pRB can either induce or suppress apoptosis, depending on the cellular context. This perspective examines Hilgendorf et al.'s identification of a novel, tumor-suppressive function of pRB as a direct activator of apoptosis at the mitochondrial membrane.
Keywords: MOMP, apoptosis, cancer, pRB, retinoblastoma protein
Abstract
The retinoblastoma tumor suppressor RB is well known for its capacity to restrict cell cycle progression at the G1/S transition of the cell cycle by controlling the transcription of cell cycle genes. In this issue of Genes & Development, Hilgendorf and colleagues (pp. 1003–1015) have identified a novel tumor suppressor function for RB independent of its role as a transcriptional regulator, in which RB directly activates the apoptosis regulator Bax at the mitochondria to promote cell death.
Early paradoxical observations on RB and cell death
The cellular mechanisms of tumor suppression include cell cycle arrest, senescence, differentiation, and cell death. When the RB tumor suppressor was first extensively studied in the 1990s, its role as an inhibitor of cell cycle progression was rapidly illuminated. Subsequent studies then indicated that active RB molecules could also contribute to the establishment and maintenance of senescence as well as to promoting differentiation. At the molecular level, these studies identified RB as a transcriptional regulator with the capacity to bind transcription factors such as E2F and recruit chromatin modifiers to the promoters of target genes involved in cell proliferation and differentiation (for review, see Harbour and Dean 2000; Burkhart and Sage 2008; Chinnam and Goodrich 2011).
The connection between RB and cell death was initially a bit surprising. Indeed, induction of cell death was primarily observed as a consequence of the absence of RB or its inactivation in cells. For instance, apoptotic cell death was found to be prominent in mouse embryos deficient for the Rb gene; genetic crosses further demonstrated that E2F and p53 are crucial for the induction of cell death downstream from RB loss (Morgenbesser et al. 1994; Symonds et al. 1994; Macleod et al. 1996; Zacksenhaus et al. 1996; Pan et al. 1998; Tsai et al. 1998; Ziebold et al. 2001). Key downstream targets of E2F in this process are the p53 family member p73 as well as genes encoding cell death machinery components such as Caspases, Apaf-1, and Bcl-2 homology 3 (BH3)-only proapoptotic members of the Bcl-2 protein family (Irwin et al. 2000; Moroni et al. 2001; Nahle et al. 2002; Hershko and Ginsberg 2004). RB may also directly bind to and inhibit proapoptotic molecules in some contexts (Adegbola and Pasternack 2005).
These observations led to a model in which cancer initiation following loss of RB—as in retinoblastoma—must happen in a cell type that is inherently resistant to the death resulting from loss of RB function (Chen et al. 2004). These studies also provided a basis for the frequent coalteration of the RB and p53 pathways in cancer cells, as RB pathway mutant tumor cells likely need to abrogate p53 proapoptotic function in order to expand during tumor progression (Sherr and McCormick 2002).
Proapoptotic role for RB
Concomitantly with these studies demonstrating that loss of RB leads to increased cell death, an increasing number of reports have shown a proapoptotic role for RB. For example, studies from the Wang laboratory (Chau et al. 2002) showed that RB is cleaved by Caspases in response to TNFα and that preventing this cleavage reduces the amount of death downstream from the TNFα receptor (but not in response to DNA damage). These data could suggest that either RB needs to be inactivated for death to be induced efficiently, in keeping with the anti-apoptotic role of RB described above, or alternatively, cleaved RB is proapoptotic.
Since these initial studies, evidence for a direct role for RB in promoting cell death has accumulated. For example, in Caenorhabditis elegans, genetic crosses have shown that the RB homolog Lin-35 collaborates with E2F to promote apoptosis during development of the anterior pharynx (Reddien et al. 2007). Lin-35 also promotes germ cell apoptosis in part by repressing the expression of ced-9, an ortholog of the anti-apoptotic factor BCL2 (Schertel and Conradt 2007). In addition, in mammalian cells, expression of a form of RB that cannot be phosphorylated on its major CDK phosphorylation sites can stimulate the apoptotic response to TNFα (Masselli and Wang 2006). RB also plays a role in the cell death induced by histone deacetylase inhibitors in colon cancer cell lines (Wagner and Roemer 2005). Furthermore, efficient induction of death by TGFβ in some cancer cells requires a complex between RB, E2F-1, and the acetyltransferase P/CAF (Korah et al. 2012). Finally, a previous report from the Lees laboratory (Ianari et al. 2009) further showed that the phosphorylated forms of RB found in proliferative cells and normally less able to bind to E2F-1 can be induced to interact with E2F-1 in response to genotoxic stress (DNA damage) or oncogenic stress (expression of E1A). In the same study, stress-induced RB/E2F-1 complexes containing P/CAF or p300 were shown to bind to and activate the transcription of cell death genes such as p73 and Casp7 (Ianari et al. 2009).
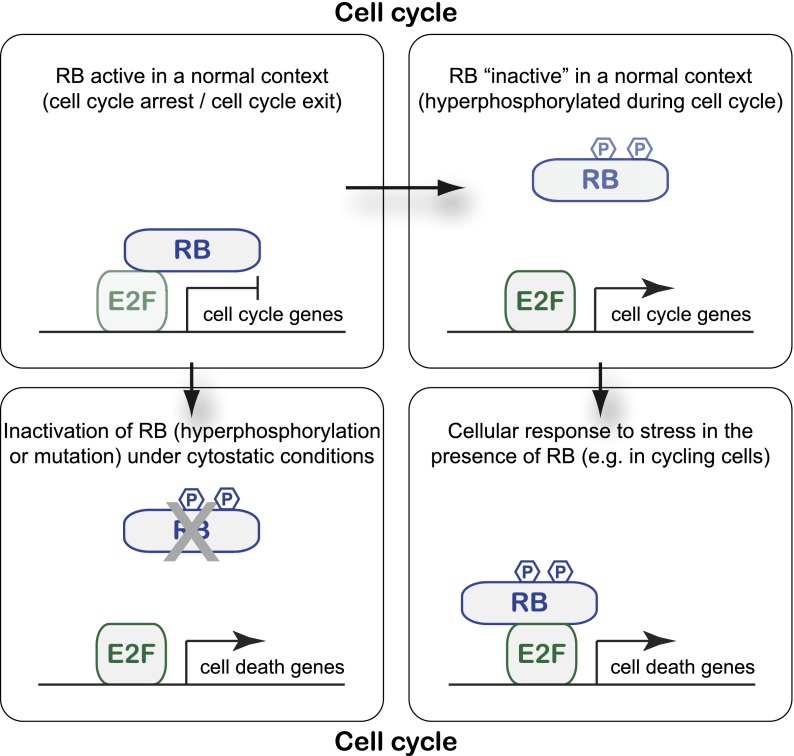
A way to reconcile these different studies is that RB has both anti-apoptotic and proapoptotic roles, depending on the cellular context: In normally slowly proliferating or quiescent cell populations, acute loss of RB function (by deletion but also by rapid hyperphosphorylation) can induce cell death in part by deregulated activity of E2F and activation of p53. This proapoptotic consequence of RB loss may, for example, help prevent cancer initiation from quiescent populations of stem cells. In cells that are already proliferating, RB molecules (including phosphorylated molecules usually thought to be “inactive”) may be able to sense genotoxic or oncogenic stresses and complex specifically with E2F-1 to activate the expression of cell death genes and promote apoptosis (Fig. 1). While the detailed mechanisms underlying the formation and activation of RB/E2F-1 proapoptotic transcriptional complexes are not fully established, the specific interaction between RB and E2F-1 relies on the C-terminal domain of RB and post-translational modifications of both proteins (Dick and Dyson 2003; Rubin et al. 2005; Julian et al. 2008; Carnevale et al. 2012). This proapoptotic stress sensor function of RB may provide a safety mechanism to eliminate cells that have committed to divide but may carry genomic alterations due to replication stress.
Figure 1.
Anti- and proapoptotic functions of the RB transcriptional regulator. (Top) During normal cell cycle progression, RB phosphorylation by Cyclin/CDK complexes results in the dissociation of RB and E2F transcription factors and activation of E2F target genes. RB's ability to restrict cell cycle progression is thought to be one of its major tumor-suppressive roles in cells. (Bottom left) When RB is inappropriately inactivated in quiescent cells or cells exiting the cell cycle—for example, by gene mutation, protein degradation, or hyperphosphorylation—activation of E2F in the wrong context may result in the activation of cell death genes (in addition to cell cycle genes), which may help suppress cancer initiation. (Bottom right) In response to certain stresses (e.g., DNA damage or oncogene activation), RB can specifically bind to E2F-1 even in cycling cells and promote the expression of cell death genes, thereby thwarting the expansion of abnormal cells and suppressing cancer development.
Direct activation of the apoptotic machinery by RB at the mitochondria
These previous studies building a case for a proapoptotic role for RB have often focused only on mechanisms linked to the transcriptional regulation by RB of genes involved in cell death. However, the fastest way to trigger cell death in cells is independent of the transcriptional machinery and involves changes in the membrane potential of mitochondria in cells (Wang 2001). In this issue of Genes & Development, Hilgendorf et al. (2013) provide compelling evidence that a fraction of RB is at the mitochondria, where it can activate Bax and promote cell death in response to TNFα treatment or other stresses. First, the investigators showed that RB can promote TNFα-induced apoptotic cell death in a translation-independent manner in RAT16 cells using treatment with cycloheximide to inhibit translation. The investigators then showed that this effect is dependent on the Bax gene and not Bak using immortalized mouse embryonic fibroblasts deficient for each of these two proapoptotic genes. Intrigued by these observations, Hilgendorf et al. (2013) sought to determine whether RB could localize to the mitochondria, where Bax normally resides. Indeed, they found that a fraction of the RB pool is constitutively mitochondrial in IMR90 human cells and mouse liver cells, including phosphorylated forms of RB. RB can directly interact with Bax, conformationally activate it, and trigger mitochondrial outer membrane permeabilization (MOMP) and release of cytochrome c, an essential step in the initiation of apoptosis. Furthermore, a truncated form of RB containing only its “small pocket” (a domain known to be essential for RB's tumor suppressor function) is sufficient to promote cell death induced by TNFα treatment in the presence of cycloheximide. Strikingly, an RB mutant lacking the nuclear localization signal but carrying a mitochondrial import signal was found to be sufficient to drive apoptosis. The remarkable ability of this mutant to suppress the growth of Rb−/−;p53−/− osteosarcoma tumor cells to an extent comparable with nuclear RB suggests that the proapoptotic action of RB at the mitochondria may be as critical as its transcriptional tumor suppressor role, at least in certain contexts, and further demonstrates that this proapoptotic role of RB is p53-independent.
Similarities and differences between RB and p53
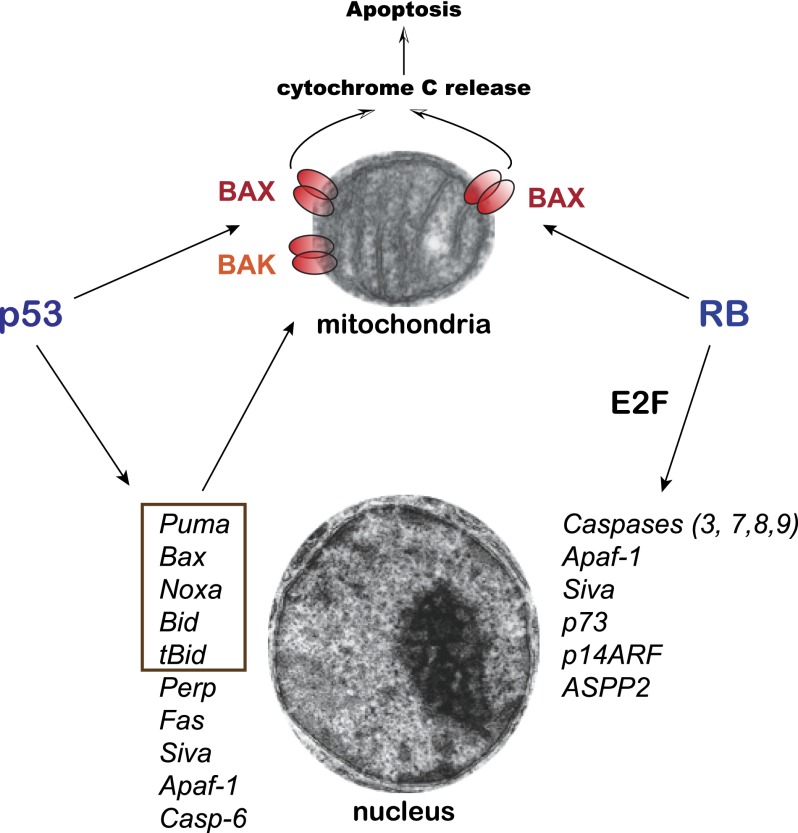
This study from the Lees laboratory (Hilgendorf et al. 2013) recalls that of RB's fellow tumor suppressor, the p53 transcription factor (Fig. 2; Brady and Attardi 2010), which, in startling findings in 1994, was discovered to induce apoptosis in the presence of transcription and translation inhibitors (Caelles et al. 1994). The story took an even more surprising twist in 2000, when p53 was reported to localize to mitochondria during apoptosis, where it acted directly to promote apoptosis (Marchenko et al. 2000). While greeted with skepticism initially, mounting evidence from different laboratories buttressed the notion of p53 action at the mitochondria, albeit through different mechanisms according to the specific model system examined. The common theme is that p53 interacts with Bcl-2 family members in each case, but the specific players vary. In the original study, p53 was found to interact with Bcl2 or Bcl-XL at the mitochondria to neutralize their activity and thus activate proapoptotic Bax and Bak proteins to drive MOMP (Mihara et al. 2003). Interestingly, tumor-derived p53 mutants fail to interact with Bcl-2 or Bcl-XL, suggesting the importance of these interactions for p53-mediated tumor suppression. Moreover, as with the RB story here, mitochondrially targeted p53 was found to be sufficient to trigger apoptosis and inhibit lymphoma growth in vivo (Marchenko et al. 2000; Talos et al. 2005). In a subsequent study, p53 was discovered to interact with Bak, freeing it from the influence of the anti-apoptotic Mcl-1 and triggering Bak oligomerization to induce apoptosis (Leu et al. 2004). Finally, p53 can bind Bcl-XL in the cytoplasm but becomes displaced upon induction of the proapoptotic Bcl-2 family member Puma, liberating p53 to transit to the mitochondrial membrane, where it promotes Bax oligomerization and apoptosis (Chipuk et al. 2004, 2005). Thus, like RB, p53 can promote Bax or Bak oligomerization/activation and membrane permeabilization Recently, the role of p53 in mitochondrial compromise was found to extend even beyond apoptosis; p53 was found to enter the mitochondrial matrix and drive necrosis, thereby contributing to stroke phenotypes (Vaseva et al. 2012). These observations highlight the similarities between p53 and RB tumor suppressor functions and further provide some possible directions for the future studies of RB's proapoptotic role at the mitochondria and the contribution of this pool of RB molecules to embryonic development, homeostasis, and cancer initiation and progression.
Figure 2.
Similarities between RB and p53 in the induction of apoptotic cell death. Both p53 (directly) and RB (with E2F1 or via E2F activity) can increase the transcription of proapoptotic genes in the nucleus. Only some of the known targets are shown, with a focus on genes encoding key apoptotic regulators and common targets between p53 and E2F. Components or regulators of the apoptotic machinery at the mitochondria are highlighted in a box. Note that Puma can disrupt the interaction between p53 and Bcl-XL, thereby allowing p53 to activate Bax and Bak. The new study from Hilgendorf et al. (2013) indicates that RB can also directly control Bax at the mitochondria, similarly to p53, which can activate Bax and Bak at the mitochondrial membrane to promote apoptotic cell death.
Conclusions
Hilgendorf et al. (2013) have identified a novel function for RB in cells. This proapoptotic role for RB at the mitochondria expands the tumor suppressor activity of an already versatile protein. Future experiments should investigate the physiological contexts in which this novel activity is most significant, including mouse models of cancer. One report in glioblastoma cells suggests that the RB family member p130 can potentiate the cell death induced by DNA damage in glioblastoma cells (Pucci et al. 2002). It would be interesting to know whether p130 and the other RB family member, p107, both of which share functional and structural similarities with RB, also share its capacity to interact with Bax and regulate apoptotic cell death directly.
The ability of two potent tumor suppressors to induce apoptosis via mitochondrial pathways raises the intriguing question of why RB and p53 evolved to participate in this alternate cell death pathway. Perhaps the ability to induce apoptosis at the mitochondria provides a means to trigger very rapid cell death in response to stresses without the need for the intermediary steps of transcription and translation. Alternatively, this mitochondrial effect may be one way to fine-tune the cellular response to stress. The double whammy imparted by apoptosis driven both at the mitochondrial membrane and in the nucleus provides a fail-safe mechanism to ensure the certain demise of the cell in the context of tumor suppressor action.
Acknowledgments
The work in J.S.'s laboratory on RB is supported by the National Institutes of Health (NIH)/National Cancer Institute (grant R01 CA114102) and California Institute for Regenerative Medicine (grant RB1-01385). J.S. is a Leukemia and Lymphoma Society scholar and the Harriet and Mary Zelencik Scientist in Children's Cancer and Blood Diseases. The work in L.D.A.'s laboratory on p53 is supported by the NIH (grants R01 CA140875 and R21 CA169673), the Department of Defense, and the Leukemia and Lymphoma Society. We apologize to our colleagues whose work was not cited due to space limitations.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.219451.113.
References
- Adegbola O, Pasternack GR 2005. Phosphorylated retinoblastoma protein complexes with pp32 and inhibits pp32-mediated apoptosis. J Biol Chem 280: 15497–15502 [DOI] [PubMed] [Google Scholar]
- Brady CA, Attardi LD 2010. p53 at a glance. J Cell Sci 123: 2527–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart DL, Sage J 2008. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer 8: 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caelles C, Helmberg A, Karin M 1994. p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature 370: 220–223 [DOI] [PubMed] [Google Scholar]
- Carnevale J, Palander O, Seifried LA, Dick FA 2012. DNA damage signals through differentially modified E2F1 molecules to induce apoptosis. Mol Cell Biol 32: 900–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau BN, Borges HL, Chen TT, Masselli A, Hunton IC, Wang JY 2002. Signal-dependent protection from apoptosis in mice expressing caspase-resistant Rb. Nat Cell Biol 4: 757–765 [DOI] [PubMed] [Google Scholar]
- Chen D, Livne-Bar I, Vanderluit JL, Slack RS, Agochiya M, Bremner R 2004. Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell 5: 539–551 [DOI] [PubMed] [Google Scholar]
- Chinnam M, Goodrich DW 2011. RB1, development, and cancer. Curr Top Dev Biol 94: 129–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR 2004. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303: 1010–1014 [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR 2005. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science 309: 1732–1735 [DOI] [PubMed] [Google Scholar]
- Dick FA, Dyson N 2003. pRB contains an E2F1-specific binding domain that allows E2F1-induced apoptosis to be regulated separately from other E2F activities. Mol Cell 12: 639–649 [DOI] [PubMed] [Google Scholar]
- Harbour JW, Dean DC 2000. Rb function in cell-cycle regulation and apoptosis. Nat Cell Biol 2: E65–E67 [DOI] [PubMed] [Google Scholar]
- Hershko T, Ginsberg D 2004. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J Biol Chem 279: 8627–8634 [DOI] [PubMed] [Google Scholar]
- Hilgendorf KI, Leshchiner ES, Nedelcu S, Maynard MA, Calo E, Ianari A, Walensky LD, Lees JA 2013. The retinoblastoma protein induces apoptosis directly at the mitochondria. Genes Dev (this issue). doi: 10.1101/gad.211326.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianari A, Natale T, Calo E, Ferretti E, Alesse E, Screpanti I, Haigis K, Gulino A, Lees JA 2009. Proapoptotic function of the retinoblastoma tumor suppressor protein. Cancer Cell 15: 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, Vousden KH, et al. 2000. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 407: 645–648 [DOI] [PubMed] [Google Scholar]
- Julian LM, Palander O, Seifried LA, Foster JE, Dick FA 2008. Characterization of an E2F1-specific binding domain in pRB and its implications for apoptotic regulation. Oncogene 27: 1572–1579 [DOI] [PubMed] [Google Scholar]
- Korah J, Falah N, Lacerte A, Lebrun JJ 2012. A transcriptionally active pRb–E2F1–P/CAF signaling pathway is central to TGFβ-mediated apoptosis. Cell Death Dis 3: e407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu JI, Dumont P, Hafey M, Murphy ME, George DL 2004. Mitochondrial p53 activates Bak and causes disruption of a Bak–Mcl1 complex. Nat Cell Biol 6: 443–450 [DOI] [PubMed] [Google Scholar]
- Macleod KF, Hu Y, Jacks T 1996. Loss of Rb activates both p53-dependent and independent cell death pathways in the developing mouse nervous system. EMBO J 15: 6178–6188 [PMC free article] [PubMed] [Google Scholar]
- Marchenko ND, Zaika A, Moll UM 2000. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem 275: 16202–16212 [DOI] [PubMed] [Google Scholar]
- Masselli A, Wang JY 2006. Phosphorylation site mutated RB exerts contrasting effects on apoptotic response to different stimuli. Oncogene 25: 1290–1298 [DOI] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM 2003. p53 has a direct apoptogenic role at the mitochondria. Mol Cell 11: 577–590 [DOI] [PubMed] [Google Scholar]
- Morgenbesser SD, Williams BO, Jacks T, DePinho RA 1994. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature 371: 72–74 [DOI] [PubMed] [Google Scholar]
- Moroni MC, Hickman ES, Lazzerini Denchi E, Caprara G, Colli E, Cecconi F, Muller H, Helin K 2001. Apaf-1 is a transcriptional target for E2F and p53. Nat Cell Biol 3: 552–558 [DOI] [PubMed] [Google Scholar]
- Nahle Z, Polakoff J, Davuluri RV, McCurrach ME, Jacobson MD, Narita M, Zhang MQ, Lazebnik Y, Bar-Sagi D, Lowe SW 2002. Direct coupling of the cell cycle and cell death machinery by E2F. Nat Cell Biol 4: 859–864 [DOI] [PubMed] [Google Scholar]
- Pan H, Yin C, Dyson NJ, Harlow E, Yamasaki L, Van Dyke T 1998. Key roles for E2F1 in signaling p53-dependent apoptosis and in cell division within developing tumors. Mol Cell 2: 283–292 [DOI] [PubMed] [Google Scholar]
- Pucci B, Claudio PP, Masciullo V, Bellincampi L, Terrinoni A, Khalili K, Melino G, Giordano A 2002. pRb2/p130 promotes radiation-induced cell death in the glioblastoma cell line HJC12 by p73 upregulation and Bcl-2 downregulation. Oncogene 21: 5897–5905 [DOI] [PubMed] [Google Scholar]
- Reddien PW, Andersen EC, Huang MC, Horvitz HR 2007. DPL-1 DP, LIN-35 Rb and EFL-1 E2F act with the MCD-1 zinc-finger protein to promote programmed cell death in Caenorhabditis elegans. Genetics 175: 1719–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin SM, Gall AL, Zheng N, Pavletich NP 2005. Structure of the Rb C-terminal domain bound to E2F1-DP1: A mechanism for phosphorylation-induced E2F release. Cell 123: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Schertel C, Conradt B 2007. C. elegans orthologs of components of the RB tumor suppressor complex have distinct pro-apoptotic functions. Development 134: 3691–3701 [DOI] [PubMed] [Google Scholar]
- Sherr CJ, McCormick F 2002. The RB and p53 pathways in cancer. Cancer Cell 2: 103–112 [DOI] [PubMed] [Google Scholar]
- Symonds H, Krall L, Remington L, Saenz-Robles M, Lowe S, Jacks T, Van Dyke T 1994. p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell 78: 703–711 [DOI] [PubMed] [Google Scholar]
- Talos F, Petrenko O, Mena P, Moll UM 2005. Mitochondrially targeted p53 has tumor suppressor activities in vivo. Cancer Res 65: 9971–9981 [DOI] [PubMed] [Google Scholar]
- Tsai KY, Hu Y, Macleod KF, Crowley D, Yamasaki L, Jacks T 1998. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol Cell 2: 293–304 [DOI] [PubMed] [Google Scholar]
- Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM 2012. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell 149: 1536–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Roemer K 2005. Retinoblastoma protein is required for efficient colorectal carcinoma cell apoptosis by histone deacetylase inhibitors in the absence of p21Waf. Biochem Pharmacol 69: 1059–1067 [DOI] [PubMed] [Google Scholar]
- Wang X 2001. The expanding role of mitochondria in apoptosis. Genes Dev 15: 2922–2933 [PubMed] [Google Scholar]
- Zacksenhaus E, Jiang Z, Chung D, Marth JD, Phillips RA, Gallie BL 1996. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev 10: 3051–3064 [DOI] [PubMed] [Google Scholar]
- Ziebold U, Reza T, Caron A, Lees JA 2001. E2F3 contributes both to the inappropriate proliferation and to the apoptosis arising in Rb mutant embryos. Genes Dev 15: 386–391 [DOI] [PMC free article] [PubMed] [Google Scholar]