p53 has a dual regulatory role in autophagy. Here, Napoli and Flores discuss Kenzelmann Broz et al.'s use of a combined RNA sequencing and ChIP sequencing analysis to generate a high-resolution, genome-wide map of p53 transcriptional targets, which reveals a novel autophagy program regulated by the entire p53 family.
Keywords: p53, ChIP-seq, RNA-seq, tumor suppression, autophagy
Abstract
Autophagy is a biological process that is crucial to maintain cellular homeostasis and is regulated by several metabolic pathways, including the p53 tumor suppressor pathway. In this issue of Genes & Development, Kenzelmann Broz and colleagues (pp. 1016–1031) show how the p53 family as a whole, including p63 and p73, collaborate in controlling autophagy to support tumor suppression.
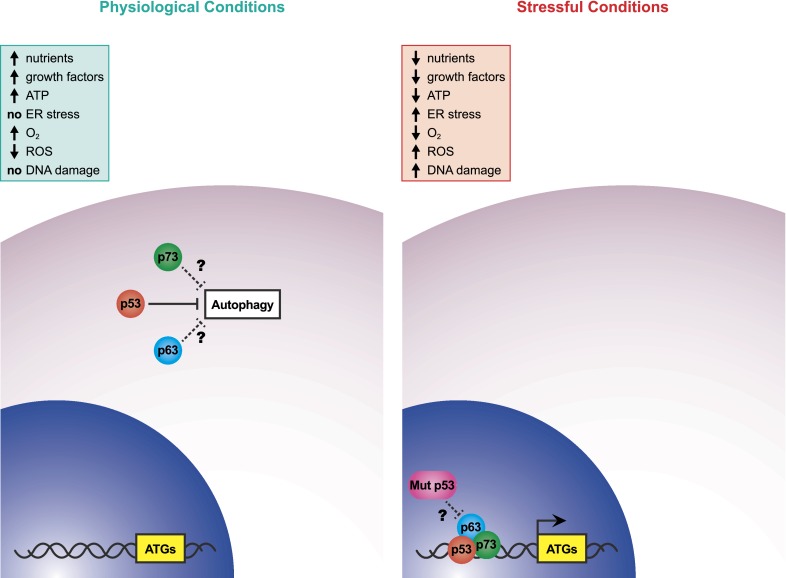
In this issue of Genes & Development, Kenzelmann Broz et al. (2013) shed new light on the complex role of the p53 tumor suppressor gene in the regulation of autophagy. By performing genome-wide transcriptional analyses on wild-type and p53−/− mouse embryo fibroblasts (MEFs) treated with doxorubicin, a commonly used chemotherapeutic agent, the investigators found that the functions of p53 extend into a larger number of biological processes than previously appreciated. Indeed, p53 is now shown to induce a plethora of genes implicated in all steps of the autophagy pathway. This transcriptional induction is triggered by p53 in both murine and human cells in response to different kinds of stimuli, such as DNA damage agents, oncogenic signaling, and genetic and pharmacological activation. Intriguingly, the investigators demonstrate that these autophagic genes are induced by the other members of the p53 family, p63 and p73, in response to doxorubicin (Fig. 1). Their crucial role is particularly evident in a p53-null background, where they can compensate for chronic p53 deficiency. In addition, the Attardi laboratory (Kenzelmann Broz et al. 2013) indicates that autophagy is essential for efficient tumor suppression induced by p53, therefore highlighting that key p53 biological responses like apoptosis and suppression of cell transformation are deeply interwoven with autophagy.
Figure 1.
Autophagy regulation by the p53 family members in cells under physiological and stressful conditions. Basal cytosolic p53 levels can inhibit autophagy, and a similar role is possible for p63 and p73 (dashed lines). Upon treatment of cells with different kinds of stress inducers, the p53 family members translocate to the nucleus, where they cooperate to trigger the expression of the ATGs. A possible dominant-negative effect by mutant p53 proteins could occur to inhibit the ability of p63 and p73 to induce autophagy.
This Perspective discusses the previously known molecular and functional connections between p53 and autophagy as well as the new contribution by Kenzelmann Broz et al. (2013) and the novel questions that it raises for the field.
The autophagic process and its physiological and pathological modulators
Exactly a half-century ago, Dr. Christian de Duve coined the term “autophagy” (from the Greek words αυτο, meaning “oneself,” and φαγɛιν, meaning “to eat”) to indicate the process through which cytoplasmic components are delivered to the lysosomes for degradation (Klionsky 2008). In particular, autophagy is characterized by the formation of double-membrane structures, called autophagosomes, engulfing organelles and soluble factors and subsequently fusing with the lysosomes to generate the autolysosomes, where the degradation of these materials occurs (Das et al. 2012).
Although involved in diverse biological phenomena, such as cellular defense from microorganisms (Zhou and Zhang 2012) and type II programmed cell death (Ouyang et al. 2012), autophagy is primarily a catabolic process mediating the degradation of long-lived proteins, protein aggregates, and damaged organelles to maintain cellular homeostasis and provide macromolecules to be recycled when starvation occurs (Rabinowitz and White 2010). This biological process is conserved throughout eukaryotic evolution (Duszenko et al. 2011), and until now, >30 autophagy-related genes (ATGs) have been discovered in yeast, and the majority of them have homologs in the human genome (Weidberg et al. 2011).
The first molecular step of the autophagic process is the generation of the preautophagosomal structure (PAS), which gives rise to the membrane isolating the organelles and soluble components to be eliminated. PAS formation is regulated by the mammalian target of rapamycin (mTOR) through two effector complexes whose hubs are the protein kinase Atg1 (ULK1, ULK2 and ULK3 in mammals) (Kamada et al. 2000) and the lipid kinase Vps34 (hVps34 in humans) (Simonsen and Tooze 2009). After PAS formation, both lipids and proteins of the autophagic core machinery continue to be accumulated, permitting the expansion of the isolation membrane, whose dynamic formation is supported by two ubiquitylation-like conjugation systems: the Atg5–Atg12–Atg16 and the Atg8 (LC3) systems (Geng and Klionsky 2008). Once these materials have been completely incorporated, the autophagosome fuses with lysosomes, and its content is degraded. The obtained products (amino acids, carbohydrates, and lipids) are then released in the cytosol to be reused by the needed anabolic processes (Rabinowitz and White 2010).
Because of its intertwined function with cellular metabolism, autophagy induction is mainly controlled by mTOR, which functionally acts as a hub integrating several upstream pathways responsible for sensing the amount of available nutrients (cAMP-dependent protein kinase A [PKA] pathway), presence of growth factors (Ras pathway), and ATP levels (the liver kinase B1 [LKB1]/5′-AMP-activated protein kinase [AMPK] pathway) (Yang and Klionsky 2010). In addition to these physiological conditions, autophagy can also be triggered as a part of stress response to diverse stimuli, such as endoplasmic reticulum (ER) stress, hypoxia, and high levels of reactive oxygen species (ROS) (Kroemer et al. 2010).
p53 as a master regulator of autophagy
Both the above-mentioned metabolic related pathways and the autophagy-inducing stressors are deeply interconnected with the tumor suppressor p53, which is considered one of the most crucial regulators of autophagy (Maiuri et al. 2010). The intriguing aspect of p53 in regulating autophagy is that this factor may act in a bivalent way toward this biological process, depending on cellular contexts. Autophagy is triggered by the transcriptional activity of p53 in response to stressful stimuli (Fig. 1). Indeed, p53 was previously reported to directly induce the expression of several proautophagic genes, such as TCS2 (tuberous sclerosis 2) and the β1 and β2 subunits of AMPK (Feng et al. 2007). In particular, upon metabolic stress conditions, AMPK can phosphorylate p53 to amplify autophagy induction (Jones et al. 2005). In response to genotoxic stress, p53 can promote the expression of PTEN (phosphatase and tensin homolog) (Stambolic et al. 2001)—which in turn curbs autophagy inhibition due to the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (PKB) pathway—and many genes coding for BH3-only proteins, which induce autophagy at least in part by releasing the proautophagic protein Beclin1 from its inhibitory partner, Bcl2 (Maiuri et al. 2007). Apart from the regulation of these genes, the most relevant contribution of p53 transcriptional activity in supporting autophagy is the up-regulation of DRAM1 (DNA damage-regulated autophagy modulator 1), coding for a lysosomal protein that is the principal mediator of p53-promoted autophagy (Crighton et al. 2006). Interestingly, DRAM1 is also required for an efficient p53-dependent apoptosis, thus representing a crucial effector of two previously unlinked p53-regulated biological processes (Crighton et al. 2007b). In addition to p53's transcriptional roles in triggering autophagy, basal p53 levels correlating with its cytoplasmic localization were demonstrated to suppress autophagy through a not yet fully characterized mechanism (Fig. 1; Tasdemir et al. 2008).
p53, p63, and p73 transcriptionally activate the same genes in autophagy
The complex role of p53 in controlling autophagy and the subsequent functional consequences are therefore not completely characterized. Kenzelmann Broz et al. (2013) now provide an essential contribution in this issue of Genes & Development by describing novel molecular insights of p53-regulated autophagy. By performing RNA sequencing (RNA-seq) in parallel with chromatin immunoprecipitation (ChIP) combined with sequencing (ChIP-seq) analyses using wild-type and p53−/− MEFs treated with doxorubicin, they discovered that several autophagy-related genes, including Atg4a, Atg4c, Atg5, Ulk1, Ulk2, and Uvrag, are direct transcriptional targets of p53 and that Atg5 is required to sustain proper p53-dependent apoptosis and suppression of cell transformation. The newly identified p53 target genes belong to different steps of the autophagic process. These include upstream regulators of autophagy (like the γ subunit of AMPK, Prkag2), components of the core autophagic machinery (such as Atg7, coding for an E1-like enzyme activating both the Atg5–Atg12–Atg16 and the LC3 complexes), and lysosomal protein-encoding genes (such as the tripeptidyl-peptidase 1 gene [TPP1]). These analyses have broadly extended our knowledge of the p53 transcriptional program that promotes autophagy, therefore increasing the number of connections between p53 and this biological process.
Intriguingly, the majority of the identified genes are induced also by the other two p53 family members, p63 and p73 (Fig. 1). Upon doxorubicin treatment in p53−/− MEFs, p63 and p73 can compensate for p53 function, suggesting that the entire p53 family cooperates in controlling cellular homeostasis through autophagy promotion similar to their collaborative induction of proapoptotic genes (Flores et al. 2002). This finding is in line with previous evidence of p73-regulated autophagy. p73 is able to induce DRAM even though p73-mediated autophagy is almost DRAM-independent, thus suggesting that the p53 family members can exploit different pathways to activate the autophagic process (Crighton et al. 2007a). In addition, in contrast to p53 that is degraded upon autophagic-inducing conditions, nutrient withdrawal and rapamycin treatment can increase p73 stability and activate a p73-dependent transcriptional program to induce autophagy, thereby implying a feedback loop between p73 and the mTOR pathway (Rosenbluth et al. 2008).
Prior to the study by the Attardi laboratory (Kenzelmann Broz et al. 2013), less information was available regarding the involvement of p63 in autophagy. ChIP-seq experiments have previously indicated p63 recruitment on the promoters of several autophagy-related genes (Kouwenhoven et al. 2010)—some of which are induced by the overexpression of ΔNp63α, one of the p63 isoforms (Su et al. 2013)—specifically upon cisplatin treatment (Huang et al. 2012).
p53, p63, and p73 regulate cellular metabolism
An indirect effect of the p53 family members in the homeostatic function of the autophagic process is provided by their role in cellular metabolism. p53 was reported to control several metabolic pathways (Maddocks and Vousden 2011). First, p53 regulates glucose metabolism by (1) repressing insulin receptor (INSR) and the glucose transporters GLUT1 and GLUT4, (2) activating TIGAR (TP53-induced glycolysis and apoptosis regulator) and HK II (hexokinase II), and (3) promoting the degradation of PGM (phosphoglycerate mutase). p53 also regulates the tricarboxylic acid (TCA) cycle by promoting the expression of GLS2 (glutaminase 2) and controls fatty acid oxidation by activating GAMT (guanidinoacetate aminotransferase). Last, p53 has been found to control mitochondrial respiration by inducing AIF (apoptosis-inducing factor) and SCO2 (synthesis of cytochrome oxidase 2).
A pivotal role in metabolism has been recently unveiled for both p63 and p73 by generating isoform-specific knockout mice with deletions in the transactivation domain containing isoforms of p63 and p73, TAp63 and TAp73, respectively. TAp63−/− mice develop insulin resistance and glucose intolerance, leading to obesity (Su et al. 2012). TAp63 can control glucose and fatty acid metabolism by inducing the expression of Sirt1 (silent information regulator T1), LKB1, and the α2 subunit of AMPK, AMPKα2 (Su et al. 2012). TAp73−/− mice also develop metabolic defects (Rufini et al. 2012). These defects are characterized by premature aging associated with impaired mitochondrial respiration and increased ROS generation, mainly due to reduced levels of the TAp73 transcriptional target gene Cox4i1 (cytochrome c oxidase subunit IV isoform 1) (Rufini et al. 2012). Taken together, this recent in vivo evidence adds further complexity to the contribution of the p53 family to the autophagic process in both physiological and pathological conditions.
Conclusions and future directions
The p53 family plays a crucial role in directly and indirectly modulating autophagy. Further research will increase our comprehension of the molecular mechanisms involved. In particular, future analyses will clarify some of the still obscure points. For example, which are the in vivo implications of p53 family-induced autophagy for tumor and metastasis suppression? What is the role of p63 and p73 in autophagy during unstressed physiological conditions? What is the contribution of the isoforms of the p53 family lacking the transactivation domain (i.e., ΔNp53, ΔNp63, and ΔNp73) in this biological process? Can the p53 mutant proteins often found in human cancers exert a dominant-negative effect on the proautophagic transcriptional activity of the p53 family members, as was shown for other biological responses like cell cycle arrest or apoptosis (Walerych et al. 2012)? These questions can now be addressed using tissues and cells from the mouse models deficient for the different p53 family members and the distinct isoforms of p63 and p73.
The answers to these and other still unanswered questions will help in completely unveiling the picture of connections between the p53 family members and autophagy and how this interdependence of the p53 family members impinge on their roles in tumor suppression.
Acknowledgments
We thank L. McCord for art production.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.219147.113.
References
- Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM 2006. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 126: 121–134 [DOI] [PubMed] [Google Scholar]
- Crighton D, O'Prey J, Bell HS, Ryan KM 2007a. p73 regulates DRAM-independent autophagy that does not contribute to programmed cell death. Cell Death Differ 14: 1071–1079 [DOI] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, Ryan KM 2007b. DRAM links autophagy to p53 and programmed cell death. Autophagy 3: 72–74 [DOI] [PubMed] [Google Scholar]
- Das G, Shravage BV, Baehrecke EH 2012. Regulation and function of autophagy during cell survival and cell death. Cold Spring Harb Perspect Biol 4: a008813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duszenko M, Ginger ML, Brennand A, Gualdron-Lopez M, Colombo MI, Coombs GH, Coppens I, Jayabalasingham B, Langsley G, de Castro SL, et al. 2011. Autophagy in protists. Autophagy 7: 127–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, Levine AJ 2007. The regulation of AMPK β1, TSC2, and PTEN expression by p53: Stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1–AKT–mTOR pathways. Cancer Res 67: 3043–3053 [DOI] [PubMed] [Google Scholar]
- Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, Jacks T 2002. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 416: 560–564 [DOI] [PubMed] [Google Scholar]
- Geng J, Klionsky DJ 2008. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: Beyond the usual suspects' review series. EMBO Rep 9: 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Guerrero-Preston R, Ratovitski EA 2012. Phospho-ΔNp63α-dependent regulation of autophagic signaling through transcription and micro-RNA modulation. Cell Cycle 11: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB 2005. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell 18: 283–293 [DOI] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 150: 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenzelmann Broz D, Spano Mello S, Bieging KT, Jiang D, Dusek RL, Brady CA, Sidow A, Attardi LD 2013. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev (this issue). doi: 10.1101/gad.212282.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ 2008. Autophagy revisited: A conversation with Christian de Duve. Autophagy 4: 740–743 [DOI] [PubMed] [Google Scholar]
- Kouwenhoven EN, van Heeringen SJ, Tena JJ, Oti M, Dutilh BE, Alonso ME, de la Calle-Mustienes E, Smeenk L, Rinne T, Parsaulian L, et al. 2010. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet 6: e1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B 2010. Autophagy and the integrated stress response. Mol Cell 40: 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks OD, Vousden KH 2011. Metabolic regulation by p53. J Mol Med 89: 237–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Criollo A, Tasdemir E, Vicencio JM, Tajeddine N, Hickman JA, Geneste O, Kroemer G 2007. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L). Autophagy 3: 374–376 [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Galluzzi L, Morselli E, Kepp O, Malik SA, Kroemer G 2010. Autophagy regulation by p53. Curr Opin Cell Biol 22: 181–185 [DOI] [PubMed] [Google Scholar]
- Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT, Liu B, Bao JK 2012. Programmed cell death pathways in cancer: A review of apoptosis, autophagy and programmed necrosis. Cell Prolif 45: 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, White E 2010. Autophagy and metabolism. Science 330: 1344–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluth JM, Mays DJ, Pino MF, Tang LJ, Pietenpol JA 2008. A gene signature-based approach identifies mTOR as a regulator of p73. Mol Cell Biol 28: 5951–5964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufini A, Niklison-Chirou MV, Inoue S, Tomasini R, Harris IS, Marino A, Federici M, Dinsdale D, Knight RA, Melino G, et al. 2012. TAp73 depletion accelerates aging through metabolic dysregulation. Genes Dev 26: 2009–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Tooze SA 2009. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol 186: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, Benchimol S, Mak TW 2001. Regulation of PTEN transcription by p53. Mol Cell 8: 317–325 [DOI] [PubMed] [Google Scholar]
- Su X, Gi YJ, Chakravarti D, Chan IL, Zhang A, Xia X, Tsai KY, Flores ER 2012. TAp63 is a master transcriptional regulator of lipid and glucose metabolism. Cell Metab 16: 511–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Chakravarti D, Flores ER 2013. p63 steps into the limelight: Crucial roles in the suppression of tumorigenesis and metastasis. Nat Rev Cancer 13: 136–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, Criollo A, Morselli E, Zhu C, Harper F, et al. 2008. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 10: 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walerych D, Napoli M, Collavin L, Del Sal G 2012. The rebel angel: Mutant p53 as the driving oncogene in breast cancer. Carcinogenesis 33: 2007–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Elazar Z 2011. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem 80: 125–156 [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ 2010. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr Opin Cell Biol 22: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XJ, Zhang H 2012. Autophagy in immunity: Implications in etiology of autoimmune/autoinflammatory diseases. Autophagy 8: 1286–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]