Formation of the anterior–posterior axis of the mouse embryo relies on reciprocal signaling between the pluripotent epiblast and its neighboring tissues, the extraembryonic ectoderm and visceral endoderm. While examining anterior visceral endoderm (AVE) induction, Nowotschin et al. found that the T-box gene Eomesodermin (Eomes) plays an essential role in AVE recruitment. Eomes function was necessary and sufficient to activate the lim homeodomain transcription factor Lhx1 in the visceral endoderm. These findings demonstrate that Eomes initiates a transcriptional program controlling anterior–posterior identity.
Keywords: Eomesodermin, Tbr2, axis formation, visceral endoderm, AVE, DVE Lhx1
Abstract
Reciprocal inductive interactions between the embryonic and extraembryonic tissues establish the anterior–posterior (AP) axis of the early mouse embryo. The anterior visceral endoderm (AVE) signaling center emerges at the distal tip of the embryo at embryonic day 5.5 and translocates to the prospective anterior side of the embryo. The process of AVE induction and migration are poorly understood. Here we demonstrate that the T-box gene Eomesodermin (Eomes) plays an essential role in AVE recruitment, in part by directly activating the homeobox transcription factor Lhx1. Thus, Eomes function in the visceral endoderm (VE) initiates an instructive transcriptional program controlling AP identity.
Formation of the anterior–posterior (AP) axis of the mouse embryo relies on precisely coordinated reciprocal signaling activities between the pluripotent epiblast and its neighboring tissues, the extraembryonic ectoderm (ExE) and visceral endoderm (VE). AP axis formation is initiated at late blastocyst stages with the specification of the distal VE (DVE), a subpopulation of primitive endoderm cells, which, a day later, around E5.5, reside at the distal tip of the post-implantation egg cylinder (Takaoka et al. 2011). Nodal signals from the epiblast lead to recruitment of additional cells—the so-termed anterior visceral endoderm (AVE)—that, together with the initial DVE pioneer population, migrate toward the embryonic/extraembryonic boundary, thereby triggering reorganization of cells within the VE epithelium (Arnold and Robertson 2009; Rossant and Tam 2009; Nowotschin and Hadjantonakis 2010). This Nodal signaling-dependent, unilateral movement of cells converts the pre-existing proximodistal (PD) axis of the egg cylinder to an AP axis (Norris et al. 2002). Cells of the AVE express secreted Nodal, Bmp, and Wnt antagonists, thereby restricting signaling to the posterior epiblast and confining the site of nascent mesoderm induction to the primitive streak.
Eomesodermin (Eomes, also referred to as Tbr2), a member of the T-box family of transcription factors, is dynamically expressed in both the embryonic and extraembryonic tissues of the early embryo. Eomes mutants exhibit defects in the trophectoderm and arrest at implantation (Russ et al. 2000), obscuring its role at later stages of development. Chimera analysis, together with epiblast-specific ablation, has uncovered essential functions for Eomes in epithelial-to-mesenchymal transition (EMT) and mesoderm delamination as well as in specification of the definitive endoderm and cardiac mesoderm (Arnold et al. 2008; Costello et al. 2011). Intriguingly, immunohistochemistry and studies of GFP reporter mice have revealed Eomes expression in the embryonic VE (EmVE) at embryonic day 5.5 (E5.5), prompting our efforts to investigate its role in this tissue (Kwon and Hadjantonakis 2007; Arnold et al. 2009).
Results and Discussion
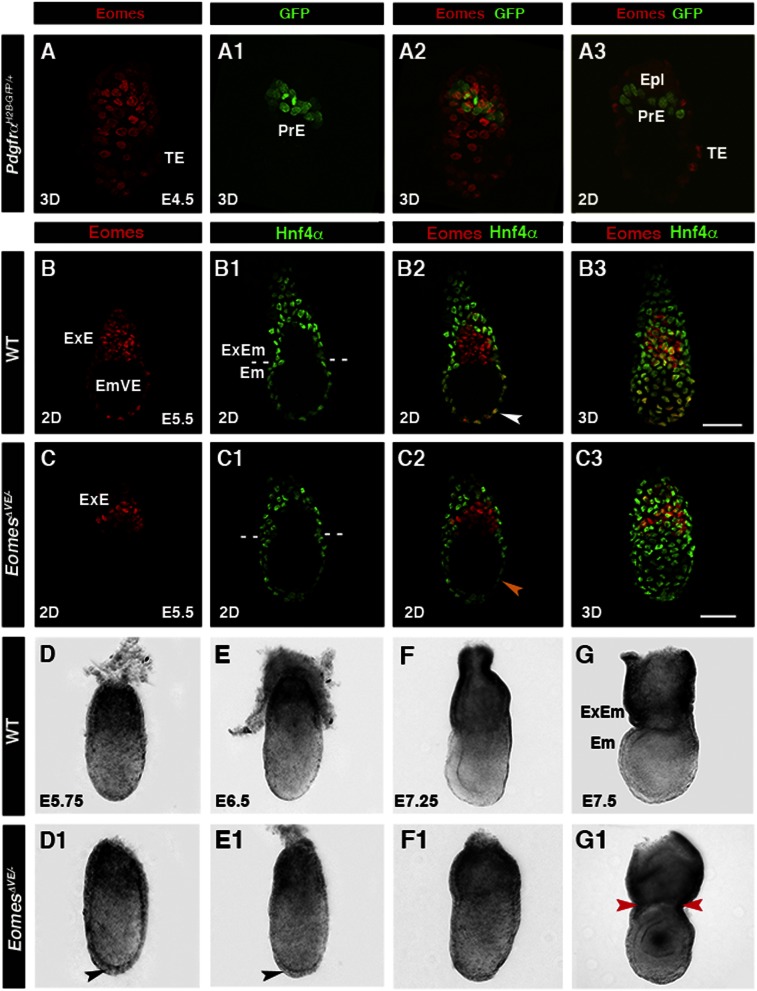
We performed immunohistochemistry to determine the onset of Eomes expression. Eomes was undetectable in the primitive endoderm of the late blastocyst (E4.5) but present in the EmVE at E5.5, as assessed by colocalization with a PdgfraH2B-GFP reporter (Plusa et al. 2008) and Hnf4α immunoreactivity (Fig. 1A–A3,B–B3; Kwon et al. 2008). Thus, Eomes is not expressed when DVE cells are initially specified in the late blastocyst but rather is induced throughout the EmVE by E5.5, coincident with AVE recruitment, specification, and migration. To bypass the requirement for Eomes in the trophectoderm, we used a Ttr-Cre transgene to selectively delete Eomes in the VE (Kwon and Hadjantonakis 2009). Ttr-Cre activity mediates complete deletion throughout the VE by E5.5 (Supplemental Fig. 1A,B; Kwon and Hadjantonakis 2009). EomesΔVE/− mutant embryos recovered at E5.5 were indistinguishable from their littermates by gross morphology. As expected, Eomes protein was present in the ExE but undetectable in the EmVE in E5.5 EomesΔVE/− mutants (Fig. 1C). Thus, VE-specific deletion of Eomes is achieved prior to AVE specification. Morphological analysis of EomesΔVE/− mutants between E5.75 and E7.5 revealed two distinct phenotypes. At E5.75, EomesΔVE/− mutants could be distinguished from the wild-type littermates by the presence of a thickening throughout the VE, which, by E6.5, appeared more pronounced at the distal tip of the embryo (Fig. 1D,D1,E,E1, black arrowheads). By E7.5, the few EomesΔVE/− mutants recovered could be distinguished from littermates due to their distorted shape (Fig. 1F,F1) and the presence of a constriction at the embryonic/extraembryonic boundary (Fig. 1G,G1, red arrowheads).
Figure 1.
Eomes is activated in the EmVE, and genetic ablation in the VE results in morphogenesis defects. (A–A3) E4.5 PdgfrαH2B-GFP embryo depicting nuclear-localized Eomes (red) in trophectoderm (TE) and absence in primitive endoderm (PrE). (B–B3) Colocalization of Eomes (red) and Hnf4α (green) in the EmVE (white arrowhead). (C–C3) Localization of Eomes and Hnf4α in E5.5 embryo with VE-specific Eomes inactivation (EomesΔVE/−). Note the lack of colocalization of Eomes and Hnf4α protein (C2; orange arrowhead). (D,D1) E5.75 EomesΔVE/− mutant embryos (D1) exhibit increased thickening of the VE at the distal tip (D1; black arrowhead) compared with wild-type (WT; D). (E,E1) At E6.5, the AVE has migrated anteriorly in wild type (E) but remains thickened distally in EomesΔVE/− mutants (E1; black arrowhead). At E7.25 (F,F1), EomesΔVE/− mutants display aberrant morphology (F1) compared with wild type (F), becoming exacerbated by E7.5 (G1). (G,G1) Note constriction at embryonic/extraembryonic junction (ExEM) (red arrowheads). (2D) Single optical section; (3D) projection of z-stack; (Em) embryonic; (Epi) epiblast; (ExE) extraembryonic ectoderm.
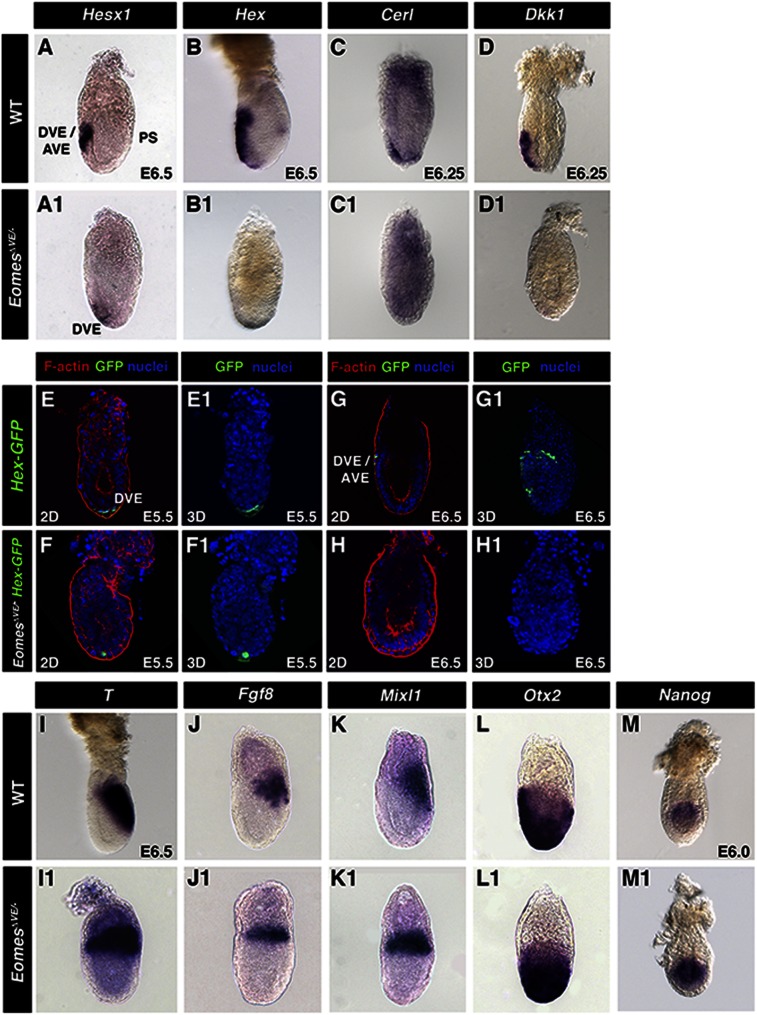
Since the persistence of a thickened VE at the distal tip of the embryo is often associated with a failure in AVE migration, we examined the expression of AVE markers in EomesΔVE/− mutant embryos. Whole-mount in situ hybridization for Hesx1, Hex, Cerl1, and Dkk1 revealed significantly reduced levels or loss of expression, localized at the distal tip of the embryo, as compared with wild-type littermates (Fig. 2A–D,A1–D1). To further investigate the dynamics of AVE specification, we used the Hex-GFP reporter strain (Rodriguez et al. 2001). Time-lapse imaging of Hex-GFPTg/+ embryos revealed GFP expression in a small group of DVE cells at the distal tip of the egg cylinder by E5.5 (Fig. 2E,E1) and an increase in fluorescence levels of the reporter over time, concomitant with the directional migration of a cohort of GFP-positive DVE/AVE cells toward the embryonic/extraembryonic junction (Fig. 2G,G1), as has been reported previously (Srinivas et al. 2004). In contrast, as compared with wild-type embryos at E5.5, EomesΔVE/−;Hex-GFPTg/+ embryos exhibited low levels of GFP fluorescence in a reduced number of distal cells (Fig. 2F,F1). Moreover, fluorescence was undetectable at later stages (Fig. 2H,H1), suggesting that loss of Eomes results in failure to correctly maintain the DVE and disrupts recruitment of further Hex-expressing cells that normally give rise to the migratory AVE population. Failure to position the AVE results in aberrant mesoderm induction, with nascent mesoderm markers T/Brachyury, Fgf8, and Mixl1 (Fig. 2I–K) observed throughout the proximal epiblast. Coincident with a perturbation of PD polarity, Otx2 was also expressed more broadly in the distal epiblast and overlying VE (Fig. 2L,L1). In wild-type embryos, Nanog expression is restricted to the proximal posterior epiblast (Fig. 2M), whereas in contrast, here we observed Nanog expressed throughout the entire epiblast in EomesΔVE/− mutants (Fig. 2M1). However, expression of the epiblast marker Oct4 was unperturbed in EomesΔVE/− mutants (data not shown).
Figure 2.
Eomes is required for maintenance of the DVE and specification and migration of the AVE. (A–D1) Whole-mount in situ hybridization of E6.5 embryos of the indicated genotypes using AVE/DVE-specific markers Hesx1, Hex, Cerl, and Dkk1. (E–H) Aberrant DVE specification (E5.5; E) and migration (E6.5; G) in EomesΔVE/− mutants (F,H) versus wild type (WT) (E,G). (Green) Hex-GFP; (red) F-actin; (blue) nuclei. (I–L) Whole-mount in situ hybridization of E6.5 embryos using posterior (T) and intermediate streak (Fgf8, Mixl1) markers (I–K) and the anterior marker Otx2 (I–L) in wild type and EomesΔVE/− mutants (I1–L1). (M,M1) Expression of Nanog in E6.0 wild-type and EomesΔVE/− mutant embryos.
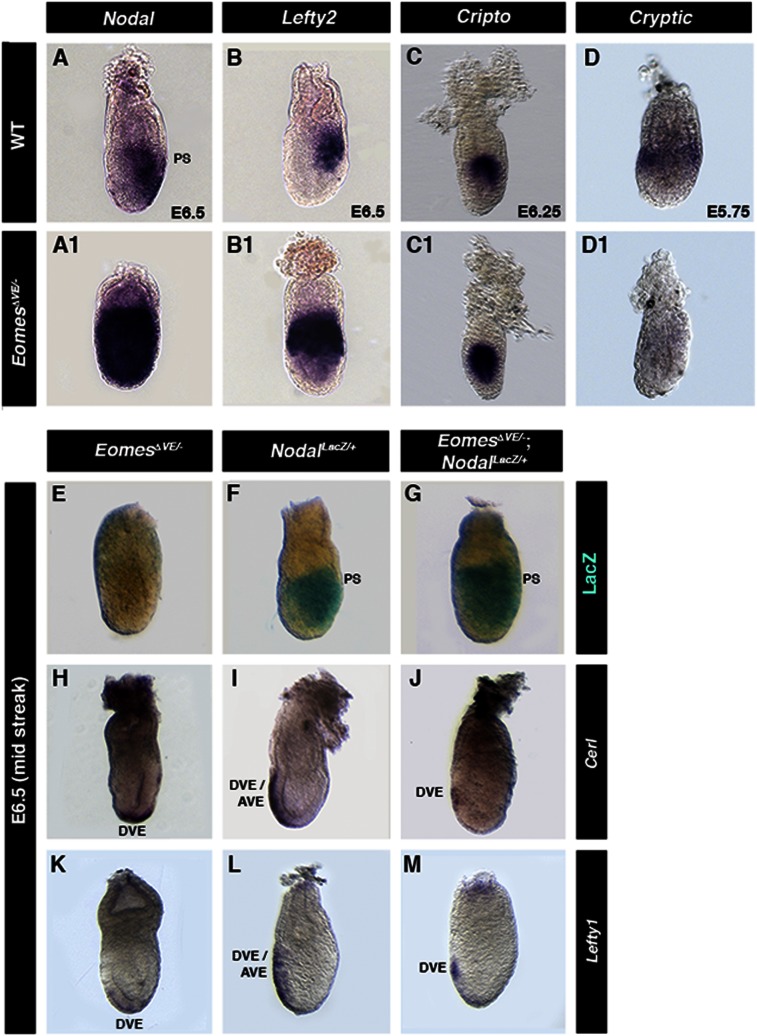
Both Wnt and Nodal signaling play essential roles in AP patterning. Next, we investigated the activity of the TOPGAL Wnt signaling reporter (DasGupta and Fuchs 1999) in EomesΔVE/− mutants. Reporter expression levels were unaffected, but expression was restricted proximally due to the failure to correctly position the primitive streak (data not shown). In contrast, Nodal expression was markedly up-regulated throughout the epiblast (Fig. 3A,A1). Additionally, expression of Lefty2, a direct target and antagonist of Nodal, as well the Nodal coreceptor Cripto became radialized in EomesΔVE/− mutants (Fig. 3B–C1). In contrast, expression of Cryptic, another Nodal coreceptor, was down-regulated in the VE of EomesΔVE/− mutants (Fig. 3D1). Cryptic expression is normally restricted to the proximal EmVE at prestreak stages (Fig. 3D). Loss of Cryptic expression strengthens the argument for defective specification of the EmVE in EomesΔVE/− mutants.
Figure 3.
AVE migration involves a Nodal-dependent role of Eomes in the EmVE. (A–D) Expression of Nodal signaling components and DVE/AVE markers in E6.5 embryos of the indicated genotypes. (A1) Nodal. (B1) Lefty2. (C1) Cripto. (D) Cryptic. (F) NodalLacZ/+ embryo exhibiting β-gal activity in the posterior epiblast. (G) A stage-matched EomesΔVE/−;NodalLacZ/+ mutant embryo lacking one copy of Nodal exhibits posteriorly restricted β-gal activity. Cerl (H–J) and Lefty1 (K–M) reveal partial anterior DVE migration in EomesΔVE/−;NodalLacZ/+ embryos (J,M) compared with distal localization in EomesΔVE/− mutants (H,K) but not as robust as in NodalLacZ/+ controls (I,L).
To test whether reducing Nodal levels could rescue some of the observed defects, we generated EomesΔVE/−;NodalLacZ/+ embryos. EomesΔVE/−;NodalLacZ/+ embryos were recovered at Mendelian ratios at midstreak (E6.5) (Fig. 3E–M) and early bud (E7.5) stages (Supplemental Fig. 2A–F). As judged by gross morphology, NodalLacZ reporter expression, and marker gene analysis, at E6.5, EomesΔVE/−;NodalLacZ/+ embryos exhibited an intermediate phenotype between wild-type embryos and EomesΔVE/− mutants (Fig. 3E–M). The VE thickening observed in the EomesΔVE/− mutants was absent in EomesΔVE/−;NodalLacZ/+ embryos. EomesΔVE/−;NodalLacZ/+ embryos express Cerl1 and Lefty1, albeit at very reduced levels (Fig. 3J,M). Consequently, the primitive streak became localized posteriorly as judged by NodalLacZ (Supplemental Fig. 2A–C) and T/Brachyury expression (Supplemental Fig. 2D–F). However, at E7.5, EomesΔVE/−;NodalLacZ/+ embryos exhibited a constriction at the embryonic/extraembryonic boundary (Supplemental Fig. 2C,F). Hence, increased Nodal signaling from the epiblast appeared to exacerbate the EomesΔVE/− mutant phenotype. Restraining Nodal signaling in EomesΔVE/− embryos was sufficient for initiation of DVE migration, as DVE formation is not defective in EomesΔVE/− mutants (Fig. 2F). However, AVE specification and migration was still impaired in EomesΔVE/−;NodalLacZ/+ embryos.
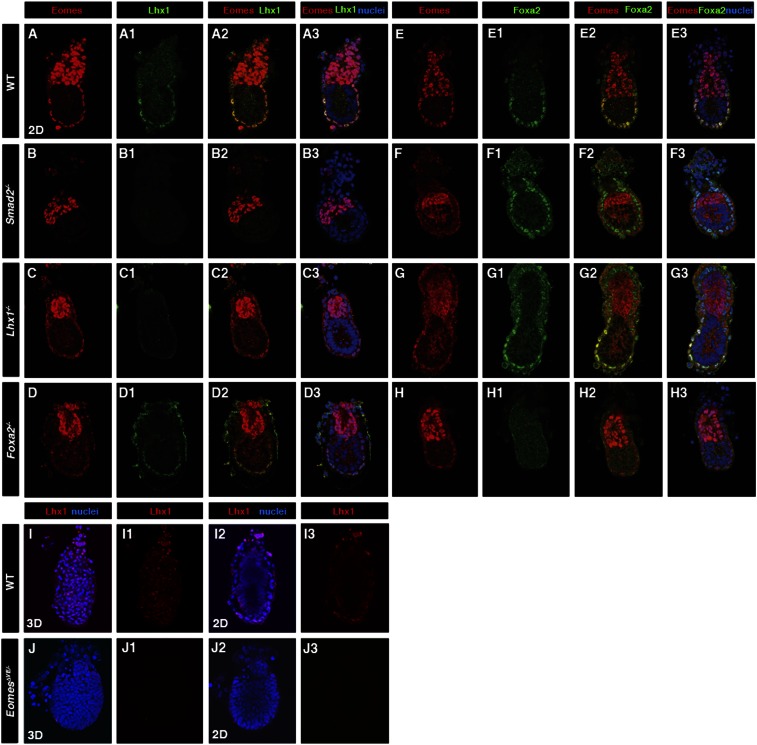
Conditional deletion of Eomes from the early EmVE prevents the correct specification and maintenance of the AVE. The simplest scenario is that Eomes acts upstream to activate the hierarchy of transcription factors governing AVE formation. Nodal signaling in the early epiblast is known to be responsible for induction of the AVE via a Smad2-dependent pathway (Brennan et al. 2001). The LIM domain factor Lhx1 and forkhead factor Foxa2 are both required for correct formation and function of the AVE. Loss of either gene results in failure to establish a normal AP axis, likely due to impaired migration of the AVE and consequent defects in primitive streak formation (Ang and Rossant 1994; Shawlot and Behringer 1995; Dufort et al. 1998; Kinder et al. 2001). Interestingly, the Lhx1;Foxa2 double-mutant phenotype closely resembles that of Smad2 mutants. Markers of the DVE/AVE are not induced, and the entire epiblast adopts a mesodermal character (Waldrip et al. 1998; Perea-Gomez et al. 1999), suggesting that these transcription factors work in parallel or cooperatively to regulate the gene regulatory network (GRN) underlying AVE specification. In wild-type E5.5 embryos, Eomes, Lhx1, and Foxa2 are expressed in the EmVE (Fig. 4A–A3,E–E3). In Smad2−/− embryos, both Eomes and Lhx1 are lost, while Foxa2 expression is still evident (Fig. 4B–B3,F–F3). In contrast, in Lhx1 mutants, Eomes and Foxa2 expression is unaffected (Fig. 4C–C3,G–G3). Foxa2 is not required for the VE to maintain Eomes or Lhx1 expression (Fig. 4D–D3,H–H3), whereas in EomesΔVE/− mutants, Lhx1 is absent in the EmVE (Fig. 4I–I3,J–J3). Hence, activation of Lhx1 depends on the Smad2–Eomes pathway, whereas Nodal signaling levels are less critical for Foxa2 expression.
Figure 4.
Smad2-dependent Eomes and Lhx1 expression in the EmVE. Localization of Eomes, Lhx1, and Foxa2 in E5.5 embryos of the indicated genotypes. (A–A3) In wild type (WT), Eomes is present in the ExE and EmVE. Lhx1 colocalized with Eomes in EmVE only. (B–B3) In Smad2−/− mutants, EmVE expression of Eomes and Lhx1 is lost, while Eomes expression in ExE is maintained. (C–C3) In the absence of Lhx1, EmVE Eomes is present. (D–D3) Foxa2 is not required for the expression of Eomes and Lhx1. (E–E3) In wild type, Foxa2 is expressed throughout the VE. Foxa2 expression is unchanged in Smad2−/− (F–F3) and Lhx1−/− (G–G3) mutants. (H–H3) Lack of Foxa2 does not alter expression of Eomes in EmVE. (I–I3,J–J3) Absence of Lhx1 in EomesΔVE/− mutant.
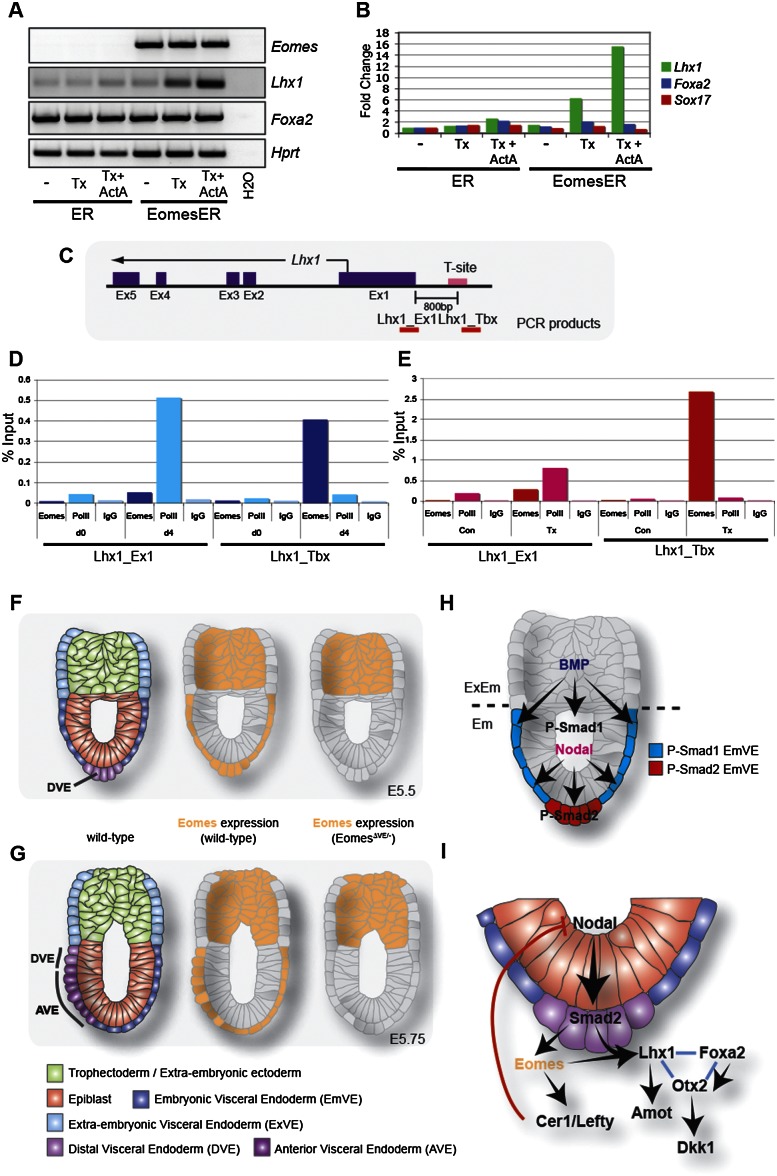
To test whether Lhx1 is a direct Eomes target, we exploited cell culture protocols that promote the formation of extraembryonic endoderm populations. First, we used forced expression of Gata6 to direct embryonic stem (ES) cells toward an extraembryonic endoderm fate (Shimosato et al. 2007). A regulatable Gata6 expression vector was stably introduced into wild-type or Smad2-deficient ES cells (Tremblay et al. 2000), in which Nodal signaling is significantly impaired. Following induction of Gata6 via addition of dexamethasone, Eomes and Lxh1 transcript levels were assessed. Wild-type cell clones expressed Eomes and Lhx1 transcripts in response to Gata6 induction, while expression of both genes was severely reduced in Smad2-deficient clones (Supplemental Fig. 3A). Thus, robust Eomes and Lhx1 induction depends on active Nodal–phospho-Smad2 signaling. Next, we examined XEN cell cultures (Kunath et al. 2005). XEN cells express low or undetectable levels of Eomes and Lhx1 as assessed by RT–PCR (Fig. 5A) and expression microarrays (Supplemental Fig. 3B). Stably transfected XEN subclones carrying an EomesER-inducible transgene were generated, and Lhx1 transcript levels were monitored following addition of either tamoxifen alone or in combination with recombinant Activin A. Nuclear translocation of EomesER resulted in a significantly elevated Lhx1 expression (Fig. 5A,B), and Lhx1 levels were further increased by Activin A treatment. As a control, Foxa2 transcription was unaffected by forced Eomes expression, providing further evidence that Foxa2 expression is Eomes-independent (Fig. 5A,B).
Figure 5.
Lhx1 is a target of Eomes. RT–PCR (A) and quantitative RT–PCR (B) analysis of XEN cells expressing a tamoxifen-inducible Eomes fusion. Translocation of Eomes into the nucleus upon tamoxifen (Tx) treatment leads to an increase in Lhx1 expression. Addition of ActivinA results in a further increase of Lhx1 expression. Expression of other VE markers (Foxa2 and Sox17) remains unchanged. (C) Schematic representation of the Lhx1 locus with putative T-site upstream of the TSS. Red bars indicate areas amplified by qPCR after ChIP. (Ex) Exon. (D) ChIP analysis of DMSO-differentiated P19Cl6 cells using antibodies for Eomes, PolII, and IgG control. qRT–PCR analysis shows binding of Eomes to the Lhx1 T-site in differentiating cells. (D0) Day 0; (d4) day 4 of differentiation. (E) P19CL6 cells expressing Eomes-ER show binding of Eomes to Lhx1 locus upon induction. (Con) Untreated cells; (Tx) day 4 tamoxifen-treated cells. (F) At E5.5, the DVE is positioned at the distal tip of the embryo. Eomes is expressed in the ExE and EmVE. In EomesΔVE/− mutants, EmVE expression is genetically ablated. (G) At E5.75, the DVE has begun to migrate to the embryonic/extraembryonic boundary, the AVE has been specified and is migrating coordinately with the DVE. In the EomesΔVE/− mutant, the AVE is not specified, and a thickening in the distal EmVE is evident. (H) BMP and Nodal signaling are essential in establishing the PD axis. AVE formation is dependent on mutually exclusive activity of Smad1 and Smad2 and occurs at the distal tip of the EmVE devoid of phospho-Smad1 and containing high levels of activated Smad2. (I) Model of the GRN controlling AVE/DVE formation.
To establish whether Lhx1 is directly regulated by Eomes, we used interspecies alignments to identify conserved T-box consensus binding sites mapping to the Lhx1 locus (Supplemental Fig. 3C). This analysis identified two T-box-binding site (T-site) motifs: one site 3′ to the locus and a second site 800 base pairs (bp) upstream of the transcriptional start site (TSS) (Supplemental Fig. 3B). We previously used DMSO-treated P19Cl6 cells together with tamoxifen-treated P19Cl6 EomesER cells to identify Mesp1/2 as direct Eomes targets (Costello et al. 2011). As expected, P19Cl6 cultures show markedly up-regulated expression of Eomes 2 d post-DMSO induction, which is followed 2 d later by robust induction of Lhx1 transcripts (Supplemental Fig. 3E). To directly evaluate Eomes occupancy adjacent to the TSS, we performed chromatin immunoprecipitation (ChIP). Both endogenous Eomes and EomesER (4 d post-differentiation) were specifically enriched at the T-site. Eomes occupancy of this site correlates with RNA polymerase II (PolII) binding at the Lhx1 TSS, consistent with induction of Lhx1 transcription (Fig. 5D,E). Since Eomes is the only T-box transcription factor known to be expressed in the EmVE (Arnold et al. 2009), these data provide compelling evidence that Lhx1 is a direct target of Eomes during AVE specification. To explore whether Eomes acts to not only induce but also maintain Lhx1 expression during migration of the AVE, we examined expression in embryos. Double-labeling experiments revealed colocalization of both transcription factors in the AVE of E6.5 of wild-type embryos (Supplemental Fig. 4).
The present study provides new insights into DVE/AVE functions in promoting anterior fates in the early post-implantation embryo. We identify a critical role for the Nodal–Smad2 pathway in activating expression of the T-box transcription factor Eomes in the EmVE population. Conditional loss of Eomes expression in the VE does not perturb DVE formation but rather blocks correct specification of the AVE. Our ChIP and epistasis experiments reveal that this is due to a requirement for Eomes in binding to and activating expression of the Lim domain transcription factor Lhx1. Amot, which regulates VE movement in the early embryo (Shimono and Behringer 2003), is regulated by Lhx1 (Shimono and Behringer 1999), and Lhx1 is also known to bind the promoter of the AVE marker Hesx1 (Chou et al. 2006). Thus, Eomes acts upstream of Lhx1 in the GRN underlying AVE specification and migration (Fig. 5I). In contrast, Foxa2 expression is Nodal-independent (Brennan et al. 2001), as loss of neither Smad2, Eomes, nor Lhx1 prevents expression of Foxa2 in the VE. Hence, both Foxa2 and Lhx1 are independently required for proper AVE function. In addition to Nodal–Smad2, a second signaling pathway must function in AVE formation. Identification of upstream regulators of Foxa2 will require additional experiments.
Signaling cues from the epiblast and ExE temporally and spatially intersect to precisely pattern the growing VE and restrict induction of the AVE to the most distal population within the EmVE. At E5.5, the EmVE uniformly expresses Eomes, Lhx1, and Foxa2, the three transcription factors identified as having critical roles in AVE formation. Anterior patterning of the early post-implantation embryo requires a continuum of opposing Nodal/Bmp signaling activities. Nodal and Bmp signals from the early epiblast and ExE, respectively, are required to distinguish the EmVE versus the extraembryonic VE (ExVE) (Mesnard et al. 2006; Yamamoto et al. 2009). As the embryo expands, the most distal VE cells become positioned increasingly farther away from the ExE, the source of BMP ligands, while retaining close apposition to the Nodal-expressing epiblast. Cell proliferation generates reciprocal PD gradients of phospho-Smad1/5 (Yamamoto et al. 2009) and phospho-Smad2. Thus, cells in which Bmp signaling is lowest give rise to AVE. Consistent with this, physical removal of the ExE results in a dramatic expansion of the AVE (Rodriguez et al. 2005; Mesnard et al. 2006). These observations, together with data presented here, provide a refined molecular model for induction and maintenance of the AVE (Fig. 5H,I). Graded phospho-Smad1/5 versus phospho-Smad2 along the PD axis likely restricts high levels of activated Smad2 to the most distal cells. Eomes is known to directly complex with Smad2 (Picozzi et al. 2009) but may only assemble efficiently in the distal-most cells, where the AVE targets—including the key growth factor antagonists Lefty1, Cerl1, and Dkk1—are directly or indirectly activated. Foxa2 expression is Nodal-independent, but the AVE phenotype of Smad2 mutants is more severe than Foxa2 mutants. This suggests that Foxa2 likely works together with Smad2-dependent partners, including Eomes and Lhx1, to robustly and efficiently activate the GRN underpinning formation, maintenance, and migration of a fully functional AVE.
Materials and methods
Mouse strains, embryo collection, and staining
Mouse strains used were as follows: Eomesflox/flox (Mao et al. 2008), Ttr-Cre (Kwon and Hadjantonakis 2009), PdgfraH2B-GFP/+ (Hamilton et al. 2003), Hex-GFP (Rodriguez et al. 2001), NodalLacZ (Collignon et al. 1996), Lhx1 (Shawlot and Behringer 1995), Foxa2 (Ang and Rossant 1994), and Smad2 (Waldrip et al. 1998). EomesVE-deleted/− embryos (referred to as EomesΔVE/−) were generated by crossing Eomes+/−;Ttr-CreTg/+ males with Eomesflox/flox females. For time-lapse imaging, embryos were cultured in 50% rat serum/50% DMEM-F12, 5% CO2, at 37°C.
E4.5 embryos were flushed from uteri in M2 (Millipore) and fixed in 4% PFA for 10 min at room temperature. Post-implantation embryos were dissected in DMEM/F12 (Gibco) and fixed in 4% PFA overnight at 4°C for in situ hybridization and for 20 min at room temperature for immunostaining.
Whole-mount in situ hybridization and LacZ staining were performed according to standard protocols (Nagy et al. 2002). Antisense riboprobes were used to detect genes of interest (Supplemental Table S1). Embryos were permeabilized in 0.5% Triton in PBS for 20 min and then washed in 0.1% Triton in PBS. Embryos were blocked in 10% goat serum, 2% BSA, and 0.3% Triton in PBS for 1 h at 4°C and then incubated with primary antibodies overnight at 4°C, followed by washes in 0.1% Triton in PBS and incubation with fluorophore-conjugated secondary antibodies (AlexaFluor, Invitrogen) overnight at 4°C, and then washed in 0.1% Triton in PBS and counterstained with Hoechst 33342 to detect nuclei (1:500; Invitrogen). Primary antibodies used were anti-Eomes (1:500; Abcam), anti-HNF4α (1:500; Santa Cruz Biotechnology), anti-Lhx1 (1:50; Santa Cruz Biotechnology), and anti-Foxa2 (1:100; Santa Cruz Biotechnology). Fluorescence intensity levels were measured on data acquired under identical imaging parameters using Imaris software’s Spot function (Bitplane).
Cell culture, ChIP, and RNA analysis
P19Cl6 and P19EoER cells were cultured as previously described (Costello et al. 2011). P19Cl6 cells were differentiated in medium containing 1% DMSO. EomesER was activated in P19EoER cells by the addition of 1 μg/mL 4-hydroxytamoxifen (4OHT). ChIP was performed as previously described (Costello et al. 2011) with the following antibodies: anti-Eomes (Abcam, ab23345), anti-PolII (Santa Cruz Biotechnology, sc-899x), and normal rabbit IgG (Santa Cruz Biotechnology, sc-2027). ChIP primer sequences are provided in Supplemental Table S2. XEN cells were cultured as previously described (Kunath et al. 2005). pCAG-EomesER-IRESPuro (Niwa et al. 2005) was introduced into XEN cells and selected with 1 μg/mL puromycin to generate EomesER expressing subclones. For EomesER activation, 1 μg/mL 4OHT was added to cultures. Activin A (R&D Systems) was added at a concentration of 50 μg/mL. Wild-type (CCE) and Smad2-null (KT-15) ES cells were cultured as previously described (Costello et al. 2011). ES cells were electroporated with pCAG-Gata6GR-IRESPuro (Shimosato et al. 2007) and selected in puromycin. Addition of 100 mM dexamethasone (Sigma) resulted in the activation of Gata6GR and conversion of cells to a XEN-like phenotype (Shimosato et al. 2007).
RNA was isolated and analyzed as previously described (Costello et al. 2011). Primer sequences are provided in Supplemental Table S3.
Wide-field images were acquired with a Zeiss Axiocam camera on a Leica M165FC. Laser-scanning confocal data were acquired on a Zeiss LSM510META or Olympus FV1000. Raw data were processed using Zeiss AIM/ZEN and Bitplane Imaris software.
Acknowledgments
We thank R. Behringer, T. Rodriquez, J.Rossant, and P. Soriano for mouse lines; H. Niwa for expression plasmids; M. Kang and N. Schrode for microarray data; and E. Bikoff and A. Perea-Gomez for advice and discussions. This work was supported by a Wellcome Trust Principal Research Fellowship to E.J.R., the NIH (HD052115 and DK084391 to A.K.H.), and the Muscular Dystrophy Association (186552 to S.N.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.215152.113.
References
- Ang SL, Rossant J 1994. HNF-3β is essential for node and notochord formation in mouse development. Cell 78: 561–574 [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Robertson EJ 2009. Making a commitment: Cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol 10: 91–103 [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Hofmann UK, Bikoff EK, Robertson EJ 2008. Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development 135: 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ, Sugnaseelan J, Groszer M, Srinivas S, Robertson EJ 2009. Generation and analysis of a mouse line harboring GFP in the Eomes/Tbr2 locus. Genesis 47: 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RS, Robertson EJ 2001. Nodal signalling in the epiblast patterns the early mouse embryo. Nature 411: 965–969 [DOI] [PubMed] [Google Scholar]
- Chou SJ, Hermesz E, Hatta T, Feltner D, El-Hodiri HM, Jamrich M, Mahon K 2006. Conserved regulatory elements establish the dynamic expression of Rpx/HesxI in early vertebrate development. Dev Biol 292: 533–545 [DOI] [PubMed] [Google Scholar]
- Collignon J, Varlet I, Robertson EJ 1996. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature 381: 155–158 [DOI] [PubMed] [Google Scholar]
- Costello I, Pimeisl IM, Drager S, Bikoff EK, Robertson EJ, Arnold SJ 2011. The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat Cell Biol 13: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta R, Fuchs E 1999. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development 126: 4557–4568 [DOI] [PubMed] [Google Scholar]
- Dufort D, Schwartz L, Harpal K, Rossant J 1998. The transcription factor HNF3β is required in visceral endoderm for normal primitive streak morphogenesis. Development 125: 3015–3025 [DOI] [PubMed] [Google Scholar]
- Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P 2003. Evolutionary divergence of platelet-derived growth factor α receptor signaling mechanisms. Mol Cell Biol 23: 4013–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinder SJ, Tsang TE, Ang SL, Behringer RR, Tam PP 2001. Defects of the body plan of mutant embryos lacking Lim1, Otx2 or Hnf3β activity. Int J Dev Biol 45: 347–355 [PubMed] [Google Scholar]
- Kunath T, Arnaud D, Uy GD, Okamoto I, Chureau C, Yamanaka Y, Heard E, Gardner RL, Avner P, Rossant J 2005. Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development 132: 1649–1661 [DOI] [PubMed] [Google Scholar]
- Kwon GS, Hadjantonakis AK 2007. Eomes:GFP—a tool for live imaging cells of the trophoblast, primitive streak, and telencephalon in the mouse embryo. Genesis 45: 208–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon GS, Hadjantonakis AK 2009. Transthyretin mouse transgenes direct RFP expression or Cre-mediated recombination throughout the visceral endoderm. Genesis 47: 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon GS, Viotti M, Hadjantonakis AK 2008. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev Cell 15: 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao CA, Kiyama T, Pan P, Furuta Y, Hadjantonakis AK, Klein WH 2008. Eomesodermin, a target gene of Pou4f2, is required for retinal ganglion cell and optic nerve development in the mouse. Development 135: 271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnard D, Guzman-Ayala M, Constam DB 2006. Nodal specifies embryonic visceral endoderm and sustains pluripotent cells in the epiblast before overt axial patterning. Development 133: 2497–2505 [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vinterstein K, Behringer R Manipulating the mouse embryo. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J 2005. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123: 917–929 [DOI] [PubMed] [Google Scholar]
- Norris DP, Brennan J, Bikoff EK, Robertson EJ 2002. The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development 129: 3455–3468 [DOI] [PubMed] [Google Scholar]
- Nowotschin S, Hadjantonakis AK 2010. Cellular dynamics in the early mouse embryo: From axis formation to gastrulation. Curr Opin Genet Dev 20: 420–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Gomez A, Shawlot W, Sasaki H, Behringer RR, Ang S 1999. HNF3β and Lim1 interact in the visceral endoderm to regulate primitive streak formation and anterior–posterior polarity in the mouse embryo. Development 126: 4499–4511 [DOI] [PubMed] [Google Scholar]
- Picozzi P, Wang F, Cronk K, Ryan K 2009. Eomesodermin requires transforming growth factor-β/activin signaling and binds Smad2 to activate mesodermal genes. J Biol Chem 284: 2397–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis AK 2008. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development 135: 3081–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez TA, Casey ES, Harland RM, Smith JC, Beddington RS 2001. Distinct enhancer elements control Hex expression during gastrulation and early organogenesis. Dev Biol 234: 304–316 [DOI] [PubMed] [Google Scholar]
- Rodriguez TA, Srinivas S, Clements MP, Smith JC, Beddington RS 2005. Induction and migration of the anterior visceral endoderm is regulated by the extra-embryonic ectoderm. Development 132: 2513–2520 [DOI] [PubMed] [Google Scholar]
- Rossant J, Tam PP 2009. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development 136: 701–713 [DOI] [PubMed] [Google Scholar]
- Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, et al. 2000. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 404: 95–99 [DOI] [PubMed] [Google Scholar]
- Shawlot W, Behringer RR 1995. Requirement for Lim1 in head-organizer function. Nature 374: 425–430 [DOI] [PubMed] [Google Scholar]
- Shimono A, Behringer RR 1999. Isolation of novel cDNAs by subtractions between the anterior mesendoderm of single mouse gastrula stage embryos. Dev Biol 209: 369–380 [DOI] [PubMed] [Google Scholar]
- Shimono A, Behringer RR 2003. Angiomotin regulates visceral endoderm movements during mouse embryogenesis. Curr Biol 13: 613–617 [DOI] [PubMed] [Google Scholar]
- Shimosato D, Shiki M, Niwa H 2007. Extra-embryonic endoderm cells derived from ES cells induced by GATA factors acquire the character of XEN cells. BMC Dev Biol 7: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Rodriguez T, Clements M, Smith JC, Beddington RS 2004. Active cell migration drives the unilateral movements of the anterior visceral endoderm. Development 131: 1157–1164 [DOI] [PubMed] [Google Scholar]
- Takaoka K, Yamamoto M, Hamada H 2011. Origin and role of distal visceral endoderm, a group of cells that determines anterior–posterior polarity of the mouse embryo. Nat Cell Biol 13: 743–752 [DOI] [PubMed] [Google Scholar]
- Tremblay KD, Hoodless PA, Bikoff EK, Robertson EJ 2000. Formation of the definitive endoderm in mouse is a Smad2-dependent process. Development 127: 3079–3090 [DOI] [PubMed] [Google Scholar]
- Waldrip WR, Bikoff EK, Hoodless PA, Wrana JL, Robertson EJ 1998. Smad2 signaling in extraembryonic tissues determines anterior–posterior polarity of the early mouse embryo. Cell 92: 797–808 [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Beppu H, Takaoka K, Meno C, Li E, Miyazono K, Hamada H 2009. Antagonism between Smad1 and Smad2 signaling determines the site of distal visceral endoderm formation in the mouse embryo. J Cell Biol 184: 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]