Specific microRNAs (miRNAs), including miR-134, localize to neuronal dendrites, where they control synaptic protein synthesis and plasticity. Bicker et al. show that the neuronal precursor-miRNA-134 (pre-miR-134) accumulates in dendrites of hippocampal neurons and at synapses in vivo. The DEAH-box helicase DHX36 mediates dendritic localization of pre-miR-134 by directly associating with its terminal loop. DHX36 is required for both miR-134-dependent inhibition of target gene expression and the control of dendritic spine size. These results identify a mechanism of miRNA transport in dendrites.
Keywords: dendritic transport, DHX36, precursor-microRNA, protein synthesis, synaptic plasticity
Abstract
Specific microRNAs (miRNAs), including miR-134, localize to neuronal dendrites, where they control synaptic protein synthesis and plasticity. However, the mechanism of miRNA transport is unknown. We found that the neuronal precursor-miRNA-134 (pre-miR-134) accumulates in dendrites of hippocampal neurons and at synapses in vivo. Dendritic localization of pre-miR-134 is mediated by the DEAH-box helicase DHX36, which directly associates with the pre-miR-134 terminal loop. DHX36 function is required for miR-134-dependent inhibition of target gene expression and the control of dendritic spine size. Dendritically localized pre-miR-134 could provide a local source of miR-134 that can be mobilized in an activity-dependent manner during plasticity.
MicroRNAs (miRNAs) are important regulators of morphological and functional plasticity in the developing and mature nervous system (Ashraf et al. 2006; Schratt et al. 2006; Rajasethupathy et al. 2009; Gao et al. 2010). Specific miRNAs control plasticity by binding to dendritic mRNAs (Schuman et al. 2006; Bramham and Wells 2007; Martin and Ephrussi 2009), thereby modulating the local synthesis of the respective proteins at synapses in an activity-dependent manner (Schratt et al. 2006; Banerjee et al. 2009; Fiore et al. 2009; Muddashetty et al. 2011). The biogenesis of miRNAs involves two subsequent RNase-dependent cleavage steps: Drosha-mediated processing of primary miRNA transcripts (pri-miRNAs) in the nucleus followed by Dicer-mediated processing of 70- to 100-nucleotide (nt) stem–loop precursors (precursor-miRNAs [pre-miRNAs]) in the cytoplasm (Kim 2005; Krol et al. 2010). miRNA biogenesis can be regulated by sequence-specific RNA-binding proteins (RBPs) that preferentially interact with the terminal loop structure of pri- and pre-miRNAs (Krol et al. 2010; Newman and Hammond 2010). Interestingly, miR-29b contains a sequence element at the 3′ end of the mature miRNA that directs its nuclear localization (Hwang et al. 2007), but nothing is known about miRNA sequence determinants responsible for localization to specific cytoplasmic compartments, such as the synapto-dendritic compartment of neurons.
We speculated that dendritic localization of specific miRNAs could occur at the pre-miRNA stage, possibly by the presence of sequence elements within pre-miRNA terminal loops that serve as binding sites for RBPs, which are in turn part of transport complexes. Dendritic localization of specific pre-miRNAs would offer the advantage of a local source of miRNAs that could be mobilized, for example, in response to synaptic activity.
Results and Discussion
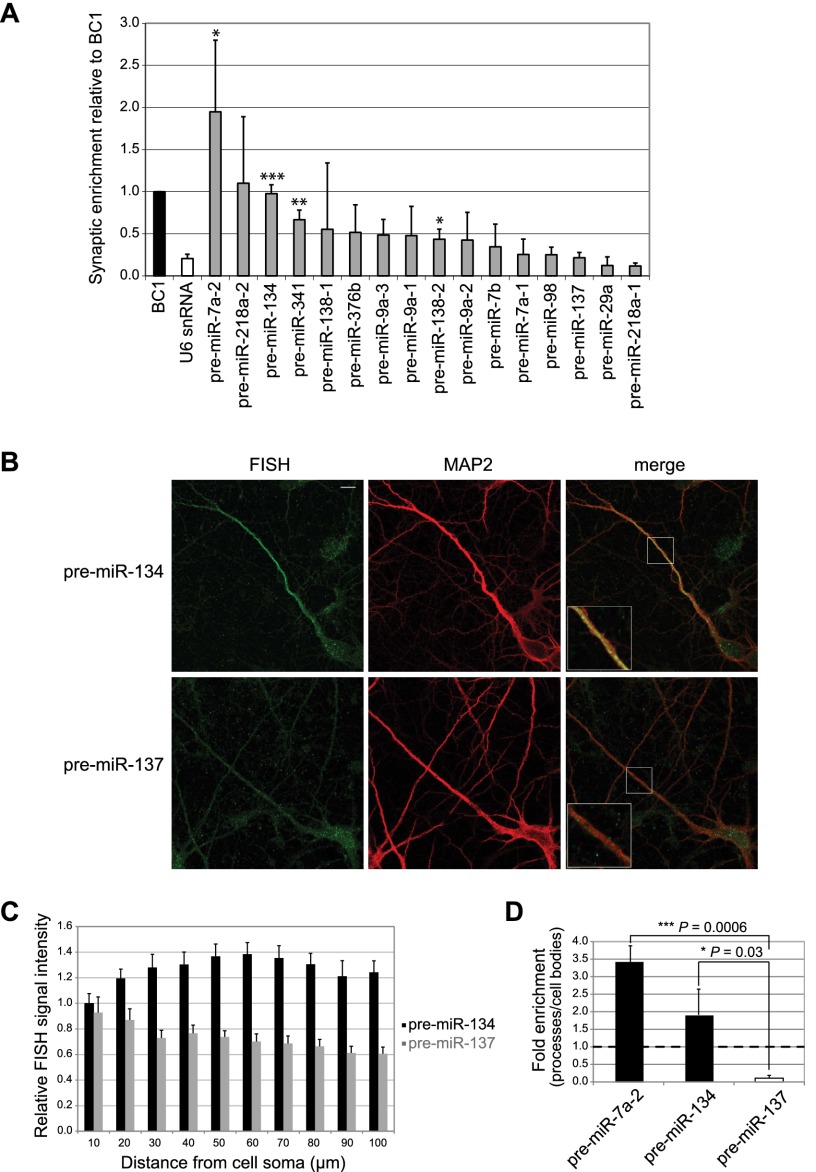
To interrogate synapto-dendritic localization of pre-miRNAs, we selected 16 previously identified dendritic miRNAs (Schratt et al. 2006; Siegel et al. 2009) and determined corresponding pre-miRNA levels in synaptosomes by quantitative RT–PCR (qRT–PCR). We found that for two of these candidates (pre-miR-7a-2 and pre-miR-134), the degree of synaptic enrichment was significantly higher compared with the nuclear U6 snRNA and at least equal to the known dendritic noncoding RNA BC1 (Brosius and Tiedge 2001), suggesting that these pre-miRNAs are actively transported into the synapto-dendritic compartment (Fig. 1A). Due to the well-documented synaptic functions of miR-134 (Schratt et al. 2006; Fiore et al. 2009; Gao et al. 2010), we decided to focus on pre-miR-134. Synaptic enrichment of pre-miR-134 was validated by Northern blot of synaptosomal and whole-brain RNA preparations (Supplemental Fig. S1; Siegel et al. 2009). Since synaptosomes contain both pre- and postsynaptic compartments, we performed locked nucleic acid (LNA)-based fluorescent in situ hybridization (FISH) in rat hippocampal neurons (7 d in vitro [DIV]) (Fig. 1B) to determine the subcellular localization of pre-miR-134. BDNF (brain-derived neurotrophic factor) treatment of neurons was used to increase pre-miR-134 expression (Fiore et al. 2009). Using a FISH probe specifically recognizing the loop sequence of pre-miR-134 (Supplemental Fig. S2), we detected a specific signal in the neuronal cell body as well as discrete punctate staining in proximal and distal parts of MAP2-positive dendrites (Fig. 1B, top panel, insert at higher magnification). In contrast, expression of pre-miR-137, which was not enriched in synaptosomes, was restricted to the cell body compartment (Fig. 1B, bottom panel). Quantification of FISH signals along the entire length of multiple dendrites confirms the specific dendritic localization of pre-miR-134 (Fig. 1C). The specificity of our FISH protocol was validated with negative (scrambled LNA oligonucleotide) and positive (LNA oligonucleotide recognizing mature miR-134) (Schratt et al. 2006) control probes (Supplemental Fig. S3). The presence of pre-miR-134 and pre-miR-7a-2, but not pre-miR-137, in processes of rat hippocampal neurons was further confirmed using a compartmentalized culture system (Fig. 1D; Supplemental Fig. S4a,b). This system also faithfully recapitulated process enrichment of mature miR-134 but not of the nondendritic miR-133b (Supplemental Fig. S4c). Together, our data obtained with synaptosomes, FISH, and compartmentalized neuron cultures support a specific localization of endogenous pre-miR-134 in dendrites of primary hippocampal neurons and at synapses in vivo.
Figure 1.
pre-miR-134 localizes to neuronal dendrites and synapses. (A) Levels of the indicated pre-miRNAs, BC1, and U6 snRNA in rat postnatal day 15 (P15) synaptosomes relative to whole forebrain measured by qRT–PCR (mean ± SD, n = 3). (*) P < 0.05; (**) P < 0.01; (***) P < 0.001). BC1 was set to one. (B, left) Representative images from FISH on BDNF-treated hippocampal neurons (7 DIV) using LNA probes directed against the terminal loop of either pre-miR-134 (top) or pre-miR-137 (bottom). (Middle) MAP2 immunostaining. (Right) Merge. Inserts at higher magnification illustrate the presence of pre-miR-134 puncta and the absence of pre-miR-137 puncta in distal dendrites. Bar, 10 μm. (C) Quantification of FISH analysis performed in B. Relative signal intensities of dendritic segments derived from 20 neurons of each condition ±SD. (D) Levels of indicated pre-miRNAs in the process compartment of hippocampal neurons relative to cell bodies measured by qRT–PCR (mean ± SD, n = 3).
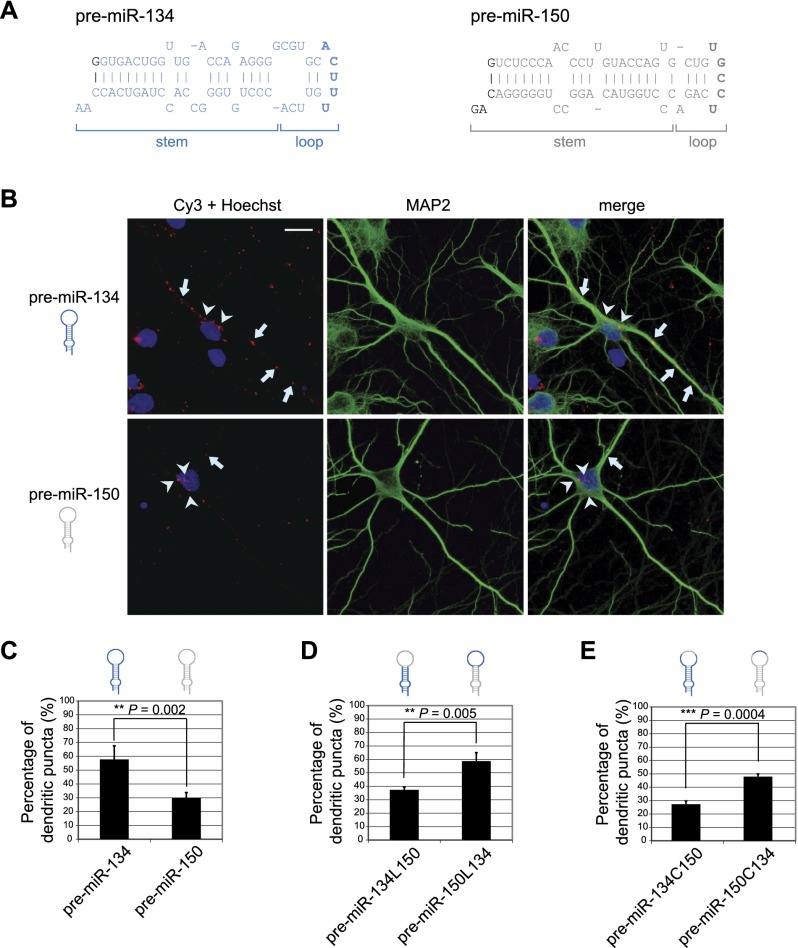
We next sought to determine the specific sequence elements required for dendritic localization of pre-miR-134 in hippocampal neurons. We transfected in vitro transcribed, fluorescently (Cy3) labeled pre-miRNAs into hippocampal neurons and analyzed dendritic localization by fluorescence confocal microscopy. All synthetic pre-miRNAs contain a 2-nt 3′ overhang, thereby resembling endogenous Drosha cleavage products (Fig. 2A). The majority of puncta formed by wild-type Cy3-pre-miR-134 localized to dendrites (Fig. 2B, top panel, arrows), demonstrating that exogenous pre-miR-134 recapitulates dendritic localization of the endogenous counterpart. The subcellular localization of Cy3-pre-miR-134 puncta was specific, since the majority of a nondendritic pre-miRNA, Cy3-pre-miR-150, localized to the neuronal cell body (Fig. 2B, bottom panel, arrowheads). Quantification of Cy3-positive puncta from at least 60 neurons for each experimental condition confirmed the specific dendritic localization of Cy3-pre-miR-134 (Fig. 2C). Only Cy3-positive puncta that were intracellularly localized were considered, as judged by the overlap with the dendritic marker protein MAP2 along the entire confocal Z-stack (Supplemental Fig. S5a). In contrast, we did not observe localization of Cy3-pre-miR-134 to TAU-1-positive axons (Supplemental Fig. S5b). Importantly, the vast majority of pre-miR-134 within these puncta was not yet processed at the time of analysis (2.5 h after transfection), as shown by FISH (Supplemental Fig. S6). The loop region is the most accessible structure within pre-miRNAs and often serves as a binding platform for regulatory protein complexes (Krol et al. 2010; Newman and Hammond 2010). Replacing the loop sequence of pre-miR-134 with the loop of the nondendritic pre-miR-150 (pre-miR-134L150) abolished dendritic accumulation of pre-miR-134 puncta (Fig. 2D; Supplemental Fig. S7a). Conversely, putting the pre-miR-134 loop sequence within the context of the pre-miR-150 stem (pre-miR-150L134) completely restored dendritic localization (Fig. 2D; Supplemental Fig. S7a). Further mutagenesis analysis revealed that exchanging the five central loop nucleotides (Fig. 2A, bold letters) within the context of pre-miR-134 (pre-miR-134C150) abolished dendritic enrichment, whereas a pre-miR-150 mutant containing the central loop nucleotides of pre-miR-134 (pre-miR-150C134) largely regained the capacity to localize to dendrites (Fig. 2E; Supplemental Fig. S7b). Together, these experiments suggest that the pre-miR-134 loop sequence is an important determinant of pre-miR-134 dendritic localization.
Figure 2.
The pre-miR-134 terminal loop is necessary and sufficient for dendritic targeting. (A) Sequences of in vitro transcribed pre-miR-134 (blue) and pre-miR-150 (gray). The five central nucleotides of the terminal loop are shown in bold. Sequence changes required for efficient in vitro transcription (G at position +1) and for correct 3′ termini (2-nt overhang) are indicated in black. (B) Representative images of hippocampal neurons transfected with indicated Cy3-pre-miRNAs. (Left) Cy3-pre-miRNA (red) and Hoechst (blue). (Middle) MAP2 (green). (Right) Merge. Arrows and arrowheads indicate pre-miRNA puncta localized in dendrites or the cell body, respectively. Bar, 10 μm. (C–E) Percentage of dendritic puncta for the indicated pre-miRNAs or mutants (mean ± SD, n = 4, 64 neurons per condition). (L) Loop; (C) central loop.
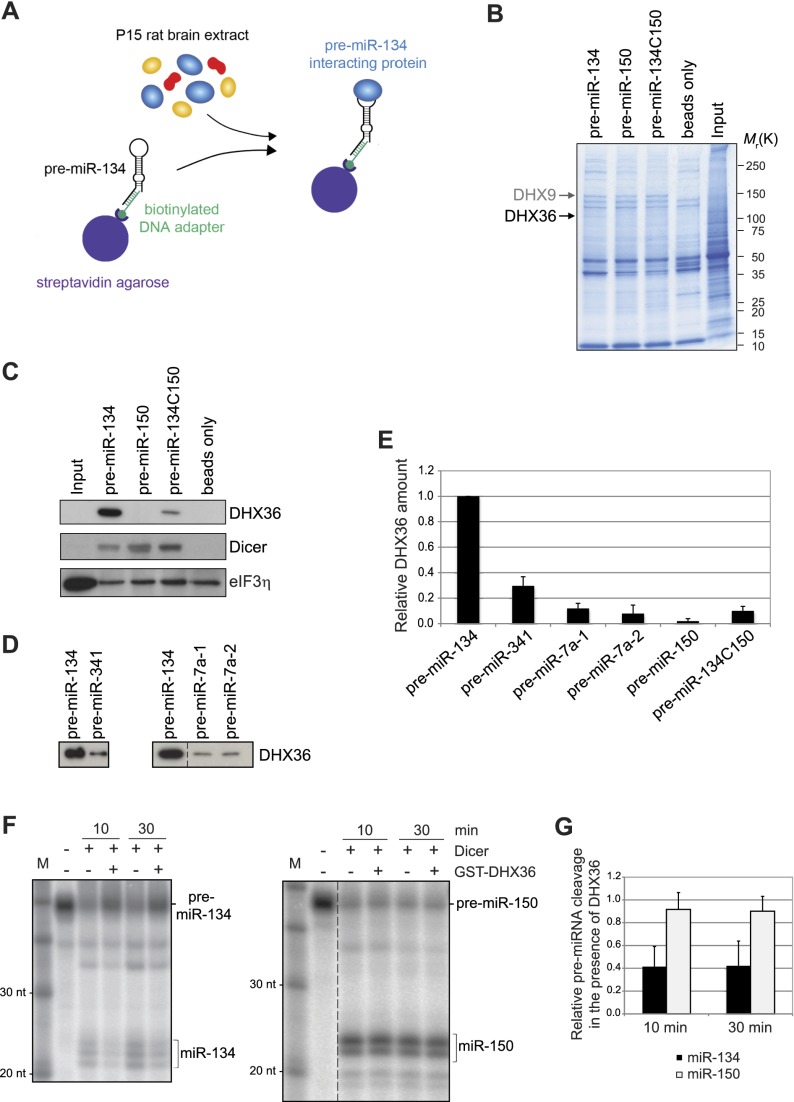
We speculated that specific RBPs that interact with the pre-miR-134 loop might be responsible for directed transport to the synapto-dendritic compartment. To identify proteins that selectively interact with the pre-miR-134 loop, we performed pull-down experiments by incubating rat forebrain protein extract with biotin-tagged pre-miR-134 or respective mutants coupled to streptavidin beads (Fig. 3A; Heo et al. 2008, 2009). Thereby, we identified one band above 100 kDa that was specifically present in eluates from pull-downs with wild-type pre-miR-134 but not with pre-miR-150 or pre-miR-134C150 (Fig. 3B, arrow). We identified this protein as DHX36 by mass spectrometry (Supplemental Table T1). In addition, we identified DHX9 (Fig. 3B, arrow) as a protein that interacts with all tested pre-miRNAs, in accordance with a described function of DHX9 within RNA-induced silencing complex (RISC) (Robb and Rana 2007). The specific interaction of pre-miR-134 with DHX36 could be confirmed by Western blot using an anti-DHX36 antibody (Fig. 3C, top panel). Importantly, Dicer interacted with all three pre-miRNAs (Fig. 3C, middle panel), demonstrating that all of them were per se competent in protein binding. An unrelated protein, eIF3η, did not specifically interact with any of the tested pre-miRNAs (Fig. 3C, bottom panel). In the case of other dendritically localized pre-miRNAs, the interaction with DHX36 was strongly reduced (Fig. 3D,E), suggesting that DHX36 preferentially interacts with pre-miR-134. Affinity-purified full-length DHX36 and the isolated DHX36 N-terminal domain, which contains an RNA-binding motif (Chalupnikova et al. 2008), both associated with radiolabeled pre-miR-134 in electrophoretic mobility shift assay (EMSA) in vitro (Supplemental Fig. S8), demonstrating a direct interaction between DHX36 and pre-miR-134. Using in vitro pre-miRNA cleavage assays (Leuschner and Martinez 2007), we found that DHX36 interfered with cleavage of synthetic pre-miR-134 by recombinant Dicer but not with Dicer-mediated pre-miR-150 cleavage (Fig. 3F,G). Together, our experiments demonstrate that DHX36 directly interacts with pre-miR-134 and reduces Dicer-mediated pre-miR-134 cleavage in vitro.
Figure 3.
DHX36 interacts with the pre-miR-134 terminal loop. (A) Scheme of the purification strategy used to isolate pre-miR-134-interacting proteins from P15 rat brain extract. (B) Coomassie gel of pull-downs using rat P15 brain extract and the indicated pre-miRNAs. Arrows point to bands that were identified by mass spectrometry as DHX9 (∼140 kDa) and DHX36 (∼110 kDa). (C) Western blot against the indicated proteins with extracts from pull-downs shown in B. (D) Western blot against DHX36 with extracts from the indicated pre-miRNA pull-downs. (E) Quantification of DHX36 binding in pre-miRNA pull-down experiments (pre-miR-134 = 1; mean ± SD, n = 3). (F) In vitro pre-miRNA cleavage assay. pre-miRNAs were incubated with recombinant Dicer for the indicated time in either the presence or absence of GST-DHX36. The position of pre-miRNAs and mature miRNAs is indicated. (G) Ratio of mature miRNA to pre-miRNA as an index for Dicer cleavage activity (mean ± SD, n = 4–5; conditions without DHX36 were set to 1 for both time points).
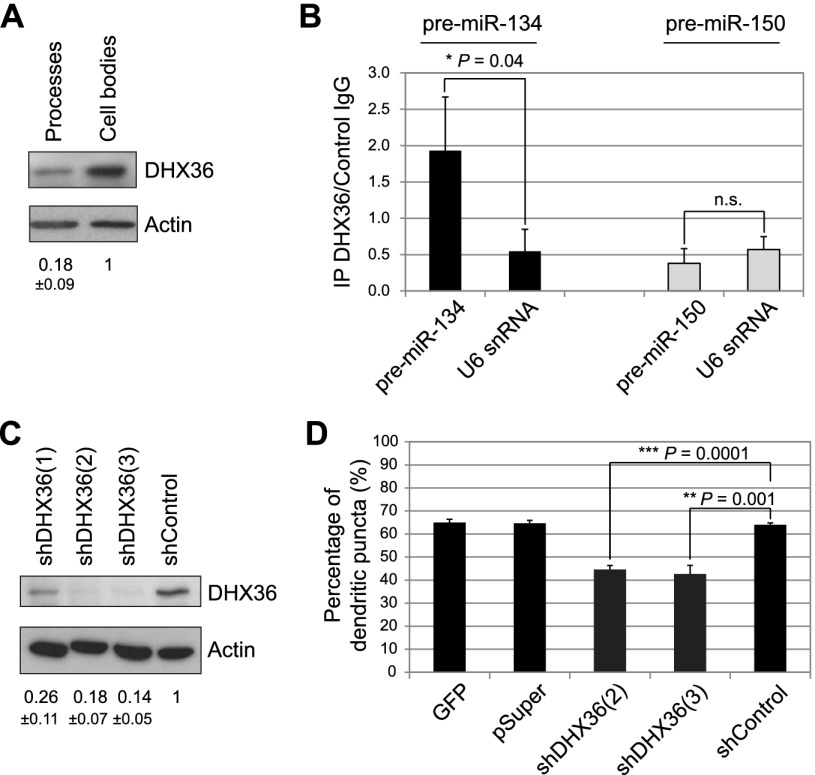
We next addressed whether DHX36 functions in dendritic localization of pre-miR-134. DHX36 was shown to resolve secondary nucleic acid structures in the nucleus (Vaughn et al. 2005) and regulate RNA metabolism in the cytoplasm in nonneuronal cells (Tran et al. 2004; Chalupnikova et al. 2008; Zhang et al. 2011). However, DHX36 has not yet been studied in neurons. Using compartmentalized primary rat hippocampal neuron cultures, we detected DHX36 expression in both the cell body and, to a lesser extent, the process compartment (Fig. 4A). In further support of a function of DHX36 in neuronal processes, ectopically expressed GFP-DHX36 fusion protein localized to dendrites and axons (Supplemental Fig. S9). To test whether pre-miR-134 interacts with DHX36 in neurons, we transfected cortical neurons at 6 DIV with either pre-miR-134 or pre-miR-150 and performed an RNA immunoprecipitation (RNA-IP) assay. We found that transfected pre-miR-134, in contrast to pre-miR-150, was specifically enriched in anti-DHX36 compared with IgG immunoprecipitates, as judged by qRT–PCR (Fig. 4B). This finding suggests that endogenous DHX36 and synthetic pre-miR-134 interact in neurons. To investigate the function of DHX36, we designed shRNA expression constructs targeting different regions of the rat DHX36 mRNA and assessed their efficiency by nucleofection (Fig. 4C). To assess the role of DHX36 in dendritic transport of pre-miR-134, we combined plasmid-based knockdown of DHX36 (3 d of expression) with short-term transfection of Cy3-labeled pre-miRNAs. To rule out potential effects of DHX36 knockdown on miR-134 processing, we decided to study subcellular distribution of synthetic Cy3-pre-miR-134 shortly after transfection. At 2.5 h after transfection, Cy3-pre-miR-134 was found to be excluded from the nucleus, and the vast majority of Cy3-pre-miR-134 was still not processed (Supplemental Fig. S6). Neurons transfected with the two most effective DHX36 shRNAs displayed a significant reduction in the percentage of dendritic Cy3-pre-miR-134 puncta (Fig. 4D; Supplemental Fig. S10a). DHX36 knockdown did not affect global levels of pre-miR-134 and mature miR-134 (Supplemental Fig. S10b,c). These results suggest that DHX36 is specifically required for the accumulation of pre-miR-134 in neuronal dendrites.
Figure 4.
DHX36 is required for dendritic localization of pre-miR-134 in hippocampal neurons. (A) Western blot against the indicated proteins with extracts from process or cell body fractions of hippocampal neurons. Quantification of band intensities is indicated below (mean ± SD, n = 3). (B) RNA-IP of cortical neurons transfected with the indicated pre-miRNAs using a DHX36 or control IgG antibody. Ratio of RNA amounts from DHX36 immunoprecipitates to control IgG immunoprecipitates as determined by qRT–PCR is shown (mean ± SD, n = 3) (C) Western blot against the indicated proteins with extracts from hippocampal neurons nucleofected with the indicated shRNA-expressing constructs. Quantification of band intensities is indicated below (mean ± SD, n = 2). (D) Percentage of dendritic Cy3-pre-miR-134 puncta observed in hippocampal neurons transfected with the indicated constructs (mean ± SD, n = 3, ANOVA: F(4;10) = 84, P < 0.001; 30 neurons per condition).
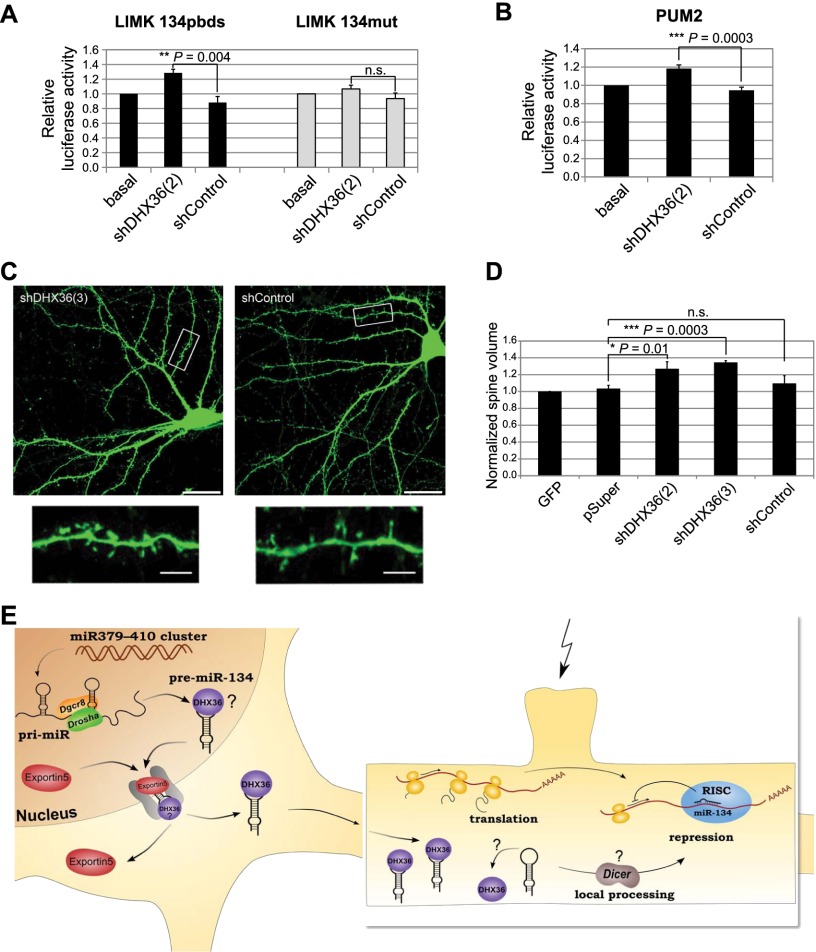
Finally, we addressed whether DHX36-dependent localization of pre-miR-134 might contribute to the activity of miR-134 in dendrites. For these experiments, we first used a luciferase reporter gene that contains a perfect miR-134-binding site in the context of the Limk1 3′ untranslated region (UTR; Limk1 134pbds) (Christensen et al. 2010). The Limk1 3′ UTR confers dendritic localization of the reporter (Schratt et al. 2006), allowing us to study the role of DHX36 in local translation. Upon DHX36 knockdown, expression of the Limk1 134pbds reporter gene was significantly increased compared with control conditions, whereas expression of a reporter gene containing a mutated miR-134-binding site was unaffected (Fig. 5A). In addition, both a Pum2 luciferase reporter gene containing a single miR-134-binding site (Fiore et al. 2009) and endogenous Limk1 protein expression were similarly induced by DHX36 knockdown in neurons (Fig. 5B; Supplemental Fig. S10d). Together, these results demonstrate that DHX36 knockdown results in an increased expression of known dendritic miR-134 target genes, possibly due to reduced dendritic pre-miR-134 accumulation. Nevertheless, based on our data, we cannot rule out that derepression of miR-134 targets in the cell body might also contribute to the observed effects of DHX36 knockdown. Finally, we interrogated whether DHX36 knockdown had an impact on neuronal function, possibly via regulation of miR-134 activity. Toward this end, we analyzed dendritic spine morphology, since we previously found that treatment of hippocampal neurons with miR-134 function-blocking antisense oligonucleotides led to elevated spine size (Schratt et al. 2006). Consistent with reduced dendritic miR-134 activity upon DHX36 knockdown, we found that neurons transfected with a DHX36 shRNA construct contained a higher fraction of large mushroom-shaped spines compared with neurons transfected with a control shRNA (Fig. 5C; Supplemental Fig. S11). Accordingly, the average spine volume of neurons transfected with two independent DHX36 shRNA constructs was significantly higher compared with control conditions (Fig. 5D). This result demonstrates that DHX36 negatively regulates dendritic spine morphogenesis in hippocampal neurons. It is further consistent with the idea that DHX36-dependent pre-miR-134 accumulation in dendrites could be important for neuronal function.
Figure 5.
DHX36 is required for miR-134-regulated neuronal functions. (A,B) Reporter gene activity in primary neurons transfected with the indicated shRNA constructs and firefly luciferase reporters containing Limk1 (A; 15 DIV hippocampal neurons) and Pum2 (B; 15 DIV cortical neurons) 3′ UTRs. Cotransfection of empty Renilla reporter served as transfection control. Relative luciferase activity represents the ratio of firefly to Renilla activity; basal condition was set to 1 (mean ± SD, n = 3, each experimental condition measured in duplicates within one experiment). (C) Representative images of hippocampal neurons transfected with either shDHX36(3) (left) or shControl (right) shRNAs. Inserts at higher magnification illustrate increased proportion of large, mushroom-shaped spines in the shDHX36 condition compared with shControl. Bars: main panels, 20 μm; inserts, 5 μm. (D) Quantification of average normalized spine volume as determined by the ratio of GFP signal in dendritic spines to total cell intensity. GFP-only condition was set to 1 (mean ± SD, n = 3; ANOVA: F(3;8) = 15,921, P = 0.001; 18 neurons per condition; 150–200 spines per neuron). (E) Working model for the role of DHX36 in pre-miR-134 dendritic transport and processing. The pre-miR-134 loop contains a cis-acting dendritic targeting element that is recognized by DHX36, resulting in pre-miR-134 transport to the dendrite. In the postsynaptic compartment, stimulation of specific dendritic spines (flash) may trigger Dicer-dependent pre-miRNA processing (possibly involving release of DHX36), thereby enabling an enhanced production of mature miR-134 and repression of miR-134 target mRNAs.
Taken together, our results show that DHX36 plays a role in dendritic localization of pre-miR-134 and spine morphogenesis, a process regulated by miR-134 (Fig. 5E). This adds subcellular localization to the growing list of functions for proteins that associate with the terminal loop of specific pre-miRNAs (Krol et al. 2010). The presumptive DHX36-binding site within the pre-miR-134 central loop is not present in the loop of other dendritic pre-miRNAs, which, together with our in vitro binding assays, suggests that distinct localization mechanisms are used by specific dendritic pre-miRNAs. Previously, we found that mature miR-134 is delivered to dendrites when transfected into neurons (Schratt et al. 2006). The underlying mechanism is unknown but could involve cotransport of miR-134 with its dendritic target mRNAs, such as Limk1. We speculate that dendritically localized pre-miR-134 might, in parallel, provide a pool of miR-134 that can be rapidly mobilized in response to activity. This local pool could be important to reinforce miR-134-mediated repression of gene expression, for example, in conditions of chronically elevated activity that are known to induce homeostatic synaptic scaling (Turrigiano 2008). The molecular mechanisms involved in a possible local control of miR-134 production in dendrites are currently unknown. Dicer has been detected in synaptosomes (Lugli et al. 2005), suggesting that pre-miR-134 could undergo local Dicer-dependent processing at synapses. Since DHX36 competes with Dicer in vitro (Fig. 3F) and Argonaute complexes that contain DHX36 are devoid of Dicer activity (Hock et al. 2007), we speculate that pre-miR-134 has to be released from DHX36 upon activity to allow efficient pre-miR-134 processing at the synapse (Fig. 5E). Future work will address the mechanism of activity-dependent local processing of pre-miR-134 and its significance for dendritic protein synthesis and synaptic plasticity.
Materials and methods
Cell culture
Primary hippocampal and cortical embryonic (embryonic day 18) neuron cultures were prepared and cultured as described (Schratt et al. 2006). For compartmentalized neuron cultures, dissociated hippocampal neurons were plated onto 1-μm pore and 30-mm diameter polyethylene tetraphthalate (PET) membrane filter inserts (Millipore) that were matrix-coated with poly-L-lysine (Sigma-Aldrich) and Laminin (BD Biosciences) on the top and bottom (Poon et al. 2006).
Transfections
Transfections of primary neurons using Lipofectamine 2000 (Invitrogen) were performed as described (Siegel et al. 2009). For validation of shRNA efficacy, primary cortical neurons of rat embryos (E18) were nucleofected using the P3 Primary Cell 4D-Nucleofector kit (Lonza), program DC-104, according to the manufacturer's instructions. Transfections of primary hippocampal or cortical neurons with in vitro transcribed pre-miRNAs (25–75 nM) were performed using siPORT NeoFX transfection agent (Ambion).
Preparation of synaptosomes
Synaptosomes were prepared from postnatal day 15 (P15) Sprague-Dawley rat pups as previously described (Schratt et al. 2004).
In vitro transcription of pre-miRNAs
Pre-miRNAs were in vitro transcribed from dsDNA templates (Supplemental Material) using the MEGAshortscript T7 kit (Ambion) according to the manufacturer's instructions, with a template concentration of 150 nM and an incubation time of 4 h at 37°C. pre-miRNAs were purified using illustra MicroSpin G-25 columns (GE Healthcare).
RNA labeling
Labeling of pre-miRNAs by Cy3 was performed using the Label IT nucleic acid labeling kit (Mirus) according to the manufacturer's instructions. For Dicer assays, pre-miRNAs were 5′ end-labeled using [γ-32P] ATP (PerkinElmer) as previously described (Leuschner and Martinez 2007).
Subcellular localization of in vitro transcribed pre-miRNAs
Rat hippocampal neurons were transfected at 6 DIV with 30 nM Cy3-labeled precursors using siPORT NeoFX transfection agent (Ambion) for 2 h. Fifteen minutes post-transfection, cells were treated with Cell Scrub buffer (Genlantis) for 5 min to remove extracellular Cy3 complexes, fixed, and stained for MAP2 to visualize dendrites. Nuclei were counterstained with Hoechst (Invitrogen). Data sets were blinded to the experimental conditions, and Z-stack images (seven consecutive optical sections per stack) were taken with an LSM 5 Pascal confocal microscope (Zeiss). ImageJ software (“analyze particles” option) was used for subsequent analysis of intracellular Cy3 puncta distribution. For quantification, the average percentage of dendritic puncta compared with the total number of intracellular puncta was determined for each pre-miRNA.
RNA pull-down
The RNA pull-down approach used in this study was based on a previously described protocol for isolation of pre-let-7-binding proteins concerning the general procedure (Heo et al. 2008) and on a protocol for FMRP immunoprecipitation concerning brain lysate preparation (Edbauer et al. 2010). See the Supplemental Material for further details.
RNA-IP
For RNA-IP, cortical neurons were transfected with in vitro transcribed pre-miRNAs and immunoprecipitated using rabbit anti-DHX36 antibody (Novus Biologicals). RNA-IP was performed using the same buffer system as described for the RNA pull-down approach. A detailed protocol can be found in the Supplemental Material.
FISH
Dissociated hippocampal neurons were treated with BDNF (PeproTech, 50 ng/mL) for 2 h before fixation. FISH was performed as previously described (Fiore et al. 2009) using FITC-labeled LNA probes (5 pmol per well in the 24-well format; Exiqon) directed against the pre-miR-134 loop, the pre-miR-137 loop, the mature miR-134 sequence, and a control probe of unrelated sequence (scramble). Signals were further amplified by a two-step antibody procedure. Neurons were costained for the dendritic protein MAP2.
Luciferase assay
Primary neurons were transfected in duplicates with 50 ng of pGL3 firefly reporter constructs and equal amounts of empty Renilla reporter as transfection control. For DHX36 knockdown, neurons were cotransfected with 20 ng of the respective pSuper construct. Luciferase assays were performed after 3 d of expression using the Dual-Luciferase reporter assay system (Promega) on the GloMax R96 Microplate Luminometer (Promega).
Spine analysis
For spine analysis, images were taken on an LSM5 Zeiss Pascal confocal microscope and analyzed with ImageJ software as previously described (Siegel et al. 2009). For each condition, spines from 18 representative hippocampal neurons (150–200 spines per neuron) derived from three independent experiments were measured.
Antibodies
Primary antibodies used in this study were as follows: mouse anti-β-actin antibody (Sigma), rabbit anti-DHX36 antibody (Novus Biologicals; immunoprecipitation, Western), goat anti-DHX36 antibody (Santa Cruz Biotechnology; Western), rabbit anti-Dicer antibody (D-349; provided by Witold Filipowicz, FMI Basel), goat anti-eIF3η antibody (Santa Cruz Biotechnology), Alexa488 rabbit anti-FITC antibody (Molecular Probes), rabbit anti-GFAP antibody (DAKO), mouse anti-MAP2 antibody (Sigma), and rabbit anti-NUP153 antibody (Abcam).
Plasmids
shRNA sequences (summarized in the Supplemental Material) were designed using the online available Dharmacon siRNA design center and cloned into pSuper (Oligoengine). Reporter plasmids pGL3 Limk 134pbds (Christensen et al. 2010), pGL3 Limk1 134mut (Schratt et al. 2006), and pGL3 Pum2(long) (Fiore et al. 2009) have been described. pAcG3XE-RHAU-His6, pGEX-2T-GST-Nter, and pEGFP-C1-DHX36 were provided by Yoshikuni Nagamine (Friedrich Miescher Institute, Basel, Switzerland).
Dicer assay
[32P] 5′ end-labeled pre-miRNAs (20,000 counts per minute [cpm]) were incubated in a processing reaction with 1 U of recombinant Dicer (Recombinant Dicer Enzyme kit, Genlantis) for 10–30 min at 37°C in either the absence or presence (preincubation for 1 h at room temperature) of 30 nM GST-DHX36 (full-length). Reaction products were resolved on a 15% denaturing PAGE and visualized by autoradiography.
Mass spectrometry analysis
MALDI was performed using an Autoflex Bruker Daltonics mass spectrometer. Data analysis was done with the MASCOT program (Matrix Science). A detailed protocol can be found in the Supplemental Material.
Statistics
For each data set, three independent biological experiments were performed if not stated otherwise. Error bars represent standard deviation. P-values were calculated with Student's t-test (two-tailed, type 2) for one-way comparisons and with ANOVA followed by post-hoc test (Student's t-test with Bonferroni correction) for multi-way comparisons.
Acknowledgments
We thank W. Filipowicz and Y. Nagamine for providing reagents; U. Beck, E. Becker, N. Braun, K. Burk, R. Gondrum, G. Jarosch, H. Kaiser, B. Kowalski, and T. Wüst for excellent technical assistance; and R. Fiore for critically reading the manuscript. This work was funded by the DFG (SFB593) and the EMBO Young Investigator Program. S.B. is a recipient of a DOC-fFORTE fellowship of the Austrian Academy of Sciences.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.211243.112.
References
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S 2006. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell 124: 191–205 [DOI] [PubMed] [Google Scholar]
- Banerjee S, Neveu P, Kosik KS 2009. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron 64: 871–884 [DOI] [PubMed] [Google Scholar]
- Bramham CR, Wells DG 2007. Dendritic mRNA: Transport, translation and function. Nat Rev Neurosci 8: 776–789 [DOI] [PubMed] [Google Scholar]
- Brosius J, Tiedge H 2001. Neuronal BC1 RNA: Intracellular transport and activity-dependent modulation. Results Probl Cell Differ 34: 129–138 [DOI] [PubMed] [Google Scholar]
- Chalupnikova K, Lattmann S, Selak N, Iwamoto F, Fujiki Y, Nagamine Y 2008. Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J Biol Chem 283: 35186–35198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen M, Larsen LA, Kauppinen S, Schratt G 2010. Recombinant adeno-associated virus-mediated microRNA delivery into the postnatal mouse brain reveals a role for miR-134 in dendritogenesis in vivo. Front Neural Circuits 3: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M 2010. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, Greenberg ME, Schratt G 2009. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J 28: 697–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH 2010. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466: 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN 2008. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell 32: 276–284 [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708 [DOI] [PubMed] [Google Scholar]
- Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G 2007. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep 8: 1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT 2007. A hexanucleotide element directs microRNA nuclear import. Science 315: 97–100 [DOI] [PubMed] [Google Scholar]
- Kim VN 2005. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol 6: 376–385 [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11: 597–610 [DOI] [PubMed] [Google Scholar]
- Leuschner PJ, Martinez J 2007. In vitro analysis of microRNA processing using recombinant Dicer and cytoplasmic extracts of HeLa cells. Methods 43: 105–109 [DOI] [PubMed] [Google Scholar]
- Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR 2005. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem 94: 896–905 [DOI] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A 2009. mRNA localization: Gene expression in the spatial dimension. Cell 136: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ 2011. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell 42: 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Hammond SM 2010. Emerging paradigms of regulated microRNA processing. Genes Dev 24: 1086–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC 2006. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci 26: 13390–13399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasethupathy P, Fiumara F, Sheridan R, Betel D, Puthanveettil SV, Russo JJ, Sander C, Tuschl T, Kandel E 2009. Characterization of small RNAs in aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron 63: 803–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb GB, Rana TM 2007. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol Cell 26: 523–537 [DOI] [PubMed] [Google Scholar]
- Schratt GM, Nigh EA, Chen WG, Hu L, Greenberg ME 2004. BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J Neurosci 24: 9366–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME 2006. A brain-specific microRNA regulates dendritic spine development. Nature 439: 283–289 [DOI] [PubMed] [Google Scholar]
- Schuman EM, Dynes JL, Steward O 2006. Synaptic regulation of translation of dendritic mRNAs. J Neurosci 26: 7143–7146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, et al. 2009. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol 11: 705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H, Schilling M, Wirbelauer C, Hess D, Nagamine Y 2004. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol Cell 13: 101–111 [DOI] [PubMed] [Google Scholar]
- Turrigiano GG 2008. The self-tuning neuron: Synaptic scaling of excitatory synapses. Cell 135: 422–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn JP, Creacy SD, Routh ED, Joyner-Butt C, Jenkins GS, Pauli S, Nagamine Y, Akman SA 2005. The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J Biol Chem 280: 38117–38120 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Kim T, Bao M, Facchinetti V, Jung SY, Ghaffari AA, Qin J, Cheng G, Liu YJ 2011. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity 34: 866–878 [DOI] [PMC free article] [PubMed] [Google Scholar]