Abstract
WASP family proteins are nucleation promoting factors that bind to and activate the Arp2/3 complex in order to stimulate nucleation of branched actin filaments. The WASP family consists of WASP, N-WASP, WAVE1-3, WASH, and the novel family members WHAMM and JMY. Each of the family members contains a C-terminus responsible for their nucleation promoting activity and unique N-termini that allow for them to be regulated in a spatiotemporal manner. Upon activation they reorganize the cytoskeleton for different cellular functions depending on their subcellular localization and regulatory protein interactions. Emerging evidence indicates that WASH, WHAMM, and JMY have functions that require the coordination of both actin polymerization and microtubule dynamics. Here, we review the mechanisms of regulation for each family member and their associated in vivo functions including cell migration, vesicle trafficking, and neuronal development.
Keywords: Arp2/3, WASP, WAVE, WASH, WHAMM, JMY
1. Overview of WASP/WAVE Signaling
1.1. Introduction
Actin reorganization in the cell is essential for muscle contractility and dynamic cell changes including endocytosis, migration, and formation of protrusive structures such as filopodia and lamellipodia. Actin monomers (G-actin) dynamically assemble into double helices [1], forming filamentous actin (F-actin) that is further organized into bundled or branched arrays. The association of three actin monomers is required for de novo actin polymerization, a critical step that is slow to occur with purified actin. In cells, this process is potently accelerated by the regulated activation of cellular nucleation factors (e.g. the Arp2/3 complex, Spire, and formins). This review focuses on recent advances in our understanding of how the Arp2/3 complex is activated to potentiate new actin filament formation. The Arp2/3 complex binds to the sides of existing actin filaments [2] and induces the nucleation of new filaments at a 70o angle [3]. This results in a dense meshwork of actin that accumulates at the leading edge of migrating cells [4] and in dendritic spines of neurons [5]. The Arp2/3 complex alone has low levels of actin nucleation activity; however the addition of nucleation promoting factors (NPFs), such as those of the WASP/WAVE family, increases the rate of actin nucleation [6].
1.2 Domain architecture and regulation
WASP (or Wiskott-Aldrich syndrome protein) was originally discovered as a mutated gene in Wiskott-Aldrich syndrome, a recessive X-linked immunodeficiency disorder [7]. Since then, an ever-increasing number of related proteins, categorized as class I NPFs, have been discovered through sequence homology studies. Class I NPFs include WASP, N-WASP, WAVE1-3, WASH, and the recently discovered WHAMM and JMY. Each of these proteins is defined by a functionally important C-terminal domain that is composed of a verprolin homology sequence, connecting sequence, and acidic sequence (VCA domain, also called WA, WCA domains). The verprolin homology sequence binds actin monomers, while the acidic sequence binds to and induces a change in the conformation of the Arp2/3 complex. This conformational switch results in a rearrangement of two actin-related subunits in the heptameric complex, Arp2 and Arp3 [8]. In combination with the actin monomer(s) held by the verprolin homology sequence, these can mimic the actin trimer that is needed to rapidly initiate cellular actin polymerization.
There is debate over whether one or two VCA domains bind to the Arp2/3 complex in order to induce activation, but evidence appears to support a dimerization model. On one hand, reconstruction and crystallography studies have proposed that only one VCA domain binds to the Arp2/3 complex, even in excess of VCA [9]. However, VCA dimerization appears to increase Arp2/3 dependent actin polymerization [10]. While numerous cross-linking studies have shown that VCA does interact with the Arp2/3 complex directly, variations on which subunits of the Arp2/3 complex are responsible for VCA interaction have been reported (reviewed in [10]). Modeling of a 2:1 VCA:Arp2/3 interaction addresses these discrepancies [10]. Also, WASP VCA dimerization has been shown to occur in vitro through sedimentation velocity analysis ultracentrifugation and light scattering experiments, as well as in vivo in HEK293 cells [11]. Any of the aforementioned inconsistencies could be due to variation between members of the WASP/WAVE family, or could be contingent upon binding of other regulatory proteins.
1.2.1 WASP
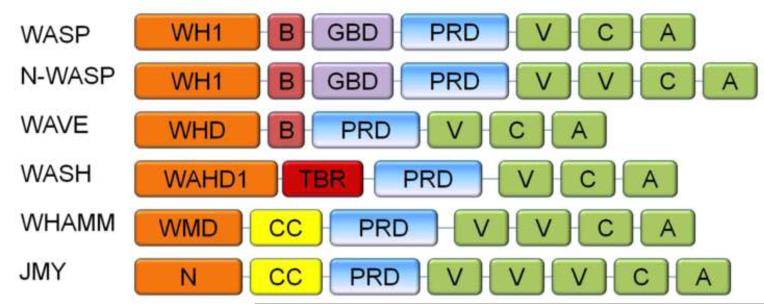
While each member of the WASP/WAVE family contains a conserved carboxy terminal sequence that potently activates the Arp2/3 complex, their amino termini specify their individuality, which endows each member with a unique ability to spatially and temporally regulate branched actin nucleation (Figure 1). For example, WASP contains several other regulatory domains including an WASP homology domain (WH1), a basic region, a GTPase-binding domain (GBD), and a proline-rich domain (PRD). These domains allow for regulation of activity both by autoinhibition and external signals.
Fig. 1. Domain structures of WASP family proteins.
All of the WASP family members exhibit a proline rich region and a VCA tail in the C-terminus, but contain unique N-termini. WH1: WASP Homology domain, B: Basic domain, GBD: GTPase binding domain, PRD: Proline rich domain, V: Verprolin homology, C: Connecting sequence, A: Acidic sequence, WHD: WAVE homology domain, WAHD1: WASH Homology domain, TBR: Tubulin binding region, WMD: WHAMM membrane-interacting domain, N: N-terminal region
Structural studies demonstrate that WASP is autoinhibited by the binding of the GBD to the VCA domain. This interaction prevents full length WASP from binding with and activating the Arp2/3 complex [12]. The GBD binds to the switch I and α5 regions of active GTP-bound Cdc42, allowing for distinction between multiple GTPases [13]. Accordingly, neither Rac nor Rho releases WASP autoinhibition [14]. Binding of Cdc42 to the GBD releases the VCA domain from the GBD and allows WASP to activate the Arp2/3 complex.
WASP autoinhibition is also modulated by phosphorylation of the GBD at tyrosine 291 [15]. In the autoinhibited structure, Y291 is inaccessible to tyrosine kinases. However, upon Cdc42 binding, the GBD is opened, allowing for phosphorylation of Y291 by SH2-containing tyrosine kinases, such as Src. Phosphorylated WASP is able to promote Arp2/3-dependent actin nucleation in vitro, independent of Cdc42 binding, suggesting phosphorylation converts WASP into a constitutively active form that may function to potentiate the duration and magnitude of its cellular signaling in vivo. Consistent with this possibility, overexpression of a phosphomimic mutant of WASP, Y291E, induces filopodium formation in macrophages [16]. Phosphorylation at Y291 also promotes WASP ubiquitination at lysine 76 and lysine 81, both within the WH1 domain [17], providing a posttranslational mechanism to negatively regulate WASP in vivo. In addition to tyrosine phosphorylation, two serines, S483 and S484, which lay between the C and A domains, may also be subject to phosphoregulation. Serine phosphorylation at these sites by casein kinase 2 results in a 7-fold increase in binding affinity between WASP and the Arp2/3 complex and significantly accelerates actin polymerization and nucleation in vitro [18].
Due to the modular domain structure of WASP, our understanding of WASP regulatory mechanisms continues to evolve and is becoming increasingly multifaceted. WIP (WASP-interacting protein) was identified in a yeast two-hybrid screen to search for novel WASP binding proteins and was confirmed to directly interact with WASP both in vitro and in vivo [19]. WIP binds to the WH1 domain and appears to functionally accelerate WASP-dependent actin polymerization, perhaps through its ability to also bind profilin, an actin monomer binding protein. WIP also stabilizes WASP and protects it from calpain-induced degradation both in vitro and in activated T and B cells [20]. Moreover, WASP expression levels are decreased in the lymphocytes of WAS patients with missense mutations in the WH1 domain, indicating that the stabilizing effect of WIP binding may be critically important for WASP expression levels in vivo.
1.2.2 N-WASP
Neural-WASP (N-WASP) shares high sequence homology and domain organization with WASP, yet it is regulated by its own specific binding partners within the N-terminus. N-WASP overexpression in COS7 cells results in formation of long spiky filopodia dependent on Cdc42 activation, linking N-WASP to Cdc42 signaling to Arp2/3 [21]. It also contains a slightly modified C-terminal tail containing two verprolin homology sequences within its VCA tail (VVCA) which increases its potency for Arp2/3 activation when compared to WASP or WAVE [22]. This suggests that the dual verprolin homology domains induce more potent changes in the actin cytoskeleton than single verprolin homology domains. N-WASP is ubiquitously expressed [23] and because of its more potent VVCA domain it must be tightly regulated to prevent unwanted abnormalities in cell morphology and motility.
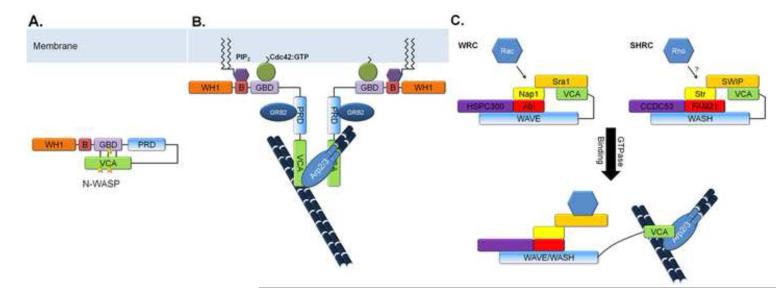
Like WASP, N-WASP is autoinhibited through interactions between the GBD and the VCA domain (Figure 2a). This regulation, however, appears to be modified by phosphatidyl-inositol (4,5)-bisphosphate (PIP2), which binds the basic domain of N-WASP in a synergistic fashion with Cdc42 [24, 25] (Figure 2b). Alone, N-WASP is in a tight autoinhibitory conformation in which both the GBD and the basic domains are partially occluded. Binding of either Cdc42 or PIP2 results in a loosening of the inhibited conformation and allows the other to bind cooperatively, amplifying and coordinating membrane and GTPase signals involved in N-WASP regulation [24]. Furthermore, in vitro cross-linking and sedimentation studies have provided evidence that, at high concentrations, N-WASP forms self-assembled dimers that can inhibit its activity in trans [26].
Fig. 2. Regulation of WASP family proteins.
(a) WASP and N-WASP are autoinhibited through intramolecular interactions between the GBD and VCA domain. (b) This autoinhibition can be released upon Cdc42 or PIP2 binding, or upon phosphorylation (phosphorylation sites marked with stars). Dimerization of VCA domains and interactions with the Arp2/3 complex leads to robust activation of branched actin nucleation. WASP can also self-dimerize in an inhibitory manner (not shown for clarity). GRB2 binds to the PRD and stabilizes the active WASP proteins. (c) WAVE and WASH proteins are sequestered in regulatory complexes, termed WRC and SHRC respectively. The VCA domain of WAVE is sequestered in a complex with Sra1. Rac1 binds to Sra1 to induce a conformational change that releases the VCA domain to allow for Arp2/3 activation. The SHRC makes up a structurally related inhibitory complex that is thought to behave in a similar manner.
N-WASP and WASP are also alike in that N-WASP is phosphorylated by Src family kinases [27]. Fyn phosphorylates N-WASP on tyrosine 253 (Y253), resulting in a PIP2- and Cdc42-independent activation mechanism. Phosphorylated N-WASP is, however, susceptible to ubiquitin-dependent proteasomal degradation. Thus, depending on the conditions of the cell, regulation of N-WASP can be both positively and negatively controlled by phosphorylation.
In addition to activation by PIP2, Cdc42, and Src family kinases, N-WASP can also be activated by WASP interacting SH3 protein (WISH) and the adaptor protein GRB2 via their SH3 domains. WISH contains a proline rich sequence that is also bound by GRB2 (which is possesses two SH3 domains), strengthening these interactions within an N-WASP complex. WISH binds to N-WASPs proline rich domain and enhances Arp2/3 complex activation, even when coexpressed with an N-WASP mutant lacking the ability to bind to Cdc42 [28]. Surprisingly, WISH retains the ability to activate the Arp2/3 complex independent of N-WASP, indicating WISH may stimulate Arp2/3 by regulating other NPFs as well. GRB2 appears to also potentiate the activation of Arp2/3-dependent actin polymerization by preferentially binding to N-WASP in its monomeric and active form, preventing the trans inhibition of N-WASP activity [26].
1.2.3 WAVE
WAVE was simultaneously discovered as a Dictyostelium discoideum homolog to WASP that acted as a suppressor of cAMP receptor signaling (termed Scar in this paper) [29] as well as a human homolog of WASP and N-WASP (WASP family Verprolin homologous protein-WAVE) that reorganized actin downstream of Rac [30]. Three isoforms of WAVE, WAVE1-3, exist that all contain a basic domain, a proline rich region, and a VCA domain as found in WASP and N-WASP. Importantly, WAVE proteins, unlike WASP, have basal actin nucleation activity [31] and possess an N-terminal WAVE homology domain (WHD) instead of the GBD [29]. Therefore, WAVE proteins are regulated in a manner that is quite distinct from WASP and N-WASP.
It is now known that WAVE is sequestered in a protein complex, the WAVE Regulatory Complex (WRC), composed of Sra1 (Specifically Rac binding protein 1, also known as CYFIP1), Nap1 (Nck-associated protein 1), HSPC300 (hematopoietic stem/progenitor cell protein 300), and Abi-1 (Abl interactor 1, also known as e3B1) [32] (Figure 2c). Whereas WASP and N-WASP are activated by Cdc42 through the GBD, WAVE is activated by Rac in an indirect mechanism through the WRC. Constitutively active Rac promotes translocation of WAVE from the cytosol to the plasma membrane in cells [30] and Sra-1 is a direct Rac effector linking active Rac and WAVE proteins. Sra-1, a 140 kDa protein, was initially identified in a screen using affinity chromatography to isolate proteins bound to Rac-GTPγS [33]. In a similar but independent screen for Rac effectors, Nap1 was isolated along with multiple other proteins including Sra-1 [34]. Both Sra-1 and Nap1 are essential in Rac-dependent lamellipodia outgrowth [35].
Sra-1, Nap1, and Abi-1 associate with WAVE in both resting and Rac-activated melanoma cells [35]. These proteins were identified in a tandem mass spectrometry identification assay of proteins associated of WAVE2, further supporting the formation of these proteins into a stable complex [36]. WAVE1 was also found to interact with Sra-1 and Nap1, along with HSPC300 [32], and evidence suggests that all three WAVE isoforms behave in a similar manner within the WRC [37]. Structurally, Nap1 and Abi-1 provide the core of the complex [38], with Abi-1 binding to the WHD of WAVE1 [36]. Sra-1 binds directly to Nap1 and HSPC300 binds to Abi-1 [38]. Rac appears to activate WAVE through the indirect interaction with WRC members, in contrast to WASP and N-WASP which directly bind Cdc42. In addition, recent evidence demonstrates that WRC is activated by Rac and Arf GTPases in a cooperative manner in vitro [39], with Arf being able to bind both Sra-1 and Nap1. Cooperative activation of WAVE proteins by Arf and Rac isoforms could be important since several lines of evidence indicate that cell migration and membrane ruffling require coordinated Arf and Rac signaling [40, 41]. Additional indirect mechanisms may also exist to regulate WAVE proteins. For example, WAVE2 can indirectly bind with Rac through insulin receptor substrate p53 (IRSp53), possibly providing a supplementary regulation mechanism [42].
The molecular mechanism by which active Rac releases WAVE inhibition by the WRC has been a topic of active research. In vitro, the WRC diminishes the ability of WAVE1 to promote actin nucleation [32], suggesting that the complex has an inhibitory effect. However, in 293T cells, expression of either constitutively active or dominant negative Rac1 does not disrupt the WRC [36], providing in vivo evidence that the complex does not dissociate upon Rac activation. In addition, experiments in Drosophila cell lines show that the complex is involved localization of WAVE within the cell and protects WAVE from degradation from the proteasome [43]. Structural analysis shows that the V and C regions of the VCA bind to Sra1, preventing WAVE from binding to actin monomers. Upon Rac activation, the complex undergoes a conformational change, releasing the VCA domain from the WRC such that it can interact with actin monomers and the Arp2/3 complex [44]. This model of WAVE regulation is supported by evidence that the WRC pentamer is inactive in vitro, but a WAVE:Abi-1:HSPC300 complex that lacks Sra-1 and Nap-1 is active [11].
The mechanism of activation of WAVE1 by Rac via the WRC is subject to an additional layer of regulation by WRP (WAVE-associated Rac-GAP protein) [45]. WRP contains a C-terminal SH3 domain that binds to the proline-rich region of WAVE1. It also contains a central Rho-family GAP domain that selectively promotes the intrinsic GTPase activity in Rac, causing Rac to hydrolyze GTP and become inactive [45]. WRP exhibits an IF-BAR domain as well [46], a phosphoinositide lipid-binding domain that remodels membrane morphology to promote outward protrusions. Thus, WRP appears directly link the regulation of signaling to WAVE1 with changes in membrane topology, allowing for tighter control of WAVE1 activation of Arp2/3 at the plasma membrane.
1.2.4 WASH
WASH (WASP and Scar Homolog) is a recently characterized addition to the family that contains a C-terminal VCA domain that also functions to activate the Arp2/3 complex. WASH also has a proline-rich region, but exhibits two N-terminal WASH homology domains that are evolutionarily conserved within WASH orthologs of other species [47]. WASH may exist in multiple subcellular sites within cells. For example, WASH colocalizes with actin in filopodia and lamellipodia in Cos7 cells [47] and interacts with the Arp2/3 complex [48, 49]. Additionally, other studies show that WASH exists in cytoplasmic puncta that colocalize with transferrin and EAA1, markers for sorting and recycling endosomes, suggesting it may modulate Arp2/3 during receptor trafficking.
Like the WAVE proteins, WASH does not appear to be regulated by an autoinhibitory mechanism. Instead it functions in a pentameric complex (the WASH Regulatory Complex, or SHRC) containing Strumpellin, FAM21, SWIP, and CCDC53 that appears to be functionally very similar to the WRC that mediates WAVE protein inhibition and activation [50] (Figure 2c). Proteomic studies also suggest that CapZ, an actin capping protein, is included in the SHRC complex, and this may be important to regulate the stability of WASH as siRNA to a CapZ subunit results in WASH degradation [51]. However, other studies suggest that the FAB21 subunit of the SHRC binds to and inhibits the actin capping property of CapZ, but that CapZ itself is dispensable for WASH regulation [30, 32]. FAM21 may be an important complex member for regulating WASH activity in cells since it affects SHRC localization through its C-terminus, potentially through interactions with phospholipids in endosomes [52].
Although the SHRC is composed of proteins that are distinct from those of the WRC, the functional and structural similarities between the two are quite remarkable. For example, electron microscopy of purified SHRC suggests the overall structural topology of the complex is quite similar to the organization of the WRC. Biochemically, SWIP and Strumpellin share a degree of sequence identity to Sra-1 and Nap1, respectively. All four of these proteins are also predicted to form helical structures, which are thought to interact with the predicted N-terminal coil-coiled structures of WASH and WAVE. In support of this notion, a WASH mutant lacking the WASH homology domains was unable to form a complex with any of the aforementioned proteins in vitro. Additionally, WASH has intrinsic actin nucleation activity that is inhibited in the SHRC in vitro [50]. This strongly suggests the SHRC may inhibit WASH until activated, much like the case for the WRC and Rac regulation of the WAVE proteins. However, more evidence is needed to confirm this function for the SHRC in vivo.
If the SHRC inhibits WASH, then how is it activated? At least in the case of Drosophila WASH, it may be activated downstream of the small GTPase Rho [48]. This interesting because it suggests the possibility that the Arp2/3 complex can be activated downstream of Cdc42 (WASP and N-WASP), Rac (WAVE1-3), and Rho (WASH), all of which are intricately involved in cytoskeletal signaling. In addition to inducing branched actin nucleation via Arp2/3 activation, Drosophila WASH can also bundle F-actin and microtubules in vitro. In vivo, Drosophila WASH appears to play a role in oogenesis and deficiencies in WASH lead to smaller eggs and sterile females. More work is needed to understand how the loss of WASH leads to these abnormalities via its effects on actin or microtubules. Additionally, it is unclear whether mammalian WASH is activated downstream of RhoA, or other as of yet unidentified GTPases [50]. Given the potential role of WASH in regulating vesicle trafficking (section 2.2) it seems possible that it will be regulated by GTPases other than the canonical Rho/Rac/Cdc42.
1.2.5 Novel WASP family members WHAMM and JMY
WHAMM (WASP homology associated with actin, membranes, and microtubules) was initially categorized as a WASP family protein through sequence homology searches for the VCA domain [53]. It is present in vertebrates, but not in C. elegans or Drosophila organisms, indicating it has evolved more recently than other family members. Interestingly, like N-WASP, WHAMM encodes two verprolin homology sequences (VVCA) as well as a proline rich domain that likely serves as a docking site for other proteins that contain proline-binding domains such as the SH3 domain. However, WHAMM also has an N-terminal domain that bears no sequence homology to any other WASP family proteins. This N-terminal domain targets WHAMM to the Golgi apparatus and is thus termed the WHAMM membrane-interacting domain (WMD). WHAMM also possess a coiled coil region that binds to microtubules and upon binding to microtubules, the C-terminal VVCA domain is hidden, preventing WHAMM from interacting with the Arp2/3 complex [54]. This suggests that WHAMM activity is directly controlled by the microtubule cytoskeleton and implicates crosstalk between the microtubule and actin cytoskeletons. .
JMY (junction-mediating and –regulatory protein) was not originally identified as a potential regulator of the actin cytoskeleton; rather it was initially found as a transcriptional cofactor for p53 [55]. However it was subsequently realized to also contain a VCA domain containing three verprolin homology sequences (VVVCA) and a proline rich domain [56], as well as two unexpected nuclear localization signals (NLS), one of which is within the VVVCA domain. Similar to WHAMM, it contains a coiled-coil domain and a unique N-terminal region. In vitro JMY assembles unbranched actin filaments, most likely by using its three verprolin homology domains to sequester actin monomers in a nucleation type organization to spur new actin filament polymerization. It can also activate the Arp2/3 complex and induce the formation of branched actin filaments [56], suggesting that JMY may either act as a multifunctional organizer of different actin structures, or be regulated in a way that would allow JMY to create branched or unbranched filaments depending on its localization and cellular conditions. JMY localization may be regulated by G-actin concentration within the cytoplasm. When cells are more motile and actin monomers are plentiful, the VVVCA domain binds actin monomers, masking the second NLS and preventing importin binding [57]. This allows JMY to become highly localized to the leading edge where its effects on actin may be mediated [56]. However, upon DNA damage, monomeric actin is sequestered in filamentous form, preventing their binding to JMY and defaulting JMY to the nucleus by importins [57].
2 Biological effects
2.1 Dynamic membrane protrusion
Probably the most well studied role of endogenous WAVE proteins is the formation of lamellipodia and Rac-induced ruffling around the leading edge of fibroblasts [30]. According to the dendritic nucleation model, WAVE is recruited and activated at the membrane, resulting in the growth of the actin meshwork that extends the membrane. This dynamic actin regulation is important in the motility of migrating cells. Chronically wounded tissue, for example, has decreased WAVE protein expression compared to cells surrounding acute wounds, presumably leaving cells unable to migrate and promote healing properly [58]. Furthermore, WASP and WAVE proteins, as well as their interactors, have been implicated in various cancers in various stages [59]. For example, in early cancer stages WAVE2 suppresses cell invasion by promoting cell-cell adhesion. However, in later cancer stages, WAVE2 promotes elongated cell morphology and motility, corresponding to an increase in metastasis.
Consistent with the deficiencies in WAVE leading to migratory defects, upsetting the balance of WAVE regulators results in similar repercussions. WRP, the WAVE-associated Rac-GAP protein, has recently been tied to proper migration of neuronal precursor cells from the ventricular region into the rostral migratory stream and olfactory bulb. Mismigration of these precursors due to genetic knockout of WRP leads to blockage of the cerebral aqueduct and, in most cases, obstructive hydrocephalus [60]. Therefore, it is likely that proper regulation of WAVE and WASP proteins may be important in migration rates and also accurate responsiveness to environmental factors that guide their directionality.
External cues can lead to other dynamic cell morphology differences. N-WASP, for example, is involved in phagocytosis that is marked by increased actin dynamics. N-WASP is recruited to the plasma membrane by Nck and, upon activation by Cdc42, induces actin polymerization so engulfment of foreign particles can occur [61]. Interestingly, N-WASP mediated signaling may continue to guide the engulfed vesicle (see section 2.2) after phagocytosis and internalization has occurred.
2.2 Internalization, endosomes, and vesicle trafficking
WASH, N-WASP, and WHAMM appear to coordinate actin polymerization with vesicle trafficking (Figure 3a). WASH, for example, is thought to play an important role in recycling and sorting endosomes. FAM21, one of the SHRC complex members, interacts with phospholipids and VPS35, a cargo-selective protein associated with retromers, and may be responsible for recruiting the SHRC to endosomes [62]. WASH associates with endosomes containing transferrin receptor and EAA1, which marks recycling and early endosomes, respectively. In addition, it colocalizes with Rab4, Rab11, and Rab5, corresponding to fast and slow recycling vesicles, and early endosomes respectively. WASH also promotes Arp2/3 dependent actin polymerization at endosomes in a microtubule binding dependent manner. When WASH is abrogated, transferrin collects in long tube-like membrane projections emanating from the endosomes. These projections are associated with microtubules and their presence delays EGF transport to late endosomes [49], suggesting that WASH may play a role in fission of recycling vesicles off of endosomes [51]. This may play a critical role in regulating important cell functions as recently, WASH and the Arp2/3 complex were found to play a positive role in trafficking of integrins to the membrane. Interestingly, WASH deficient cells showed decreased adhesion and a corresponding increased cell motility in wound healing assays [63].
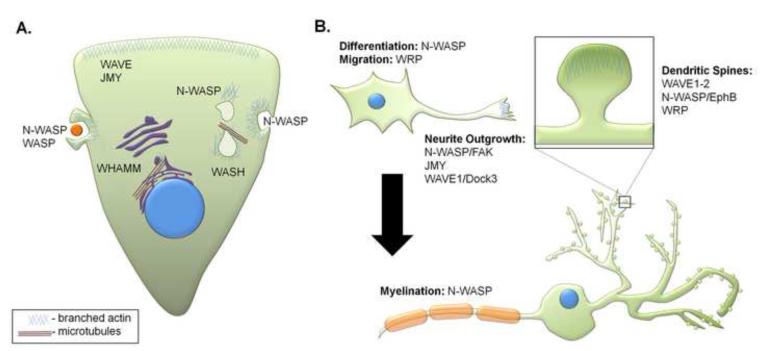
Fig. 3. Cellular roles of WASP family proteins.
(a) WASP family proteins coordinate membrane and cytoskeletal events, such as migration, endocytosis, and vesicle transport. WAVE and JMY play roles in cellular motility at the leading edge. WASP and N-WASP are involved in phagocytosis and endocytosis. N-WASP also leads to endosomal “rocketing”. WASH is responsible for pinching recycling vesicles off of endosomes, and WHAMM is essential for proper anterograde transport from the ER to the Golgi; both of these processes require interactions with microtubules. (b) WASP family proteins are involved in several aspects of neuronal development, including differentiation, migration, neurite outgrowth and myelination. In mature neurons, WASP and WAVE proteins are involved in the development and morphology of dendritic spines.
Interestingly, N-WASP appears to enhance clathrin-mediated endocytosis in addition to also regulating endosome trafficking. In yeast there is good evidence that actin polymerization is required for endocytosis, yet the role of actin in mammalian cells is less clear [64]. Despite this, actin and Arp2/3 localize to clathrin-coated pits in mammalian cells, and loss of N-WASP impairs the endocytosis of receptors such as epidermal growth factor receptor [65]. Additionally, like WASH, N-WASP colocalizes with motile endosomes in vivo. However, instead of affecting endosome shape in a microtubule-dependent manner, N-WASP activates Arp2/3 dependent actin nucleation in conjunction with adaptor proteins Nck and Grb2 and creates an actin comet tail that “rockets” endosome vesicles through the cytoplasm. This activity is induced by elevated levels of PIP2 at the endosome membrane that may bind the basic region of N-WASP, but the function (if any) of Cdc42 at the endosome is unclear [66].
While WASH and N-WASP have been shown to affect endosome transport and maturation, WHAMM may play a role in ER/Golgi transport [53]. WHAMM associates peripherally with the Golgi apparatus and induces actin polymerization in an Arp2/3 complex dependent manner. Overexpression or depletion of WHAMM in Cos7 and HeLa cells leads to disruption, dispersal, and abnormal morphology and function of the Golgi apparatus. This suggests that tight control of WHAMM expression may be required for proper Golgi morphology and function. WHAMM colocalizes and assists with the movement of tubular structures out of the endoplasmic reticulum during anterograde transport, contingent upon actin and microtubule dynamics. However, Arp2/3 complex activity only appears necessary for stabilizing membrane tubulation during transport and is not essential for transport of smaller vesicles. Further work is needed to elucidate exactly how WHAMM functions mechanistically in vesicular transport. Studies on the mechanism of WHAMM in this process may shed light on the complexity of cytoskeletal regulation between actin, microtubules, and membranes in the process of endosomal and vesicle trafficking.
2.3 Neuronal development
Dendritic spines serve as the primary postsynaptic structure for excitatory neurotransmission. They are incredibly small structures whose cytoskeleton is almost exclusively composed of actin filaments. They develop from filopodia that initially emerge from the dendritic shaft during the first weeks after birth and mature upon contact with pre-synaptic axons. Regulation of actin is not only critical for initial spine development and maturation, but is also thought to underlie the mechanisms of synaptic plasticity important for brain function. Synapses respond to neurotransmitters to positively and negatively modulate synaptic strength, and their morphology changes as the strength of their synaptic efficacy is modified. These can be long-lasting changes and are termed long term potentiation (LTP) and long term depression (LTD) [67]. LTD is dependent on actin depolymerization and results in a decrease in spine volume and a functional weakening of the synapse, whereas LTP induction stimulates actin polymerization and results in rapid increase in spine volume and strengthening of the synapse.
To further understand how synaptic plasticity takes place, we must first understand the resting state of a dendritic spine. Dendritic spines are dynamic structures [68], suggesting that the spine is not quiescent at resting state, but instead is regulating actin signaling pathways to keep the spine primed for action. Fluorescence recovery after photobleaching (FRAP) analysis in spines expressing GFP-actin shows that nearly 85% of the actin in dendritic spines is dynamic, and that the faction of “stable” actin filaments is relatively low [69]. Supporting this, the motility of spines is rapidly decreased upon treatment with the actin polymerization inhibitor cytochalasin D [70]. This suggests that dendritic spine motility is due to constant actin remodeling and polymerization.
Actin treadmilling, the depolymerization of linear actin filaments at one end at approximately the same rate as polymerization at the other, does not seem like a plausible explanation for the high dynamics of the spine membrane. The treadmilling model does not fully account for the force that would be necessary to push out a membrane as seen in the leading edges of cells [71]. However, severing and nucleation of actin filaments seems to be plausible as a meshwork of actin would be much more stable than individual filaments. Thus, the regulation of the WASP family and their ability to activate Arp2/3 dependent actin polymerization is likely to be critical for proper neuronal development.
Several lines of experimental evidence support this likelihood (Figure 3b). For example, WRP is disrupted in a balanced chromosomal translocation in a patient with 3p-syndrome, which is characterized by several cognitive impairments and mental retardation [72]. Neurons transfected with WRP show decreased neurite outgrowth [45] and WAVE1 knockout neurons exhibit abnormalities in growth cone morphology [73]. WRP knockout mice show decreased dendritic filopodia and excitatory spine density [46]. However, WRP does not seem to be required for spine maintenance. Accordingly, WRP knockout mice show cognition impairments in the novel object test, Morris water maze reversal, and passive avoidance.
Analogous to the neurological problems resulting from WRP deficiencies, WAVE-1 knockout mice also have motor and learning/memory disabilities compared to wildtype mice [74]. This may arise from multiple synaptic deficits associated with the loss of WAVE1. Neurons from WAVE-1 knockout mice, as well as knock in mice expressing a WRP-binding deficient mutant of WAVE1, exhibit decreased spine density in the hippocampus and cortex, decreased LTD, increased LTP, and increased NMDAR:AMPAR ratios [73]. These results suggest that WRP and WAVE1 signaling complex is important for multiple aspects of excitatory synapse function.
Although WAVE is clearly regulated downstream of Rac signaling, other signaling mechanisms may also be at play to influence its ability to modulate dendritic spines. For example, WAVE-1 also interacts with Cdk5/p35 and its ability to activate the Arp2/3 complex may be inhibited by Cdk5 through phosphorylation. Cdk5 overexpression led to decreased spine density in cultured neurons [75]. Additionally, WAVE1 also appears to be regulated by forming complexes with GEFs. In response to BDNF, WAVE1 is recruited to the neuronal plasma membrane by Dock3, a Rac-GEF, and upon Rac activation Dock3 is phosphorylated and released from WAVE1. This spatiotemporal regulation of WAVE1 is thought to induce BDNF-dependent axonal outgrowth [76]. The finding that WAVE1 can be regulated by both GAP (WRP) and GEF (Dock3) complexes is interesting and suggests that the regulation of Rac locally in the vicinity of WAVE1 may be tightly coordinated with WAVE1 activation. WAVE2, Dysbindin-1 (a protein implicated in schizophrenia), and Abi1 have also been found to act in a complex to regulate dendritic spine morphology, and knockdown of dysbindin-1 leads to abnormal spine morphology [77].
N-WASP, which is also highly expressed in neuronal tissues, has been implicated in several aspects of neuronal development. For example, the activation of the EphB receptor leads to a signaling complex formation consisting of intersectin, a Cdc42 GEF, and N-WASP. EphB activity can induce synaptogenesis, and inhibition of N-WASP expression leads to decreased spine formation in the cultured neurons [78]. N-WASP is also phosphorylated by FAK (focal adhesion kinase) in developing hippocampal neurons to promote neurite outgrowth [79], and is localized by Nck1 and Cdc42 to the growth cone to promote expansion [80]. In addition to regulating neurite outgrowth, N-WASP also appears to be involved the differentiation of neural stem cells by promoting increased filopodia [81]. Furthermore, N-WASP has been found to affect myelination outgrowth from Schwann cells, which wrap around axons by using a giant lamellipodial sheet-like structure [82]. JMY has recently been shown to localize to the cytoplasm and negatively regulate neurite outgrowth in a neuronal cell line, and this regulation is dependent partially on its ability to interact with the Arp2/3 complex [83]. Collectively, this evidence suggests that NPFs are highly regulated in and critical for multiple stages of brain development.
3 Future studies
Although the NPF field began with realization that WASP and WAVE family proteins could reorganize actin by activating the Arp2/3 complex downstream of Rho-family GTPases, new connections between many of the new WASP family members and microtubules or other GTPases is likely to drive the field into new directions. Understanding the interplay between various cytoskeletal elements within the cell will provide a more holistic picture of how various motile processes within the cell, from neuronal development to vesicle transport, work together and are regulated so precisely with respect to each other. Insights into the integration of signaling pathways involving newer members of the family, including JMY, WHAMM, and WASH will likely reveal new insights into these mechanisms. Future work should further reveal new roles for WASP family proteins in cell physiology and how those pathways may be interlinked. We are slowly beginning to recognize the patterns in actin signaling; however, the complexity in these pathways due to regulation in time and space are still unfolding.
Highlights.
WASP family proteins include WASP, N-WASP, WAVE1-3, WASH, WHAMM, and JMY.
Each family member regulates actin remodeling in specific cellular contexts.
Their unique regulation specifies their individual and diverse cellular functions.
They are involved in processes such as vesicle trafficking and neural development.
Acknowledgements
This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. 1106401 (L.E.B.) and National Institutes of Health Grant RO1-NS059957 (S.H.S.)
Abbreviations
- NPF
nucleation promoting factor
- VCA
verprolin homology
- connecting sequence
acidic sequence containing domain
- WRC
WAVE regulatory complex
- SHRC
WASH regulatory complex
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Fowler WE, Aebi U. A consistent picture of the actin filament related to the orientation of the actin molecule. The Journal of cell biology. 1983;97:264–9. doi: 10.1083/jcb.97.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mullins RD, Stafford WF, Pollard TD. Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba. The Journal of cell biology. 1997;136:331–43. doi: 10.1083/jcb.136.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:6181–6. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. The Journal of cell biology. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Korobova F, Svitkina T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Molecular biology of the cell. 2010;21:165–76. doi: 10.1091/mbc.E09-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Welch MD, Rosenblatt J, Skoble J, Portnoy DA, Mitchison TJ. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–8. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- [7].Derry JM, Ochs HD, Francke U. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell. 1994;78:635–44. doi: 10.1016/0092-8674(94)90528-2. [DOI] [PubMed] [Google Scholar]
- [8].Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, et al. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–84. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- [9].Gaucher JF, Mauge C, Didry D, Guichard B, Renault L, Carlier MF. Interactions of isolated C-terminal fragments of Neural Wiskott-Aldrich Syndrome Protein (N-WASP) with actin and Arp2/3 complex. The Journal of biological chemistry. 2012 doi: 10.1074/jbc.M112.394361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 complex is bound and activated by two WASP proteins. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E472–9. doi: 10.1073/pnas.1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Padrick SB, Cheng HC, Ismail AM, Panchal SC, Doolittle LK, Kim S, et al. Hierarchical regulation of WASP/WAVE proteins. Molecular cell. 2008;32:426–38. doi: 10.1016/j.molcel.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404:151–8. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- [13].Abdul-Manan N, Aghazadeh B, Liu GA, Majumdar A, Ouerfelli O, Siminovitch KA, et al. Structure of Cdc42 in complex with the GTPase-binding domain of the ‘Wiskott-Aldrich syndrome’ protein. Nature. 1999;399:379–83. doi: 10.1038/20726. [DOI] [PubMed] [Google Scholar]
- [14].Symons M, Derry JM, Karlak B, Jiang S, Lemahieu V, McCormick F, et al. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–34. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- [15].Torres E, Rosen MK. Contingent phosphorylation/dephosphorylation provides a mechanism of molecular memory in WASP. Molecular cell. 2003;11:1215–27. doi: 10.1016/s1097-2765(03)00139-4. [DOI] [PubMed] [Google Scholar]
- [16].Cory GO, Garg R, Cramer R, Ridley AJ. Phosphorylation of tyrosine 291 enhances the ability of WASp to stimulate actin polymerization and filopodium formation. Wiskott-Aldrich Syndrome protein. The Journal of biological chemistry. 2002;277:45115–21. doi: 10.1074/jbc.M203346200. [DOI] [PubMed] [Google Scholar]
- [17].Reicher B, Joseph N, David A, Pauker MH, Perl O, Barda-Saad M. Ubiquitylation-dependent negative regulation of WASp is essential for actin cytoskeleton dynamics. Molecular and cellular biology. 2012;32:3153–63. doi: 10.1128/MCB.00161-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cory GO, Cramer R, Blanchoin L, Ridley AJ. Phosphorylation of the WASP-VCA domain increases its affinity for the Arp2/3 complex and enhances actin polymerization by WASP. Molecular cell. 2003;11:1229–39. doi: 10.1016/s1097-2765(03)00172-2. [DOI] [PubMed] [Google Scholar]
- [19].Ramesh N, Anton IM, Hartwig JH, Geha RS. WIP, a protein associated with wiskott-aldrich syndrome protein, induces actin polymerization and redistribution in lymphoid cells. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:14671–6. doi: 10.1073/pnas.94.26.14671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].de la Fuente MA, Sasahara Y, Calamito M, Anton IM, Elkhal A, Gallego MD, et al. WIP is a chaperone for Wiskott-Aldrich syndrome protein (WASP) Proceedings of the National Academy of Sciences of the United States of America. 2007;104:926–31. doi: 10.1073/pnas.0610275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, et al. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–31. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- [22].Yamaguchi H, Miki H, Suetsugu S, Ma L, Kirschner MW, Takenawa T. Two tandem verprolin homology domains are necessary for a strong activation of Arp2/3 complex-induced actin polymerization and induction of microspike formation by N-WASP. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:12631–6. doi: 10.1073/pnas.190351397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Miki H, Miura K, Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. The EMBO journal. 1996;15:5326–35. [PMC free article] [PubMed] [Google Scholar]
- [24].Prehoda KE, Scott JA, Mullins RD, Lim WA. Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science. 2000;290:801–6. doi: 10.1126/science.290.5492.801. [DOI] [PubMed] [Google Scholar]
- [25].Rohatgi R, Ho HY, Kirschner MW. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4, 5-bisphosphate. The Journal of cell biology. 2000;150:1299–310. doi: 10.1083/jcb.150.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Carlier MF, Nioche P, Broutin-L’Hermite I, Boujemaa R, Le Clainche C, Egile C, et al. GRB2 links signaling to actin assembly by enhancing interaction of neural Wiskott-Aldrich syndrome protein (N-WASp) with actin-related protein (ARP2/3) complex. The Journal of biological chemistry. 2000;275:21946–52. doi: 10.1074/jbc.M000687200. [DOI] [PubMed] [Google Scholar]
- [27].Suetsugu S, Hattori M, Miki H, Tezuka T, Yamamoto T, Mikoshiba K, et al. Sustained activation of N-WASP through phosphorylation is essential for neurite extension. Developmental cell. 2002;3:645–58. doi: 10.1016/s1534-5807(02)00324-6. [DOI] [PubMed] [Google Scholar]
- [28].Fukuoka M, Suetsugu S, Miki H, Fukami K, Endo T, Takenawa T. A novel neural Wiskott-Aldrich syndrome protein (N-WASP) binding protein, WISH, induces Arp2/3 complex activation independent of Cdc42. The Journal of cell biology. 2001;152:471–82. doi: 10.1083/jcb.152.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bear JE, Rawls JF, Saxe CL., 3rd SCAR, a WASP-related protein, isolated as a suppressor of receptor defects in late Dictyostelium development. The Journal of cell biology. 1998;142:1325–35. doi: 10.1083/jcb.142.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. The EMBO journal. 1998;17:6932–41. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, et al. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:3739–44. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–3. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- [33].Kobayashi K, Kuroda S, Fukata M, Nakamura T, Nagase T, Nomura N, et al. p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. The Journal of biological chemistry. 1998;273:291–5. doi: 10.1074/jbc.273.1.291. [DOI] [PubMed] [Google Scholar]
- [34].Kitamura Y, Kitamura T, Sakaue H, Maeda T, Ueno H, Nishio S, et al. Interaction of Nck-associated protein 1 with activated GTP-binding protein Rac. The Biochemical journal. 1997;322(Pt 3):873–8. doi: 10.1042/bj3220873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Steffen A, Rottner K, Ehinger J, Innocenti M, Scita G, Wehland J, et al. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. The EMBO journal. 2004;23:749–59. doi: 10.1038/sj.emboj.7600084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Innocenti M, Zucconi A, Disanza A, Frittoli E, Areces LB, Steffen A, et al. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nature cell biology. 2004;6:319–27. doi: 10.1038/ncb1105. [DOI] [PubMed] [Google Scholar]
- [37].Stovold CF, Millard TH, Machesky LM. Inclusion of Scar/WAVE3 in a similar complex to Scar/WAVE1 and 2. BMC cell biology. 2005;6:11. doi: 10.1186/1471-2121-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gautreau A, Ho HY, Li J, Steen H, Gygi SP, Kirschner MW. Purification and architecture of the ubiquitous Wave complex. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4379–83. doi: 10.1073/pnas.0400628101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Koronakis V, Hume PJ, Humphreys D, Liu T, Horning O, Jensen ON, et al. WAVE regulatory complex activation by cooperating GTPases Arf and Rac1. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14449–54. doi: 10.1073/pnas.1107666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhang Q, Calafat J, Janssen H, Greenberg S. ARF6 is required for growth factor- and rac-mediated membrane ruffling in macrophages at a stage distal to rac membrane targeting. Molecular and cellular biology. 1999;19:8158–68. doi: 10.1128/mcb.19.12.8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Santy LC, Casanova JE. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. The Journal of cell biology. 2001;154:599–610. doi: 10.1083/jcb.200104019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Miki H, Yamaguchi H, Suetsugu S, Takenawa T. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature. 2000;408:732–5. doi: 10.1038/35047107. [DOI] [PubMed] [Google Scholar]
- [43].Kunda P, Craig G, Dominguez V, Baum B. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Current biology : CB. 2003;13:1867–75. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- [44].Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, et al. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468:533–8. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Soderling SH, Binns KL, Wayman GA, Davee SM, Ong SH, Pawson T, et al. The WRP component of the WAVE-1 complex attenuates Rac-mediated signalling. Nature cell biology. 2002;4:970–5. doi: 10.1038/ncb886. [DOI] [PubMed] [Google Scholar]
- [46].Carlson BR, Lloyd KE, Kruszewski A, Kim IH, Rodriguiz RM, Heindel C, et al. WRP/srGAP3 facilitates the initiation of spine development by an inverse F-BAR domain, and its loss impairs long-term memory. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:2447–60. doi: 10.1523/JNEUROSCI.4433-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Linardopoulou EV, Parghi SS, Friedman C, Osborn GE, Parkhurst SM, Trask BJ. Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS genetics. 2007;3:e237. doi: 10.1371/journal.pgen.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Liu R, Abreu-Blanco MT, Barry KC, Linardopoulou EV, Osborn GE, Parkhurst SM. Wash functions downstream of Rho and links linear and branched actin nucleation factors. Development. 2009;136:2849–60. doi: 10.1242/dev.035246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Duleh SN, Welch MD. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton. 2010;67:193–206. doi: 10.1002/cm.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Jia D, Gomez TS, Metlagel Z, Umetani J, Otwinowski Z, Rosen MK, et al. WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10442–7. doi: 10.1073/pnas.0913293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Developmental cell. 2009;17:712–23. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- [52].Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Developmental cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Campellone KG, Webb NJ, Znameroski EA, Welch MD. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 2008;134:148–61. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Shen QT, Hsiue PP, Sindelar CV, Welch MD, Campellone KG, Wang HW. Structural insights into WHAMM-mediated cytoskeletal coordination during membrane remodeling. The Journal of cell biology. 2012;199:111–24. doi: 10.1083/jcb.201204010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Shikama N, Lee CW, France S, Delavaine L, Lyon J, Krstic-Demonacos M, et al. A novel cofactor for p300 that regulates the p53 response. Molecular cell. 1999;4:365–76. doi: 10.1016/s1097-2765(00)80338-x. [DOI] [PubMed] [Google Scholar]
- [56].Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nature cell biology. 2009;11:451–9. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Zuchero JB, Belin B, Mullins RD. Actin binding to WH2 domains regulates nuclear import of the multifunctional actin regulator JMY. Molecular biology of the cell. 2012;23:853–63. doi: 10.1091/mbc.E11-12-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Jiang WG, Ye L, Patel G, Harding KG. Expression of WAVEs, the WASP (Wiskott-Aldrich syndrome protein) family of verprolin homologous proteins in human wound tissues and the biological influence on human keratinocytes. Wound repair and regeneration : official publication of the Wound Healing Society [and] the European Tissue Repair Society. 2010;18:594–604. doi: 10.1111/j.1524-475X.2010.00630.x. [DOI] [PubMed] [Google Scholar]
- [59].Kurisu S, Takenawa T. WASP and WAVE family proteins: friends or foes in cancer invasion? Cancer science. 2010;101:2093–104. doi: 10.1111/j.1349-7006.2010.01654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kim IH, Carlson BR, Heindel CC, Kim H, Soderling SH. Disruption of Wave Associated Rac-GAP (Wrp) Leads to Abnormal Adult Neural Progenitor Migration Associated with Hydrocephalus. The Journal of biological chemistry. 2012 doi: 10.1074/jbc.M112.398834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Dart AE, Donnelly SK, Holden DW, Way M, Caron E. Nck and Cdc42 co-operate to recruit N-WASP to promote FcgammaR-mediated phagocytosis. Journal of cell science. 2012;125:2825–30. doi: 10.1242/jcs.106583. [DOI] [PubMed] [Google Scholar]
- [62].Jia D, Gomez TS, Billadeau DD, Rosen MK. Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Molecular biology of the cell. 2012;23:2352–61. doi: 10.1091/mbc.E11-12-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Duleh SN, Welch MD. Regulation of integrin trafficking, cell adhesion, and cell migration by WASH and the Arp2/3 complex. Cytoskeleton. 2012 doi: 10.1002/cm.21069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Engqvist-Goldstein AE, Drubin DG. Actin assembly and endocytosis: from yeast to mammals. Annual review of cell and developmental biology. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- [65].Benesch S, Polo S, Lai FP, Anderson KI, Stradal TE, Wehland J, et al. N-WASP deficiency impairs EGF internalization and actin assembly at clathrin-coated pits. Journal of cell science. 2005;118:3103–15. doi: 10.1242/jcs.02444. [DOI] [PubMed] [Google Scholar]
- [66].Benesch S, Lommel S, Steffen A, Stradal TE, Scaplehorn N, Way M, et al. Phosphatidylinositol 4,5-biphosphate (PIP2)-induced vesicle movement depends on N-WASP and involves Nck, WIP, and Grb2. The Journal of biological chemistry. 2002;277:37771–6. doi: 10.1074/jbc.M204145200. [DOI] [PubMed] [Google Scholar]
- [67].Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- [68].Dailey ME, Smith SJ. The dynamics of dendritic structure in developing hippocampal slices. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1996;16:2983–94. doi: 10.1523/JNEUROSCI.16-09-02983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Star EN, Kwiatkowski DJ, Murthy VN. Rapid turnover of actin in dendritic spines and its regulation by activity. Nature neuroscience. 2002;5:239–46. doi: 10.1038/nn811. [DOI] [PubMed] [Google Scholar]
- [70].Fischer M, Kaech S, Knutti D, Matus A. Rapid actin-based plasticity in dendritic spines. Neuron. 1998;20:847–54. doi: 10.1016/s0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- [71].Theriot JA, Mitchison TJ. Actin microfilament dynamics in locomoting cells. Nature. 1991;352:126–31. doi: 10.1038/352126a0. [DOI] [PubMed] [Google Scholar]
- [72].Endris V, Wogatzky B, Leimer U, Bartsch D, Zatyka M, Latif F, et al. The novel Rho-GTPase activating gene MEGAP/ srGAP3 has a putative role in severe mental retardation. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11754–9. doi: 10.1073/pnas.162241099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Soderling SH, Guire ES, Kaech S, White J, Zhang F, Schutz K, et al. A WAVE-1 and WRP signaling complex regulates spine density, synaptic plasticity, and memory. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:355–65. doi: 10.1523/JNEUROSCI.3209-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Soderling SH, Langeberg LK, Soderling JA, Davee SM, Simerly R, Raber J, et al. Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:1723–8. doi: 10.1073/pnas.0438033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Kim Y, Sung JY, Ceglia I, Lee KW, Ahn JH, Halford JM, et al. Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature. 2006;442:814–7. doi: 10.1038/nature04976. [DOI] [PubMed] [Google Scholar]
- [76].Namekata K, Harada C, Taya C, Guo X, Kimura H, Parada LF, et al. Dock3 induces axonal outgrowth by stimulating membrane recruitment of the WAVE complex. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7586–91. doi: 10.1073/pnas.0914514107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ito H, Morishita R, Shinoda T, Iwamoto I, Sudo K, Okamoto K, et al. Dysbindin-1, WAVE2 and Abi-1 form a complex that regulates dendritic spine formation. Molecular psychiatry. 2010;15:976–86. doi: 10.1038/mp.2010.69. [DOI] [PubMed] [Google Scholar]
- [78].Irie F, Yamaguchi Y. EphB receptors regulate dendritic spine development via intersectin, Cdc42 and N-WASP. Nature neuroscience. 2002;5:1117–8. doi: 10.1038/nn964. [DOI] [PubMed] [Google Scholar]
- [79].Chacon MR, Navarro AI, Cuesto G, Del Pino I, Scott R, Morales M, et al. Focal adhesion kinase regulates actin nucleation and neuronal filopodia formation during axonal growth. Development. 2012;139:3200–10. doi: 10.1242/dev.080564. [DOI] [PubMed] [Google Scholar]
- [80].Shekarabi M, Moore SW, Tritsch NX, Morris SJ, Bouchard JF, Kennedy TE. Deleted in colorectal cancer binding netrin-1 mediates cell substrate adhesion and recruits Cdc42, Rac1, Pak1, and N-WASP into an intracellular signaling complex that promotes growth cone expansion. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:3132–41. doi: 10.1523/JNEUROSCI.1920-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Liebau S, Steinestel J, Linta L, Kleger A, Storch A, Schoen M, et al. An SK3 channel/nWASP/Abi-1 complex is involved in early neurogenesis. PloS one. 2011;6:e18148. doi: 10.1371/journal.pone.0018148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Novak N, Bar V, Sabanay H, Frechter S, Jaegle M, Snapper SB, et al. N-WASP is required for membrane wrapping and myelination by Schwann cells. The Journal of cell biology. 2011;192:243–50. doi: 10.1083/jcb.201010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Firat-Karalar EN, Hsiue PP, Welch MD. The actin nucleation factor JMY is a negative regulator of neuritogenesis. Molecular biology of the cell. 2011;22:4563–74. doi: 10.1091/mbc.E11-06-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]