Abstract
Protein biomarkers are critical for diagnosis, prognosis, and treatment of disease. The transition from protein biomarker discovery to verification can be a rate limiting step in clinical development of new diagnostics. Liquid chromatography-selected reaction monitoring mass spectrometry (LC-SRM MS) is becoming an important tool for biomarker verification studies in highly complex biological samples. Analyte enrichment or sample fractionation is often necessary to reduce sample complexity and improve sensitivity of SRM for quantitation of clinically relevant biomarker candidates present at the low ng/mL range in blood. In this paper, we describe an alternative method for sample preparation for LC-SRM MS, which does not rely on availability of antibodies. This new platform is based on selective enrichment of proteotypic peptides from complex biological peptide mixtures via isoelectric focusing (IEF) on a digital ProteomeChip (dPC™) for SRM quantitation using a triple quadrupole (QQQ) instrument with an LC-Chip (Chip/Chip/SRM). To demonstrate the value of this approach, the optimization of the Chip/Chip/SRM platform was performed using prostate specific antigen (PSA) added to female plasma as a model system. The combination of immunodepletion of albumin and IgG with peptide fractionation on the dPC, followed by SRM analysis, resulted in a limit of quantitation of PSA added to female plasma at the level of ~1–2.5 ng/mL with a CV of ~13%. The optimized platform was applied to measure levels of PSA in plasma of a small cohort of male patients with prostate cancer (PCa) and healthy matched controls with concentrations ranging from 1.5 to 25 ng/mL. A good correlation (r2 = 0.9459) was observed between standard clinical ELISA tests and the SRM-based-assay. Our data demonstrate that the combination of IEF on the dPC and SRM (Chip/Chip/SRM) can be successfully applied for verification of low abundance protein biomarkers in complex samples.
Keywords: Isoelectric focusing, IEF, digital ProteomeChip, dPC, selected reaction monitoring, SRM, prostate specific antigen, PSA, QQQ, LC-Chip
INTRODUCTION
The field of clinical proteomics has the goal of the discovery, verification and validation of disease-specific protein biomarkers that can ultimately be applied as blood tests for diagnostic, prognostic or theranostic purposes. The discovery process typically involves comparative studies of a limited number of clinical samples using fractionation and shotgun proteomics to identify a panel of proteins whose concentrations are found to change in diseased blood relative to control samples1. It is generally assumed that a number of clinically relevant candidate biomarkers will be present in blood at the low ng/mL level2, 3 and that, after discovery, these candidate biomarkers will need to be further verified and then validated in large patient sample sets. The assay of such low level proteins must have the capability of high throughput, sensitivity, and the ability to measure multiple targets within a single run, i.e., multiplexing1, 4, 5. The shortcomings of ELISA (high cost, long lead time, high failure rate, difficulty to multiplex, etc.) or the absence of specific and well characterized antibodies for newly discovered targets have prompted the development of alternative methods for targeted quantitation of proteins in highly complex plasma samples.
Liquid chromatography-selected reaction monitoring (LC-SRM) coupled with stable isotope dilution has been shown to be a potential effective method for biomarker verification and possibly validation of novel protein biomarker candidates6–8. In addition to speed, sensitivity, and multiplexing capability, the SRM assay can be used to discriminate between closely related protein isoforms9, a characteristic that cannot be easily achieved with an ELISA assay. The SRM assay is also relatively easy to implement and optimize. However, when SRM is applied directly to unfractionated plasma, the method is generally not sufficiently sensitive, being limited to detection of proteins at the low µg/mL level10–13. The recent introduction of a dual ion funnel interface has enhanced the sensitivity of SRM by approximately 10-fold, and detection of proteins ranging from 40 to 80 ng/mL within unfractionated mouse plasma was achieved14. Nonetheless, analyte enrichment at the protein and/or peptide level is generally necessary to reduce sample complexity and improve sensitivity for quantitating low abundant biomarkers at only a few ng/mL concentration in blood or lower.
Strategies for lowering the detection limit of the SRM assay have focused on reducing sample complexity prior to the SRM analysis. These approaches include enrichment of proteins or peptides using antibodies9, 15, 16 and depletion of the most abundant plasma proteins in combination with peptide fractionation using charge- or hydrophobicity-based separations6, 17, 18. Although immunoextraction using stable isotope standards and capture by anti-peptide antibodies (SISCAPA)7, 19 has been shown to reach low ng/mL protein sensitivities, the approach requires well characterized and effective antibodies. Comparable limits of quantitation have been achieved using a combination of immunodepletion and strong cation exchange (SCX) fractionation18
Isoelectric focusing (IEF) has been employed for separation and enrichment of low abundant proteins derived from cell lysates20, tissues21, and plasma22–24 prior to MS analysis. To date, IEF, however, has not been exploited as a peptide enrichment method in combination with SRM25. Various approaches to IEF, such as capillary isoelectric focusing (cIEF), immobilized pH gradient isoelectric focusing (IPG-IEF) and Off-Gel™ IEF, have been applied for separating complex peptide mixtures prior to MS. However, for sample pretreatment prior to SRM, the technology remains problematic. For example, capillary isoelectric focusing shows high resolving power, but the low loading capacity and the fact that the system requires complicated switching valves and capillary traps, makes the routine use of this system difficult26. Other disadvantages of using liquid-based IEF may include moderate resolving power and the presence of large concentrations of carrier ampholytes that can interfere with MS27–30. Immobilized pH gradient isoelectric focusing offers high capacity, wide dynamic range, and high-resolution, but it is a low throughput method31, 32. The Off-Gel™ IEF approach has been successful in-solution recovery of peptides and high capacity, but it is also a low throughput method, with separations taking as long as 24 hours33, 34. Furthermore, it uses high sample volumes that require further sample processing steps when employed with capillary LC systems.
We previously reported on the use of digital ProteomeChip (dPC™) technology (Cell Biosciences, Santa Clara, CA) for fractionation and enrichment of medium to low abundant proteins from human plasma35. In this study, we have utilized isoelectric focusing on the digital ProteomeChip (dPC™) technology (Cell Biosciences, Santa Clara, CA) for selective enrichment of proteotypic peptides from plasma samples for SRM analysis using a triple quadrupole (QQQ) instrument with high-performance liquid chromatography (HPLC)-Chip microfluidic system. The dPC consists of discrete pH gel plugs for pH intervals to trap proteins or peptides in respect to their pI (See Figure S1 in the Supplement for a description of the system). Isoelectric focusing using the dPC offers several potential advantages for SRM analysis. Separation is fast (less than 1 hour), ampholyte-free, and capable of fractionating high loads of proteins and/or peptides per run, which are then recovered into low volumes. Moreover, the development and optimization of both dPC and SRM methods can be rapid. This paper demonstrates that IEF on the dPC in combination with SRM can be a general approach for verification of low abundant biomarker candidates in plasma.
To evaluate the platform, prostate specific antigen (PSA), added to female plasma, was chosen as a model sample. As the sample was used previously17, 18, 36–39, we could compare our approach to that of others who employed alternative sample clean-up and enrichment methods17, 18. In the present work, the combination of immunodepletion of albumin and IgG along with peptide fractionation on the dPC, resulted in the reproducible quantitation of PSA at levels as low as 1.0 ng/mL in plasma. Furthermore, PSA levels in male prostate cancer (PCa) patient plasmas determined with the platform were well correlated with ELISA analyses of PSA. The IEF Chip/LC-Chip-SRM (termed Chip/Chip/SRM) methodology combines two powerful low volume separation technologies: (1) dPC rapidly resolves and concentrates peptides into individual pI fractions, each in a separate gel plug; (2) the LC-Chip allows highly efficient, sensitive, and reproducible LC separation. The final SRM analysis is conducted on a triple quadrupole mass spectrometer in a conventional approach.
EXPERIMENTAL PROCEDURES
Materials
Bovine sequencing grade modified trypsin was purchased from Promega (Madison, WI). Formic acid (FA), acetonitrile (MeCN), Optima LC/MS grade water, and other buffer reagents were from Thermo Fisher Scientific (Fair Lawn, NJ). The isotopically labeled peptides were synthesized and quantitated (amino acid analysis) by AnaSpec (San Jose, CA). The unlabeled versions of peptides were synthesized by GenScript Corp. (Piscataway, NJ) and quantitated (amino acid analysis) at Dana Farber Cancer Institute Core Facilities (Boston, MA). Human prostate-specific antigen (PSA) was purchased from Scripps Laboratories (San Diego, CA) and submitted for amino acid analysis to Dana Farber. After appropriate Institutional Review Board approval from Northeastern University, both matched control and prostate cancer (PCa) plasma samples were purchased from ProteoGenex (Culver City, CA).
Removal of albumin and IgG from plasma
CaptureSelect® affinity ligand with specificity for albumin (BAC B.V., The Netherlands) was immobilized on POROS® AL 20 µm Self Pack® media (Life Technologies, Inc., Carlsbad, CA) and packed into a PEEK column (Isolation Technologies, Hopedale, MA) to prepare the albumin depletion column (4.6 mm × 100 mm)40. Protein A media (POROS® MabCapture™ A, Life Technologies, Inc.), packed into a 4.6 mm × 50 mm PEEK column, was used for depletion of IgG. Depletion was performed on a Shimadzu LC-20AD HPLC instrument (Shimadzu Scientific Instruments, Columbia, MD). The packed columns were connected in series and equilibrated with 1X PBS prior to plasma depletion. Plasma sample (100 µL) was diluted 5-fold with 1X PBS and loaded onto the IgG and albumin depletion columns at 0.5 mL/min for 12 min. The flow rate was then increased to 2 mL/min for an additional 3 min. The flow-through fraction was collected into 4 mL Amicon 5 kDa MWCO filter (Millipore, MA) and concentrated to 50 µL at 2,500 × g using a bench-top centrifuge (model 5804R Eppendorf North America, Westbury, NY). Samples were removed and filters washed with 50 µL of denaturing buffer (6 M guanidine-HCl in 0.1 M ammonium bicarbonate, pH 8.0); the wash was then combined with the concentrated samples. The bound albumin and IgG were eluted with 18 mL of elution buffer (100 mM glycine, pH 2.5) at 3 mL/min. Following elution, the columns were regenerated with 12 mL of neutralization buffer (500 mM Tris, 1 M NaCl, 0.05% sodium NaN3, pH 7.4) and then re-equilibrated with 25 mL of 1X PBS at 5 mL/min. Total protein concentration was determined by the Bradford protein assay.
PSA test samples
Prostate specific antigen (PSA) test samples were prepared by adding PSA protein prior to albumin and IgG depletion of human plasma from a healthy female donor at 0, 0.25, 0.5, 1.0, 2.5, 5.0, 25, and 50 ng/mL. A second group of PSA test samples was prepared by adding PSA at 0, 0.25, 0.5, 1.0, 2.5, and 5.0 ng/mL after immunoaffinity depletion of albumin and IgG from the female plasma.
Human plasma sample preparation for dPC isoelectric focusing
The isotopically labeled internal standard peptides were added just prior to trypsin digestion at a concentration of 3.8 ng/mL. Tryptic digests of spiked depleted plasma samples were prepared as follows: samples were denatured with 6 M guanidine-HCl in 0.1 M ammonium bicarbonate, pH 8.0, and then reduced by addition of 0.5 M TCEP solution to a final concentration of 20 mM. The mixture was incubated at room temperature for 15 min; 0.5 M iodoacetamide was added to a final concentration of 40 mM, and the sample was incubated at room temperature for 30 min in the dark. The reaction was quenched with 10 mM DTT for 5 min at room temperature. Guanidine hydrochloride concentration was diluted 6-fold with 0.1 M ammonium bicarbonate, pH 8.0, prior to overnight digestion at 37°C with trypsin using a 1:25 (w/w) enzyme to substrate ratio. Digests were terminated with formic acid to a final concentration of 1% formic acid and guanidine hydrochloride, and excess reagents were removed using Oasis HLB 1cc (30 mg) cartridges (Waters, Milford, MA) according to the following procedure: (1) wash cartridge with 3 × 500 µL 80% acetonitrile in 0.1% formic acid; (2) equilibrate with 3 × 500 µL 0.1% formic acid; (3) load sample digest; (4) wash 4 × 500 µL 0.1% formic acid; (5) elute peptides with 2 × 500 µL 0.1% formic acid. Washing and elution were carried out using a Vac Elut 20 vacuum manifold (Varian, Palo Alto, CA). Eluates were frozen at −80°C and concentrated to a volume of 10 µL via vacuum centrifugation.
dPC IEF Chip enrichment/separation of peptides
In order to determine the dPC focusing pH region and peak maxima for the signature peptides, 1 µg of each of the peptide standards was dissolved in 600 µL of cathode focusing buffer for the dPC chip in the range of pH 3.00 – 5.00 (the information on the composition and concentration of the cathode buffer is proprietary). The dPC was placed between the anode and cathode chambers. After loading the sample, isoelectric focusing was performed for 1 hour at 50 V. Triplicate samples were separated by dPC. After focusing, each dPC was briefly rinsed with water, and all 41 gel plugs, in pools of 4 per fraction (i.e. 0.2 pH units), were manually ejected into microcentrifuge tubes using a dull end needle device supplied by the manuracturer. Target peptides were extracted from specific gel plugs with 50 µL of 0.2% formic acid in 50% acetonitrile at 37°C for 1 hour. Peptides were collected in fresh microcentrifuge tubes, concentrated to a volume of 20 µL via vacuum centrifugation, and analyzed by LC-Chip-SRM (see below). The dPC focusing profile for each of the peptides was constructed by integrating the SRM peak area at the m/z value of the selected transition ion used for quantitation, as described in SRM assay optimization. Actual digested plasma samples were reconstituted in the corresponding dPC running buffer and processed using the above isoelectric focusing conditions. Pools of eight gel plugs (0.4 pH units) per fraction were generated according to the dPC focusing profile of the signature peptides from above. The previous conditions were for extraction and concentration of PSA fractions at a volume of 10 µL. Extracts were quantitatively analyzed by the LC-Chip-SRM described in the following.
LC-Chip-SRM
Nano-flow liquid chromatography was performed using a 1200 Series LC system connected to an LC-Chip Cube interfaced to a 6460 Triple Quadrupole (QQQ) LC-MS/MS (Agilent Technologies, Santa Clara, CA). Agilent MassHunter software (version B.02.01) was used for data acquisition and processing. The LC separation of peptides was carried out on a Protein ID Chip consisting of a 160 nL trapping column and a 75 µm × 150 mm separation column packed with Zorbax 300SB-C18 5-µm (Agilent). Processed samples or peptide standards were loaded onto the enrichment column using an Agilent 1260 autosampler. Solid-phase extraction (SPE) of the analytes prior to gradient separation was performed by the capillary LC pump delivering a mixture of 97% water/0.1% formic acid (mobile phase A) and 3% acetonitrile/0.1% formic acid at 4 µL/min. The sample was eluted in the backflush mode from the enrichment column and transferred to the analytical column by automatic switching of the LC-Chip nanorotary valve. Peptides were separated at a flow rate of 600 nL/min by a nanopump delivering a linear gradient of 5–70% mobile phase B in 7 min, followed by 2 min for column re-equilibration. The analyses were performed in the positive ionization mode with a capillary voltage set at 1850 V and an electron multiplier voltage (Delta EMV) at 200 V. The drying gas flow rate was 1.8 L nitrogen/min with an interface heater temperature of 325°C. The MS fragmentor voltage was fixed at 135 V. SRM transition dwell times were 100 ms, with Q1 set to “wide” and Q3 to “unit”.
Proteotypic peptide selection and SRM assay optimization
To select the proteotypic peptides, we used the enhanced signature peptide (ESP) predictor41. Additionally, the ESP predictor was utilized to generate a list of SRM transition candidates for each peptide consisting of only y ions. The theoretical pI values of the predicted peptides were calculated using the pI function of L’Atelier BioInformatique de Marseille (A.B.I.M) (http://www.up.univ-mrs.fr/~wabim/). The most predominant charge state of selected peptides was verified using the 6460 QQQ with LC-MS (MS2Scan) acquisition in the 200–1400 m/z range with a scan time of 500 ms. Using MassHunter Optimizer software for peptides, the collision energy (CE) voltage was automatically optimized for the SRM transition candidates. Briefly, the QQQ was set to operate in a targeted fashion whereby only molecular ions corresponding to the most dominant charge state [M + 2]2+ of selected peptides were transmitted through Q1, and SRM transition candidates were monitored in Q3. Both Q1 and Q3 resolution were set to “unit,” and a default dwell time of 5 ms was used.
PSA ELISA procedure
The human PSA ELISA kit was obtained from Alpha Diagnostics Intl. Inc. (San Antonio, TX), and the assays were conducted following the recommended procedures. Briefly, an ELISA plate was washed with 300 µL of 1X wash buffer. During the assay, 25 µL of standards, controls, and plasma samples, together with 100 µL of an antibody-enzyme conjugate, were added to each well which was coated with a primary antibody. The plate was incubated at room temperature for 30 min. After that time, solution in each well was removed, and the wells were washed four times with 300 µL of 1X wash buffer. An aliquot (100 µL) of 3,3’,5,5’-tetramethylbenzidine (TMB) was added to each well, and the plate was incubated at room temperature for 15 min. The reaction was ended by adding 50 µL of sulfuric acid solution. The absorbance of individual wells was measured at 450 nm using SpectraMax 340PC384 Absorbance Microplate Reader (Molecular Devices, Sunnyvale, CA). A standard curve was obtained by plotting the absorbance and the concentrations of the protein standards.
RESULTS AND DISCUSSION
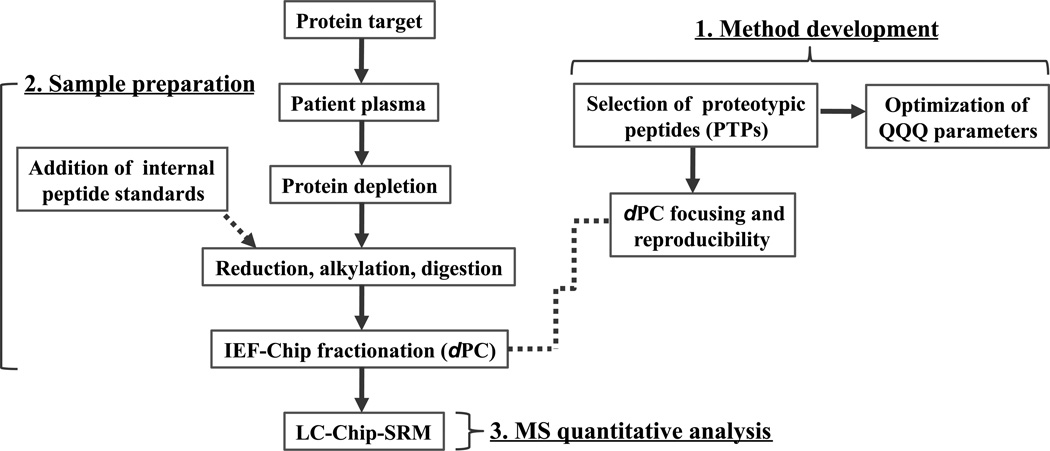
A workflow chart of the Chip/Chip/SRM platform is presented in Figure 1. The approach consisted of three steps: (1) method development – selection of proteotypic peptides, determination of IEF profile, and optimization of the SRM assay; (2) sample preparation – proteolysis of immunodepleted plasma and separation of peptides on the IEF Chip; and (3) LC Chip/SRM quantitative analysis. To evaluate the platform, we first optimized the method using PSA in female plasma as a test sample. Next, data from prostate cancer patient plasma samples were obtained and compared with that of a corresponding ELISA analysis on the same samples.
Figure 1.
Workflow for analysis of plasma samples using isoelectric focusing on the dPC and SRM for selective enrichment and quantitation of signature peptides derived from target proteins.
Selection of proteotypic peptides for PSA and the SRM assay
Currently, a variety of databases of MS experimental data42, 43 or computational tools44–47 are available to aid in the selection of proteotypic peptides for protein targets, without the need for initial MS discovery experiments. In this study, the selection of proteotypic peptides was conducted using the ESP predictor, a computational approach trained to predict high-responding peptides in an ESI-MS experiment41. The selection criteria were primarily based not only on peptide-response calculated by the ESP predictor, but also on pI values that fall within the pH range of the currently available dPC chips (pH 3.00–5.00, pH 4.20–6.20, and pH 6.00–8.00). Peptides IVGGWECEK and LSEPAELTDAVK with calculated ESP/pI values of 0.36/4.3 and 0.84/3.9, respectively, were selected as proteotypic peptides for the PSA protein. These peptides are identical to those used by others utilizing PSA for development of SRM platforms for clinical applications (without consideration of pI values)17, 18, 36, 37. The stable isotope labeled standards of the selected peptides were commercially synthesized with a single, uniformly labeled [13C6] leucine or [13C5] valine amino acid. The SRM assay was optimized for each peptide by determining appropriate precursor and product ion transitions in order to maximize sensitivity and specificity. The results of this SRM optimization are shown in Table 1.
Table 1.
Isoelectric focusing and mass spectrometric properties of proteotypic peptides for prostate specific antigen (PSA).
| Protein | MW (kDa) |
Proteotypic Peptidesa | ESP Valuec |
pI | dPC | MH+ (mono) |
Charge state, x, (Q1) |
Precursor ion (m/z), Q1 |
Product ions (m/z), Q3 |
Collision Energy (V) |
Quantitation transition (m/z) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Prostate specific antigen |
29 | LSEPAELTDAVK | 0.84 | 3.9 | 3.0–5.0 | 1272.7 | 2 | 636.8 | 775.4, 846.5, 943.5 | 26, 22, 14 | 636.8 → 943.5 |
| LSEPAEL[13C6]TDAVK | 1278.7 | 2 | 640.8 | 781.4, 852.5, 949.5 | 26, 22, 14 | 640.8 → 949.5 | |||||
| IVGGWECamcEKb | 0.36 | 4.3 | 3.0–5.0 | 1078.5 | 2 | 539.7 | 809.3, 866.3, 965.4 | 15, 15, 15 | 539.7 → 866.3 | ||
| IV[13C6]GGWECamcEKb | 1083.5 | 2 | 542.2 | 809.3, 866.3, 971.4 | 15, 15, 15 | 542.2 → 866.3 |
The stable isotope labeled amino acids are indicated in bold.
Carbamidomethyl group were introduced during peptide synthesis.
ESP value – probability value.
LC-Chip-SRM assay for PSA quantitative analysis
We first evaluated LC-Chip-SRM analytical performance, independent of the dPC chip, to assess the analytical method. For this purpose, plasma, depleted of albumin and IgG, was digested and then spiked with both unlabeled and stable isotopic labeled peptide standards. A number of analytical characteristics were determined, including linearity (r2), limit of detection (LOD, 3X signal-to-noise ratio (S/N); S/N = peak intensity at the apex/noise level prior to and following the analyte peak), limit of quantitation (LOQ, 10X S/N), accuracy (percent relative error, %RE), and precision (% CV = (mean calculated concentration (n = 4)/standard deviation) × 100). The isotopically labeled peptide internal standards were prepared at 3.8 ng/mL using immunodepleted human female plasma digest (diluted 40-fold) as sample matrix. The sample matrix and internal standard samples were found to have no target analytes as determined by SRM analysis. During each analysis, solutions containing 0.06, 0.12, 0.24, 0.94, 3.8, 15, 60, and 240 ng/mL of the unlabeled peptides were constructed in the presence of a fixed amount, 3.8 ng/mL, of the isotopically labeled peptides. Four replicates of 4 µL aliquots of each working standard solution were injected into the LC-Chip for SRM analysis. Using MassHunter quantitative analysis software, we obtained linearity plots and evaluated accuracy and precision. The highest intensity transition was used to quantitate the peptides, and two other significant transitions were monitored for assessment of the specificity and selectivity of the SRM assay. The responses of the quantitation transitions for IVGGWECEK (m/z 539.7 → 866.3) and LSEPAELTDAVK (m/z 636.8 → 943.5) peptides derived from PSA were found to be linear (r2 = 0.9851 and 0.9941, respectively) over the concentration range examined (Figure S2 in the Supplement), while the accuracy of the measurements ranged from −7.8 to 10% and −9.4 to 1.0%, respectively. The CVs for both of the target peptides were within_+ 16%. The LC-SRM LOQ for both peptides was approximately 0.12 ng/mL, and the LOD was ≤ 0.06 ng/mL.
dPC fractionation
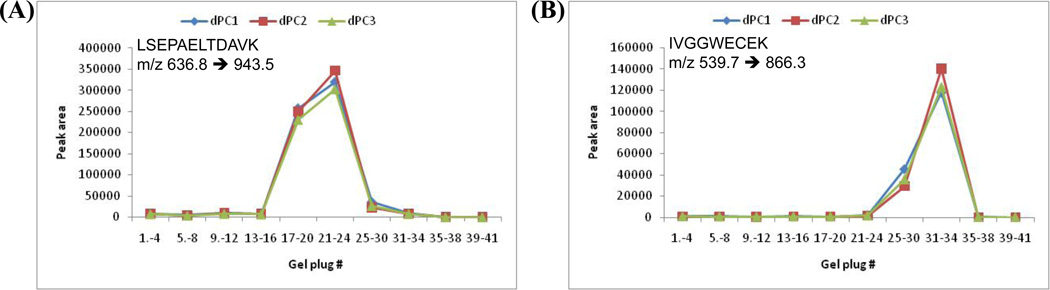
The dPC chip consisted of 41 discrete gel plugs with pH intervals of 0.05 over a pH range of 2, which, when placed in an electric field, trapped proteins or peptides from a complex mixture according to the respective pI values of the species. The operating principles of the dPC are shown in Figure S1 in the Supplement. The electrophoretic focusing of signature peptides was investigated in order to determine the trapping pH region as well as the repeatability of the IEF process. Figure 2 illustrates the distribution of PSA peptide standards in dPC gel plugs throughout the 3.00–5.00 pH chips. Both peptides focused consistently over 8 gel plugs corresponding to 0.4 pH units. A narrower electrofocusing peak could be obtained on a dPC with greater pH differences between the gel plugs (e.g., 0.2 pH unit); these chips are not commercially available at the present time, but have been made in prototype. Based on the results of Figure 2, the gel plugs were pooled into one fraction per peptide (two fractions total). Eight gel plugs (0.4 pH units) per fraction were used for LC-Chip-SRM analysis of PSA peptides from plasma samples.
Figure 2.
dPC electrofocusing and reproducibility for PSA standard peptides. SRM analysis of ~1 µg of standard peptide solution of peptide LSEPAELTDAVK, pI 3.9 (A) and peptide IVGGWECEK, pI 4.3 (B) after IEF on the pH 3.0–5.0 dPC.
Development of Chip/Chip/SRM platform for plasma samples
In order for Chip/Chip/SRM to be of utility for biomarker analysis, the platform must be, as noted earlier, capable of precise and accurate measurements of clinically relevant proteins present in plasma at the low ng/mL level48. Prostate specific antigen (PSA) added to female plasma represents a good model system for the development of new workflows for LC-SRM assay, as has been used by others17, 18, 36, 37. The study began with nondepleted human female plasma, to which the PSA protein standard was added at 50 ng/mL. This sample was subjected to digestion with trypsin and IEF fractionation on the dPC, followed by LC-SRM analysis, targeting both proteotypic peptides derived from PSA. The detection of PSA in the highly complex, nondepleted plasma that was fractionated on the dPC was not observed, even at 50 ng/mL of PSA added. The conclusion from this experiment was that pre-fractionation of the plasma sample was required prior to fractionation on the dPC.
Albumin and IgG, the two highest concentration proteins in plasma were removed from the human female plasma to which the protein standard was added. The performance of the Chip/Chip/SRM platform was evaluated using six immunodepleted plasma samples containing PSA at concentrations ranging from 0 to 50 ng/mL. The results, derived from duplicate LC-Chip-SRM analyses for two different samples (n = 4), are presented in Table 2 (i). The LOQ for PSA peptide IVGGWECEK was between 1.0 and 2.5 ng/mL, with an estimated average recovery of ~ 36% and a CV of ~13%. Recoveries were determined by comparing the SRM signal of the peptide to the internal calibration plot generated using the synthetic peptide. Similar quantitative data for the second signature peptide, LSEPAELTDAVK, derived from PSA was also obtained.
Table 2.
Results of quantitative Chip/Chip/SRM measurements for PSA protein added to human female plasma (i) prior to immunodepletion of albumin and IgGa with LOQ indicated in bold; (ii) recovery of PSA after immunodepletion step only as determined by ELISA; and (iii) Chip/Chip/SRM measurements for PSA protein added to female plasma after immunodepletion with the LOQ specified in bold.
| Target ng/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Protein | Proteotypic peptide |
Q1 → Q3 | Quantitative measurements |
0.25 | 0.50 | 1.0 | 2.5 | 5.0 | 25 | 50 |
| (i) | PSA added prior to immunodepletion/dPC | |||||||||
| PSA | IVGGWECamcEK | 539.7 → 866.3 | Calculated ng/mL | NSD | 0.10 | 0.29 | 1.1 | 2.6 | 19 | 35 |
| % CV | NSD | 137 | 13 | 18 | 12 | 8.2 | 10 | |||
| % Recoveryb | NSD | 20 | 29 | 42 | 53 | 76 | 70 | |||
| PSA | LSEPAELTDAVK | 636.8 → 943.5 | Calculated ng/mL | NSD | 0.13 | 0.36 | 1.2 | 2.7 | 15 | 32 |
| % CV | NSD | 132 | 19 | 14 | 18 | 11 | 12 | |||
| % Recoveryb | NSD | 20 | 36 | 47 | 55 | 60 | 64 | |||
| (ii) | PSA after immunodepletion (without dPC), % recovery measured by ELISA (n = 2) | |||||||||
| Total PSA measured | N/A | N/A | N/A | 41 | 65 | 71 | 65 | |||
| (iii) | PSA added after immunodepletion/dPC | |||||||||
| PSA | IVGGWECamcEK | 539.7 → 866.3 | Calculated ng/mL | 0.11 | 0.21 | 1.0 | 2.8 | 4.1 | N/A | N/A |
| % CV | 122 | 2.8 | 10.0 | 9.9 | 9.6 | |||||
| % Recoveryb | 44 | 41 | 101 | 110 | 81 | |||||
| PSA | LSEPAELTDAVK | 636.8 → 943.5 | Calculated ng/mL | 0.20 | 0.24 | 1.0 | 2.6 | 4.1 | N/A | N/A |
| % CV | 115 | 22 | 16 | 13 | 11 | |||||
| % Recoveryb | 80 | 48 | 98 | 103 | 82 | |||||
Sample Preparations and SRM experiments were performed in duplicate for a total of four replicates (n = 4).
% Recovery = (calculated (ng/mL)/target (ng/mL) × 100.
N/A = sample were not investigated.
NSD = no single detected.
Over the whole PSA concentration range, protein recoveries ranged from 29 to 76% and the overall coefficients of variation ranged from 8.2 to 18%. The recoveries and precision improved at higher concentrations of PSA, as expected. The sensitivity and precision values from our platform compare well to those obtained by Fortin et. al.17 (LOQ 1.5 ng/mL and CV ~10%) in a study with PSA added to human female plasma prior to albumin depletion and solid-phase extraction (SPE) fractionation coupled to SRM. As will be shown below, the lower recovery obtained for the 1.0 ng/mL sample in the present work is mainly associated with immunodepletion and not fractionation in the dPC. To determine linearity of the Chip/Chip/SRM platform, calibration plots were generated by measuring the peak area ratio (sample peptide/isotopically labeled peptide internal standard) versus protein concentration in the range of 1.0 – 50 ng/mL. A good linear correlation was established between the peak area ratio and protein concentration with a coefficient of linearity of r2 = 0.9812, see Figure S3 in the Supplement.
Specificity of the method was examined by processing two samples of depleted female blank plasma on the dPC and analysis by SRM. Although SRM signals were detected at retention times close to those of both PSA proteotypic peptides, the detection was nonspecific due to the lack of all three transitions monitored with peak area never greater than 20% of the LOQ.
Effect of immunodepletion on PSA recovery
The major concern associated with removal of abundant proteins from plasma is the potential loss of proteins of interest. The loss of PSA during the immunodepletion step (including sample concentration via ultrafiltration) was investigated using ELISA for plasma analysis with PSA added in the range of 2.5 – 50 ng/mL. The ELISA results shown in Table 2 (ii) reveal that losses associated with high abundant protein removal are a major contributor to the lower overall recoveries of the platform at low PSA concentrations in plasma. We were not able to study the 1.0 ng/mL sample due to the ELISA’s limit of quantitation (1.5 ng/mL). However, we found that at 2.5 ng/mL, the depletion of albumin and IgG resulted in only ~ 40% recovery of PSA. The losses could be related to both non-specific binding of PSA to the depletion column and the downstream filtration/concentration step. Recoveries of proteins in the lower concentration range could be likely improved with optimization of the depletion step. The recovery for samples containing PSA at or above 5.0 ng/mL was ~ 65% and remained constant across all examined concentrations above this level.
Efficiency of trypsin digestion and dPC fractionation
The completeness of trypsin digestion and its effect on protein recovery was difficult to evaluate due to the lack of a stable isotopically labeled protein standard. However, the recent introduction of such reagents will enable SRM absolute quantitation in the future49. In order to explore digestion and dPC losses of PSA at low concentrations, samples containing PSA between 1.0 and 5.0 ng/mL were prepared by adding the PSA protein standard to female plasma, after the immunodepletion and ultrafiltration steps. Further fractionation on the dPC allowed analysis of PSA by the SRM method, resulting in approximately 100% (81–110%) recoveries at concentrations ≥1.0 ng/mL. These values indicate high efficiency of trypsin digestion and minimal losses during sample de-salting, dPC fractionation and extraction prior to SRM analysis. These results allowed us to test lower concentrations (0.5 and 0.25 ng/mL), leading to LOQ value of 0.5 ng/mL with recovery and CV values of 41% and 2.8%, respectively, Table 2 (iii). Using the LOQ value obtained for PSA when added to plasma after depletion of albumin and immunoglobulin allowed a comparison of the present platform to a study by Keshishian et. al18, in which PSA was added after depletion of the twelve most abundant plasma proteins, followed by fractionation using strong cation exchange prior to SRM analyses. The LOQ for PSA using this latter approach was reported to be 2.2 ng/mL, approximately 5 fold higher than the platform of the current paper.
Enrichment with the dPC
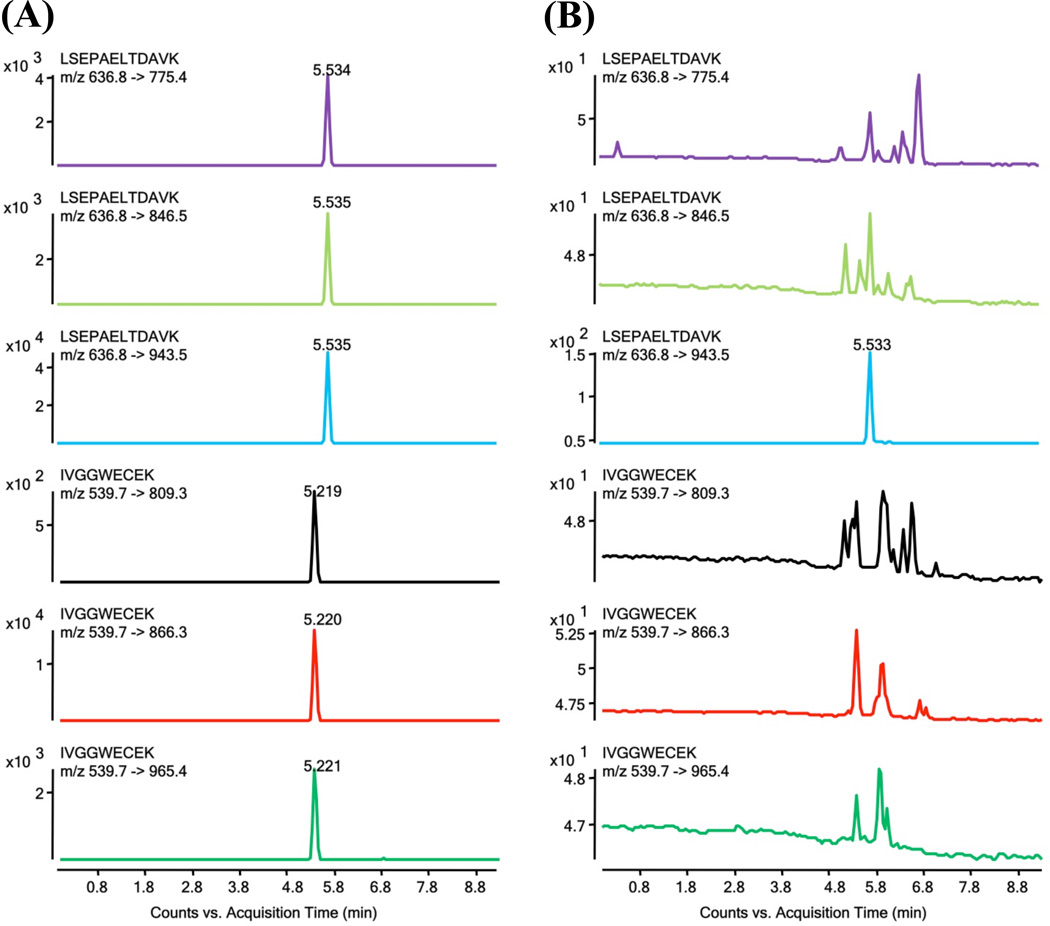
We investigated the enrichment of the PSA peptides from the dPC step by comparing the SRM response of a PSA plasma sample at 25 ng/mL processed using the platform with the response of the same sample omitting the dPC step. As illustrated in Figure 3, a 200-fold enhancement of the PSA peptide ion signal was achieved by the dPC. The differences in signal intensity for the target analyte are likely a result of reduction in ion suppression due to focusing over a narrow pH range (0.4 pH units) that helped to eliminate other sample digest peptide components in the plasma. Fractionation on the dPC reduces sample complexity and concentrates the target proteins diluted in plasma down to low volumes. The enrichment is attributed to the parallel isoelectric focusing incorporated into the dPC and the fast analyte transport between different gel plugs allowing for facile collection efficiency of the target peptides50.
Figure 3.
Enrichment capability of the dPC - comparison of PSA ion signals with and without enrichment on the dPC. Comparison of XICs acquired on all three SRM transitions for LSEPAELTDAVK (elution time 5.5 min) and IVGGWECEK (elution time 5.2 min) peptides derived from PSA standard protein added to plasma at 25 ng/mL and enriched (A) with the dPC and (B) without the dPC.
Chip/Chip/SRM versus ELISA for quantitation of PSA in plasma patient samples
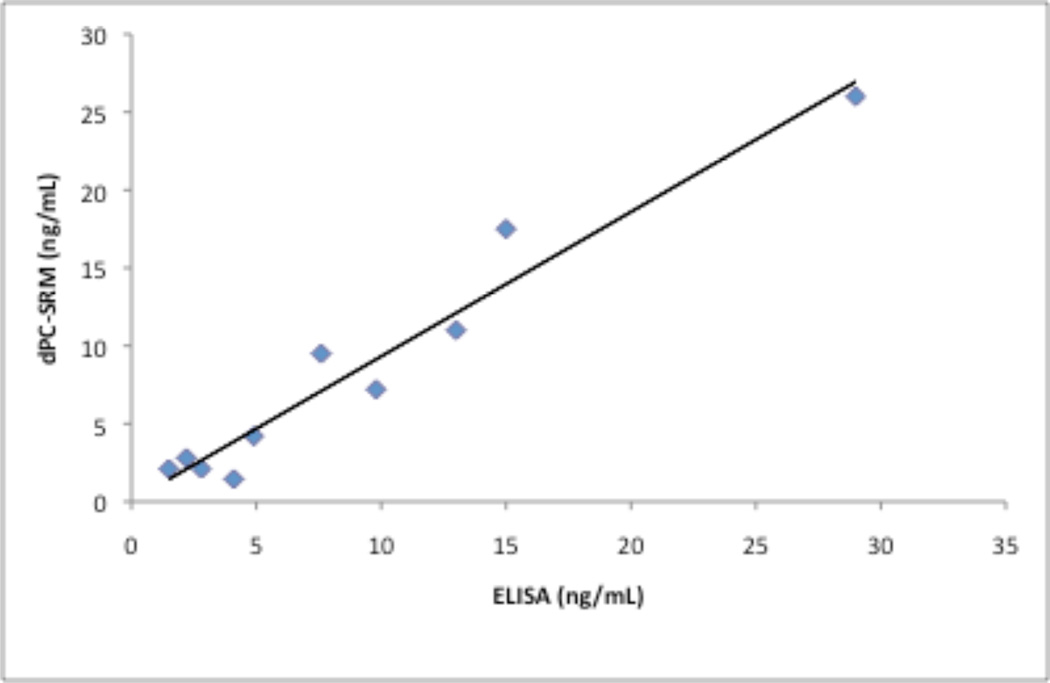
We next compared concentrations of PSA measured by ELISA to the values obtained using the Chip/Chip/SRM method. We used plasma samples collected from five control and five PCa male patients between the ages of 67 and 80 years. Each clinical sample was analyzed in duplicate by LC-Chip-SRM. The quantitation was performed by interpolation from the Chip/Chip/SRM calibration plots obtained for PSA added to plasma prior to removal of albumin and IgG (Figure S2 in the Supplement). The calibrated linear range for the Chip/Chip/SRM assay was from 1.0 to 50 ng/mL (higher levels were not investigated), and 1.5 to 80 ng/mL for the ELISA. Levels of PSA measured by the ELISA and the Chip/Chip/SRM showed good agreement, r2 = 0.9459 (Figure 4). The PSA concentrations measured in each clinical sample using both methods are listed in Table S1 in the Supplement. Figure S4 in the Supplement shows the comparison of the XICs acquired on the three transitions, 539.7 → 809.3, 539.7 → 866.3, and 539.7 → 965.4 for the IVGGWECEK analyte peptide in a patient with PSA level quantitated at 1.5 ng/mL (A) and the LOQ for PSA protein spiked into plasma prior to depletion of albumin and IgG (B). The results demonstrate that the present platform is successful in analyzing PSA in plasma in clinical samples down to the low ng/mL level.
Figure 4.
Correlation between PSA levels in plasma of patients with PCa assayed by ELISA and the Chip/Chip/SRM assay (r2 = 0.9459). The PSA concentration from the analysis of SRM transition 539.7 → 866.3 for IVGGWECEK peptide (y axis) was plotted versus the ELISA results (x axis).
CONCLUSION
Biomarker discovery has been a major by-product of the post-genomic era. Application of genomic and proteomic technologies has allowed for the discovery of far more candidates than can possibly be verified in a timely fashion.
The overall goal of our study was to combine isoelectric focusing on the dPC and SRM for selective enrichment of proteotypic peptides from plasma peptide mixtures for sensitive, quantitative, and reproducible MS-based assay for routine measurement of low abundance protein biomarkers in clinical samples. Using PSA as the model protein, we have demonstrated the high sensitivity and analytical performance of the platform. Our data compare well with other approaches tested on PSA. Low ng/mL sensitivity and high precision comparable to the requirements of a clinical assay were obtained. Further experiments such as inter-day accuracy and precision, stability of targeted peptides, in solution stability of internal standards, and multiplexing capabilities need to be performed to determine the feasibility of the proposed method in a clinical setting. It is likely that sensitivity, accuracy, and precision of this assay could be further enhanced by the use of the dual ion funnel interface14 or multiple reaction monitoring cubed (MRM3)39. Most importantly, we were able to accurately measure clinically relevant quantities of PSA in patient samples. The low ng/mL amounts of PSA measured by the Chip/Chip/SRM across the different patients were well correlated to those measured by a commercial ELISA test.
Although the described protocol is of low sample throughput, one of the biggest advantages of the Chip/Chip/SRM platform over other contemporary peptide fractionation approaches using charge- or hydrophobicity-based separations6, 7, 18 is the short assay development time and. The assay can practically be deployed for routine testing in a matter of a week even with the use of current experimental conditions and setup. Currently, the sample preparation and analysis takes approximately less than 24 hours; however, it can be performed more rapidly by shortening and optimizing the protein depletion, trypsin digestion, and peptide extraction steps prior to the dPC IEF chip. This can be accomplished through the use of high throughput automated technologies for sample preparation. For example, automated high throughput methods for removal of abundant proteins are commercially available (SepproTip system, Precision System Science, Pleasanton, CA); minimizing sample handling, can potentially lead to better sample recoveries for this step. Also, trypsin digestion, simultaneous sample clean-up and sample concentration, which are compatible with high throughput 96-microtiter well formats have been previously developed51 and can be easily adapted to work in parallel with the Chip/Chip/SRM platform. Furthermore, an automated trypsin digestion work station (Digilab ProGest Protein Digestion Station) is available as a commercial instrument (Ark Scientific Pte Ltd, Singapore). Other advantages of the Chip/Chip/SRM platform include good reproducibility and enrichment power due to the “digital” design of the chip. The Chip/Chip/SRM assay eliminates issues related to availability of antibodies as raising highly specific antibodies to a novel protein or peptide target can be a daunting task even if cost is not a concern. The assay is a general method for verification of protein biomarker candidates in biological samples. We postulate that it can be applied to a large number of human proteins, especially targets, for which antibodies are not available or while such antibodies are being developed and tested.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grants RO1 GM 15847 (BLK) and 1RO1 CA122591 (WSH). The authors would like to thank Drs. R. Garlick, W. Skea, and D. Argoti, for continued support and discussions. Contribution #960 from the Barnett Institute of Chemical and Biological Analysis.
Footnotes
SUPPORTING INFORMATION
Supplemental tables and figures. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Anderson NL. The roles of multiple proteomic platforms in a pipeline for new diagnostics. Mol Cell Proteomics. 2005;4(10):1441–1444. doi: 10.1074/mcp.I500001-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 3.Schiess R, Wollscheid B, Aebersold R. Targeted proteomic strategy for clinical biomarker discovery. Mol Oncol. 2009;3(1):33–44. doi: 10.1016/j.molonc.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJ, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM, Hiltke T, Jackson A, Johansen EB, Kinsinger CR, Li J, Mesri M, Neubert TA, Niles RK, Pulsipher TC, Ransohoff D, Rodriguez H, Rudnick PA, Smith D, Tabb DL, Tegeler TJ, Variyath AM, Vega-Montoto LJ, Wahlander A, Waldemarson S, Wang M, Whiteaker JR, Zhao L, Anderson NL, Fisher SJ, Liebler DC, Paulovich AG, Regnier FE, Tempst P, Carr SA. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27(7):633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keshishian H, Addona T, Burgess M, Mani DR, Shi X, Kuhn E, Sabatine MS, Gerszten RE, Carr SA. Quantification of cardiovascular biomarkers in patient plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2009;8(10):2339–2349. doi: 10.1074/mcp.M900140-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhn E, Addona T, Keshishian H, Burgess M, Mani DR, Lee RT, Sabatine MS, Gerszten RE, Carr SA. Developing multiplexed assays for troponin I and interleukin-33 in plasma by peptide immunoaffinity enrichment and targeted mass spectrometry. Clin Chem. 2009;55(6):1108–1117. doi: 10.1373/clinchem.2009.123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whiteaker JR, Zhao L, Zhang HY, Feng LC, Piening BD, Anderson L, Paulovich AG. Antibody-based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers. Anal Biochem. 2007;362(1):44–54. doi: 10.1016/j.ab.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez MF, Rezai T, Sarracino DA, Prakash A, Krastins B, Athanas M, Singh RJ, Barnidge DR, Oran P, Borges C, Nelson RW. Selected reaction monitoring-mass spectrometric immunoassay responsive to parathyroid hormone and related variants. Clin Chem. 2010;56(2):281–290. doi: 10.1373/clinchem.2009.137323. [DOI] [PubMed] [Google Scholar]
- 10.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5(4):573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Janecki DJ, Bemis KG, Tegeler TJ, Sanghani PC, Zhai L, Hurley TD, Bosron WF, Wang M. A multiple reaction monitoring method for absolute quantification of the human liver alcohol dehydrogenase ADH1C1 isoenzyme. Anal Biochem. 2007;369(1):18–26. doi: 10.1016/j.ab.2007.06.043. [DOI] [PubMed] [Google Scholar]
- 12.Kuzyk MA, Smith D, Yang J, Cross TJ, Jackson AM, Hardie DB, Anderson NL, Borchers CH. Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol Cell Proteomics. 2009;8(8):1860–1877. doi: 10.1074/mcp.M800540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams DK, Muddiman DC. Absolute quantification of C-reactive protein in human plasma derived from patients with epithelial ovarian cancer utilizing protein cleavage isotope dilution mass spectrometry. J Proteome Res. 2009;8(2):1085–1090. doi: 10.1021/pr800922p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hossain M, Kaleta DT, Robinson EW, Liu T, Zhao R, Page JS, Kelly RT, Moore RJ, Tang K, Camp DG, 2nd, Qian WJ, Smith RD. Enhanced Sensitivity for Selected Reaction Monitoring Mass Spectrometry-based Targeted Proteomics Using a Dual Stage Electrodynamic Ion Funnel Interface. Mol Cell Proteomics. 2011;10(2) doi: 10.1074/mcp.M000062-MCP201. M000062MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicol GR, Han M, Kim J, Birse CE, Brand E, Nguyen A, Mesri M, FitzHugh W, Kaminker P, Moore PA, Ruben SM, He T. Use of an immunoaffinity-mass spectrometry-based approach for the quantification of protein biomarkers from serum samples of lung cancer patients. Mol Cell Proteomics. 2008;7(10):1974–1982. doi: 10.1074/mcp.M700476-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Zhen E, Berna MJ, Jin Z, Pritt ML, Watson DE, Ackermann BL, Hale JE. Quantification of heart fatty acid binding protein as a biomarker for drug-induced cardiac and musculoskeletal necroses. Proteomics Clinical Applications. 2007;1:661–671. doi: 10.1002/prca.200700006. [DOI] [PubMed] [Google Scholar]
- 17.Fortin T, Salvador A, Charrier JP, Lenz C, Lacoux X, Morla A, Choquet-Kastylevsky G, Lemoine J. Clinical quantitation of prostate-specific antigen biomarker in the low nanogram/milliliter range by conventional bore liquid chromatography-tandem mass spectrometry (multiple reaction monitoring) coupling and correlation with ELISA tests. Mol Cell Proteomics. 2009;8(5):1006–1015. doi: 10.1074/mcp.M800238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6(12):2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) J Proteome Res. 2004;3(2):235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 20.Chen X, Simon ES, Xiang Y, Kachman M, Andrews PC, Wang Y. Quantitative proteomics analysis of cell cycle-regulated Golgi disassembly and reassembly. J Biol Chem. 285(10):7197–7207. doi: 10.1074/jbc.M109.047084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Rudnick PA, Evans EL, Li J, Zhuang Z, Devoe DL, Lee CS, Balgley BM. Proteome analysis of microdissected tumor tissue using a capillary isoelectric focusing-based multidimensional separation platform coupled with ESI-tandem MS. Anal Chem. 2005;77(20):6549–6556. doi: 10.1021/ac050491b. [DOI] [PubMed] [Google Scholar]
- 22.Gygi SP, Corthals GL, Zhang Y, Rochon Y, Aebersold R. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc Natl Acad Sci U S A. 2000;97(17):9390–9395. doi: 10.1073/pnas.160270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuroda Y, Yukinaga H, Kitano M, Noguchi T, Nemati M, Shibukawa A, Nakagawa T, Matsuzaki K. On-line capillary isoelectric focusing-mass spectrometry for quantitative analysis of peptides and proteins. J Pharm Biomed Anal. 2005;37(3):423–428. doi: 10.1016/j.jpba.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 24.Lilley KS, Razzaq A, Dupree P. Two-dimensional gel electrophoresis: recent advances in sample preparation, detection and quantitation. Curr Opin Chem Biol. 2002;6(1):46–50. doi: 10.1016/s1367-5931(01)00275-7. [DOI] [PubMed] [Google Scholar]
- 25.Huttenhain R, Malmstrom J, Picotti P, Aebersold R. Perspectives of targeted mass spectrometry for protein biomarker verification. Curr Opin Chem Biol. 2009;13(5–6):518–525. doi: 10.1016/j.cbpa.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Lee CS, Shen Y, Smith RD, Baehrecke EH. Integration of capillary isoelectric focusing with capillary reversed-phase liquid chromatography for two-dimensional proteomics separation. Electrophoresis. 2002;23(18):3143–3148. doi: 10.1002/1522-2683(200209)23:18<3143::AID-ELPS3143>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 27.Baczek T. Fractionation of peptides and identification of proteins from Saccharomyces cerevisiae in proteomics with the use of reversed-phase capillary liquid chromatography and pI-based approach. J Pharm Biomed Anal. 2004;35(4):895–904. doi: 10.1016/j.jpba.2004.02.032. [DOI] [PubMed] [Google Scholar]
- 28.Baczek T. Fractionation of peptides in proteomics with the use of pI-based approach and ZipTip pipette tips. J Pharm Biomed Anal. 2004;34(5):851–860. doi: 10.1016/j.jpba.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Moritz RL, Ji H, Schutz F, Connolly LM, Kapp EA, Speed TP, Simpson RJ. A proteome strategy for fractionating proteins and peptides using continuous free-flow electrophoresis coupled off-line to reversed-phase high-performance liquid chromatography. Anal Chem. 2004;76(16):4811–4824. doi: 10.1021/ac049717l. [DOI] [PubMed] [Google Scholar]
- 30.Xiao Z, Conrads TP, Lucas DA, Janini GM, Schaefer CF, Buetow KH, Issaq HJ, Veenstra TD. Direct ampholyte-free liquid-phase isoelectric peptide focusing: application to the human serum proteome. Electrophoresis. 2004;25(1):128–133. doi: 10.1002/elps.200305700. [DOI] [PubMed] [Google Scholar]
- 31.Cargile BJ, Sevinsky JR, Essader AS, Stephenson JL, Jr, Bundy JL. Immobilized pH gradient isoelectric focusing as a first-dimension separation in shotgun proteomics. J Biomol Tech. 2005;16(3):181–189. [PMC free article] [PubMed] [Google Scholar]
- 32.Essader AS, Cargile BJ, Bundy JL, Stephenson JL., Jr A comparison of immobilized pH gradient isoelectric focusing and strong-cation-exchange chromatography as a first dimension in shotgun proteomics. Proteomics. 2005;5(1):24–34. doi: 10.1002/pmic.200400888. [DOI] [PubMed] [Google Scholar]
- 33.Heller M, Ye M, Michel PE, Morier P, Stalder D, Junger MA, Aebersold R, Reymond F, Rossier JS. Added value for tandem mass spectrometry shotgun proteomics data validation through isoelectric focusing of peptides. J Proteome Res. 2005;4(6):2273–2282. doi: 10.1021/pr050193v. [DOI] [PubMed] [Google Scholar]
- 34.Horth P, Miller CA, Preckel T, Wenz C. Efficient fractionation and improved protein identification by peptide OFFGEL electrophoresis. Mol Cell Proteomics. 2006;5(10):1968–1974. doi: 10.1074/mcp.T600037-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Zeng Z, Hincapie M, Pitteri SJ, Hanash S, Schalkwijk J, Hogan JM, Wang H, Hancock WS. A Proteomics Platform Combining Depletion, Multi-lectin Affinity Chromatography (M-LAC), and Isoelectric Focusing to Study the Breast Cancer Proteome. Anal Chem. 2011;83(12):4845–4854. doi: 10.1021/ac2002802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulasingam V, Smith CR, Batruch I, Buckler A, Jeffery DA, Diamandis EP. "Product ion monitoring" assay for prostate-specific antigen in serum using a linear ion-trap. J Proteome Res. 2008;7(2):640–647. doi: 10.1021/pr7005999. [DOI] [PubMed] [Google Scholar]
- 37.Barnidge DR, Goodmanson MK, Klee GG, Muddiman DC. Absolute quantification of the model biomarker prostate-specific antigen in serum by LC-Ms/MS using protein cleavage and isotope dilution mass spectrometry. J Proteome Res. 2004;3(3):644–652. doi: 10.1021/pr049963d. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Tian Y, Rezai T, Prakash A, Lopez MF, Chan DW, Zhang H. Simultaneous analysis of glycosylated and sialylated prostate-specific antigen revealing differential distribution of glycosylated prostate-specific antigen isoforms in prostate cancer tissues. Anal Chem. 2011;83(1):240–245. doi: 10.1021/ac102319g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fortin T, Salvador A, Charrier JP, Lenz C, Bettsworth F, Lacoux X, Choquet-Kastylevsky G, Lemoine J. Multiple reaction monitoring cubed for protein quantification at the low nanogram/milliliter level in nondepleted human serum. Anal Chem. 2009;81(22):9343–9352. doi: 10.1021/ac901447h. [DOI] [PubMed] [Google Scholar]
- 40.Kullolli M, Hancock WS, Hincapie M. Automated platform for fractionation of human plasma glycoproteome in clinical proteomics. Anal Chem. 2010;82(1):115–120. doi: 10.1021/ac9013308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fusaro VA, Mani DR, Mesirov JP, Carr SA. Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat Biotechnol. 2009;27(2):190–198. doi: 10.1038/nbt.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J Proteome Res. 2004;3(6):1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 43.Deutsch EW, Lam H, Aebersold R. PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 2008;9(5):429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, Kuster B, Aebersold R. Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol. 2007;25(1):125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- 45.Sanders WS, Bridges SM, McCarthy FM, Nanduri B, Burgess SC. Prediction of peptides observable by mass spectrometry applied at the experimental set level. BMC Bioinformatics. 2007;8(Suppl 7):S23. doi: 10.1186/1471-2105-8-S7-S23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang H, Arnold RJ, Alves P, Xun Z, Clemmer DE, Novotny MV, Reilly JP, Radivojac P. A computational approach toward label-free protein quantification using predicted peptide detectability. Bioinformatics. 2006;22(14):e481–e488. doi: 10.1093/bioinformatics/btl237. [DOI] [PubMed] [Google Scholar]
- 47.Webb-Robertson BJ, Cannon WR, Oehmen CS, Shah AR, Gurumoorthi V, Lipton MS, Waters KM. A support vector machine model for the prediction of proteotypic peptides for accurate mass and time proteomics. Bioinformatics. 2008;24(13):1503–1509. doi: 10.1093/bioinformatics/btn218. [DOI] [PubMed] [Google Scholar]
- 48.Surinova S, Schiess R, Huttenhain R, Cerciello F, Wollscheid B, Aebersold R. On the development of plasma protein biomarkers. J Proteome Res. 2011;10(1):5–16. doi: 10.1021/pr1008515. [DOI] [PubMed] [Google Scholar]
- 49.OriGene Technologies launches over 5000 heavy isotope labeled full-length human proteins as quantitative internal standards for SRM/MRM Mass spectrometry. [accessed March 12, 2011]; www.origene.com;
- 50.Zilberstein G, Korol L, Bukshpan S, Baskin E. Parallel isoelectric focusing chip. Proteomics. 2004;4(9):2533–2540. doi: 10.1002/pmic.200300794. [DOI] [PubMed] [Google Scholar]
- 51.Nissum M, Schneider U, Kuhfuss S, Obermaier C, Wildgruber R, Posch A, Eckerskorn C. In-gel digestion of proteins using a solid-phase extraction microplate. Anal Chem. 2004;76(7):2040–2045. doi: 10.1021/ac035165f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.