Abstract
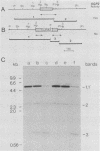
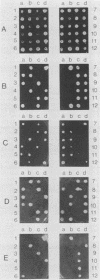
Loss of function of GPA1, which encodes a guanine-nucleotide-binding protein, arrests the cell at the G1 phase and allows it to mate, suggesting that the gpa1 mutation spontaneously exerts an intracellular signal that mimics the action of mating factor. We have cloned the SGP2 gene, which was first identified as a secondary mutation that allowed a gpa1::HIS3 mutant to grow and to show a non-cell-type-specific sterile phenotype. Disruption of SGP2 confers temperature-sensitive growth and a-specific sterile phenotypes, characteristics similar to those conferred by the dpr1 (ram) mutation, a suppressor of RAS2Val-19. The following observations indicate that SGP2 and DPR1 are in fact identical. (i) The cloned SGP2 complements both the temperature-sensitive growth and the a-specific sterility of the dpr1 mutant and can be integrated into the chromosomal DPR1 locus. (ii) The cloned DPR1, in turn, complements the ability of sgp2 to suppress the lethality of gpa1::HIS3. (iii) The dpr1 mutation suppresses the growth defect of gpa1::HIS3, and the dpr1 gpa1::HIS3 strain shows a non-cell-type-specific sterile phenotype. (iv) sgp2 is closely linked to the dpr1 locus. The DPR1 product has been shown to be responsible for processing and fatty acid acylation of a-factor and RAS proteins at their carboxyl termini. Therefore, the SGP2 (DPR1) product may be involved in membrane localization of an essential component in the mating-factor signaling pathway.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Bender A., Sprague G. F., Jr Yeast peptide pheromones, a-factor and alpha-factor, activate a common response mechanism in their target cells. Cell. 1986 Dec 26;47(6):929–937. doi: 10.1016/0092-8674(86)90808-1. [DOI] [PubMed] [Google Scholar]
- Betz R., Crabb J. W., Meyer H. E., Wittig R., Duntze W. Amino acid sequences of a-factor mating peptides from Saccharomyces cerevisiae. J Biol Chem. 1987 Jan 15;262(2):546–548. [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Burkholder A. C., Hartwell L. H. The yeast alpha-factor receptor: structural properties deduced from the sequence of the STE2 gene. Nucleic Acids Res. 1985 Dec 9;13(23):8463–8475. doi: 10.1093/nar/13.23.8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschenes R. J., Broach J. R. Fatty acylation is important but not essential for Saccharomyces cerevisiae RAS function. Mol Cell Biol. 1987 Jul;7(7):2344–2351. doi: 10.1128/mcb.7.7.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel C., Kurjan J. The yeast SCG1 gene: a G alpha-like protein implicated in the a- and alpha-factor response pathway. Cell. 1987 Sep 25;50(7):1001–1010. doi: 10.1016/0092-8674(87)90166-8. [DOI] [PubMed] [Google Scholar]
- Dixon R. A., Kobilka B. K., Strader D. J., Benovic J. L., Dohlman H. G., Frielle T., Bolanowski M. A., Bennett C. D., Rands E., Diehl R. E. Cloning of the gene and cDNA for mammalian beta-adrenergic receptor and homology with rhodopsin. Nature. 1986 May 1;321(6065):75–79. doi: 10.1038/321075a0. [DOI] [PubMed] [Google Scholar]
- Gilman A. G. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- Hagen D. C., McCaffrey G., Sprague G. F., Jr Evidence the yeast STE3 gene encodes a receptor for the peptide pheromone a factor: gene sequence and implications for the structure of the presumed receptor. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1418–1422. doi: 10.1073/pnas.83.5.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J Cell Biol. 1980 Jun;85(3):811–822. doi: 10.1083/jcb.85.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. B., Fong H. K., Teplow D. B., Dreyer W. J., Simon M. I. Isolation and characterization of a cDNA clone for the gamma subunit of bovine retinal transducin. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6948–6952. doi: 10.1073/pnas.81.22.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahng K. Y., Ferguson J., Reed S. I. Mutations in a gene encoding the alpha subunit of a Saccharomyces cerevisiae G protein indicate a role in mating pheromone signaling. Mol Cell Biol. 1988 Jun;8(6):2484–2493. doi: 10.1128/mcb.8.6.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenness D. D., Burkholder A. C., Hartwell L. H. Binding of alpha-factor pheromone to yeast a cells: chemical and genetic evidence for an alpha-factor receptor. Cell. 1983 Dec;35(2 Pt 1):521–529. doi: 10.1016/0092-8674(83)90186-1. [DOI] [PubMed] [Google Scholar]
- Kubo T., Fukuda K., Mikami A., Maeda A., Takahashi H., Mishina M., Haga T., Haga K., Ichiyama A., Kangawa K. Cloning, sequencing and expression of complementary DNA encoding the muscarinic acetylcholine receptor. Nature. 1986 Oct 2;323(6087):411–416. doi: 10.1038/323411a0. [DOI] [PubMed] [Google Scholar]
- Mackay V., Manney T. R. Mutations affecting sexual conjugation and related processes in Saccharomyces cerevisiae. II. Genetic analysis of nonmating mutants. Genetics. 1974 Feb;76(2):273–288. doi: 10.1093/genetics/76.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima I., Nakafuku M., Nakayama N., Brenner C., Miyajima A., Kaibuchi K., Arai K., Kaziro Y., Matsumoto K. GPA1, a haploid-specific essential gene, encodes a yeast homolog of mammalian G protein which may be involved in mating factor signal transduction. Cell. 1987 Sep 25;50(7):1011–1019. doi: 10.1016/0092-8674(87)90167-x. [DOI] [PubMed] [Google Scholar]
- Miyajima I., Nakayama N., Nakafuku M., Kaziro Y., Arai K., Matsumoto K. Suppressors of a gpa1 mutation cause sterility in Saccharomyces cerevisiae. Genetics. 1988 Aug;119(4):797–804. doi: 10.1093/genetics/119.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakafuku M., Itoh H., Nakamura S., Kaziro Y. Occurrence in Saccharomyces cerevisiae of a gene homologous to the cDNA coding for the alpha subunit of mammalian G proteins. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2140–2144. doi: 10.1073/pnas.84.8.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama N., Kaziro Y., Arai K., Matsumoto K. Role of STE genes in the mating factor signaling pathway mediated by GPA1 in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Sep;8(9):3777–3783. doi: 10.1128/mcb.8.9.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama N., Miyajima A., Arai K. Common signal transduction system shared by STE2 and STE3 in haploid cells of Saccharomyces cerevisiae: autocrine cell-cycle arrest results from forced expression of STE2. EMBO J. 1987 Jan;6(1):249–254. doi: 10.1002/j.1460-2075.1987.tb04746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama N., Miyajima A., Arai K. Nucleotide sequences of STE2 and STE3, cell type-specific sterile genes from Saccharomyces cerevisiae. EMBO J. 1985 Oct;4(10):2643–2648. doi: 10.1002/j.1460-2075.1985.tb03982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J., Hogness D. S. Isolation, sequence analysis, and intron-exon arrangement of the gene encoding bovine rhodopsin. Cell. 1983 Oct;34(3):807–814. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- Nogi Y., Matsumoto K., Toh-e A., Oshima Y. Interaction of super-repressible and dominant constitutive mutations for the synthesis of galactose pathway enzymes in Saccharomyces cerevisiae. Mol Gen Genet. 1977 Apr 29;152(3):137–144. doi: 10.1007/BF00268810. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov YuA Rhodopsin and bacteriorhodopsin: structure-function relationships. FEBS Lett. 1982 Nov 8;148(2):179–191. doi: 10.1016/0014-5793(82)80805-3. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sprague G. F., Jr, Blair L. C., Thorner J. Cell interactions and regulation of cell type in the yeast Saccharomyces cerevisiae. Annu Rev Microbiol. 1983;37:623–660. doi: 10.1146/annurev.mi.37.100183.003203. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wilson K. L., Herskowitz I. STE16, a new gene required for pheromone production by a cells of Saccharomyces cerevisiae. Genetics. 1987 Mar;115(3):441–449. doi: 10.1093/genetics/115.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]