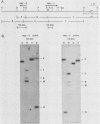
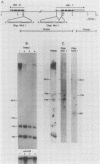
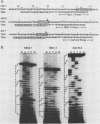
Abstract
We have cloned and analyzed the Caenorhabditis elegans regulatory myosin light-chain genes. C. elegans contains two such genes, which we have designated mlc-1 and mlc-2. The two genes are separated by 2.6 kilobases and are divergently transcribed. We determined the complete nucleotide sequences of both mlc-1 and mlc-2. A single, conservative amino acid substitution distinguishes the sequences of the two proteins. The C. elegans proteins are strongly homologous to regulatory myosin light chains of Drosophila melanogaster and vertebrates and weakly homologous to a superfamily of eucaryotic calcium-binding proteins. Both mlc-1 and mlc-2 encode abundant mRNAs. We mapped the 5' termini of these transcripts by using primer extension sequencing of mRNA templates. mlc-1 mRNAs initiate within conserved hexanucleotides at two different positions, located at -28 and -38 relative to the start of translation. The 5' terminus of mlc-2 mRNA is not encoded in the 4.8-kilobase genomic region upstream of mlc-2. Rather, mlc-2 mRNA contains at its 5' end a short, untranslated leader sequence that is identical to the trans-spliced leader sequence of three C. elegans actin genes.
Full text
PDF





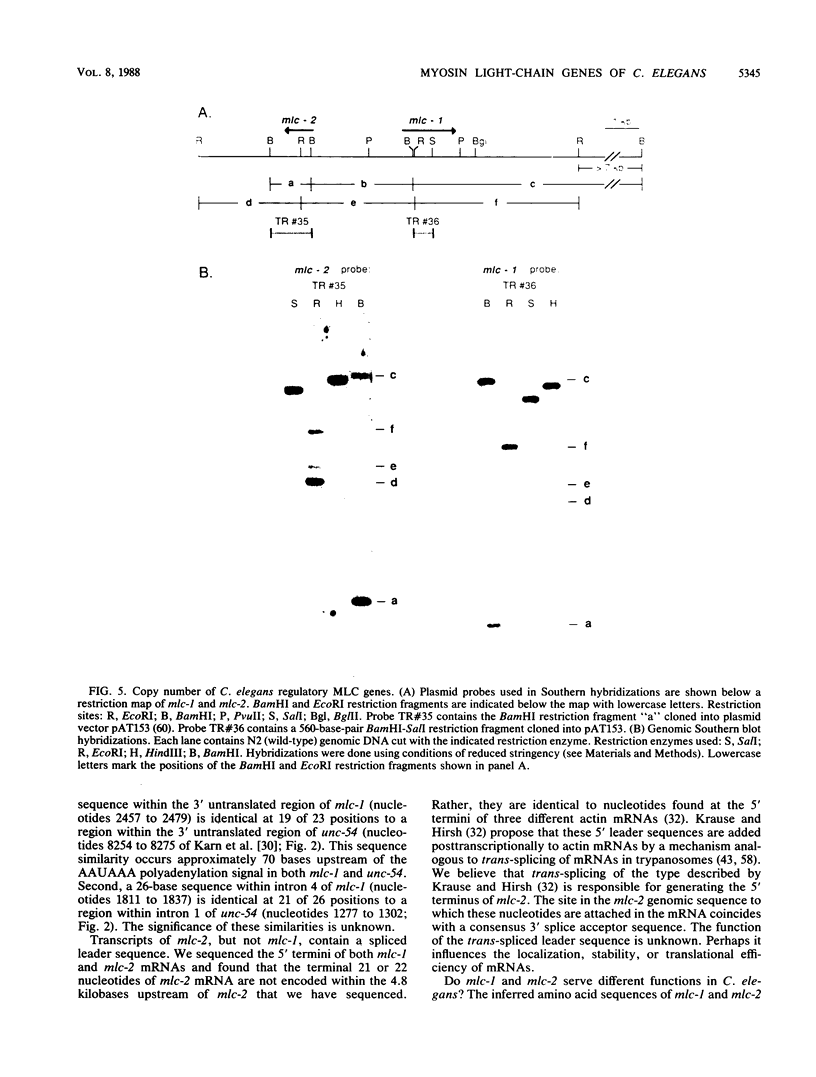



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adelstein R. S., Eisenberg E. Regulation and kinetics of the actin-myosin-ATP interaction. Annu Rev Biochem. 1980;49:921–956. doi: 10.1146/annurev.bi.49.070180.004421. [DOI] [PubMed] [Google Scholar]
- Bagshaw C. R., Kendrick-Jones J. Characterization of homologous divalent metal ion binding sites of vertebrate and molluscan myosins using electron paramagnetic resonance spectroscopy. J Mol Biol. 1979 May 25;130(3):317–336. doi: 10.1016/0022-2836(79)90544-8. [DOI] [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974 May;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm R. L., Rushforth A. M., Pollenz R. S., Kuczmarski E. R., Tafuri S. R. Dictyostelium discoideum myosin: isolation and characterization of cDNAs encoding the essential light chain. Mol Cell Biol. 1988 Feb;8(2):794–801. doi: 10.1128/mcb.8.2.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. H. Homology of myosin DTNB light chain with alkali light chains, troponin C and parvalbumin. Nature. 1976 Feb 26;259(5545):699–700. doi: 10.1038/259699a0. [DOI] [PubMed] [Google Scholar]
- Cooke R. The mechanism of muscle contraction. CRC Crit Rev Biochem. 1986;21(1):53–118. doi: 10.3109/10409238609113609. [DOI] [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Cochran M. D., Dobkin C., Reiser J., Weissmann C. Three regions upstream from the cap site are required for efficient and accurate transcription of the rabbit beta-globin gene in mouse 3T6 cells. Cell. 1983 Mar;32(3):695–706. doi: 10.1016/0092-8674(83)90055-7. [DOI] [PubMed] [Google Scholar]
- Eide D., Anderson P. The gene structures of spontaneous mutations affecting a Caenorhabditis elegans myosin heavy chain gene. Genetics. 1985 Jan;109(1):67–79. doi: 10.1093/genetics/109.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E., Hill T. L. Muscle contraction and free energy transduction in biological systems. Science. 1985 Mar 1;227(4690):999–1006. doi: 10.1126/science.3156404. [DOI] [PubMed] [Google Scholar]
- Ellis R. E., Sulston J. E., Coulson A. R. The rDNA of C. elegans: sequence and structure. Nucleic Acids Res. 1986 Mar 11;14(5):2345–2364. doi: 10.1093/nar/14.5.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein H. F., Waterston R. H., Brenner S. A mutant affecting the heavy chain of myosin in Caenorhabditis elegans. J Mol Biol. 1974 Dec 5;90(2):291–300. doi: 10.1016/0022-2836(74)90374-x. [DOI] [PubMed] [Google Scholar]
- Falkenthal S., Parker V. P., Mattox W. W., Davidson N. Drosophila melanogaster has only one myosin alkali light-chain gene which encodes a protein with considerable amino acid sequence homology to chicken myosin alkali light chains. Mol Cell Biol. 1984 May;4(5):956–965. doi: 10.1128/mcb.4.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A. Integrative transformation of Caenorhabditis elegans. EMBO J. 1986 Oct;5(10):2673–2680. doi: 10.1002/j.1460-2075.1986.tb04550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Goodman M., Pechère J. F., Haiech J., Demaille J. G. Evolutionary diversification of structure and function in the family of intracellular calcium-binding proteins. J Mol Evol. 1979 Nov;13(4):331–352. doi: 10.1007/BF01731373. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Identification of regulatory sequences in the prelude sequences of an H2A histone gene by the study of specific deletion mutants in vivo. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1432–1436. doi: 10.1073/pnas.77.3.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington W. F., Rodgers M. E. Myosin. Annu Rev Biochem. 1984;53:35–73. doi: 10.1146/annurev.bi.53.070184.000343. [DOI] [PubMed] [Google Scholar]
- Harris H. E., Epstein H. F. Myosin and paramyosin of Caenorhabditis elegans: biochemical and structural properties of wild-type and mutant proteins. Cell. 1977 Apr;10(4):709–719. doi: 10.1016/0092-8674(77)90105-2. [DOI] [PubMed] [Google Scholar]
- Harris H. E., Tso M. Y., Epstein H. F. Actin and myosin-linked calcium regulation in the nematode Caenorhabditis elegans. Biochemical and structural properties of native filaments and purified proteins. Biochemistry. 1977 Mar 8;16(5):859–865. doi: 10.1021/bi00624a008. [DOI] [PubMed] [Google Scholar]
- Henry G. D., Trayer I. P., Brewer S., Levine B. A. The widespread distribution of alpha-N-trimethylalanine as the N-terminal amino acid of light chains from vertebrate striated muscle myosins. Eur J Biochem. 1985 Apr 1;148(1):75–82. doi: 10.1111/j.1432-1033.1985.tb08809.x. [DOI] [PubMed] [Google Scholar]
- Hibberd M. G., Trentham D. R. Relationships between chemical and mechanical events during muscular contraction. Annu Rev Biophys Biophys Chem. 1986;15:119–161. doi: 10.1146/annurev.bb.15.060186.001003. [DOI] [PubMed] [Google Scholar]
- Hicks J., Fink G. R. Identification of chromosomal location of yeast DNA from hybrid plasmid p Yeleu 10. Nature. 1977 Sep 15;269(5625):265–267. doi: 10.1038/269265a0. [DOI] [PubMed] [Google Scholar]
- Hohn B. In vitro packaging of lambda and cosmid DNA. Methods Enzymol. 1979;68:299–309. doi: 10.1016/0076-6879(79)68021-7. [DOI] [PubMed] [Google Scholar]
- Karn J., Brenner S., Barnett L. Protein structural domains in the Caenorhabditis elegans unc-54 myosin heavy chain gene are not separated by introns. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4253–4257. doi: 10.1073/pnas.80.14.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M., Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987 Jun 19;49(6):753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretsinger R. H. Structure and evolution of calcium-modulated proteins. CRC Crit Rev Biochem. 1980;8(2):119–174. doi: 10.3109/10409238009105467. [DOI] [PubMed] [Google Scholar]
- Landel C. P., Krause M., Waterston R. H., Hirsh D. DNA rearrangements of the actin gene cluster in Caenorhabditis elegans accompany reversion of three muscle mutants. J Mol Biol. 1984 Dec 15;180(3):497–513. doi: 10.1016/0022-2836(84)90024-x. [DOI] [PubMed] [Google Scholar]
- Lehman W., Szent-Györgyi A. G. Regulation of muscular contraction. Distribution of actin control and myosin control in the animal kingdom. J Gen Physiol. 1975 Jul;66(1):1–30. doi: 10.1085/jgp.66.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman W. Thick-filament-linked calcium regulation in vertebrate striated muscle. Nature. 1978 Jul 6;274(5666):80–81. doi: 10.1038/274080a0. [DOI] [PubMed] [Google Scholar]
- Mackenzie J. M., Jr, Schachat F., Epstein H. F. Immunocytochemical localization of two myosins within the same muslce cells in Caenorhabditis elegans. Cell. 1978 Oct;15(2):413–419. doi: 10.1016/0092-8674(78)90010-7. [DOI] [PubMed] [Google Scholar]
- Matsuda G., Suzuyama Y., Maita T., Umegane T. The L-2 light chain of chicken skeletal muscle myosin. FEBS Lett. 1977 Dec 1;84(1):53–56. doi: 10.1016/0014-5793(77)81055-7. [DOI] [PubMed] [Google Scholar]
- Meinkoth J., Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984 May 1;138(2):267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Moerman D. G., Benian G. M., Barstead R. J., Schriefer L. A., Waterston R. H. Identification and intracellular localization of the unc-22 gene product of Caenorhabditis elegans. Genes Dev. 1988 Jan;2(1):93–105. doi: 10.1101/gad.2.1.93. [DOI] [PubMed] [Google Scholar]
- Murphy W. J., Watkins K. P., Agabian N. Identification of a novel Y branch structure as an intermediate in trypanosome mRNA processing: evidence for trans splicing. Cell. 1986 Nov 21;47(4):517–525. doi: 10.1016/0092-8674(86)90616-1. [DOI] [PubMed] [Google Scholar]
- Nabeshima Y., Fujii-Kuriyama Y., Muramatsu M., Ogata K. Alternative transcription and two modes of splicing results in two myosin light chains from one gene. Nature. 1984 Mar 22;308(5957):333–338. doi: 10.1038/308333a0. [DOI] [PubMed] [Google Scholar]
- Nabeshima Y., Nabeshima Y., Nonomura Y., Fujii-Kuriyama Y. Nonmuscle and smooth muscle myosin light chain mRNAs are generated from a single gene by the tissue-specific alternative RNA splicing. J Biol Chem. 1987 Aug 5;262(22):10608–10612. [PubMed] [Google Scholar]
- Nudel U., Calvo J. M., Shani M., Levy Z. The nucleotide sequence of a rat myosin light chain 2 gene. Nucleic Acids Res. 1984 Sep 25;12(18):7175–7186. doi: 10.1093/nar/12.18.7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Parker V. P., Falkenthal S., Davidson N. Characterization of the myosin light-chain-2 gene of Drosophila melanogaster. Mol Cell Biol. 1985 Nov;5(11):3058–3068. doi: 10.1128/mcb.5.11.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periasamy M., Strehler E. E., Garfinkel L. I., Gubits R. M., Ruiz-Opazo N., Nadal-Ginard B. Fast skeletal muscle myosin light chains 1 and 3 are produced from a single gene by a combined process of differential RNA transcription and splicing. J Biol Chem. 1984 Nov 10;259(21):13595–13604. [PubMed] [Google Scholar]
- Pulliam D. L., Sawyna V., Levine R. J. Calcium sensitivity of vertebrate skeletal muscle myosin. Biochemistry. 1983 May 10;22(10):2324–2331. doi: 10.1021/bi00279a004. [DOI] [PubMed] [Google Scholar]
- Reinach F. C., Nagai K., Kendrick-Jones J. Site-directed mutagenesis of the regulatory light-chain Ca2+/Mg2+ binding site and its role in hybrid myosins. Nature. 1986 Jul 3;322(6074):80–83. doi: 10.1038/322080a0. [DOI] [PubMed] [Google Scholar]
- Robert B., Daubas P., Akimenko M. A., Cohen A., Garner I., Guenet J. L., Buckingham M. A single locus in the mouse encodes both myosin light chains 1 and 3, a second locus corresponds to a related pseudogene. Cell. 1984 Nov;39(1):129–140. doi: 10.1016/0092-8674(84)90198-3. [DOI] [PubMed] [Google Scholar]
- Ross J. A precursor of globin messenger RNA. J Mol Biol. 1976 Sep 15;106(2):403–420. doi: 10.1016/0022-2836(76)90093-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slightom J. L., Blechl A. E., Smithies O. Human fetal G gamma- and A gamma-globin genes: complete nucleotide sequences suggest that DNA can be exchanged between these duplicated genes. Cell. 1980 Oct;21(3):627–638. doi: 10.1016/0092-8674(80)90426-2. [DOI] [PubMed] [Google Scholar]
- Spieth J., Denison K., Zucker E., Blumenthal T. The nucleotide sequence of a nematode vitellogenin gene. Nucleic Acids Res. 1985 Oct 11;13(19):7129–7138. doi: 10.1093/nar/13.19.7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Brenner S. The DNA of Caenorhabditis elegans. Genetics. 1974 May;77(1):95–104. doi: 10.1093/genetics/77.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton R. E., Boothroyd J. C. Evidence for trans splicing in trypanosomes. Cell. 1986 Nov 21;47(4):527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffenetti J., Mischke D., Pardue M. L. Isolation and characterization of the gene for myosin light chain two of Drosophila melanogaster. J Cell Biol. 1987 Jan;104(1):19–28. doi: 10.1083/jcb.104.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Waterston R. H., Fishpool R. M., Brenner S. Mutants affecting paramyosin in Caenorhabditis elegans. J Mol Biol. 1977 Dec 15;117(3):679–697. doi: 10.1016/0022-2836(77)90064-x. [DOI] [PubMed] [Google Scholar]
- Waterston R. H., Hirsh D., Lane T. R. Dominant mutations affecting muscle structure in Caenorhabditis elegans that map near the actin gene cluster. J Mol Biol. 1984 Dec 15;180(3):473–496. doi: 10.1016/0022-2836(84)90023-8. [DOI] [PubMed] [Google Scholar]
- Winter B., Klapthor H., Wiebauer K., Delius H., Arnold H. H. Isolation and characterization of the chicken cardiac myosin light chain (L-2A) gene. Evidence for two additional N-terminal amino acids. J Biol Chem. 1985 Apr 10;260(7):4478–4483. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Kent J. R., Cech T. R. A labile phosphodiester bond at the ligation junction in a circular intervening sequence RNA. Science. 1984 May 11;224(4649):574–578. doi: 10.1126/science.6200938. [DOI] [PubMed] [Google Scholar]