The retinoblastoma protein gene RB-1 is mutated in one-third of human tumors. Hilgendorf et al. now identify a nonnuclear role for retinoblastoma protein (pRB) in apoptosis induction. A fraction of endogenous pRB was constitutively associated with mitochondria, and recombinant pRB was sufficient to trigger BAX-dependent apoptosis. A mutant that targets pRB to mitochondria retained the ability to promote apoptosis in vitro as well as block further tumor development in Rb−/−;p53−/− tumors. These results suggest a nontranscriptional, tumor-suppressive role for pRB in the direct activation of BAX and mitochondrial apoptosis.
Keywords: MOMP, apoptosis, cancer, pRB, retinoblastoma protein
Abstract
The retinoblastoma protein gene RB-1 is mutated in one-third of human tumors. Its protein product, pRB (retinoblastoma protein), functions as a transcriptional coregulator in many fundamental cellular processes. Here, we report a nonnuclear role for pRB in apoptosis induction via pRB's direct participation in mitochondrial apoptosis. We uncovered this activity by finding that pRB potentiated TNFα-induced apoptosis even when translation was blocked. This proapoptotic function was highly BAX-dependent, suggesting a role in mitochondrial apoptosis, and accordingly, a fraction of endogenous pRB constitutively associated with mitochondria. Remarkably, we found that recombinant pRB was sufficient to trigger the BAX-dependent permeabilization of mitochondria or liposomes in vitro. Moreover, pRB interacted with BAX in vivo and could directly bind and conformationally activate BAX in vitro. Finally, by targeting pRB specifically to mitochondria, we generated a mutant that lacked pRB's classic nuclear roles. This mito-tagged pRB retained the ability to promote apoptosis in response to TNFα and also additional apoptotic stimuli. Most importantly, induced expression of mito-tagged pRB in Rb−/−;p53−/− tumors was sufficient to block further tumor development. Together, these data establish a nontranscriptional role for pRB in direct activation of BAX and mitochondrial apoptosis in response to diverse stimuli, which is profoundly tumor-suppressive.
Regulation of retinoblastoma protein (pRB) is perturbed in most, if not all, cancers (Sherr and McCormick 2002). pRB functions in many cellular processes and is a key regulator of the cell cycle by interacting with and inhibiting E2F transcription factors (van den Heuvel and Dyson 2008). Upon mitogenic signaling, cdk/cylin complexes phosphorylate pRB, resulting in release of E2Fs and cell cycle progression (van den Heuvel and Dyson 2008). Notably, the two-thirds of human tumors that are RB-1 wild type typically carry mutations in upstream regulators of pRB (p16Inka, cyclinD, or cdk4) that promote cdk/cylin activation and thus pRB phosphorylation (Sherr and McCormick 2002). Since these mutations all inactivate pRB's anti-proliferative function, little attention has been paid to the status of RB-1 in considering tumor treatment. However, we note that pRB also functions as a transcriptional coregulator of differentiation, senescence, and apoptosis genes (Ianari et al. 2009; Calo et al. 2010; Gordon and Du 2011; Viatour and Sage 2011). In addition, a portion of the pRB protein exists in the cytoplasm (Jiao et al. 2006, 2008; Roth et al. 2009; Fulcher et al. 2010) and even at mitochondria (Ferecatu et al. 2009), but no known roles have been assigned to these species.
This current study concerns the role of pRB in the regulation of apoptosis. It is already well established that pRB can either promote or suppress apoptosis through both direct and indirect transcriptional mechanisms (Ianari et al. 2009; Gordon and Du 2011). The earliest evidence for the anti-apoptotic role of pRB emerged from the characterization of the Rb-null mouse, which exhibits increased levels of apoptosis in the nervous system, lens, and skeletal muscle (Jacks et al. 1992). Subsequent studies showed that this phenotype is mostly non-cell-autonomous, resulting from the deregulation of cell cycle genes and the consequent overproliferation of placental tissues that disrupts its normal architecture and vascularization and causes hypoxia in embryonic tissues (de Bruin et al. 2003; Wu et al. 2003; Wenzel et al. 2007). Loss of pRB in mouse embryonic fibroblasts (MEFs) results in increased sensitivity to genotoxic stress (Knudsen and Knudsen 2008). However, this is thought to be an indirect consequence of failure to prevent cell cycle entry in the absence of pRB as well as increased chromosomal instability (Knudsen et al. 2000; Bosco et al. 2004; Burkhart and Sage 2008; Manning and Dyson 2012). In contrast and more consistent with a tumor suppressor role, pRB can also act in a proapoptotic manner in highly proliferative cells (Knudsen et al. 1999; Araki et al. 2008; Ianari et al. 2009; Milet et al. 2010; Carnevale et al. 2012). In this context, pRB and also hyperphosphorylated pRB contribute directly to apoptosis by functioning in a transcriptionally active pRB:E2F1 complex that promotes expression of proapoptotic genes, such as caspase 7 and p73, in response to DNA damage (Ianari et al. 2009). Taken together, these studies suggest that the ability of pRB to promote or repress apoptosis, at least in response to genotoxic stress, may be dictated by the cellular context.
The proapoptotic role of pRB has been primarily investigated in the context of DNA damage. The findings in this current study followed from our analysis of pRB's apoptotic function in response to another apoptotic stimulus, TNFα. TNFα can promote apoptosis via both the extrinsic and mitochondrial/intrinsic pathways (Jin and El-Deiry 2005). The extrinsic pathway involves direct activation of the caspase cascade. In contrast, the intrinsic pathway depends on activation of the Bcl-2 protein family members BAX and BAK, which trigger mitochondrial outer membrane permeabilization (MOMP) and release of proapoptotic factors, such as cytochrome c, that lead to effector caspase activation (Jin and El-Deiry 2005; Brunelle and Letai 2009; Chipuk et al. 2010; Wyllie 2010; Martinou and Youle 2011). Notably, in addition to its proapoptotic response, TNFα also induces a NFκB-mediated, proinflammatory response, which inhibits apoptosis (Karin and Lin 2002). Thus, to study its apoptotic function, TNFα is commonly used in conjunction with a factor that abrogates the proinflammatory response, such as the proteasome inhibitor MG-132 or the translational inhibitor cycloheximide (CHX) (Traenckner et al. 1994; Karin and Lin 2002).
In this current study, we show that pRB enhances apoptosis in response to TNFα and can exert this effect in the presence of translation inhibition. This finding led us to discover a novel, nonnuclear function for the pRB protein. Specifically, pRB is associated with mitochondria and can induce MOMP by directly binding and activating BAX. By localizing pRB specifically to the mitochondria, we showed that this mitochondrial apoptosis function is capable of yielding potent tumor suppression in the absence of pRB's canonical nuclear functions. Importantly, mitochondrial pRB can respond to a wide variety of proapoptotic signals, suggesting that this represents a general and potent tumor-suppressive mechanism. Additionally, we found that cdk-phosphorylated pRB is present at the mitochondria, and pRB's mitochondrial apoptosis function remains intact in the presence of tumorigenic events, such as p16 inactivation, that promote pRB phosphorylation. Thus, we believe that it may be possible to exploit pRB's mitochondrial apoptosis role as a therapeutic treatment in the majority of human tumors that retain wild-type Rb-1.
Results
pRB is proapoptotic in response to TNFα treatment even in the presence of an inhibitor of translation
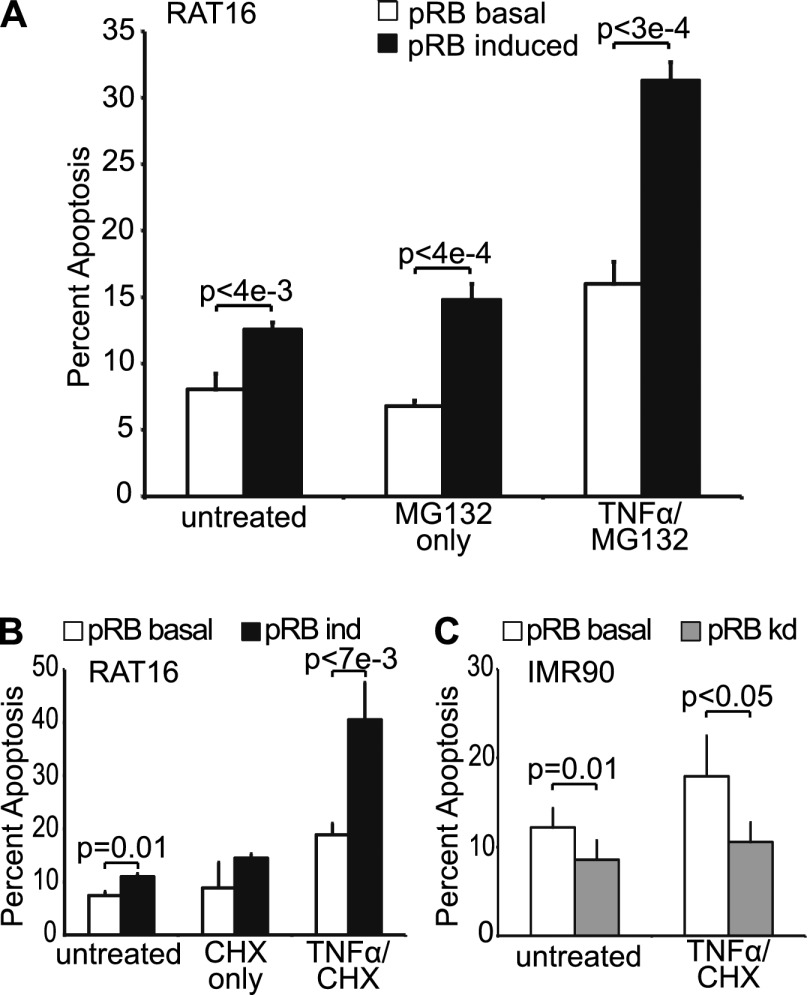
Our initial interest in exploring pRB's role in TNFα-induced apoptosis stemmed from prior reports that a phosphorylation site mutant version of pRB played a proapoptotic role in the TNFα response (Masselli and Wang 2006). We began our studies using a stable variant of an immortalized rat embryonic fibroblast cell line, RAT16, in which doxycycline withdrawal induces ectopic pRB expression. RAT16 cells with basal or induced pRB expression were treated with TNFα in concert with the proteasome inhibitor MG132 to block the TNFα-induced activation of NFκB and its proinflammatory response. We found that ectopic pRB expression significantly enhanced TNFα-induced apoptosis as measured by AnnexinV staining (Fig. 1A). Additionally, we note that pRB also caused a subtle increase in apoptosis even in the absence of TNFα treatment (Fig. 1A). We further confirmed these data by analysis of cleaved effector caspases 3 and 7 protein levels (Supplemental Fig. S1A). We wanted to confirm that this was not simply a consequence of pRB overexpression and thus also assessed the role of the endogenous pRB using knockdown cell lines. Notably, even partial knockdown of endogenous pRB in RAT16 cells was sufficient to significantly impair TNFα-induced apoptosis (Supplemental Fig. S1B) without any detectable disruption of cell cycle phasing (Supplemental Fig. S1C). Similar results were observed in a second cell line, human primary IMR90 fibroblasts, in which pRB knockdown also suppressed TNFα-induced apoptosis (Supplemental Fig. S1D). Thus, modulation of pRB levels by either overexpression or knockdown is sufficient to enhance or depress the apoptotic response to TNFα.
Figure 1.
pRB promotes TNFα-induced apoptosis in a transcription-independent manner. (A) RAT16 cells with and without induced expression of pRB for 24 h were treated with TNFα and MG132 for 48 h and analyzed for apoptosis by AnnexinV staining. Induced expression of pRB resulted in increased levels of apoptosis without TNFα treatment and greatly enhanced TNFα/MG132-induced apoptosis. (B) Induced expression of pRB for 24 h in RAT16 cells increased apoptosis resulting from 24 h of TNFα and CHX treatment. (C) Stable knockdown of pRB in IMR90 cells decreased apoptosis induced by 24 h of treatment with TNFα and CHX. (A–C) Graph bars represent the average of at least three independent experiments (±SD).
We anticipated that pRB's contribution to the proapoptotic TNFα response would reflect its ability to transcriptionally activate proapoptotic genes, as observed in our prior DNA damage studies (Ianari et al. 2009). However, we did not detect any significant changes in the levels of apoptotic, inflammatory, or autophagic mRNAs, including known pRB:E2F1 targets, in response to pRB induction in our RAT16-TNFα experiments (Supplemental Fig. S1E). This led us to consider that perhaps pRB might be acting independently of transcription. To explore this possibility, we took advantage of the fact that CHX can be used instead of MG132 to block activation of NFκB (Traenckner et al. 1994; Karin and Lin 2002). Since CHX acts to block translation, this would preclude any effects that required gene expression changes. Strikingly, induction of pRB in RAT16 cells was able to potentiate apoptosis in response to TNFα and CHX (Fig. 1B), with control experiments verifying translation inhibition by CHX (Supplemental Fig. S2). Moreover, pRB knockdown in IMR90s impaired TNFα/CHX-induced apoptosis (Fig. 1C). Thus, taken together, our data show that pRB synergizes with TNFα to promote apoptosis in both primary and immortalized human and rodent cell lines in the absence of translation. This strongly suggests that pRB can act through a previously unappreciated mechanism.
pRB activates mitochondrial apoptosis in a BAX-dependent manner
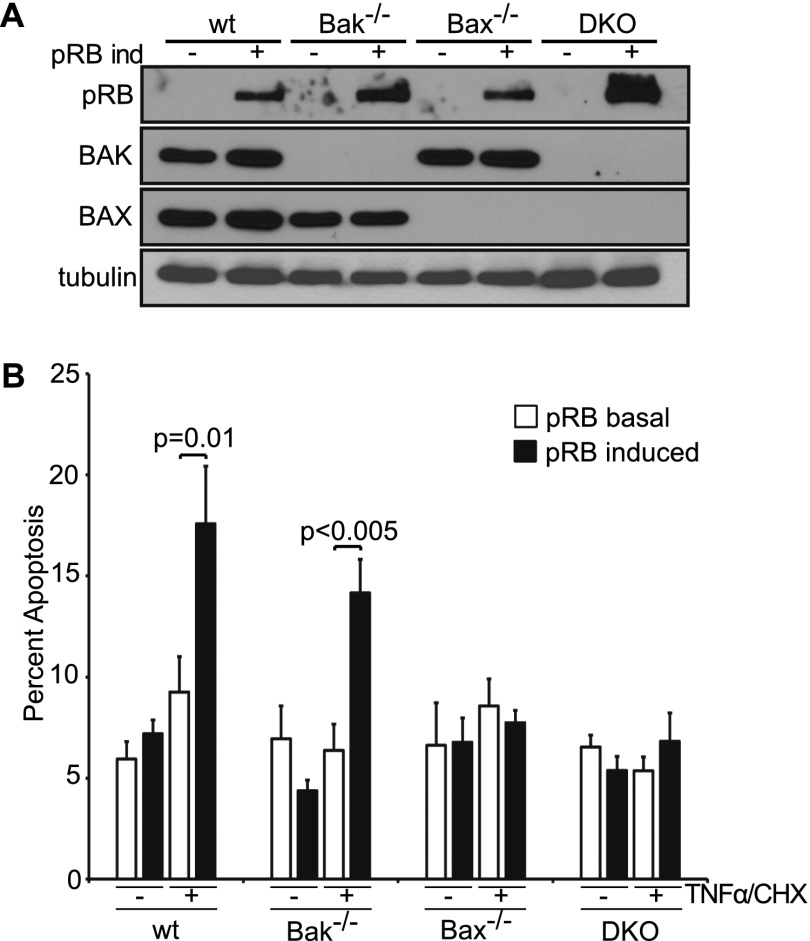
TNFα can induce apoptosis through both the extrinsic and mitochondrial pathways. To further pinpoint pRB's role, we exploited the fact that the mitochondrial pathway is highly dependent on BAX and/or BAK (Chipuk et al. 2010; Martinou and Youle 2011). Specifically, we generated stable pools of immortalized wild-type, Bak−/−, Bax−/−, or Bax−/−;Bak−/− MEFs that would allow for doxycycline-inducible pRB expression (Fig. 2A) and assayed for apoptosis in the absence and presence of TNFα/CHX treatment. To ensure that we were assaying the BAX/BAK dependence of any pRB effect rather than a general difference in the apoptotic potential of the various Bax/Bak genotypes, we conducted these experiments using treatment conditions that yielded minimal response to TNFα/CHX in the uninduced (basal pRB) cells (Fig. 2B). Remarkably, pRB induction enhanced TNFα-induced apoptosis in both wild-type (P = 0.01) and Bak−/− (P < 0.005) MEFs but had no effect in either Bax−/− or Bax−/−;Bak−/− MEFs (Fig. 2B). This analysis yielded two important conclusions. First, pRB exerts its effect on TNFα-induced apoptosis by acting specifically on the mitochondrial pathway. Second, pRB requires BAX, but not BAK, for this activity.
Figure 2.
pRB promotes mitochondrial apoptosis in a BAX-dependent manner. (A) pRB expression was induced for 24 h in stable pools of wild-type, Bak−/−, Bax−/−, and Bax−/−;Bak−/− immortalized MEFs by doxycycline addition and confirmed by Western blotting using antibodies against pRB, BAK, BAX, and tubulin. (B) Wild-type, Bak−/−, Bax−/−, and Bax−/−;Bak−/− immortalized MEFs with or without 24 h of pRB expression were left untreated or treated with TNFα and CHX for 10 h and analyzed for apoptosis by AnnexinV staining. Induction of pRB in wild-type and Bak−/−, but not Bax−/− or Bax−/−;Bak−/−, MEFs sensitized to TNFα/CHX-induced apoptosis. Each MEF variant was independently generated twice. Graph bars represent the average of three representative, independent experiments (±SD).
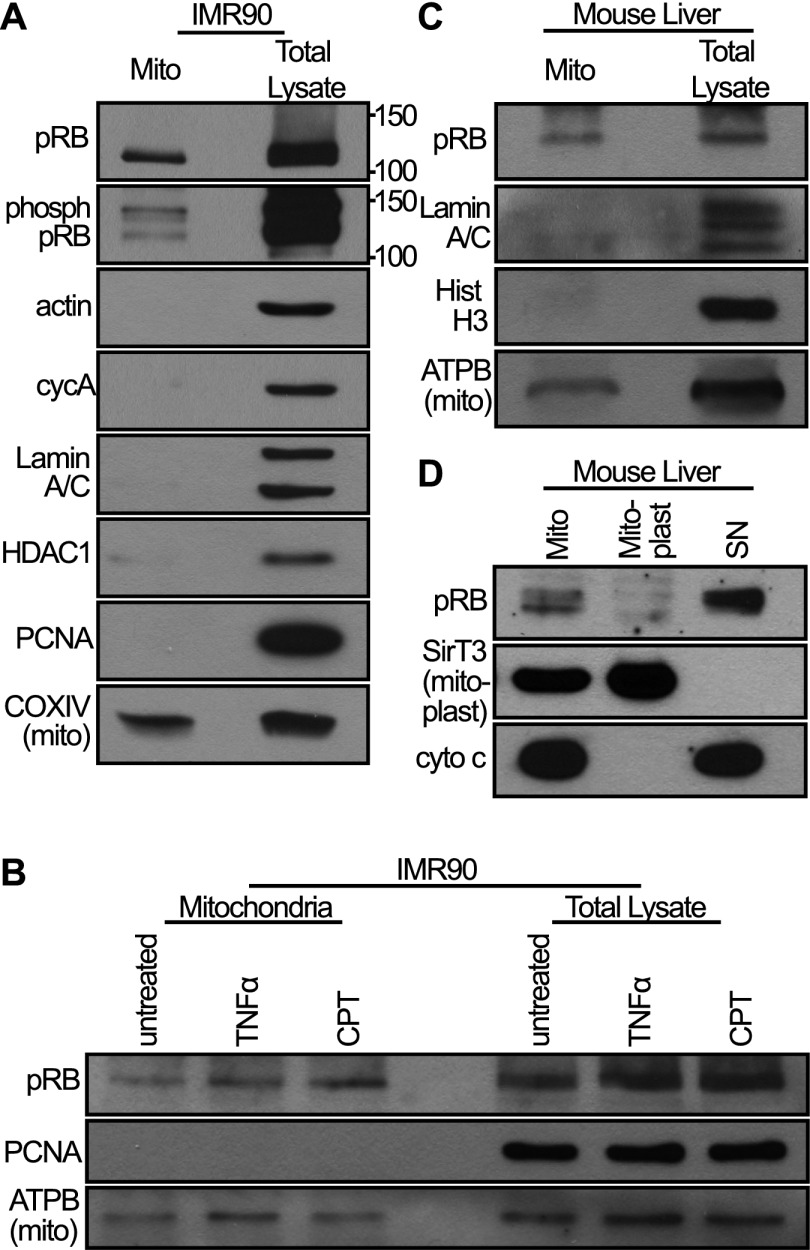
pRB is thought of primarily as a nuclear protein but also exists in the cytoplasm (Jiao et al. 2006, 2008; Roth et al. 2009; Fulcher et al. 2010). Indeed, pRB has even been reported at mitochondria (Ferecatu et al. 2009), albeit without any known function. Thus, we speculated that pRB might act directly at mitochondria. To validate and further explore the mitochondrial localization of pRB, we first fractionated IMR90 cells. We note that our goal was not to necessarily recover all mitochondria but rather to obtain a mitochondrial fraction free of nuclear and cytoplasmic contamination, as confirmed by Western blotting for various nuclear, cytoplasmic, and mitochondrial markers (Fig. 3). Intriguingly, we detected a portion of endogenous pRB in the isolated mitochondria and found that this exists even in the absence of treatment with TNFα or genotoxic agents (Fig. 3A,B). Furthermore, using phospho-specific pRB antibodies, we showed that this mitochondrial pRB includes the cdk-phosphorylated species (Fig. 3A). Based on the relative loading and Western blot signals, we estimate that ∼5% of the total cellular pRB is present in the mitochondrial fraction. Since we optimized the mitochondrial fractionation for purity, not completeness, this is likely a conservative estimate of the level of mitochondrial pRB.
Figure 3.
A fraction of pRB constitutively localizes to mitochondria. (A) IMR90 cell mitochondria were fractionated, and equal amounts (in micrograms) of mitochondrial fraction (mito) versus total lysate were analyzed by Western blotting using an antibody against pRB and a cocktail of antibodies against phosphorylated pRB. A fraction of pRB, including phospho-pRB, localizes to mitochondria. The purity of the mitochondrial fraction was verified using control nuclear and cytoplasmic markers. (B) Mitochondria of IMR90 cells treated with camptothecin (CPT; 5 h) or TNFα (24 h) were isolated and analyzed by Western blotting. The levels of mitochondrial pRB are unaffected by treatment with these drugs. (C) Mitochondria were isolated from mouse livers and analyzed by Western blotting. A fraction of pRB is present at mouse liver mitochondria. (D) Mouse liver mitochondria were subfractionated into mitoplast and nonmitoplast (SN). Mitochondrial pRB localizes outside the mitoplast. (A–D) Data are representative of at least three independent experiments.
To further validate the mitochondrial localization of pRB in vivo, we next examined mitochondria from mouse livers. Again, Western blotting showed that a fraction of pRB existed within the mitochondrial fraction (Fig. 3C). Finally, subfractionation of mouse liver mitochondria localized pRB outside of the mitoplast (inner mitochondrial membrane and enclosed matrix) (Fig. 3D), consistent with the notion that it is associated with the outer mitochondrial membrane and therefore coincident with the site of BAX/BAK action.
pRB directly activates Bax and induces cytochrome c release from isolated mitochondria
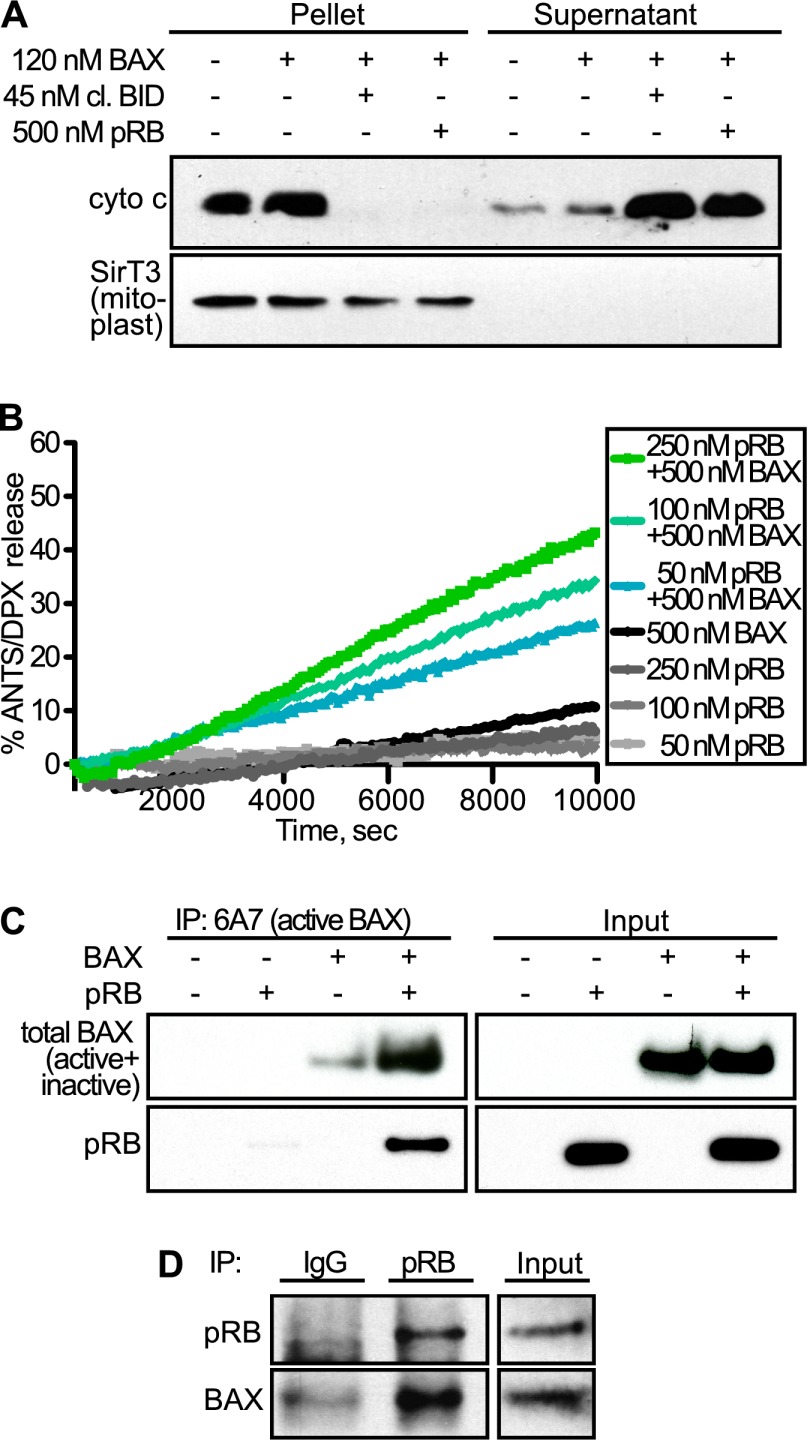
Having established that pRB is associated with mitochondria in vivo, we hypothesized that pRB might act to trigger MOMP. To test this, we performed in vitro cytochrome c release assays. First, we isolated mitochondria from wild-type mouse livers and added recombinant monomeric BAX because BAX is not associated with mitochondria in unstressed conditions (Walensky et al. 2006). As expected, addition of BAX alone did not result in cytochrome c release (Fig. 4A). Remarkably, coaddition of purified, baculovirus-expressed human pRB and monomeric BAX was sufficient to induce cytochrome c release (Fig. 4A). This was comparable with the effect of cleaved BID, a BH3-only member of the BCL-2 family that is a physiologic trigger of MOMP. Thus, recombinant pRB can induce MOMP in isolated mitochondria supplemented with monomeric BAX.
Figure 4.
pRB induces MOMP by directly activating BAX. (A) Mouse liver mitochondria were isolated; supplemented with recombinant, monomeric BAX; and incubated with and without recombinant cleaved BID (positive control) or baculovirus-expressed, recombinant human pRB. Cytochrome c release was assessed following incubation and centrifugation by Western blotting of pellet versus supernatant. Addition of either pRB or cleaved BID was sufficient to release cytochrome c into the supernatant. (B) ANTS/DPX-loaded liposomes were incubated with 50, 100, and 250 nM recombinant pRB in the presence or absence of recombinant, monomeric BAX. ANTS/DPX release was assessed over time. pRB yielded dose-dependent liposome permeabilization in a BAX-dependent manner. (C) In vitro binding assay using recombinant pRB and recombinant, monomeric BAX. Activated BAX was immunoprecipitated using an active conformation-specific BAX antibody (6A7), and binding was assessed by Western blotting using an antibody against total BAX (inactive + active) and pRB. When coincubated with inactive BAX, pRB bound to (bottom panels) and stimulated formation of (top panels) conformationally active BAX. (D) Coimmunoprecipitation experiment using IMR90 cells treated with TNFα/CHX for 3 h. Endogenous pRB was immunoprecipitated from cell extracts, and endogenous BAX association was assessed by Western blotting. pRB and BAX form an endogenous complex in vivo in TNFα/CHX-treated cells. (A–D) Data are representative of at least three independent experiments.
Next, we wanted to investigate the BAX/BAK dependence of this activity. As isolated mitochondria contain monomeric BAK, we repeated this assay using mitochondria isolated from Alb-crepos;Baxf/−;Bak−/− mouse livers (Walensky et al. 2006). Consistent with our cellular studies, pRB only induced cytochrome c release when these mitochondria were supplemented with BAX (Supplemental Fig. S3). Thus, pRB induces MOMP in both a dose- and BAX-dependent manner. To further explore this BAX specificity, we performed a liposome release assay in which freshly prepared ANTS/DPX-loaded liposomes were incubated with increasing concentrations of purified pRB in the absence or presence of recombinant, monomeric BAX (Fig. 4B). As expected, addition of either BAX or pRB (up to 250 nM) alone was insufficient to yield ANTS/DPX release. In contrast, upon coaddition, pRB was sufficient to trigger BAX-dependent liposome permeabilization in a dose- and time-dependent manner.
We wanted to further investigate pRB's ability to directly activate BAX. It is well established that BAX undergoes a conformational change upon activation that can be detected using an antibody (6A7) specific for the active conformation of BAX (Hsu and Youle 1997). Thus, we performed an in vitro binding assay in which recombinant pRB and/or monomeric BAX were incubated separately or together in the absence of mitochondria or liposomes. We then screened for BAX activation by immunoprecipitation with 6A7, and assayed pRB association by Western blotting (Fig. 4C). Notably, we found that addition of pRB promoted formation of the active BAX conformation (Fig. 4C, upper panels). Moreover, pRB coimmunoprecipitated with this active BAX species (Fig. 4C, lower panels). Thus, pRB can both associate with BAX and trigger its conformational activation. Since the liposome and coimmunoprecipitation assays were both conducted in the absence of any other proteins, we can conclude that pRB is sufficient to directly activate BAX and induce BAX-mediated membrane permeabilization.
We next sought to validate the pRB:BAX interaction in the in vivo context and with endogenous proteins. Endogenous interactions between BAX and proapoptotic proteins, including BH3-only proteins, are notoriously difficult to observe due to the dynamic, “hit and run” nature of these interactions (Eskes et al. 2000; Perez and White 2000; Walensky and Gavathiotis 2011). Thus, to enable detection of such transient interactions, we treated primary human IMR90 fibroblasts with TNFα/CHX and then cross-linked using the amine-reactive cross-linking agent DSP. Using this approach, we were able to detect BAX within immunoprecipitates of the endogenous pRB protein (Fig. 4D). Thus, taken together, our in vitro and in vivo experiments show that pRB can bind directly to BAX and induce it to adopt the active conformation that triggers MOMP and cytochrome c release.
The mitochondrial function of pRB induces apoptosis even in the absence of pRB's nuclear functions
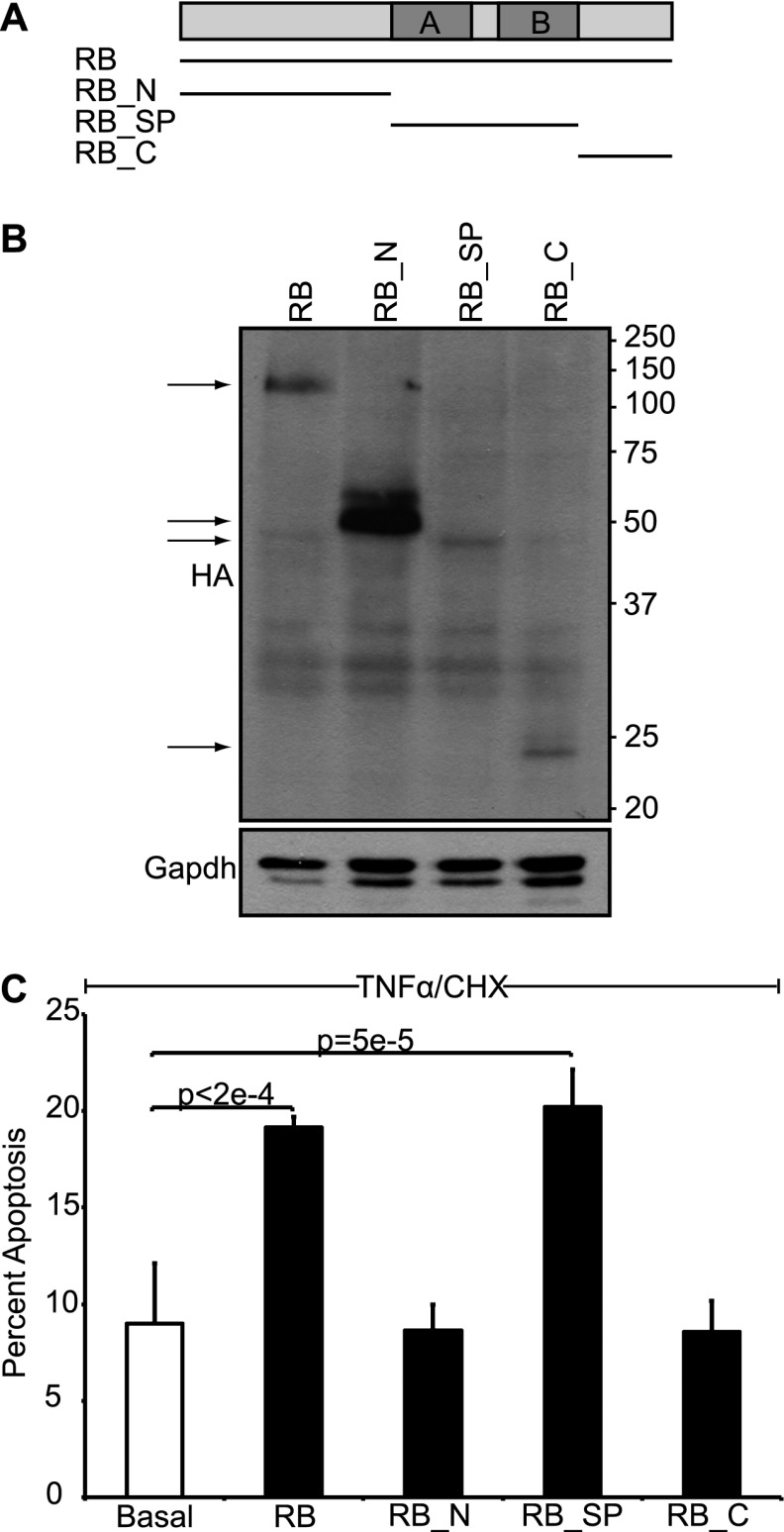
Our data have identified a mitochondrial function for pRB. We next wanted to address how this function contributes to pRB's tumor-suppressive activity. First, we asked which region of pRB is required for transcription-independent apoptosis. The tumor suppressor pRB is traditionally divided up into three domains (Fig. 5A): the N terminus (residues 1–372), the small pocket domain (residues 373–766), and the C terminus (residues 767–928). The small pocket is required for pRB's known biological functions, and most of the mutations found in human tumors disrupt this domain. We expressed the three domains (RB_N, RB_SP, and RB_C) in an inducible manner using stable RAT16 cell lines (Fig. 5A,B). Notably, expression of the small pocket of pRB, but not the N-terminal or C-terminal domains, promoted apoptosis in response to TNFα and CHX (Fig. 5C). Thus, pRB's ability to induce transcription-independent apoptosis mapped to the small pocket domain. Since this domain is essential for pRB's tumor-suppressive activity, these data are consistent with functional significance. Unfortunately, the small pocket is also required for most other pRB functions.
Figure 5.
The small pocket of pRB is sufficient to induce transcription-independent apoptosis. (A) Schematic of pRB domains: the N terminus (RB_N; residues 1–372), the small pocket domain (RB_SP; residues 373–766), and the C terminus (RB_C; residues 767–928). (B) Stable variants of RAT16 cells allowing for inducible expression of HA-tagged full-length pRB, RB_N, RB_SP, and RB_C by doxycycline withdrawal were generated, and expression was verified after 24 h by Western blotting using an HA-antibody. (C) RAT16 cells with and without expression of pRB, RB_N, RB_SP, and RB_C for 24 h were treated with TNFα and CHX for 24 h, and levels of apoptosis were assessed by AnnexinV staining. Induced expression of full-length pRB and RB_SP, but not RB_N or RB_C, sensitized to TNFα/CHX-induced apoptosis. (B,C) Each RAT16 variant was independently generated three times. Graph bars represent the average of three representative, independent experiments (±SD).
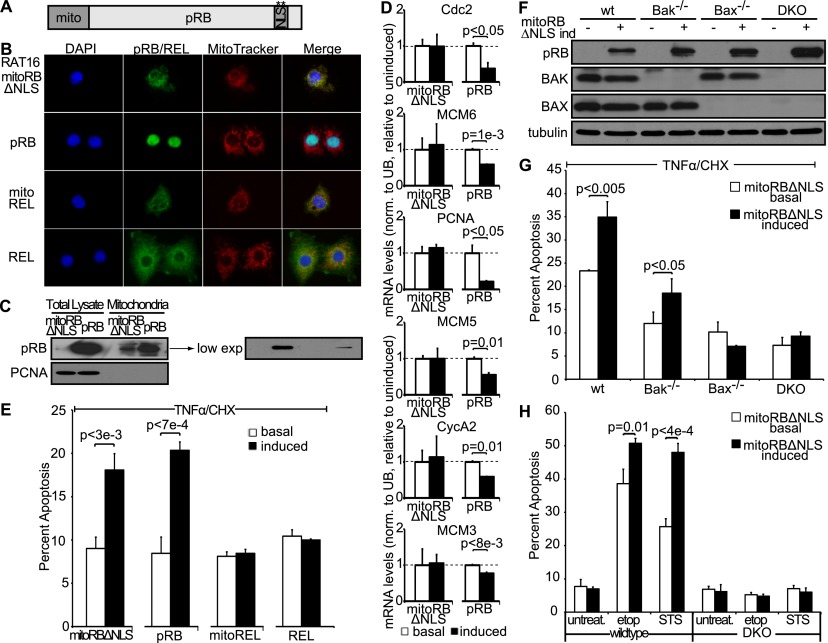
As an alternative approach, we sought to study the impact of the mitochondrial pRB activity in the absence of pRB's canonical nuclear functions by targeting pRB specifically to the mitochondria. Mutation of the NLS and direct targeting to the outside of the mitochondria using the transmembrane domain of Bcl2 proved insufficient for exclusive mitochondrial localization (data not shown), and instead we combined NLS mutation with fusion to the mitochondrial leader peptide of ornithine transcarbamylase (mitoRBΔNLS) (Fig. 6A). We note that this leader peptide targets to the matrix of mitochondria but has been used successfully to study protein function at the outer mitochondrial membrane, presumably by allowing resorting to other mitochondrial compartments (Marchenko et al. 2000). We confirmed that expression of mitoRBΔNLS did not cause general mitochondrial cytotoxicity, as judged by the absence of increased reactive oxygen species (ROS) production (data not shown). We also localized a control protein, REL, to mitochondria to further control for any nonspecific effect of mitochondrial targeting. Stable, inducible RAT16 cell lines were used to express mitoRBΔNLS, wild-type pRB, or mitoREL upon doxycycline withdrawal. Importantly, we confirmed specific targeting of mitoRBΔNLS and the control mitoREL protein to mitochondria (Fig. 6B,C). Consistent with its restricted localization, mitoRBΔNLS was unable to perform nuclear pRB functions, as judged by its inability to mediate the transcriptional repression of cell cycle genes that are known pRB targets (cdc2, mcm3, mcm5, mcm6, PCNA, and cyclinA2), in stark contrast to the wild-type pRB protein (Fig. 6D). Importantly, mitoRBΔNLS retained the ability to enhance TNFα/CHX-induced apoptosis (Fig. 6E). This was not a nonspecific effect of mitochondrial targeting, as the mitoREL control did not modulate apoptosis (Fig. 6E). We note that wild-type pRB was expressed at much higher levels than the mitoRBΔNLS protein even when considering only the mitochondrially localized fraction (Fig. 6C). Nevertheless, mitoRBΔNLS was almost as efficient as wild-type pRB at promoting TNFα/CHX-induced apoptosis. As an additional control against nonspecific mitochondrial cytotoxicity, we wanted to verify that mitoRBΔNLS works through the same directed mechanism as wild-type pRB. Thus, we assessed the BAX/BAK dependence of this protein by generating stable variants of immortalized wild-type, Bak−/−, Bax−/−, or Bax−/−;Bak−/− MEFs to allow doxycycline-inducible mitoRBΔNLS expression (Fig. 6F). Expression of mitoRBΔNLS in wild-type and Bak−/−, but not Bak−/− or Bax−/−;Bak−/−, MEFs enhanced TNFα-induced apoptosis (Fig. 6G). Thus, mitoRBΔNLS promotes apoptosis in a BAX-dependent manner, exactly like wild-type pRB. Targeting pRB directly to the mitochondria therefore successfully generates a separation-of-function mutant that is deficient for pRB's classic nuclear functions but retains the ability to induce apoptosis in a transcription-independent, BAX-dependent manner in response to TNFα.
Figure 6.
Mitochondria targeted pRB is deficient for pRB's nuclear function but induces apoptosis in response to various apoptotic stimuli. (A) Schematic of mitoRBΔNLS construct. pRB was targeted to mitochondria by fusion to the mitochondrial leader peptide of ornithine transcarbamylase and mutation of the NLS. (B) Stable variants of RAT16 cells allowing for inducible expression of mitoRBΔNLS, wild-type pRB, mitoREL, and wild-type REL were generated, and cellular localization following 24 h of induction was assessed by immunofluorescence using antibodies against pRB and REL. Mitochondria were visualized using MitoTracker. mitoRBΔNLS, and mitoREL localized to mitochondria. (C) Western blotting showing induced expression levels of mitoRBΔNLS and wild-type pRB at mitochondria and total lysate. mitoRBΔNLS expressed at lower levels than wild-type pRB even when considering the mitochondrial fraction. (D) pRB, but not mitoRBΔNLS, repressed E2F target genes cdc2, mcm3, mcm5, mcm6, PCNA, and cycA2 as measured by RT-qPCR and normalized to ubiquitin. Average of two independent experiments (±SD). (E) Induced expression of mitoRBΔNLS and wild-type pRB, but not mitoREL or REL, sensitized to apoptosis induced by 24 h of treatment with TNFα and CHX. (B–E) Each RAT16 variant was independently generated three times. Graph bars represent the average of three representative, independent experiments (±SD). (F) mitoRBΔNLS expression was induced by doxycycline addition for 24 h in stable variants of wild-type, Bak−/−, Bax−/−, and Bax−/−;Bak−/− immortalized MEFs and confirmed by Western blotting using antibodies against pRB, BAK, BAX, and tubulin. (G) Induction of mitoRBΔNLS in wild-type and Bak−/−, but not Bax−/− or Bax−/−;Bak−/−, MEFs sensitized to 10 h of treatment with TNFα and CHX. (H) Wild-type and Bax−/−;Bak−/− MEFs with and without induced expression of mitoRBΔNLS were treated with STS (1 μM) for 6 h or etoposide (25 μM) for 12 h. Induction of mitoRBΔNLS in wild-type, but not Bax−/−;Bak−/−, MEFs enhanced apoptosis in response to STS and etoposide. (F–H) Each MEF variant was independently generated twice. Graph bars represent the average of three representative, independent experiments (±SD).
In our earlier experiments, induction of wild-type pRB and RB_SP consistently yielded a small but significant increase in apoptosis even in the absence of TNFα treatment. This led us to hypothesize that mitochondrial pRB may function in a broader manner in apoptosis. We therefore evaluated the effect of mitoRBΔNLS expression on apoptosis induced by apoptotic factors other than TNFα. Immortalized wild-type or Bax−/−;Bak−/− MEFs with and without mitoRBΔNLS expression were treated with two drugs, etoposide and staurosporine (STS), that work via distinct mechanisms. Notably, mitoRBΔNLS expression enhanced both etoposide and STS-induced apoptosis (Fig. 6H). Thus, mitochondrial pRB can be activated in response to a variety of apoptotic stimuli, including both intrinsic and extrinsic stimuli. This argues that mitochondrial pRB function may contribute to pRB-induced apoptosis in various settings.
Mito targeted pRB suppresses tumor growth in vivo
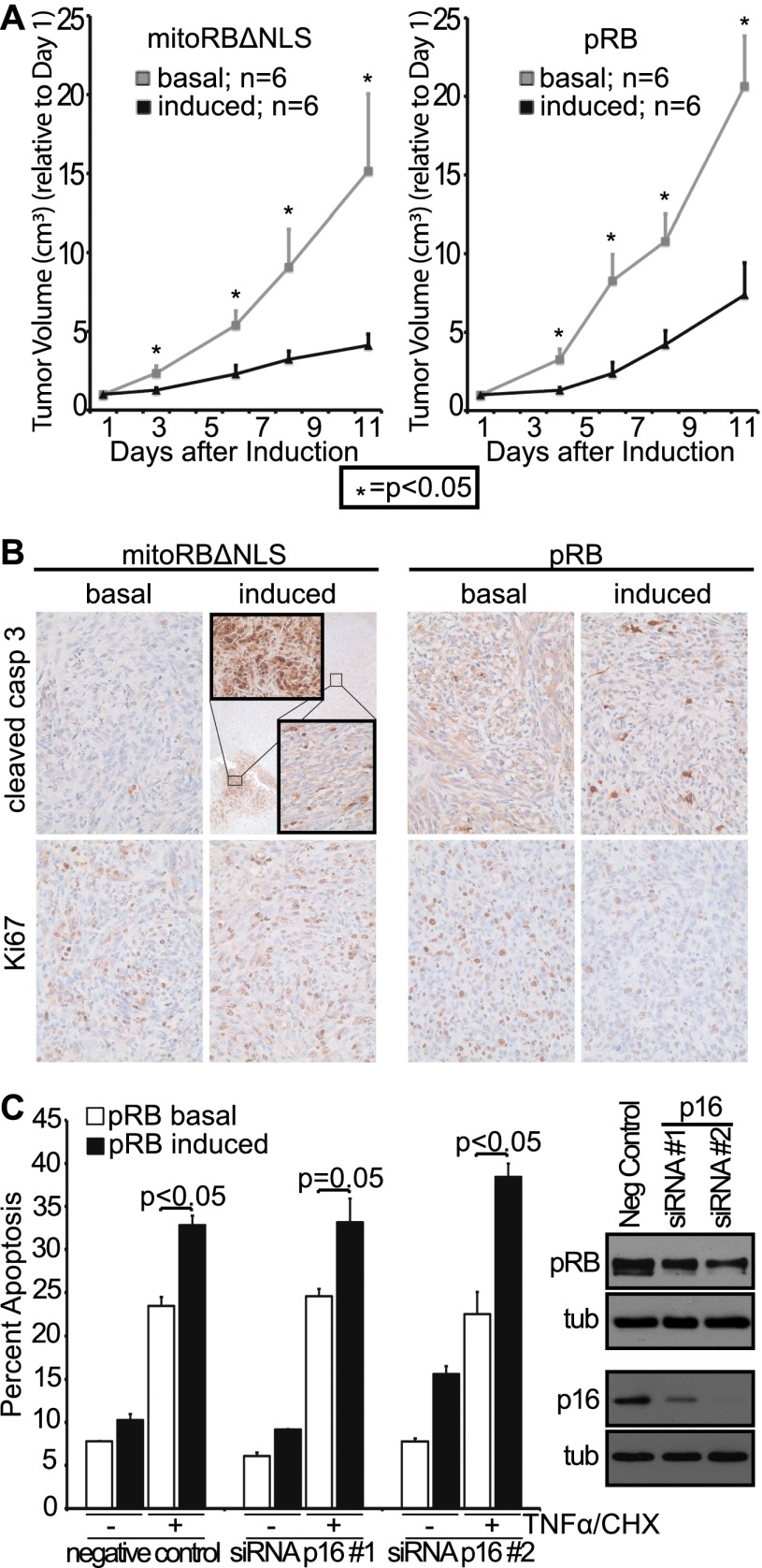
We next wanted to evaluate whether the mitochondrial function of pRB can contribute to tumor suppression in vivo. For this analysis, we used a murine Rb−/−;p53−/− osteosarcoma cell line (DKO-OS) that is capable of forming xenografts in mice. We generated stable variants of DKO-OS cells that would allow doxycycline-inducible expression of mitoRBΔNLS or control proteins (Supplemental Fig. S4A,B). Importantly, mitoRBΔNLS localized specifically to mitochondria in these tumor cells and, unlike mitoREL, promoted apoptosis in either the presence or absence of TNFα treatment (Supplemental Fig. S4C; data not shown). We injected DKO-OS-mitoRBΔNLS or DKO-OS-wtRb cells into immunocompromised mice (12 injections per cell line) and switched half of the animals to doxycycline once the tumor volume reached ∼0.05 cm3 to yield tumors without (referred to here as basal) or with mitoRBΔNLS and wild-type pRB induction. We note that these tumor cells were not treated with apoptotic stimuli but presumably were subjected to oncogenic stress. Control experiments (with parental DKO-OS cells) showed that the doxycycline treatment itself was not tumor-suppressive (Supplemental Fig. S4D). Strikingly, expression of wild-type pRB, versus mitoRBΔNLS, was similarly efficient in blocking further tumor expansion (Fig. 7A). Specifically, after 11 d, the tumor volume with basal expression was threefold greater than the tumor volume with either mitoRBΔNLS or pRB expression (Fig. 7A). Importantly, despite this equivalent impact on tumor growth, examination of tumor sections confirmed that there were clear differences in the cellular response to mitoRBΔNLS versus pRB expression (Fig. 7B). Ki67, a proliferation marker and pRB repression target, was down-regulated in tumors expressing wild-type pRB but completely unaffected by mitoRBΔNLS (Fig. 7B, bottom panels), in accordance with mitoRBΔNLS's inability to perform nuclear functions. Both pRB proteins promoted apoptosis, as judged by cleaved caspase 3 staining, but we observed qualitative differences in this response (Fig. 7B, top panels). Tumors with wild-type pRB showed a uniform increase in apoptosis (2.5-fold increase relative to basal, P < 0.01). Tumors with mitoRBΔNLS had regions comparable with the wild-type pRB response (2.3-fold increase relative to basal, P < 0.001) but also included smaller regions (Fig. 7B, inset box) that were profoundly apoptotic. Thus, wild-type pRB promotes both cell cycle arrest and apoptosis, while mitoRBΔNLS is solely apoptotic. Importantly, despite its restricted biological activity, mitoRBΔNLS was as efficient as, if not more efficient than, wild-type pRB at mediating tumor suppression in vivo.
Figure 7.
Mitochondria targeted pRB induces apoptosis in vivo and is tumor-suppressive. (A) Rb−/−;p53−/− osteosarcoma cell variants allowing for doxycycline-inducible expression of mitoRBΔNLS or wild-type pRB were injected into the flanks of nude mice (two injection sites per mouse; 12 injections per cell line variant). Once a tumor volume of ∼0.05 cm3 was reached, mitoRBΔNLS or wild-type pRB expression was induced in half the mice, and tumor volume was monitored for 11 d (n = 6 per condition, ±SEM). Induced expression of mitoRBΔNLS or wild-type pRB suppressed growth of xenografted tumors. Tumor volume normalized to day 1. (B, top) Induced expression of mitoRBΔNLS or wild-type pRB resulted in increased levels of apoptosis as measured by immunohistochemistry using a cleaved caspase 3 antibody. (Inset) Tumors expressing mitoRBΔNLS also contained small areas with very high levels of apoptosis. (Bottom) Induced expression of wild-type pRB, but not mitoRBΔNLS, decreased proliferation as measured by Ki67 staining. Images representative of six respective tumor sections. (C) Murine p16 was knocked down in immortalized wild-type MEF variants that allow for doxycycline-inducible expression of pRB. Phosphorylation status of pRB (as judged by mobility) and knockdown of p16 using two distinct siRNAs were confirmed by Western blotting. Induced expression of pRB enhances TNFα/CHX-induced apoptosis in the presence and absence of p16, and this activity is not inactivated by pRB phosphorylation. Bars indicate the average of two independent experiments (±SD).
These observations raise the possibility that the mitochondrial function of pRB could be employed as a tumor-suppressive mechanism even in p53-deficient cells, at least when re-expressed in Rb-null cells. This is very intriguing because approximately two-thirds of human tumors are wild type for the Rb-1 gene but instead carry alterations in upstream regulators of pRB (p16Ink4A, cyclin D, or cdk4) that promote its cdk phosphorylation. Thus, we wanted to determine whether pRB can mediate its mitochondrial apoptosis function in the presence of such tumorigenic changes. This question was further spurred by our finding that cdk-phosphorylated pRB is observed in the mitochondrial fractions (Fig. 3A). To address the influence of pRB phosphorylation on mitochondrial pRB function, we used two distinct siRNAs to knock down p16 in our immortalized MEF populations that allow for doxycycline-inducible expression of pRB. We confirmed that these siRNAs yielded near-complete p16 knockdown and promoted hyperphosphorylation of the doxycycline-induced pRB (Fig. 7C). Importantly, pRB enhanced TNFα/CHX-induced apoptosis with similar efficiency in MEFs with p16 knockdown versus those transfected with a negative control duplex (Fig. 7C). These data strongly suggest that cdk-phosphorylated pRB is competent to induce mitochondrial apoptosis and that this function will persist in the majority of human tumors that contain wild-type, constitutively phosphorylated pRB.
Discussion
We have known for more than two decades that pRB is localized in both the nucleus and cytoplasm. However, prior studies have largely focused on pRB's nuclear functions, particularly its widespread transcriptional roles. To our knowledge, no biological function has been assigned to cytoplasmic pRB. In fact, it has been suggested that this species is essentially inactive, and its existence simply reflects the loss of nuclear tethering of cdk-phosphorylated pRB (Mittnacht and Weinberg 1991; Templeton 1992). By following the interplay between pRB and TNFα-induced apoptosis, we uncovered clear evidence for a biological role for pRB at mitochondria in various cellular settings, including normal, immortalized, and tumor cells. Initially, we found that pRB not only enhanced TNFα-induced apoptosis, but could do so even in the presence of CHX. This argued that pRB could act in a novel manner to promote apoptosis. We then discovered that this pRB function was BAX-dependent, indicating cross-talk to the mitochondrial/intrinsic apoptotic pathway. Importantly, additional observations argue that pRB plays a direct and broadly applicable role in this pathway. First, a fraction of endogenous pRB, including some cdk-phosphorylated pRB, is constitutively localized on the outside of mitochondria. Second, recombinant pRB is able to induce MOMP and liposome permeabilization in vitro. Third, pRB can directly bind and activate BAX in vitro in the absence of any other proteins, and we confirm an interaction between the endogenous pRB and BAX proteins in vivo. Fourth, targeting pRB to mitochondria generates a separation-of-function mutant that is deficient for pRB's nuclear functions but able to induce apoptosis in response to various stimuli, including TNFα, etoposide, STS, and, presumably (since it is active in tumor cells), oncogenic stress. Finally and most importantly, this mitochondrially tethered pRB is sufficient to suppress tumorigenesis. Given these findings, we believe that endogenous, mitochondrial pRB acts nontranscriptionally and in a broadly engaged manner to promote apoptosis by activating BAX directly and inducing MOMP. To our knowledge, this is the first reported nontranscriptional and nonnuclear function for pRB.
Of course, we are not arguing that pRB is essential for mitochondrial apoptosis; this process is consistently impaired, but not ablated, by pRB deficiency. Instead, we conclude that pRB is one of a growing list of proteins that are able to modulate the activity of core apoptotic regulators. We suspect that pRB acts at the mitochondria to fine-tune the apoptotic threshold because its effects are dose-dependent (both in vitro and with overexpression/knockdown in vivo), it is constitutively localized to mitochondria, and it can potentiate many different proapoptotic signals. pRB's proapoptotic role is highly reproducible, but the fold change in our cell studies could be judged as relatively modest (often twofold to threefold). However, we note that these experiments sample only one snapshot in time. Indeed, in the context of the in vivo xenograft experiments, mitoRBΔNLS yielded the same modest increase in the frequency of apoptotic cells within tumor sections (twofold to threefold), but we can now see the cumulative effects of pRB action, and it is profoundly tumor-suppressive.
As described above, our data provide some insight into the underlying mechanism of mitochondrial pRB action; it requires BAX, but not BAK, both in vivo and in vitro, and pRB can directly bind to and activate formerly inactive monomeric BAX. Clearly, additional questions remain regarding this activity. The first concerns the precise nature of the pRB–BAX interaction. Given pRB's preference for BAX over BAK, we conclude that the pRB-binding site either is specific to BAX, and not BAK, or represents a shared domain that is somehow masked in the BAK protein. We note that we recently identified a novel and unique activation site on the BAX protein (Gavathiotis et al. 2008, 2010) and thus are interested to learn whether this might mediate pRB binding. In the case of pRB, we showed here that the small pocket domain is both necessary and sufficient for transcription-independent apoptosis and thus likely contains sequences essential for BAX binding. This is gratifying because the small pocket is essential for pRB's tumor-suppressive activity. However, this region is still relatively large and mediates interactions with many other known pRB-associated proteins. Thus, additional analysis will be required to further define the critical BAX-interacting sequences and determine how this might compete with the binding of other pRB targets. The second key question concerns the mechanisms by which proapoptotic signals trigger mitochondrial pRB to activate BAX. One obvious candidate is post-translational modifications. Our data show that cdk-phosphorylated pRB is present at mitochondria and capable of promoting mitochondrial apoptosis, based on the retention of mitochondrial pRB activity in p16 knockdown MEFs. However, we have yet to understand whether phosphorylation by cdks (or other kinases) is simply permissive for, or actively enables, pRB's mitochondrial apoptosis role. We also note that it has been previously reported that TNFα treatment induces cleavage of pRB at the C terminus, releasing a 5-kDa fragment (Tan et al. 1997; Huang et al. 2007). We were intrigued by the possibility that this cleavage might be responsible for the activation of mitochondrial pRB function by TNFα, presumably by exposing/activating the required small pocket domain. However, our exploratory analyses of both precleaved (i.e., Δ 5 kDa) and uncleavable forms of pRB in stable inducible cell lines were inconsistent with the notion that cleavage is necessary and sufficient for pRB activation (data not shown). Thus, it remains an open question how proapoptotic signals, including TNFα, trigger the mitochondrial pRB response.
Regardless of the remaining mechanistic questions, our findings considerably expand our appreciation of pRB's role in apoptosis. It is already clear that pRB can either suppress apoptosis by enforcing cell cycle arrest or promote DNA damage-induced apoptosis by transcriptionally coactivating proapoptotic genes (Ianari et al. 2009). Here, we show that in addition to these transcriptional mechanisms, pRB can also promote apoptosis directly at mitochondria. We believe that pRB's overall proapoptotic function is likely a result of the combined effect of nuclear and mitochondrial pRB. The relative extent to which these two functions contribute to apoptosis is likely context-specific. However, our mito-tagged pRB experiments clearly showed that the mitochondrial function of pRB can contribute to apoptosis in response to a broad range of stimuli, including the oncogenic context of tumor cells. Most importantly, our in vivo xenograft studies establish the tumor-suppressive potential of mitochondrial pRB in the absence of classic, nuclear pRB functions. Given these observations, we conclude that mitochondrial apoptosis represents a novel and bona fide mechanism of tumor suppression for pRB. This adds to a growing list of ways in which pRB has the potential to be tumor-suppressive, including its classic cell cycle function (Burkhart and Sage 2008); transcriptional coregulation of apoptosis, autophagy, and metabolic genes (Tracy et al. 2007; Blanchet et al. 2011; Ciavarra and Zacksenhaus 2011; Viatour and Sage 2011; Takahashi et al. 2012); and ability to control fate commitment by modulating the transcriptional activity of core differentiation regulators (Calo et al. 2010; Viatour and Sage 2011). The extent to which each of these mechanisms of pRB action contributes to overall tumor suppression remains to be fully elucidated. The ability to localize pRB specifically to the mitochondria allowed us to study pRB's mitochondrial role in the absence of all other known pRB functions. Remarkably, at least in this context, this mito-specific pRB was as efficient as, if not more efficient than, wild-type pRB at suppressing tumorigenesis. This unequivocally establishes the potential potency of pRB's mitochondrial apoptosis function. However, it does not disavow the potential contribution of other pRB functions. It remains an open question whether the myriad roles of pRB collaborate consistently in tumor suppression or whether, in specific contexts, one or more functions have more physiological relevance than others. It is also interesting to note that the multiple functions of pRB are highly reminiscent of those of the p53 tumor suppressor. p53 was also initially linked to cell cycle arrest and then shown to play a central role in promoting apoptosis both through the transcriptional activation of proapoptotic genes and by directly inducing MOMP at the mitochondria in a transcription-independent manner (Mihara et al. 2003; Chipuk et al. 2004; Leu et al. 2004; Vousden and Prives 2009; Speidel 2010). Our data now suggest that the direct promotion of mitochondrial apoptosis is a general mechanism of tumor suppression.
Finally, we believe that our findings have significant implications for therapeutic treatment in the approximately two-thirds of human tumors that retain wild-type pRB but instead carry mutations that promote cdk/cyclin activation and pRB phosphorylation (Sherr and McCormick 2002). Historically, little attention has been paid to Rb-1 status in chemotherapeutic response because the absence of pRB or the presence of phospho-pRB similarly inactivates pRB-mediated G1 arrest. However, we previously found that phospho-pRB can activate transcription of proapoptotic genes (Ianari et al. 2009). In this current study, we show that the endogenous mitochondrial pRB includes the cdk-phosphorylated form and retains its proapoptotic role in highly proliferative tumor cells and after inactivation of the cdk inhibitor p16. Moreover, both the transcriptional and the mitochondrial proapoptotic functions of pRB occur independent of p53. Thus, we believe that it should be possible to develop chemotherapeutic strategies for the majority of human tumors that retain wild-type RB-1, which engage phospho-pRB and promote apoptosis through both transcriptional and mitochondrial mechanisms.
Materials and methods
Cell culture and drug treatment
RAT16 and IMR90 cells were grown in MEM with Earle's salts and 10% FBS, penicillin–streptomycin, L-glutamine, sodium pyruvate, and NEAA. All other cell lines were grown in DMEM with 10% FBS and penicillin–streptomycin. For TNFα treatments, 50 ng/mL recombinant mouse TNFα (Sigma) was used with 0.1 μM MG132 (Calbiochem) for 48 h or 0.5 μg/mL CHX (Sigma) for 24 h unless noted otherwise. MEFs were treated with TNFα/CHX for 10 h, 1 μM STS (Sigma) for 6 h, or 25 μM etoposide (Sigma) for 12 h. TET System-approved FBS (Clontech) and 1 μg/mL doxycycline (Clontech) were used for inducible cell lines unless noted otherwise. Overexpression was induced for 24 h prior to TNFα treatment. Doxycycline was used at 20 μg/mL and 15 μg/mL in wild-type, Bax−/−, and Bak−/− MEFs to induce expression of pRB and mitoRBΔNLS, respectively. Doxycycline (5 μg/mL) was used in Bax−/−;Bak−/− MEFs to adjust for expression levels.
FACS apoptosis analysis
Cell suspensions were stained with AnnexinV-FITC or APC (Becton Dickinson) and propidium iodide (Sigma) or 7AAD (Becton Dickinson). Total apoptotic cells were assessed by gating for AnnexinV-positive using a FACScan or FACSCalibur system (Becton Dickinson). Similar trends were observed for all experiments when only early apoptotic (AnnexinV+;PI/7AAD−) cells were considered (data not shown). For cell cycle profiling, cell suspensions were processed as previously described (Ianari et al. 2009) and analyzed by FACScan and FlowJo.
Plasmid and stable cell line generation
Human pRB knockdown was performed as described previously (Chicas et al. 2010), and the rodent Rb target sequence was TATAATGGAATCAAACTCCTC, and the Luc control was GAGCTCCCGTGAATTGGAATCC. siRNAs (10 μM) (IDT, murine p16) were transfected into MEFs using RNAiMax (Invitrogen) according to the manufacturer's recommendations. The lentiviral vector pCW22 (tet-on) was used for expression studies. Since RAT16 cells already contained tTA (tet-off), the rtTA was removed by AgeI and XmaI digestion. mitoRBΔNLS was generated by amplifying the mitochondrial leader peptide of ornithine transcarbamylase (CAAAAGCGCTATGCTGTTTAATCTGAGGA and GGAAGGCGCCTGCACTTTATTTTGTAG) and human RB (CTATGGCGCCCAAAACCCCCC and GATTTTAATTAATCATTTCTCTTCCTTGT). The NLS was mutated using GCAAAACTAAGCTTTGATATTGAAGG and TATCAAAGCTTAGTTTTGCCAGTGG, followed by HindIII digestion. mitoREL was generated by amplifying the mitochondrial leader peptide of ornithine transcarbamylase (CAAAGTTAACATGCTGTTTAATCTGAGGA and GTTTCCGGAGGCCTGCACTTTATTTTGTAG) and human REL (tGGCCTCCGGAGCGTATAACCC and CAAATTAATTAACTTATACTTGAAAAAATTCATATG). pCW22 3HA was generated using 3HA peptide (IDT) and ligated to RB_N (CAAAGTCGACATGCCGCCCAAAAC and GATTTTAATTAATCAGTGTGGAGGAATTACATTCACCT), RB_SP (CAAAGTCGACACTCCAGTTAGGA and GATTTTAATTAATCATGTTTTCAGTCTCTGCATG), and RB_C (CAAAGTCGACAATATTTTGCAGTATGC and GATTTTAATTAATCATTTCTCTTCCTTGT). Cells infected with the inducible construct underwent blasticidin (Invitrogen) selection. All RAT16 variants were independently generated three times, and all MEF variants were independently generated twice.
Mitochondrial fractionations
IMR90 mitochondria were fractionated as follows: Cells were resuspended in cold buffer A (250 mM sucrose, 20 mM HEPES at pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, protease inhibitors [Roche]) and homogenized using 20 strokes in a 0.25-in cylinder cell homogenizer (H&Y Enterprises), 0.1558-in ball. Nuclei and unlysed cells were removed by low-speed centrifugation and mitochondria, and 10% of the initial cell suspension (for total lysate) was lysed using RIPA buffer (0.5% sodium deoxycholate, 50 mM Tris HCl at pH 7.6, 1% NP40, 0.1% SDS,140 mM NaCl, 5 mM EDTA, 100 mM NaF, 2 mM NaPPi, protease inhibitors) and quantified using BCA protein assay reagent (Pierce).
Mitochondria were isolated from livers of 2- to 6-mo-old mice maintained on a mixed C57Bl/6x129sv background as follows: Livers were minced in buffer A (0.3 M Mannitol, 10 mM HEPES/K at pH 7.4, 0.1% BSA, 0.2 mM EDTA/Na at pH 8.0) and homogenized using three strokes in a Teflon dounce homogenizer. A small fraction was removed for total lysate. Nuclei and unlysed cells were removed from the remaining supernatant by low-speed centrifugation, and mitochondria were washed in buffer B (0.3 M Mannitol, 10 mM HEPES/K at pH 7.4, 0.1% BSA). For whole-mitochondria analysis, mitochondria and total lysate were lysed in RIPA buffer. For subfractionation, the mitochondrial pellet was resuspended at 2 mg/mL in hypotonic buffer (10 mM KCl, 2 mM HEPES/K at pH 7.9), incubated for 20 min on ice, and centrifuged at 14000 rpm. The pellet was washed twice with wash buffer (150 mM KCl, 2 mM HEPES/K at pH 7.9), and all three supernatants combined yielded the nonmitoplast fraction. The mitoplast was resuspended in hypotonic/wash buffer, and equal fractions were analyzed by SDS-PAGE and Western blotting. Mitochondria were isolated from livers of Alb-crepos;Baxf/−;Bak−/− mice as reported previously (Walensky et al. 2006).
In vitro cytochrome c release assay
The assay was performed as previously described (Chipuk et al. 2005). Briefly, wild-type mitochondria were resuspended in mitochondrial isolation buffer (200 mM Mannitol, 68 mM sucrose, 10 mM HEPES/K at pH 7.4, 100 mM KCl, 1 mM EDTA, 1 mM EGTA, 0.1% BSA) to a concentration of 1 μg/μL and incubated with pRB (Sigma), GST-Bax (Sigma), or cl. BID (R&D Systems) for 1 h at 37°C. DKO mitochondria were resuspended in experimental buffer (125 mM KCl, 10 mM Tris-MOPS at pH 7.4, 5 mM glutamate, 2.5 mM malate, 1 mM KPO4, 10 μM EGTA-Tris at pH 7.4) to a concentration of 1.5 mg/mL, and incubated with monomeric BAX (purified as previously described; Gavathiotis et al. 2008) and pRB (ProteinOne) for 45 min at room temperature. Samples were centrifuged at 5500g, and pellets versus supernatants were analyzed by SDS-PAGE and immunoblotting.
Liposomal release assay
The liposomal release assay was performed as described previously (Lovell et al. 2008; LaBelle et al. 2012). Briefly, large unilamellar vesicles (LUVs) were generated from a lipid mixture of 48% phosphatidylcholine, 28% phosphatidylethanolamine, 10% phosphatidylinositol, 10% dioleoyl phosphatidylserine, and 4% tetraoleoyl cardiolipin as chloroform stocks (Avanti Polar Lipids). The lipid mixture was dried in glass test tubes under nitrogen gas and then under vacuum for 15 h. The fluorescent dye ANTS (6.3 mg) and the quencher DPX (19.1 mg) were added to 1 mg of dry lipid film, and the mixture resuspended in assay buffer (200 mM KCl, 1 mM MgCl2, 10 mM HEPES at pH 7.0). After five freeze–thaw cycles, the lipid mixtures were extruded through a 100-nm nucleopore polycarbonate membrane (Whatman) using a miniextruder (Avanti). Liposomes were isolated by gravity flow SEC using a cross-linked Sepharose CL-2B column (Sigma Aldrich). LUVs (5 μL) were treated with the indicated concentrations of BAX and pRB in 384-well format (Corning) in a total reaction volume of 30 μL. After time-course fluorescence measurement on a Tecan Infinite M1000 spectrophotometer (excitation 355 nm, emission 520 nm), Triton X-100 was added to a final concentration of 0.2% (v/v) to determine maximal release.
Immunoprecipitation
For the in vitro binding assay, recombinant, monomeric BAX (1 μM) and pRB (1 μM) were mixed in 10 μL of TBS (50 mM Tris, 150 mM NaCl) and preincubated for 30 min at room temperature. The samples were diluted with 1% BSA in TBS to 80 μL and incubated with pre-equilibrated Protein A/G-agarose beads (Santa Cruz Biotechnology) and 5 μL 6A7 antibody (Santa Cruz Biotechnology, sc-23959) with rotation for 1 h at room temperature. Beads were collected and washed three times with 0.5 mL of 1% (w/v) BSA/TBS buffer.
For the endogenous interaction study, IMR90 cells were treated with 50 ng/mL TNFα and 0.5 μg/mL CHX for 3 h total and cross-linked with 1 mg/mL DSP (Pierce) for 1 h. Proteins were extracted using an NP40-based buffer (50 mM HEPES at pH 7.9, 10% glycerol, 150 mM NaCl, 1% NP40, 1 mM NaF, 10 mM B-glycerophosphate, protease inhibitors). The following antibodies were used: pRB (Cell Signaling, 9309) and normal rabbit IgG (Santa Cruz Biotechnology).
Western blotting
Samples were loaded in SDS lysis buffer (8% SDS, 250 mM TrisHCl at pH 6.6, 40% glycerol, 5% 2-mercaptoethanol, bromophenol blue), separated by SDS-PAGE, transferred to a nitrocellulose membrane, and blocked in 5% nonfat milk. The following antibodies were used in 2.5% nonfat milk: human pRB (Cell Signaling, 9309), rodent pRB (Becton Dickinson, 554136), phospho-pRB (Cell Signaling, 2181, 9301, 9307, and 9308), procaspase 7 (Cell Signaling, 9492), cleaved caspase 7 (Cell Signaling, 9491), procaspase 3 (Cell Signaling, 9662), cleaved caspase 3 (Cell Signaling, 9661), actin (Santa Cruz Biotechnology, SC1616 HRP), tubulin (Sigma, T9026), BAX (Cell Signaling, 2772; Santa Cruz Biotechnology, sc-493), BAK (Cell Signaling, 3814), cyclin A (Santa Cruz Biotechnology, sc594), HDAC1 (Upstate Biotechnology, 05-614), PCNA (Abcam, ab29), Lamin A/C (Cell Signaling, 2032), COXIV (Cell Signaling, 4850), Histone H3 (Santa Cruz Biotechnology, sc8654), ATPB (Abcam, ab5432), SirT3 (Cell Signaling, 5490), cytochrome c (Becton Dickinson, 556433), p16 (Santa Cruz Biotechnology, sc74401), HA.11 (Covance), and GAPDH (Ambion, 4300). Secondary HRP-conjugated antibodies (Santa Cruz Biotechnology) were used at 1:5000 in 1% nonfat milk.
Real-time PCR
RNA was isolated using RNAeasy kit (Qiagen) and reverse-transcribed using SuperScript III reverse transcriptase (Invitrogen). Real-time PCR reactions were performed with SYBR Green (Applied Biosystems) on the ABI Prism 7000 sequence detection system and analyzed using the 7000 SDS software. The following primers were used: Cdc2 (CTGGCCAGTTCATGGATTCT and ATCAAACTGGCAGATTTCGG), Cyclin A2 (GAGAATGTCAACCCCGAAAA and ATAAACGATGAGCACGTCCC), Mcm3 (GTACGAGGAGTTCTACATAG and TCTTCTTAGTAGCAGGACAG), Mcm5 (GAGGACCAGGAGATGCTGAG and CTTTACCGCCTCAAGTGAGC), Mcm6 (CACGATTTGGAGGGAAAGAA and TCCACGGCAATGATGAAGTA), PCNA (TCCCAGACAAGCAATGTTGA and TTATTTGGCTCCCAAGATCG), and UB (TTCGTGAAGACCCTGACC and ACTCTTTCTGGATGTTGTAGTC).
Additional primers are described in Supplemental Table S1.
Xenograft model
All animal procedures followed protocols approved by MIT's Committee on Animal Care. Nude/SCID mice (Taconic) were injected subcutaneously with 107 tumor cells per site, two sites per mouse. After euthanasia, tumors were removed, fixed overnight in formalin followed by overnight incubation in 70% ethanol, and subjected to histological processing.
Immunofluorescence and immunohistochemistry
Cells were plated at low density on coverslips, and protein expression was induced for 48 h. Mitochondria were labeled using MitoTracker Deep Red (Invitrogen) at 100 nM for 45 min at 37°C. Cells were fixed in 4% formaldehyde (Thermo Scientific), permeabilized with 0.25% Triton X-100/PBS, and blocked with 5% goat serum in 0.2% Tween 20/PBS for 30 min at 37°C. The following antibodies were used at 1:200 for 1 h at room temperature: pRB (Cell Signaling, 9309) and c-REL (Cell Signaling, 4727). Alexa Fluor 488 (Invitrogen) was used at 1:1000 for 1 h at room temperature, and slides were mounted using SlowFade Gold anti-fade reagent with DAPI (Invitrogen) and observed under a fluorescence microscope (Zeiss).
Ki67 and cleaved caspase 3 immunohistochemistry was performed with a modified citric acid unmasking protocol. Briefly, paraffin was removed from slides, followed by incubation in 0.5% H2O2/methanol for 15 min and antigen retrieval using citrate buffer (pH 6.0) in a microwave for 15 min. Slides were blocked for 1 h at room temperature in 2% normal horse serum (Ki67) or 10% goat serum (cc3) in PBS. Ki67 (1:50; Becton Dickinson, 550609) and cleaved caspase 3 (1:200; Cell Signaling, 9661) antibodies were used in 0.15% Triton/PBS overnight at 4°C. Secondary antibodies (Vector Laboratories) were used at 1:200 in PBS with 0.4% normal horse serum (Ki67) or 2% goat serum (cc3), detected using a DAB substrate kit (Vector Laboratories), and counterstained with haematoxylin.
Acknowledgments
We thank E. Knudsen for providing the RAT16 parental and LP-RB-inducible cell lines, A. Letai for wild-type and Bax−/−/Bak−/− MEFs, S. Lowe for human Rb shRNA, and M. Hemann for rodent Rb shRNA. We also thank the members of the Lees laboratory for input during the study and manuscript preparation. This work was supported by an NCI/NIH grant to J.A.L., who is a Ludwig Scholar at MIT. K.I.H. was supported by an NSF predoctoral fellowship and D.H. Koch Graduate Fellowship.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.211326.112.
References
- Araki K, Ahmad SM, Li G, Bray DA Jr, Saito K, Wang D, Wirtz U, Sreedharan S, O'Malley BW Jr, Li D 2008. Retinoblastoma RB94 enhances radiation treatment of head and neck squamous cell carcinoma. Clin Cancer Res 14: 3514–3519 [DOI] [PubMed] [Google Scholar]
- Blanchet E, Annicotte J-Sb, Lagarrigue S, Aguilar V, Clapé C, Chavey C, Fritz V, Casas Fo, Apparailly F, Auwerx J, et al. 2011. E2F transcription factor-1 regulates oxidative metabolism. Nat Cell Biol 13: 1146–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco EE, Mayhew CN, Hennigan RF, Sage J, Jacks T, Knudsen ES 2004. RB signaling prevents replication-dependent DNA double-strand breaks following genotoxic insult. Nucleic Acids Res 32: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelle J, Letai A 2009. Control of mitochondrial apoptosis by the Bcl-2 family. J Cell Sci 122: 437–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart DL, Sage J 2008. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer 8: 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Quintero-Estades JA, Danielian PS, Nedelcu S, Berman SD, Lees JA 2010. Rb regulates fate choice and lineage commitment in vivo. Nature 466: 1110–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevale J, Palander O, Seifried LA, Dick FA 2012. DNA damage signals through differentially modified E2F1 molecules to induce apoptosis. Mol Cell Biol 32: 900–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A, Wang X, Zhang C, McCurrach M, Zhao Z, Mert O, Dickins R, Narita M, Zhang M, Lowe S 2010. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell 17: 376–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk J, Kuwana T, Bouchier-Hayes L, Droin N, Newmeyer D, Schuler M, Green D 2004. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303: 1010–1014 [DOI] [PubMed] [Google Scholar]
- Chipuk J, Bouchier-Hayes L, Kuwana T, Newmeyer D, Green D 2005. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science 309: 1732–1737 [DOI] [PubMed] [Google Scholar]
- Chipuk J, Moldoveanu T, Llambi F, Parsons M, Green D 2010. The BCL-2 family reunion. Mol Cell 37: 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciavarra G, Zacksenhaus E 2011. Direct and indirect effects of the pRb tumor suppressor on autophagy. Autophagy 7: 544–546 [DOI] [PubMed] [Google Scholar]
- de Bruin A, Wu L, Saavedra HI, Wilson P, Yang Y, Rosol TJ, Weinstein M, Robinson ML, Leone G 2003. Rb function in extraembryonic lineages suppresses apoptosis in the CNS of Rb-deficient mice. Proc Natl Acad Sci 100: 6546–6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskes R, Desagher S, Antonsson B, Martinou JC 2000. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol Cell Biol 20: 929–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferecatu I, Le Floch N, Bergeaud M, Rodriguez-Enfedaque A, Rincheval V, Oliver L, Vallette FM, Mignotte B, Vayssiere JL 2009. Evidence for a mitochondrial localization of the retinoblastoma protein. BMC Cell Biol 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher A, Dias M, Jans D 2010. Binding of p110 retinoblastoma protein inhibits nuclear import of simian virus SV40 large tumor antigen. J Biol Chem 285: 17744–17797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavathiotis E, Suzuki M, Davis ML, Pitter K, Bird GH, Katz SG, Tu HC, Kim H, Cheng EH, Tjandra N, et al. 2008. BAX activation is initiated at a novel interaction site. Nature 455: 1076–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavathiotis E, Reyna DE, Davis ML, Bird GH, Walensky LD 2010. BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol Cell 40: 481–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon G, Du W 2011. Conserved RB functions in development and tumor suppression. Protein Cell 2: 864–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YT, Youle RJ 1997. Nonionic detergents induce dimerization among members of the Bcl-2 family. J Biol Chem 272: 13829–13834 [DOI] [PubMed] [Google Scholar]
- Huang X, Masselli A, Frisch SM, Hunton IC, Jiang Y, Wang JY 2007. Blockade of tumor necrosis factor-induced Bid cleavage by caspase-resistant Rb. J Biol Chem 282: 29401–29413 [DOI] [PubMed] [Google Scholar]
- Ianari A, Natale T, Calo E, Ferretti E, Alesse E, Screpanti I, Haigis K, Gulino A, Lees JA 2009. Proapoptotic function of the retinoblastoma tumor suppressor protein. Cancer Cell 15: 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA 1992. Effects of an Rb mutation in the mouse. Nature 359: 295–300 [DOI] [PubMed] [Google Scholar]
- Jiao W, Datta J, Lin H-M, Dundr M, Rane S 2006. Nucleocytoplasmic shuttling of the retinoblastoma tumor suppressor protein via Cdk phosphorylation-dependent nuclear export. J Biol Chem 281: 38098–38108 [DOI] [PubMed] [Google Scholar]
- Jiao W, Lin HM, Datta J, Braunschweig T, Chung JY, Hewitt S, Rane S 2008. Aberrant nucleocytoplasmic localization of the retinoblastoma tumor suppressor protein in human cancer correlates with moderate/poor tumor differentiation. Oncogene 27: 3156–3220 [DOI] [PubMed] [Google Scholar]
- Jin Z, El-Deiry WS 2005. Overview of cell death signaling pathways. Cancer Biol Ther 4: 139–163 [DOI] [PubMed] [Google Scholar]
- Karin M, Lin A 2002. NF-κB at the crossroads of life and death. Nat Immunol 3: 221–227 [DOI] [PubMed] [Google Scholar]
- Knudsen E, Knudsen K 2008. Tailoring to RB: Tumour suppressor status and therapeutic response. Nat Rev Cancer 8: 714–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen KE, Weber E, Arden KC, Cavenee WK, Feramisco JR, Knudsen ES 1999. The retinoblastoma tumor suppressor inhibits cellular proliferation through two distinct mechanisms: Inhibition of cell cycle progression and induction of cell death. Oncogene 18: 5239–5245 [DOI] [PubMed] [Google Scholar]
- Knudsen KE, Booth D, Naderi S, Sever-Chroneos Z, Fribourg AF, Hunton IC, Feramisco JR, Wang JY, Knudsen ES 2000. RB-dependent S-phase response to DNA damage. Mol Cell Biol 20: 7751–7763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBelle JL, Katz SG, Bird GH, Gavathiotis E, Stewart ML, Lawrence C, Fisher JK, Godes M, Pitter K, Kung AL, et al. 2012. A stapled BIM peptide overcomes apoptotic resistance in hematologic cancers. J Clin Invest 122: 2018–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu J, Dumont P, Hafey M, Murphy M, George D 2004. Mitochondrial p53 activates Bak and causes disruption of a Bak–Mcl1 complex. Nat Cell Biol 6: 443–493 [DOI] [PubMed] [Google Scholar]
- Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B, Andrews DW 2008. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell 135: 1074–1084 [DOI] [PubMed] [Google Scholar]
- Manning A, Dyson N 2012. RB: Mitotic implications of a tumour suppressor. Nat Rev Cancer 12: 220–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchenko N, Zaika A, Moll U 2000. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem 275: 16202–16214 [DOI] [PubMed] [Google Scholar]
- Martinou J-C, Youle R 2011. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev Cell 21: 92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masselli A, Wang JY 2006. Phosphorylation site mutated RB exerts contrasting effects on apoptotic response to different stimuli. Oncogene 25: 1290–1298 [DOI] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM 2003. p53 has a direct apoptogenic role at the mitochondria. Mol Cell 11: 577–590 [DOI] [PubMed] [Google Scholar]
- Milet C, Rincheval-Arnold A, Mignotte B, Guenal I 2010. The Drosophila retinoblastoma protein induces apoptosis in proliferating but not in post-mitotic cells. Cell Cycle 9: 97–103 [DOI] [PubMed] [Google Scholar]
- Mittnacht S, Weinberg RA 1991. G1/S phosphorylation of the retinoblastoma protein is associated with an altered affinity for the nuclear compartment. Cell 65: 381–393 [DOI] [PubMed] [Google Scholar]
- Perez D, White E 2000. TNF-α signals apoptosis through a bid-dependent conformational change in Bax that is inhibited by E1B 19K. Mol Cell 6: 53–63 [PubMed] [Google Scholar]
- Roth D, Harper I, Pouton C, Jans D 2009. Modulation of nucleocytoplasmic trafficking by retention in cytoplasm or nucleus. J Cell Biochem 107: 1160–1167 [DOI] [PubMed] [Google Scholar]
- Sherr CJ, McCormick F 2002. The RB and p53 pathways in cancer. Cancer Cell 2: 103–112 [DOI] [PubMed] [Google Scholar]
- Speidel D 2010. Transcription-independent p53 apoptosis: An alternative route to death. Trends Cell Biol 20: 14–38 [DOI] [PubMed] [Google Scholar]
- Takahashi C, Sasaki N, Kitajima S 2012. Twists in views on RB functions in cellular signaling, metabolism and stem cells. Cancer Sci 103: 1182–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Martin SJ, Green DR, Wang JY 1997. Degradation of retinoblastoma protein in tumor necrosis factor- and CD95-induced cell death. J Biol Chem 272: 9613–9616 [DOI] [PubMed] [Google Scholar]
- Templeton DJ 1992. Nuclear binding of purified retinoblastoma gene product is determined by cell cycle-regulated phosphorylation. Mol Cell Biol 12: 435–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF 2007. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol 27: 6229–6242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traenckner EB, Wilk S, Baeuerle PA 1994. A proteasome inhibitor prevents activation of NF-κB and stabilizes a newly phosphorylated form of I κB-α that is still bound to NF-κB. EMBO J 13: 5433–5441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ 2008. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 9: 713–724 [DOI] [PubMed] [Google Scholar]
- Viatour P, Sage J 2011. Newly identified aspects of tumor suppression by RB. Dis Model Mech 4: 581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden K, Prives C 2009. Blinded by the light: The growing complexity of p53. Cell 137: 413–444 [DOI] [PubMed] [Google Scholar]
- Walensky LD, Gavathiotis E 2011. BAX unleashed: The biochemical transformation of an inactive cytosolic monomer into a toxic mitochondrial pore. Trends Biochem Sci 36: 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky LD, Pitter K, Morash J, Oh KJ, Barbuto S, Fisher J, Smith E, Verdine GL, Korsmeyer SJ 2006. A stapled BID BH3 helix directly binds and activates BAX. Mol Cell 24: 199–210 [DOI] [PubMed] [Google Scholar]
- Wenzel PL, Wu L, de Bruin A, Chong JL, Chen WY, Dureska G, Sites E, Pan T, Sharma A, Huang K, et al. 2007. Rb is critical in a mammalian tissue stem cell population. Genes Dev 21: 85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, de Bruin A, Saavedra HI, Starovic M, Trimboli A, Yang Y, Opavska J, Wilson P, Thompson JC, Ostrowski MC, et al. 2003. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature 421: 942–947 [DOI] [PubMed] [Google Scholar]
- Wyllie A 2010. ‘Where, O death, is thy sting?’ A brief review of apoptosis biology. Mol Neurobiol 42: 4–13 [DOI] [PMC free article] [PubMed] [Google Scholar]