+ state. In the EPR spectral region around g = 1.97, at which the sharp proton-superhyperfine split NR Mo5+ signal is best detected, neither holo-MCP nor apo-NR exhibits EPR spectral features upon the addition of nitrate and NADPH. But after transfer of the Moco from holo-MCP to apo-NR, the presence of Moco with a microenvironment identical to holo-NR could unmistakenly be inferred from the characteristic EPR signal. Based on the amplitude in comparison to holo-NR, at least 34% of apo-NR could be activated by holo-MCP. This value compares favorably with the activity data as shown in Fig. 8.
The Sequence of Prosthetic Group Insertion into Neurospora NR
The timing of prosthetic group insertion into some prokaryotic molybdenum enzymes is known to be crucial for their maturation (42). To test for a mandatory sequence of prosthetic group insertion into NR, we first analyzed a NR protein defective in heme binding. Therefore, the NR variant H654A/H677A (13) was created by site-directed mutagenesis. The mutant protein was purified from the Moco-accumulating bacterial strain TP1000. CD spectroscopy showed no negative effects of the introduced mutations on protein secondary structure in comparison to the wild type protein (data not shown).
Upon chromogenic-based detection, no heme was detectable in H654A/H677A protein preparations, documenting the successful construction of a heme-free NR. The Moco/MPT content of NR H654A/H677A purified from TP1000 (determined HPLC-based) was found to be 0.81 molecules of Moco ± 0.04 per monomer (Fig. 10C), thus resembling the value quantified for the wild type NR. The molybdenum content as determined by ICP-MS revealed on average >80% saturation. Therefore, we conclude that exclusively Moco is bound to NR H654A/H677A. These findings closely resemble the Moco binding properties of the NR wild type protein.
FIGURE 10.
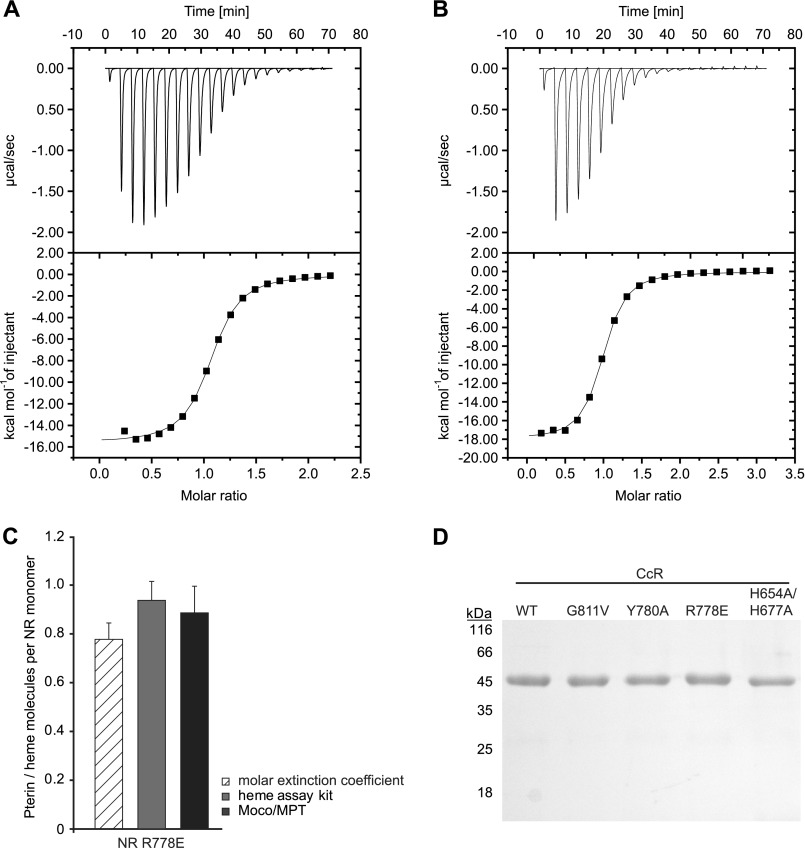
Analyses of FAD binding constant to CcR fragments using ITC. The figures show the titration data of CcR fragments (A, wild type; B, H654A/H677A) with FAD. The upper panel shows the heat released per second during the addition of FAD into the ITC cell containing CcR fragment. Lower panel, squares represent the integrated binding isotherm at each injection and normalization for the molar concentration. The solid line represents the non-linear fitting curve of the one set of sites binding model. C, stoichiometry of cofactors bound to NR variant R778E is shown. D, SDS-PAGE analysis of wild type CcR fragment and CcR variants used for FAD binding analysis by ITC.
Consequently we reason that Moco insertion into NR occurs independently from the presence of heme. In reverse, wild type-like heme binding was observed for Moco/MPT-free NR (Fig. 2E), thus documenting that NR heme binding is independent from Moco/MPT binding.
As a next step the influence of FAD on the insertion of Moco and heme, respectively, was determined. NR has a ferredoxin reductase-type FAD binding domain (43). Therefore, FAD binding mutants were constructed considering known FAD binding mutants of other enzymes with ferredoxin reductase-type FAD binding sites as well as FAD binding mutants of N. crassa and Arabidopsis thaliana NR (4, 44). Accordingly, N. crassa NR was mutated, yielding FAD binding mutants R778E, Y780A, and G811V, respectively. To show that these NR variants do not bind FAD, we quantified the FAD binding properties of an N-terminal-truncated NR variant consisting of the FAD and heme binding domain (thus forming the NR CcR fragment). Expression und purification of each CcR variant was successful, yielding the highest amounts of pure protein as documented by SDS-PAGE analysis (Fig. 10D). In the following we determined the effects of the introduced mutations in the FAD domain on the protein secondary structure using CD spectroscopy. No differences as compared with the CcR wild type CD-spectra were detectable (data not shown). Subsequently, the FAD binding properties of NR CcR fragments were quantified using ITC. By this, a FAD Kd value of 0.61 ± 0.05 μm was revealed for wild type CcR (Fig. 10A). However, ITC-based no FAD binding was detectable for CcR R778E and Y780A variants (data not shown). Next we quantified FAD binding properties of the heme-free CcR variant H654A/H677A, revealing an FAD Kd value of 0.52 ± 0.03 μm (Fig. 10B). Therefore, FAD binding to the NR CcR fragment is independent from heme binding.
In the inverse experiment we asked whether or not heme binding to NR is independent from FAD binding. Therefore, the amount of heme co-purified with NR variant R778E being unable to bind FAD was quantified. In comparison to wild type protein, NR R778E had a very similar heme content with a binding stoichiometry of 0.78 ± 0.07 molecules heme per monomer (extinction coefficient-based determination) or 0.94 ± 0.08 molecules heme per monomer (chromogenic-based determination) (Fig. 10C). Therefore, heme binding to NR is independent of FAD binding. Finally we wanted to know if Moco binding to NR is independent of FAD binding. Therefore, the amount of Moco bound to NR R778E upon purification from the Moco-accumulating E. coli strain TP1000 was quantified. NR variant R778E was co-purified with 0.89 ± 0.11 molecules of Moco per monomer (Fig. 10C), and the ICP-MS-based molybdenum quantification revealed >80% molybdenum saturation (data not shown). Therefore, NR R778E binds essentially the same amount of Moco as does the wild type protein, thus demonstrating that Moco binding to NR is independent from FAD binding.
DISCUSSION
In the past eukaryotic NR has been characterized intensively, revealing the multidomain character of this complex metalloenzyme. Although detailed knowledge is available for the holoenzyme (3–5, 28), little is known about its redox cofactor assembly. This gap of knowledge is mainly due to insufficient amounts of well defined stable apo-enzyme proteins available for analysis.
However, 40 years ago (45) a stable apo-NR was identified in the whole cell extract of the N. crassa nit-1 mutant. nit-1 extracts were found to be a valuable tool for Moco research (11) as the addition of physiologically active Moco results in formation of active NR, thus representing the only enzymatic assay system available for detecting physiologically active Moco.
Structural elucidation of the eukaryotic NR (46) revealed that Moco is deeply buried within the protein, at the end of the substrate funnel. This finding led to the questions of (i) how Moco is inserted, and (ii) considering that besides Moco, heme and FAD are also NR redox cofactors, is there a mandatory sequence for NR redox cofactor assembly? In this study we addressed both questions using the N. crassa NR as model enzyme. In a first set of experiments we characterized the Moco insertion process using recombinant Moco-free apo-NR and found that Moco is sufficient to induce NR dimer formation in this fully defined in vitro system. Consistently, cofactor-induced dimerization was already previously observed in the undefined whole cell extracts of the nit-1 system (45, 47, 48), but its sole dependence on Moco was not provable in this system.
How can Moco-induced NR dimer formation be explained? To address this question we built a structural model of N. crassa NR on the basis of our data set for the molybdenum domain of Pichia NR and analyzed its dimer interface. By this, we identified a small domain that provides ∼25% of the buried interface area.
We speculate that this domain is flexible and that it triggers dimerization upon Moco binding. In the absence of Moco it adopts a conformation unfit for dimerization, and as a result, the monomer becomes the abundant species. In the presence of Moco, the all-helical domain becomes part of the interface, and therefore, the equilibrium shifts toward the dimer. Once formed, the dimer is kinetically locked, thus precluding accessibility to the Moco binding site.
We postulate that the monomer is also kinetically locked until dimerization is triggered. Due to the abundance of Moco under natural conditions, NR is usually found to be Moco-loaded and, hence, dimeric. The emergence of both forms could result from NR overexpression in a Moco-free E. coli strain. It cannot further be ruled out that certain E. coli metabolites with weak affinity occupy the Moco binding site and hence trigger dimerization.
Analytical ultracentrifugation revealed that monomeric apo-NR is not in equilibrium with dimeric apo-NR, thus substantiating our model of NR dimerization. We were not able to verify the proposed model because as yet apo-NR crystallization failed. Furthermore, solely monomeric apo-NR is reconstitution competent, whereas dimeric apo-NR is not, which is best explained by missing accessibility of the Moco binding site.
Monomeric apo-NR binds substoichiometric Moco amounts (∼35% Moco saturation) and is, therefore, different from recombinant holo-NR that is co-purified with an approximately equimolar Moco amount (∼81% Moco saturation). EPR studies with reconstituted NR revealed no differences in the molybdenum micro environment as compared with the recombinant holoenzyme, documenting that NR active site formation is an autonomous process. How can we explain the substoichiometric Moco binding upon NR reconstitution? To ensure stoichiometric Moco binding to NR, a yet unidentified factor(s) (e.g. an enzyme-specific chaperone) may be required. However, hitherto molybdenum enzyme-specific chaperons are only known for the prokaryotic molybdenum enzymes xanthine dehydrogenase, trimethylamine oxidoreductase, and NR A (NarGHI) (42). Efficient maturation of these enzymes essentially depends on the cognate chaperones XDHC, TorD, and NarJ, respectively. However, Ilbert et al. (49) report that activation of TorA does not completely depend on the presence of the chaperone TorA in vitro but is significantly enhanced by it.
We speculate that likewise Moco insertion into NR is enhanced by a yet unknown factor(s). Because recombinant NR contains equimolar amounts of Moco, we expect these factors either to be evolutionary conserved, as is Moco biosynthesis (1), or to be functionally redundant in the expression host E. coli. Our defined in vitro reconstitution system will be a valuable tool for the identification and characterization of this factor(s).
Likewise, as compared with the undefined N. crassa nit-1 system, the in vitro system is a more suitable tool for the detection of physiologically active Moco. Hitherto, detection and quantification of physiologically active Moco was constrained, as the whole-cell nit-1 system also contains other proteins with a Moco binding site, either involved in Moco biosynthesis, Moco transfer, or Moco usage. These error sources are not present in the defined in vitro system containing solely Moco and apo-NR. Functional NR is a homo-dimeric enzyme consisting of two monomers that are built up from three distinct cofactor binding domains. This finding suggests that Moco insertion needs to be synchronized with the insertion of the remaining redox cofactors. For the bacterial molybdenum enzyme NR A (NarGHI) an obligatory sequence of metal cofactor insertion was shown (50), documenting its complex and intricate assembly process. However, we did not find any indication for a mandatory sequence of eukaryotic NR cofactor assembly, which allows us to conclude that the three NR redox cofactors are bound autonomously by their respective domains. Furthermore, we cannot exclude that NR cofactor assembly is intimately connected to protein translation, thus defining the sequence of cofactor insertion and preventing the formation of cofactor-free NR protein.
Acknowledgments
We thank Lidia Litz (from the Institute for Biophysical Chemistry, Hannover Medical School) and Anke Oelbermann (from the Department of Plant Biology, Braunschweig University of Technology) for excellent technical assistance and Dr. Joachim Greipel (from the Institute for Biophysical Chemistry, Hannover Medical School) for valuable discussions.
This work was supported by the Deutsche Forschungsgemeinschaft.
- NR
- nitrate reductase
- Moco
- molybdenum cofactor
- MPT
- molybdopterin
- CbR
- cytochrome b reducing fragment
- CcR
- cytochrome c reducing fragment
- ICP-MS
- inductively coupled plasma mass spectrometry
- ITC
- isothermal titration calorimetry
- SEC
- size-exclusion chromatography
- MCP
- Moco carrier protein.
REFERENCES
- 1. Mendel R. R., Kruse T. (2012) Cell biology of molybdenum in plants and humans. Biochim. Biophys. Acta 1823, 1568–1579 [DOI] [PubMed] [Google Scholar]
- 2. Schwarz G., Mendel R. R., Ribbe M. W. (2009) Molybdenum cofactors, enzymes, and pathways. Nature 460, 839–847 [DOI] [PubMed] [Google Scholar]
- 3. Campbell W. H. (2001) Structure and function of eukaryotic NAD(P)H:nitrate reductase. Cell. Mol. Life Sci. 58, 194–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Campbell W. H. (1999) Nitrate reductase structure, function, and regulation. Bridging the gap between biochemistry and physiology. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 277–303 [DOI] [PubMed] [Google Scholar]
- 5. Campbell W. H., Kinghorn K. R. (1990) Functional domains of assimilatory nitrate reductases and nitrite reductases. Trends Biochem. Sci. 15, 315–319 [DOI] [PubMed] [Google Scholar]
- 6. Lillo C., Meyer C., Lea U. S., Provan F., Oltedal S. (2004) Mechanism and importance of post-translational regulation of nitrate reductase. J. Exp. Bot. 55, 1275–1282 [DOI] [PubMed] [Google Scholar]
- 7. Nussaume L., Vincentz M., Meyer C., Boutin J. P., Caboche M. (1995) Post-transcriptional regulation of nitrate reductase by light is abolished by an N-terminal deletion. Plant Cell 7, 611–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poll A. M., Cherel I., Gonneau M., Leydecker M. T. (1991) Biochemical characterization of cnx nitrate reductase-deficient mutants from Nicotiana plumbaginifolia. Plant Sci. 76, 201–209 [Google Scholar]
- 9. McDonald D. W., Coddington A. (1974) Properties of the assimilatory nitrate reductase from Aspergillus nidulans. Eur. J. Biochem. 46, 169–178 [DOI] [PubMed] [Google Scholar]
- 10. De Vries S. E., Dirks R., Mendel R. R., Schaart J. G., Feenstra W. J. (1986) Biochemical characterization of some nitrate reductase deficient mutants of Nicotiana plumbaginifolia. Plant Science 44, 105–110 [Google Scholar]
- 11. Nason A., Lee K. Y., Pan S. S., Ketchum P. A., Lamberti A., DeVries J. (1971) In vitro formation of assimilatory reduced nicotinamide adenine dinucleotide phosphate:nitrate reductase from a Neurospora mutant and a component of molybdenum enzymes. Proc. Natl. Acad. Sci. U.S.A. 68, 3242–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mertens J. A., Shiraishi N., Campbell W. H. (2000) Recombinant expression of molybdenum reductase fragments of plant nitrate reductase at high levels in Pichia pastoris. Plant Physiol. 123, 743–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Okamoto P. M., Fu Y. H., Marzluf G. A. (1991) Nit-3, the structural gene of nitrate reductase in Neurospora crassa. Nucleotide sequence and regulation of mRNA synthesis and turnover. Mol. Gen. Genet. 227, 213–223 [DOI] [PubMed] [Google Scholar]
- 14. Galagan J. E., Calvo S. E., Borkovich K. A., Selker E. U., Read N. D., Jaffe D., FitzHugh W., Ma L. J., Smirnov S., Purcell S., Rehman B., Elkins T., Engels R., Wang S., Nielsen C. B., Butler J., Endrizzi M., Qui D., Ianakiev P., Bell-Pedersen D., Nelson M. A., Werner-Washburne M., Selitrennikoff C. P., Kinsey J. A., Braun E. L., Zelter A., Schulte U., Kothe G. O., Jedd G., Mewes W., Staben C., Marcotte E., Greenberg D., Roy A., Foley K., Naylor J., Stange-Thomann N., Barrett R., Gnerre S., Kamal M., Kamvysselis M., Mauceli E., Bielke C., Rudd S., Frishman D., Krystofova S., Rasmussen C., Metzenberg R. L., Perkins D. D., Kroken S., Cogoni C. (2003) The genome sequence of the filamentous fungus Neurospora crassa. Nature 422, 859–868 [DOI] [PubMed] [Google Scholar]
- 15. Borkovich K. A., Alex L. A., Yarden O., Freitag M., Turner G. E., Read N. D., Seiler S., Bell-Pedersen D., Paietta J., Plesofsky N., Plamann M., Goodrich-Tanrikulu M., Schulte U., Mannhaupt G., Nargang F. E., Radford A., Selitrennikoff C., Galagan J. E., Dunlap J. C., Loros J. J., Catcheside D., Inoue H., Aramayo R., Polymenis M., Selker E. U., Sachs M. S., Marzluf G. A., Paulsen I., Davis R., Ebbole D. J., Zelter A., Kalkman E. R., O'Rourke R., Bowring F., Yeadon J., Ishii C., Suzuki K., Sakai W., Pratt R. (2004) Lessons from the genome sequence of Neurospora crassa. Tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68, 1–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCluskey K. (2003) The Fungal Genetics Stock Center. From molds to molecules. Adv. Appl. Microbiol. 52, 245–262 [DOI] [PubMed] [Google Scholar]
- 17. Stewart V., MacGregor C. H. (1982) Nitrate reductase in Escherichia coli K-12. Involvement of chlC, chlE, and chlG loci. J. Bacteriol. 151, 788–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Palmer T., Santini C.-L., Iobbi-Nivol C., Eaves D. J., Boxer D. H., Giordano G. (1996) Involvement of the narJ and mob gene products in distinct steps in the biosynthesis of the molybdoenzyme nitrate reductase in Escherichia coli. Mol. Microbiol. 20, 875–884 [DOI] [PubMed] [Google Scholar]
- 19. Sreerama N., Woody R. W. (2004) Computation and analysis of protein circular dichroism spectra. Methods Enzymol. 383, 318–351 [DOI] [PubMed] [Google Scholar]
- 20. Zhao H., Ghirlando R., Piszczek G., Curth U., Brautigam C. A., Schuck P. (2013) Recorded scan times can limit the accuracy of sedimentation coefficients in analytical ultracentrifugation. Anal. Biochem., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Laue M. T., Shah B. D., Ridgeway T. M., Pelletier S. L. (1991) Analytical Ultracentrifugation in Biochemistry and Polymer Science, pp. 90–125, Cambridge, UK [Google Scholar]
- 23. Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwarz G., Boxer D. H., Mendel R. R. (1997) Molybdenum cofactor biosynthesis. The plant protein Cnx1 binds molybdopterin with high affinity. J. Biol. Chem. 272, 26811–26814 [DOI] [PubMed] [Google Scholar]
- 25. Redinbaugh M. G., Campbell W. H. (1985) Quaternary structure and composition of squash NADH:nitrate reductase. J. Biol. Chem. 260, 3380–3385 [PubMed] [Google Scholar]
- 26. Evans H. J, Nason A. (1952) The effect of reduced triphosphopyridine nucleotide on nitrate reduction by purified nitrate reductase. Arch. Biochem. Biophys. 39, 234–235 [DOI] [PubMed] [Google Scholar]
- 27. Garrett R. H., Nason A. (1967) Involvement of a B-type cytochrome in the assimilatory nitrate reductase of Neurospora crassa. Proc. Natl. Acad. Sci. U.S.A. 58, 1603–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fischer K., Llamas A., Tejada-Jimenez M., Schrader N., Kuper J., Ataya F. S., Galvan A., Mendel R. R., Fernandez E., Schwarz G. (2006) Function and structure of the molybdenum cofactor carrier protein from Chlamydomonas reinhardtii. J. Biol. Chem. 281, 30186–30194 [DOI] [PubMed] [Google Scholar]
- 29. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 30. Macheroux P. (1999) UV-visible spectroscopy as a tool to study flavoproteins. Methods Mol. Biol. 131, 1–7 [DOI] [PubMed] [Google Scholar]
- 31. Pan S. S., Nason A. (1978) Purification and characterization of homogeneous assimilatory reduced nicotinamide adenine dinucleotide phosphate-nitrate reductase from Neurospora crassa. Biochim. Biophys. Acta 523, 297–313 [DOI] [PubMed] [Google Scholar]
- 32. Minagawa N., Yoshimoto A. (1982) Purification and characterization of the assimilatory NADPH-nitrate reductase of Aspergillus nidulans. J. Biochem. 91, 761–774 [DOI] [PubMed] [Google Scholar]
- 33. Barbier G. G., Joshi R. C., Campbell E. R., Campbell W. H. (2004) Purification and biochemical characterization of simplified eukaryotic nitrate reductase expressed in Pichia pastoris. Protein Expr. Purif. 37, 61–71 [DOI] [PubMed] [Google Scholar]
- 34. Garrett R. H., Nason A. (1969) Further purification and properties of Neurospora nitrate reductase. J. Biol. Chem. 244, 2870–2882 [PubMed] [Google Scholar]
- 35. Lebowitz J., Lewis M. S., Schuck P. (2002) Modern analytical ultracentrifugation in protein science. A tutorial review. Protein Sci. 11, 2067–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web. A case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 37. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 38. Gutteridge S., Bray R. C., Notton B. A., Fido R. J., Hewitt E. J. (1983) Studies by electron paramagnetic resonance spectroscopy of the molybdenum centre of spinach (Spinacia oleracea) nitrate reductase. Biochem. J. 213, 137–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kay C. J., Barber M. J., Solomonson L. P., Kau D., Cannons A. C., Hipkin C. R. (1990) Spectroscopic, thermodynamic, and kinetic properties of Candida nitratophila nitrate reductase. Biochem. J. 272, 545–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kay C. J., Barber M. J. (1989) EPR and kinetic analysis of the interaction of halides and phosphate with nitrate reductase. Biochemistry 28, 5750–5758 [DOI] [PubMed] [Google Scholar]
- 41. Orme-Johnson W. H. (1980) Molybdenum and molybdenum-containing enzymes (Coughlan M. P., ed.) pp. 327–344, Pergamon Press, New York [Google Scholar]
- 42. Magalon A., Fedor J. G., Walburger A., Weiner J. H. (2011) Molybdenum enzymes in bacteria and their maturation. Coord. Chem. Rev. 255, 1159–1178 [Google Scholar]
- 43. Dym O., Eisenberg D. (2001) Sequence-structure analysis of FAD-containing proteins. Protein Sci. 10, 1712–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. González C., Brito N., Marzluf G. A. (1995) Functional analysis by site-directed mutagenesis of individual amino acid residues in the flavin domain of Neurospora crassa nitrate reductase. Mol. Gen. Genet. 249, 456–464 [DOI] [PubMed] [Google Scholar]
- 45. Nason A., Antoine A. D., Ketchum P. A., Frazier W. A., 3rd, Lee D. K. (1970) Formation of assimilatory nitrate reductase by in vitro inter-cistronic complementation in Neurospora crassa. Proc. Natl. Acad. Sci. U.S.A. 65, 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fischer K., Barbier G. G., Hecht H. J., Mendel R. R., Campbell W. H., Schwarz G. (2005) Structural basis of eukaryotic nitrate reduction. Crystal structures of the nitrate reductase active site. Plant Cell 17, 1167–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ketchum P. A., Cambier H. Y., Frazier W. A., 3rd, Madansky C. H., Nason A. (1970) In vitro assembly of Neurospora assimilatory nitrate reductase from protein subunits of a Neurospora mutant and the xanthine oxidizing or aldehyde oxidase systems of higher animals. Proc. Natl. Acad. Sci. U.S.A. 66, 1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ketchum P. A., Sevilla C. L. (1973) In vitro formation of nitrate reductase using extracts of the nitrate reductase mutant of Neurospora crassa, nit-1, and Rhodospirillum rubrum. J. Bacteriol. 116, 600–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ilbert M., Méjean V., Giudici-Orticoni M. T., Samama J. P., Iobbi-Nivol C. (2003) Involvement of a mate chaperone (TorD) in the maturation pathway of molybdoenzyme TorA. J. Biol. Chem. 278, 28787–28792 [DOI] [PubMed] [Google Scholar]
- 50. Lanciano P., Vergnes A., Grimaldi S., Guigliarelli B., Magalon A. (2007) Biogenesis of a respiratory complex is orchestrated by a single accessory protein. J. Biol. Chem. 282, 17468–17474 [DOI] [PubMed] [Google Scholar]
- 51. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X Version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 52. Horner R. D. (1983) Purification and comparison of nit-1 and wild-type NADPH:nitrate reductases of Neurospora crassa. Biochim. Biophys. Acta 744, 7–15 [Google Scholar]
- 53. Dolinsky T. J., Nielsen J. E., McCammon J. A., Baker N. A. (2004) PDB2PQR. An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schrödinger, LLC; The PyMOL Molecular Graphics System, Version 1.5.0.4. [Google Scholar]