ABSTRACT
A novel dissimilatory perchlorate-reducing bacterium (DPRB), Arcobacter sp. strain CAB, was isolated from a marina in Berkeley, CA. Phylogenetically, this halophile was most closely related to Arcobacter defluvii strain SW30-2 and Arcobacter ellisii. With acetate as the electron donor, strain CAB completely reduced perchlorate (ClO4−) or chlorate (ClO3−) [collectively designated (per)chlorate] to innocuous chloride (Cl−), likely using the perchlorate reductase (Pcr) and chlorite dismutase (Cld) enzymes. When grown with perchlorate, optimum growth was observed at 25 to 30°C, pH 7, and 3% NaCl. Transmission electron microscopy (TEM) and scanning electron microscopy (SEM) preparations were dominated by free-swimming straight rods with 1 to 2 polar flagella per cell. Strain CAB utilized a variety of organic acids, fructose, and hydrogen as electron donors coupled to (per)chlorate reduction. Further, under anoxic growth conditions strain CAB utilized the biogenic oxygen produced as a result of chlorite dismutation to oxidize catechol via the meta-cleavage pathway of aerobic catechol degradation and the catechol 2,3-dioxygenase enzyme. In addition to (per)chlorate, oxygen and nitrate were alternatively used as electron acceptors. The 3.48-Mb draft genome encoded a distinct perchlorate reduction island (PRI) containing several transposases. The genome lacks the pcrC gene, which was previously thought to be essential for (per)chlorate reduction, and appears to use an unrelated Arcobacter c-type cytochrome to perform the same function.
IMPORTANCE
The study of dissimilatory perchlorate-reducing bacteria (DPRB) has largely focused on freshwater, mesophilic, neutral-pH environments. This study identifies a novel marine DPRB in the genus Arcobacter that represents the first description of a DPRB associated with the Campylobacteraceae. Strain CAB is currently the only epsilonproteobacterial DPRB in pure culture. The genome of strain CAB lacks the pcrC gene found in all other DPRB tested, demonstrating a new variation on the (per)chlorate reduction pathway. The ability of strain CAB to oxidize catechol via the oxygenase-dependent meta-cleavage pathway in the absence of external oxygen by using the biogenic oxygen produced from the dismutation of chlorite provides a valuable model for understanding the anaerobic degradation of a broad diversity of xenobiotics which are recalcitrant to anaerobic metabolism but labile to oxygenase-dependent mechanisms.
Introduction
Perchlorate (ClO4−), a stable, water-soluble, toxic oxyanion of chlorine, has been widely produced through anthropogenic processes for use in rocket fuel, pyrotechnics, lubricants, and paints (1–3). Due to unregulated disposal of ClO4− before 1997, contamination is now widespread (3, 4), and it poses a significant health threat, as ClO4− inhibits the uptake of iodine by the thyroid gland and may lead to hypothyroidism (1, 5, 6). Recent geochemical studies indicate that extant (per)chlorate is far more prevalent than originally perceived (7–9). Although (per)chlorate has been considered to be primarily of anthropogenic origin, the widespread discovery of (per)chlorate in pristine environments (7–10), including waters and sediments from Mono Lake, CA (J. D. Coates, unpublished data); the Antarctic Dry Valleys (11); and, recently, Martian regoliths (12), has indicated a more complex and possibly ancient natural geochemical origin. This is supported by stable isotope analysis of the chlorine and oxygen isotopes of perchlorate, which allow for the distinction between anthropogenic and natural sources (13). These findings have identified the existence of an active biogeochemical redox cycle of chlorine on neoteric Earth and hint at its ancient origin.
Since the first dissimilatory perchlorate-reducing bacterium (DPRB) was isolated in 1996 (14, 15), more than 50 strains that use (per)chlorate as a metabolic electron acceptor are now known (16). All known DPRB respire perchlorate completely to chloride (Cl−) using the key enzymes perchlorate reductase (Pcr), which can reduce both perchlorate and chlorate, and chlorite dismutase (Cld). The genes encoding Cld and Pcr, cld and pcrABCD, respectively, have recently been shown to be part of a perchlorate reduction island (PRI) (17) that is horizontally transferred across diverse phylogenetic boundaries. This genomic island also contains conserved gene families predicted to be involved in regulation, electron transport, and cofactor biosynthesis (17). In contrast, a recent study by Liebensteiner et al. (18) describes a mechanism of (per)chlorate reduction in the archaeon Archaeoglobus fulgidus that involves an interplay of biotic and abiotic reactions that do not involve Cld.
Thus far, the study of microbial (per)chlorate reduction has been limited mostly to isolates (e.g., Dechloromonas, Azospira, and Ideonella) found in freshwater, mesophilic, neutral-pH environments (1, 19–25). Exceptions are the Gram-positive species Moorella perchloratireducens and Sporomusa strain An4, both of which were isolated from an underground gas storage facility (26, 27), and the hyperthermophilic archaeon Archaeoglobus fulgidus (18). The current study expands the diversity of DPRB by reporting on an isolate obtained from a marine environment. The isolate, designated strain CAB, is a marine epsilonproteobacterium obtained from a marina in Berkeley, CA. It exhibits physiological and genomic features that make it unique among both existing DPRB and previously known members of the Arcobacter genus. Although previous studies demonstrated dissimilatory perchlorate reduction by the epsilonproteobacterium Wolinella succinogenes strain HAP-1 (15), cultures of that bacterium are no longer available. Therefore, strain CAB is currently the only epsilonproteobacterial DPRB available in pure culture and the first Arcobacter known to reduce (per)chlorate.
RESULTS
Enrichment, isolation, and phylogeny.
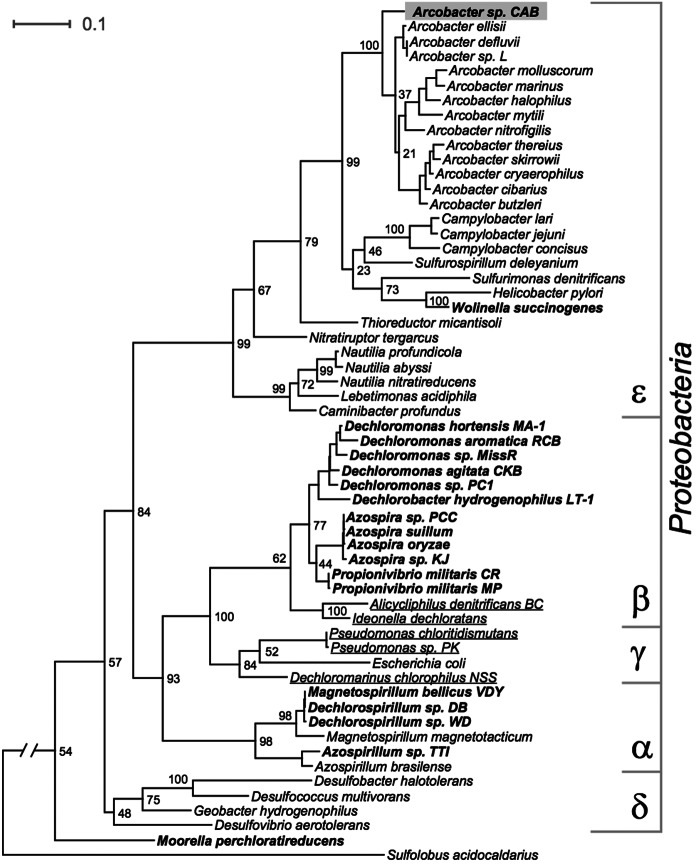
Culture enrichment was performed for 2 weeks in medium containing 3% NaCl inoculated with sediment from the Berkeley Marina. The enrichment was transferred into fresh medium twice prior to isolation. Pink colonies appeared in agar plugs after 5 to 7 days, and an isolate designated CAB was obtained using acetate and perchlorate as the sole electron donor and acceptor, respectively. No other isolates were obtained. Phylogenetic analysis of the 16S rRNA gene identified strain CAB as a previously unknown epsilonproteobacterium most closely affiliated with the genus Arcobacter (Fig. 1). The closest relatives to Arcobacter strain CAB were Arcobacter defluvii (94% 16S rRNA sequence identity), isolated from a waste treatment plant in Spain, and Arcobacter ellisii (94% 16S rRNA sequence identity), isolated from mussels in the Ebro Delta of Spain (28, 29).
FIG 1 .
Maximum likelihood tree based on 16S rRNA sequences showing the phylogenetic position of Arcobacter sp. strain CAB (gray box) both within the genus Arcobacter and within known perchlorate (bold)- and chlorate (underlined)-reducing bacteria. Bootstrap values are based on 1,000 replications and are shown at the nodes of the tree. The scale bar represents 0.1 expected change per site.
Morphology.
Strain CAB is a Gram-negative bacterium with most cells appearing to be straight rods usually 0.3 to 0.8 µm wide by 1 to 4 µm long (Fig. 2A). Strain CAB cells are highly motile and possess one or two flagella at one of the poles (Fig. 2B). In comparison, members of the genus Arcobacter typically appear as motile, slightly curved, rod-shaped cells 0.2 to 0.9 µm wide and 1 to 4 µm long with one or two polar flagella (30). Although some cells of strain CAB are slightly curved, most cells lack the characteristic curvature usually associated with the Arcobacter genus and from which the genus derives its name. Rarely, cells appear as filaments (8 to 10 µm) or in chains (Fig. 2A).
FIG 2 .
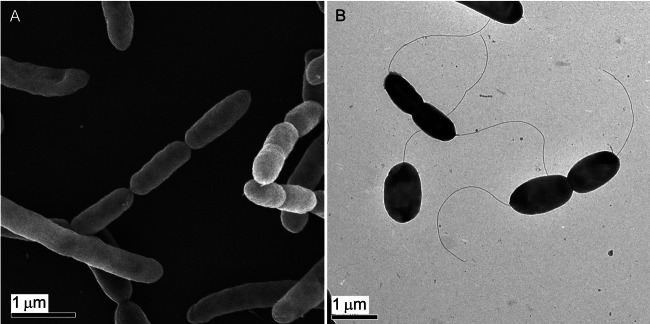
Scanning electron microscopy (A) and transmission electron microscopy (B) of Arcobacter sp. strain CAB.
Growth on acetate and perchlorate.
Arcobacter sp. strain CAB grows by coupling the oxidation of acetate to the reduction of perchlorate (Fig. 3). Strain CAB has a doubling time of 4 to 5 h and reaches a maximum optical density at 600 nm (OD600) of 0.45 when grown at 30°C with 16 mM (each) acetate and perchlorate. A 16.8 mM concentration of acetate and a 10.5 mM concentration of perchlorate were consumed over the 50-h incubation period, indicating that approximately 6.3 mM acetate (or 37%) was assimilated into biomass, which is typical for DPRB (21).
FIG 3 .
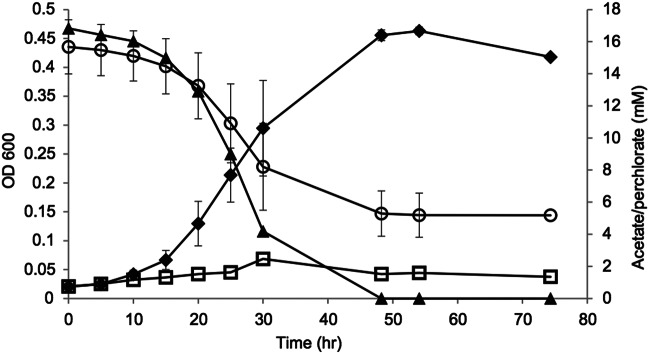
Growth curve of Arcobacter sp. strain CAB on 16 mM (each) acetate and perchlorate. Cell number increase was monitored by OD600. Diamonds, cell density (16 mM acetate-perchlorate); circles, perchlorate concentration (mM); triangles, acetate concentration (mM); squares, cell density for the negative control without perchlorate. Error bars represent standard deviations of the average of triplicate samples.
Optimum pH, temperature, and salinity.
Optimum growth pH, temperature, and salinity (percent NaCl) experiments were carried out using acetate as the electron donor and perchlorate as the electron acceptor. Strain CAB grew over the tested pH range (6 to 7.5) with an optimum of pH 7 (see Fig. S1 in the supplemental material). The optimum growth temperature was 25 to 30°C, although good growth was observed between 20 and 30°C (see Fig. S2). Furthermore, growth also occurred at 15°C, which is characteristic of the genus Arcobacter and differentiates this genus from the closely related but exclusively pathogenic genus Campylobacter (31, 32). No growth, either aerobically or anaerobically with perchlorate, was observed at 37°C. Unlike many DPRB, which cannot tolerate levels of NaCl higher than 2% (1), strain CAB is halophilic and grew best at 3% NaCl (see Fig. S3).
Fatty acid composition.
The fatty acid compositions of strain CAB cells grown aerobically on acetate or anaerobically on acetate and perchlorate were analyzed and are shown in Table 1. The predominant fatty acids under both conditions were hexadecanoic acid (C16:0), cis-9-hexadecanoic acid (C16:1ω7c), and cis-11-octadecanoic acid (C18:1ω7c) (Table 1). The presence of cis-11-hexadecanoic acid (C16:1ω5c) and cis-5-dodecanoic acid (C12:1ω7c), as well as the absence of dodecanoic acid (C12:0), distinguishes strain CAB from other recognized members of the genus Arcobacter (33).
TABLE 1 .
Whole-cell fatty acid composition of Arcobacter sp. strain CAB and other Arcobacter speciesa
| Fatty acid | % of total fatty acids in organism: |
||||||
|---|---|---|---|---|---|---|---|
|
Arcobacter sp. strain CAB |
A. marinus | A. halophilus | A. nitrofigilis | A. butzleri | A. skirrowii | ||
| Aerobic | Anaerobic | ||||||
| C10:0 | ND | 1 | ND | ND | ND | ND | ND |
| C12:0 | ND | ND | 6.6 | 8.2 | 7.2 | 6.8 | 9.1 |
| C14:0 | 3.92 | 3.03 | 5 | 5 | 4.8 | 3.2 | 2.4 |
| C16:0 | 19.86 | 25.53 | 26.2 | 19.3 | 32 | 19.6 | 22.2 |
| C18:0 | ND | ND | 1.6 | ND | ND | ND | ND |
| C12:1ω7c | 3.23 | 2.79 | ND | ND | ND | ND | ND |
| C14:1ω6c | ND | ND | ND | ND | ND | 4.9 | 1.4 |
| C14:1ω7c | 1.49 | ND | ND | ND | ND | ND | ND |
| C16:1ω5c | 1.56 | 1.29 | ND | ND | ND | ND | ND |
| C16:1ω7c | 50.19 | 46.77 | 28.4 | 37.9 | 30.9 | 19.2 | 22.8 |
| C16:1ω6t | ND | ND | ND | ND | ND | 13.8 | 7.8 |
| C18:1ω9c | ND | ND | 2.1 | ND | ND | ND | ND |
| C18:1ω7c | 12.39 | 14.51 | 22.3 | 21.7 | 12.8 | 11.6 | 19.7 |
| C12:0 3-OH | 1.58 | 1.25 | 2.9 | 2.7 | 5.5 | ND | ND |
| Iso-C16:1 I, C14:0 3-OH, and/or C12:0 alde | 1.83 | 2.42 | 2 | 2.1 | 5.3 | 17.4 | 9.2 |
Data for all species excluding strain CAB are from reference 33. ND, not detected or <1%.
Metabolic diversity.
In addition to perchlorate, strain CAB utilized both chlorate and oxygen as alternative electron acceptors, as is typical of the DPRB (Table 2). The ability of strain CAB to grow in atmospheric partial pressures of oxygen is consistent with most known DPRB and further supports its phylogenetic placement within the genus Arcobacter, as members of the genus Campylobacter are unable to grow aerobically (31, 32).
TABLE 2 .
Compounds tested as electron donors in the presence of perchlorate (10 mM) or tested as electron acceptors in the presence of acetate (10 mM)
| Compound | Concn (mMa) |
|---|---|
| Electron donors | |
| Utilized | |
| Acetate | 10 |
| Propionate | 10 |
| Citrate | 10 |
| Succinate | 1 |
| Lactate | 10 |
| Yeast extract | 1 g/liter |
| Fumarate | 10 |
| Malate | 10 |
| Fructose | 10 |
| Catechol | 1 |
| Hydrogen | ~50 kPa |
| Tested but not utilized | |
| Isobutyrate | 10 |
| Benzoate | 1, 2.5 |
| Butyrate | 10 |
| Valerate | 10 |
| Methanol | 5 |
| Ethanol | 5 |
| Glucose | 10 |
| Methane | ~25 kPa |
| Electron acceptors | |
| Utilized | |
| Perchlorate | 10 |
| Chlorate | 10 |
| Oxygen | ~25 kPa |
| Nitrate | 10 |
| Tested but not utilized | |
| Ferric citrate | 10 |
| Sulfate | 10 |
| Sulfite | 2 |
| Thiosulfate | 10 |
| Arsenate | 2.5 |
| Selenate | 2.5 |
| Malate | 10 |
| Nitrite | 2 |
Unless otherwise indicated.
Like most members of the genus Arcobacter, strain CAB can utilize nitrate as an electron acceptor (29). However, strain CAB does not grow robustly by this metabolism and accumulates significant concentrations of nitrite (4.65 mM) when given 10 mM acetate and 8 mM nitrate (see Fig. S4 in the supplemental material). Unexpectedly, the genome of strain CAB contains a complete set of genes encoding the reduction of nitrate and nitrite to ammonium. The genome encodes copies of the periplasmic nitrate reductase Nap (napA-G-H-B-F-L-D) (locus tags CAB_00029540, CAB_00029550, CAB_00029560, CAB_00029570, CAB_00029580, CAB_00029590, CAB_00029600, and CAB_00029610), the periplasmic nitrite reductase Nrf (nrfH-A-I-hyp-hyp) (locus tags CAB_00023420, CAB_00023410, CAB_00023400, CAB_00023390, and CAB_00023380), the nitrous oxide reductase Nos (nosZ-hyp-D-G-C1-C2-H-F-L) (locus tags CAB_00024950, CAB_00024960, CAB_00024970, CAB_00024980, CAB_00024990, CAB_00025000, CAB_00025010, CAB_00025020, and CAB_00025030), and the nitric oxide reductase Nor (nor-Q-D-B-C) (locus tags CAB_00034190, CAB_00034200, CAB_00034210, and CAB_00034220). The presence and the arrangement of the Nap, Nrf, and Nos clusters in strain CAB are very similar to those of the ammonifying epsilonproteobacterium Wolinella succinogenes (34). Furthermore, these clusters contain typical signatures of nitrate reduction pathways in the epsilonproteobacteria, such as the lack of napC in the Nap gene cluster and the nos-G-C1-C2-H gene arrangement that has, to date, been found exclusively in the epsilonproteobacteria. Like other members of the Arcobacter genus (35), the genome of strain CAB also includes the Nif operon, suggesting that strain CAB can fix N2, though this has not been empirically tested.
As shown in Table 2, strain CAB oxidizes a variety of organic acids coupled to the reduction of perchlorate. Hydrogen is also utilized as an electron donor, and the genome of strain CAB contains genes encoding several hydrogenases (CAB_00033260 to CAB_00033310, CAB_00000010 to CAB_00000060, and CAB_00000130 to CAB_00000180). However, the genome of strain CAB lacks genes necessary for CO2 fixation, and as such, strain CAB appears to require organic carbon for biomass synthesis in the presence of hydrogen as an electron donor.
Catechol degradation.
In contrast to previously described DPRB and members of the Arcobacter genus (28), strain CAB oxidized fructose and catechol using perchlorate as an electron acceptor (Table 2; Fig. 4). In the case of catechol, the oxidation of 1 mM catechol resulted in the reduction of 2.38 mM perchlorate (data not shown). According to the equation C6H6O2 + 3.25ClO4− → 6CO2 + 3.25Cl− + 3H2O, the theoretical ratio of catechol oxidized to perchlorate reduced is 1:3.25, implying that approximately 0.27 mM (or 26.8%) catechol was assimilated into biomass. Cultures of strain CAB grown on 1 mM catechol and 5 mM perchlorate were bright yellow, which is indicative of the production of 2-hydroxymuconate semialdehyde (2-HMS), a known intermediate of the meta-cleavage pathway of aerobic catechol oxidation (36). Absorbance at 375 nm, which is specific to 2-HMS, was indicative of the production of this compound during growth with catechol and perchlorate (Fig. 4), but not if catechol was replaced with acetate as an electron donor (data not shown). Spectrophotometric scans (300 to 800 nm) of the filtrates show a maximum absorbance peak at 375 nm (see Fig. S5 in the supplemental material), consistent with the previously published spectrum of 2-HMS (37). No maximum absorbance peak at 375 nm was seen in cultures grown with acetate instead of catechol (data not shown). The spectrum analysis could not be directly validated because of the unavailability of commercial 2-HMS. However, using the known extinction coefficient for 2-HMS of ε375 = 36,000 M−1 cm−1 (38), the measured absorbances were converted to 2-HMS concentrations. During growth of CAB on 1 mM catechol with perchlorate, the 2-HMS peak indicated a maximum concentration of 7.0 µM at t = 36 h, which declined to 1.8 µM at t = 556 hours (see Fig. S6). In order to further support the finding of 2-HMS during anaerobic oxidation of catechol with perchlorate as an electron acceptor, a separate culture of strain CAB that obtained an unusually high absorbance at 375 nm (36 µM calculated 2-HMS concentration) was subjected to liquid chromatography-mass spectrometry (LC-MS) analysis. An ion of m/z 141.0192 was observed, consistent with (M-H)– of 2-HMS (theoretical m/z 141.0193) (see Fig. S7). Strain CAB did not oxidize catechol with nitrate as an electron acceptor (data not shown).
FIG 4 .
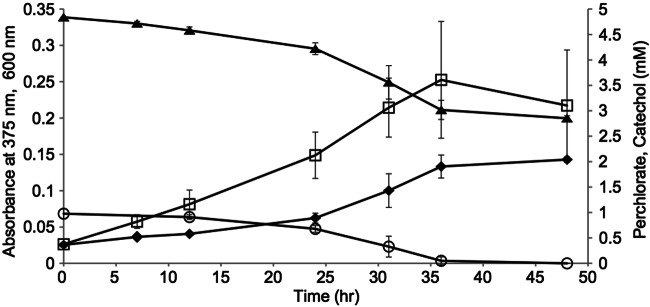
Growth curve of Arcobacter sp. strain CAB on 1 mM catechol and 5 mM perchlorate. Cell number increase was monitored by OD600. Diamonds, cell density; squares, absorbance at 375 nm; circles, catechol concentration (mM); triangles, perchlorate concentration (mM). Error bars represent standard deviations of the average of triplicate samples.
The genome of strain CAB encodes copies of all the proteins necessary for the meta-cleavage pathway of catechol degradation (38). These proteins include catechol 2,3-dioxygenase (CAB_00005480), 2-hydroxymuconate semialdehyde dehydrogenase (CAB_00005500), 2-hydroxymuconate semialdehyde hydrolase (CAB_00005570), 4-oxalocrotonate tautomerase (CAB_00005550), 4-oxalocrotonate decarboxylase (CAB_00005540), 2-oxopent-4-enoate hydratase (CAB_00005510), and 4-hydroxy-2-oxovalerate aldolase (CAB_00005530). Reverse transcription-PCR (RT-PCR) of cultures grown in the presence of acetate and perchlorate shows no or very little expression of genes in this pathway, whereas cultures grown with catechol and perchlorate have much higher expression of all these genes (see Fig. S8 in the supplemental material). The presence and transcription of these genes under these conditions are consistent with metabolic observations of catechol consumption and transient production of 2-HMS, demonstrating the unique ability of DPRB to utilize oxygenase-dependent pathways under anoxic (per)chlorate-reducing conditions.
Draft genome and perchlorate reduction island.
The draft genome of strain CAB was 3.48 Mb, which is one of the largest sequenced genomes in the epsilonproteobacteria to date (39, 40). The genome contained 367 contigs ranging in size from 201 bp to 85,599 bp. The average contig length was 9,478 bp. The GC content was low (28.16%) but is characteristic of the family Campylobacteraceae and the genus Arcobacter (30).
The genome revealed the presence of a unique perchlorate reduction island (PRI) containing striking differences from those in other DPRB (Fig. 5). As is typical of DPRB, no genes encoding a separate chlorate reductase were found. The PRI contains copies of cld (locus tag CAB_00027480), pcrABD (locus tags CAB_00027550, CAB_00027540, and CAB_00027530), and transposases (locus tags CAB_00027490, CAB_00027500, CAB_00027510, and CAB_00027560). Of particular note was the lack of the pcrC gene in the genome. This gene was considered an essential component of the PRI and is present in all other sequenced DPRB (17). The pcrC gene encodes a periplasmic multiheme c-type cytochrome that putatively mediates electron transport from the cytoplasmic membrane to the periplasmic functional PcrAB protein (16). Instead, the PRI of strain CAB contained a c-type monoheme cytochrome (locus tag CAB_00027520) (2). BLAST results revealed that this cytochrome is similar to those known from other Arcobacter genomes, including A. butzleri RM4018, A. butzleri ED-1, and A. nitrofigilis (41). Additionally, in all previously sequenced organisms the cld was located either upstream or downstream of the pcr operon on the same strand. In contrast, in Arcobacter sp. strain CAB, it was located much further away from the pcr genes and on the opposite strand (17). Between pcr and cld, there were three transposase genes, suggesting a possible mechanism for the aberrant PRI architecture relative to those of previously sequenced DPRB.
FIG 5 .
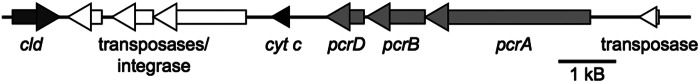
Arcobacter sp. strain CAB perchlorate reduction island. White, transposases and integrases; black, monoheme cytochrome c; dark gray, cld; light gray, pcrABD. Bar, 1 kB. Arrows indicate direction of transcription.
DISCUSSION
In this study, we describe a novel organism capable of dissimilatory (per)chlorate reduction. Although more than 50 DPRB have been isolated since 1996, Arcobacter sp. strain CAB is only the second DPRB belonging to the epsilonproteobacteria to be isolated and is currently the only one in pure culture (1). Further, nearly all isolated DPRB do not tolerate NaCl concentrations higher than 2%, whereas strain CAB is a halophile and grows best at 3% NaCl (1). As such, it provides a unique opportunity to study respiratory (per)chlorate reduction in a different phylogenetic and environmental context (1). Respiration of (per)chlorate has not been reported previously in any members of the genus Arcobacter, and the few sequenced genomes (40, 42, 43) of this genus do not carry copies of cld or pcr. As such, the existence of strain CAB suggests a novel ecological niche among members of the genus Arcobacter.
Although Arcobacter is not exclusively a pathogenic genus, members of the Arcobacter genus are most often studied because of their pathogenicity. The genus Arcobacter, which was first described in 1991 to accommodate 2 aerotolerant Campylobacter species, currently comprises 13 named members (29) plus Candidatus “Arcobacter sulfidicus,” which has not yet been fully described (44). Arcobacter species exhibit broad environmental diversity that can be divided between free-living species, usually found in marine environments, and pathogenic species known to infect birds and mammals, including humans (31). Strain CAB fails to grow at 37°C either aerobically or anaerobically with perchlorate, suggesting that strain CAB is unlikely to be a mammalian pathogen. Based on its environmental source and metabolism, strain CAB fits into the marine subclade of the Arcobacter genus. The relatively low (94%) 16S rRNA gene sequence agreement between strain CAB and its nearest relatives A. ellisii and A. defluvii indicates that strain CAB is a new species in the Arcobacter genus.
Strain CAB displays high metabolic versatility. In addition to the discovery that strain CAB is capable of reducing (per)chlorate, it was also found to oxidize fructose and catechol. Neither of these activities has been reported previously in members of the genus Arcobacter, and the inability to catabolize carbohydrates was considered diagnostic of the genus (30). These activities are also unusual among DPRB, where the majority are unable to use carbohydrates, catechol, and citrate (45).
Although (per)chlorate-reducing bacteria have been shown to degrade aromatics such as benzene in the presence of (per)chlorate, the mechanism by which this happens has not yet been completely elucidated (46–48). It has been suggested that the molecular oxygen produced by the dismutation of chlorite is internally recycled and used as a cosubstrate by oxygenases to carry out aerobic processes in anoxic environments (48, 49). Strain CAB oxidized catechol coupled to perchlorate reduction. RT-PCR and genomic data point toward a dioxygenase-mediated process in which the molecular oxygen produced from chlorite dismutation is used as a cosubstrate by the catechol 2,3-dioxygenase enzyme to convert catechol into 2-HMS.
The genome of strain CAB reveals the presence of an 11-kb contig containing a unique transposase-laden PRI that carries cld and pcrABD but lacks pcrC. This finding is surprising, as pcrC, which encodes a c-type cytochrome, was thought to be an essential part of the perchlorate reduction machinery. Strain CAB may be replacing the shuttling function of pcrC with another cytochrome, possibly the monoheme c-type cytochrome encoded in the PRI between the cld and pcr operons. If true, this would suggest that the perchlorate reduction pathway is more flexible than previously thought.
The relatively low 16S rRNA gene sequence identity, the frequent absence of curved cells, and the presence of metabolic pathways previously unknown among members of the Arcobacter distinguish strain CAB from other Arcobacter species and expand the known diversity of the genus. Further, the existence of strain CAB significantly extends the range of known DPRB. The only previously isolated DPRB from the epsilonproteobacteria is Wolinella succinogenes strain HAP-1, for which all cultures are now believed to have been lost. Though DPRB are well known among the Proteobacteria, especially within the Betaproteobacteria subclass, epsilonproteobacteria are only distantly related to known DPRB. As such, strain CAB will contribute to our understanding of the evolutionary history and diversity of the perchlorate reduction pathway.
MATERIALS AND METHODS
Culture conditions, enrichments, and isolation.
Sediment samples were obtained from the Berkeley Marina in Berkeley, CA (latitude 37.8629 N; longitude 122.3132 W), using 50-ml Falcon tubes. Sample salinity was ~1.5%, and the sample contained no detectable perchlorate. The medium used for the enrichment, isolation, and culturing contained the following (g/liter): NaCl (30), KCl (0.67), NaHCO3 (2.5), 10 ml vitamins, 10 ml minerals, and 20 ml RST minerals (21). The RST minerals contained the following (g/liter): NaCl (40), NH4Cl (50), KCl (5), KH2PO4 (5), MgSO4 ⋅ 7H2O (10), and CaCl2 ⋅ 2H2O (1). The medium was boiled for 30 to 60 s, cooled on ice, and degassed under an N2-CO2 headspace (80/20). The pH was ~6.8. Each anaerobic tube (Bellco, Vineland, NJ) received 8 ml of this medium and was autoclaved. After autoclaving, 0.5 ml of MgCl2 ⋅ 6H2O (21.2 g/100 ml distilled water [dH2O]) and 0.5 ml of CaCl2 ⋅ 2H2O (3.04 g/100 ml dH2O) were added to each tube from sterile aqueous stock solutions.
For the enrichment, 1 g of sediment sample was mixed with 9 ml of the described medium and 10 mM (each) acetate and perchlorate were used as the electron donor and acceptor, respectively. The enrichments were maintained at 30°C for 2 weeks and then transferred into fresh medium twice prior to isolation. Agar deeps (with acetate and perchlorate) (21) and streak plates (with acetate and oxygen) were used for isolation.
Optimum pH, temperature, and salinity.
Acetate and perchlorate were used to test the optimum growth temperature, pH, and salinity. The temperatures tested were 15°C, 20°C, 25°C, 30°C, 33°C, and 37°C. For salinity, the amount of NaCl added to the medium was varied from 0% to 6% in 0.5% increments. For pH, the medium described above was used, except that 10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer was substituted for NaHCO3. The medium was adjusted to the desired pH (6, 6.5, 7, or 7.5) using HCl or NaOH. All analyses were performed on triplicate cultures.
Alternate electron acceptors and electron donors.
For the electron donor profile, perchlorate was used as the sole electron acceptor (10 mM). Electron donors were added from sterile aqueous solutions to give the following final concentrations (mM, unless otherwise specified): acetate (10), propionate (10), isobutyrate (10), butyrate (10), valerate (10), methanol (5), ethanol (5), benzoate (1 and 2.5), pyruvate (5), citrate (10), succinate (1), lactate (10), glucose (10), fructose (10), yeast extract (1 g/liter), fumarate (10), malate (10), hydrogen (~50 kPa), catechol (1), and methane (~25 kPa). The tubes containing hydrogen also contained 1 mM acetate as a carbon source. To determine if strain CAB oxidized the tested electron donor coupled to perchlorate reduction, a sample was taken at t = 0 and at t = 11 days and analyzed for a decrease in the concentration of perchlorate by ion chromatography (see below). Each donor was tested in triplicate, and negative controls included (also in triplicate) a no-donor control and a no-donor-plus-1 mM acetate control. Growth was considered positive when perchlorate was consumed together with an increase in biomass and no loss of electron acceptor in the no-donor control.
For the electron acceptor profile, acetate (10 mM) was used as the sole electron donor and carbon source. Electron acceptors were added from sterile aqueous stocks in the following concentrations (mM unless otherwise specified): perchlorate (10), chlorate (10), nitrate (10), nitrite (2), sulfate (10), sulfite (2), thiosulfate (10), arsenate (2.5), selenate (2.5), ferric citrate (10), malate (10), and oxygen (~25 kPa). To determine if strain CAB reduced the tested acceptor coupled to the oxidation of acetate, a sample was taken at t = 0 and at t = 11 days and analyzed for a decrease in the concentration of acetate by high-performance liquid chromatography (HPLC) as described below. Each acceptor was tested in triplicate, and the experiment included a no-acceptor control (also in triplicate). Growth was considered positive when there was an acetate loss together with an increase of biomass and no loss of electron donor in the no-acceptor control.
Cell growth profiles.
Cell growth was measured spectrophotometrically at 600 nm. All experimental analyses were performed in triplicate, and the results are expressed as the average of these determinations.
Analytical methods.
Perchlorate concentration was measured via ion chromatography using a Dionex IonPac AS 16 (4- by 250-mm) column (Thermo Fisher Scientific, Sunnyvale, CA) with a 35 mM NaOH mobile phase at a flow rate of 1.0 ml/min. The eluting perchlorate was detected by conductivity suppressed with a Dionex ASRS operating in recycle mode. The suppressor controller was set at 100 mA for the analysis. The injection volume was 25 µl. Acetate concentrations were measured by HPLC (Dionex; model LC20), using a UV-visible (UV-Vis) detector (Dionex; AD20) at a wavelength of 210 nm and a Bio-Rad Aminex HPX-87H column with a mobile phase of 0.016 N H2SO4 (flow rate of 0.9 ml/min).
Catechol and 2-HMS were measured via liquid chromatography-mass spectrometry. For determination of catechol concentration, 2 µl of filtered sample was injected at 30°C onto a Rapid Resolution column (2.1 by 30 mm, 3.5 µm; Agilent Technologies, Santa Clara, CA) and analyzed on an Agilent 1100 series liquid chromatography system equipped with a degasser (G1379B), binary pump (G132B), autosampler (G1329B), and column compartment (G1316B) coupled to an Agilent 6510 quadrupole time of flight (QTOF) mass spectrometer equipped with a dual-spray electrospray ionization (ESI) source. Compounds were eluted with a flow rate of 0.5 ml/min and a mobile phase program of 1 min of isocratic flow of 98% A (0.1% formic acid) to 2% B (acetonitrile with 0.1% [vol/vol] formic acid) and then 1 min of linear flow to 10% B, 2 min of linear flow to 95% B, 2 min of isocratic flow, and an immediate return to 2% B with 3 min of equilibration. MS settings were as following: negative-mode ion polarity, 325°C gas temperature, 7-liter/min drying gas, 45-lb/in2 nebulizer, Vcap (capillary voltage) of 3,500 V, scanning from 50 to 1,100 m/z (1.41 spectra/s), and internal mass calibration ions m/z 112.985587 and m/z 1,033.988109. For analysis of 2-HMS, 10 µl of filtered sample was analyzed using the same method.
Scanning electron microscopy.
Silicon wafers were washed with ethanol for 30 s and air dried. A 20-µl poly-l-lysine drop was placed on the silicon wafers for 1 min and then withdrawn. The wafers were then rinsed with ultraclean water. A drop of concentrated CAB cells fixed with 2% glutaraldehyde in 0.1 M sodium cacodylate buffer was added to the silicon wafers and allowed to settle for 1 h. The silicon wafers were then processed as described in reference 24 and visualized using a Hitachi S-5000 scanning electron microscope at 20 kV.
Transmission electron microscopy.
Flagella were visualized using the negative-stain technique. Copper Formvar- and carbon-coated grids (400 mesh) were glow discharged just before use to increase hydrophilicity. A suspension (5 µl) of CAB cells fixed with 2% glutaraldehyde in 0.1 M sodium cacodylate buffer was deposited on the grids. This was sufficient to cover the grid. After 2 min, the suspension was removed by touching the ragged torn edges of filter paper to the sample until the grid surface was nearly dry. The grid was then washed 3 times on water droplets, and excess water was wicked off using filter paper as described above. A drop of 1% aqueous uranyl acetate was then added to the grid. After 2 min, the grid was dried with filter paper. Samples were examined the same day using a Tecnai transmission electron microscope at 120 kV.
Whole-cell fatty acid analysis.
Fifty milliliters of strain CAB cells was grown aerobically using 20 mM acetate and anaerobically using 20 mM (each) acetate and perchlorate in the medium described above. The cells were then harvested via centrifugation and submitted to MIDI Labs (MIDI, Inc., Newark, DE) for whole-cell fatty acid analysis.
Genomic and phylogenetic analysis.
Genomic DNA was extracted from strain CAB as described previously (50). The genomic DNA was then submitted to Eureka Genomics (Hercules, CA) for Illumina sequencing and partial assembly. The genome of Arcobacter sp. strain CAB is available on the Integrated Microbial Genomes (IMG) system of the Joint Genome Institute.
The Silva database and aligner (51) were used to align 16S rRNA gene sequences, and a maximum likelihood phylogenetic tree was constructed with 1,000 bootstrap values using RAxML-HPC (52). Accession numbers for the microorganisms used in the tree are provided in Text S1 in the supplemental material. Sulfolobus acidocaldarius was used as an outgroup.
RNA extraction and RT-PCR.
Forty milliliters of triplicate cultures growing either with 1 mM catechol and 5 mM perchlorate or with 5 mM acetate and 5 mM perchlorate was centrifuged at 7,000 × g, 4°C, for 10 min. The pellets were resuspended in 1 ml RNAwiz reagent (Ambion, Grand Island, NY), and RNA was extracted according to the manufacturer’s protocol. The Turbo DNA-free kit (Applied Biosystems, Grand Island, NY) was used to remove any contaminating DNA according to the manufacturer’s protocol. cDNA was made with 200 ng RNA using the Verso cDNA kit (Thermo Fisher Scientific, Sunnyvale, CA) according to the manufacturer’s protocol. The RNA was tested for DNA contamination using the universal primers for 16S rRNA amplification 27F (5′ AGAGTTTGATCMTGGCTCAG 3′) and 1492R (5′ GGTTACCTTGTTACGACTT 3′).
To determine if the genes in the 4-oxalocrotonate pathway of catechol oxidation were expressed when strain CAB was grown using catechol as an electron donor instead of acetate, RT-PCRs were performed using cDNA. The PCR primers used can be found in the supplemental material. Chlorite dismutase primers were used as a control. For primer pairs OEH 1F/1R, OXD 5F/5R, Cld 5F/5R, HOA 1F/1R, and CAT 1F/1R, PCR conditions were 95°C for 5 min; 30 cycles of 95°C for 45 s, 61°C for 45 s, and 72°C for 45 s; and 10 min at 72°C. For primer pair HMSD 4F/4R, the conditions were 95°C for 5 min; 30 cycles of 95°C for 45 s, 60°C for 45 s, and 72°C for 1 min; and 10 min at 72°C. For primer pair OXT 3F/3R, the conditions were 95°C for 5 min; 30 cycles of 95°C for 45 s, 58°C for 45 s, and 72°C for 30 s; and 10 min at 72°C. For primer pair HSH 4F/4R, the conditions were 95°C for 5 min; 30 cycles of 95°C for 45 s, 65°C for 45 s, and 72°C for 30 s; and 10 min at 72°C.
SUPPLEMENTAL MATERIAL
Accession numbers for microorganisms used in phylogenetic tree and RT-PCR primers used. Download
Effect of pH on the growth rate of strain CAB grown in medium containing 10 mM (each) acetate and perchlorate. Error bars represent standard deviations of the average of triplicate samples. Download
Effect of temperature on the growth rate of strain CAB grown in medium containing 10 mM (each) acetate and perchlorate. Error bars represent standard deviations of the average of triplicate samples. Download
Effect of salinity on the growth rate of strain CAB grown in medium containing 10 mM (each) acetate and perchlorate. Error bars represent standard deviations of the average of triplicate samples. Download
Growth curve of Arcobacter sp. strain CAB on 10 mM acetate and 8 mM nitrate. Cell number increase was monitored by optical density at 600 nm. Diamonds, cell density; squares, nitrite concentration (mM); circles, nitrate concentration (mM); triangles, cell density for no-acceptor control (mM). Error bars represent standard deviations of the average of triplicate samples. Download
Spectrophotometric scans (300 to 800 nm) of filtrates from triplicate cultures of strain CAB grown with 1 mM catechol and 5 mM perchlorate. Download
2-HMS and perchlorate concentration of Arcobacter sp. strain CAB grown on 1 mM catechol and 5 mM perchlorate. Squares, perchlorate concentration (mM); diamonds, 2-HMS concentration (μM). Error bars represent standard deviations of the average of triplicate samples. Download
LC/QTOF-MS of Arcobacter sp. strain CAB culture grown on 1 mM catechol and 5 mM perchlorate. Calculated 2-HMS concentration is 35.95 µM (based on absorbance at 375 nm). An ion of m/z 141.0192 was observed, consistent (accuracy, 0.71 ppm) with (M-H)− of 2-HMS (theoretical m/z 141.0193). Download
(A) RT-PCR of catechol-grown (lanes 3 to 5) and acetate-grown (lane 7) cultures of strain CAB. Lane 1 is a PCR negative control, and lane 2 is a genomic DNA positive control. Lane 6 is an acetate-grown culture of strain CAB that was transferred from a stock grown on catechol and perchlorate. (B) Proposed pathway of catechol oxidation in strain CAB. Red text, enzymes identified in the genome; black text, intermediates of the pathway. Download
ACKNOWLEDGMENTS
Funding supporting research on microbial (per)chlorate metabolism in the lab of J. D. Coates is provided by the Energy Biosciences Institute, CA. C. Carlström is grateful for funding support from the National Science Foundation’s Graduate Research Fellowship Program.
We acknowledge Robert Rohde for assisting with sample collection and Susanna Repo for guidance with RAxML.
Footnotes
Citation Carlström CI, Wang O, Melnyk RA, Bauer S, Lee J, Engelbrektson A, Coates JD. 2013. Physiological and genetic description of dissimilatory perchlorate reduction by the novel marine bacterium Arcobacter sp. strain CAB. mBio 4(3):e00217-13. doi:10.1128/mBio.00217-13.
REFERENCES
- 1. Coates JD, Achenbach LA. 2004. Microbial perchlorate reduction: rocket-fueled metabolism. Nat. Rev. Microbiol. 2:569–580 [DOI] [PubMed] [Google Scholar]
- 2. Motzer WE. 2001. Perchlorate: problems, detection, and solutions. Environ. Forensics 2:301–311 [Google Scholar]
- 3. Urbansky ET. 2002. Perchlorate as an environmental contaminant. Environ. Sci. Pollut. Res. 9:187–192 [DOI] [PubMed] [Google Scholar]
- 4. Hogue C. 2003. Rocket-fueled river. Chem. Eng. News 81:37–46 [Google Scholar]
- 5. Stanbury JB, Wyngaarden JB. 1952. Effect of perchlorate on the human thyroid gland. Metab. Clin. Exp. 1:533–539 [PubMed] [Google Scholar]
- 6. Wolff J. 1998. Perchlorate and the thyroid gland. Pharmacol. Rev. 50:89–105 [PubMed] [Google Scholar]
- 7. Rajagopalan S, Anderson T, Cox S, Harvey G, Cheng Q, Jackson WA. 2009. Perchlorate in wet deposition across North America. Environ. Sci. Technol. 43:616–622 [DOI] [PubMed] [Google Scholar]
- 8. Rajagopalan S, Anderson TA, Fahlquist L, Rainwater KA, Ridley M, Jackson WA. 2006. Widespread presence of naturally occurring perchlorate in high plains of Texas and New Mexico. Environ. Sci. Technol. 40:3156–3162 [DOI] [PubMed] [Google Scholar]
- 9. Rao B, Anderson TA, Orris GJ, Rainwater KA, Rajagopalan S, Sandvig RM, Scanlon BR, Stonestrom DA, Walvoord MA, Jackson WA. 2007. Widespread natural perchlorate in unsaturated zones of the southwest United States. Environ. Sci. Technol. 41:4522–4528 [DOI] [PubMed] [Google Scholar]
- 10. Scanlon BR, Reedy RC, Jackson WA, Rao B. 2008. Mobilization of naturally occurring perchlorate related to land-use change in the southern high plains, Texas. Environ. Sci. Technol. 42:8648–8653 [DOI] [PubMed] [Google Scholar]
- 11. Kounaves SP, Stroble ST, Anderson RM, Moore Q, Catling DC, Douglas S, McKay CP, Ming DW, Smith PH, Tamppari LK, Zent AP. 2010. Discovery of natural perchlorate in the Antarctic dry valleys and its global implications. Environ. Sci. Technol. 44:2360–2364 [DOI] [PubMed] [Google Scholar]
- 12. Hecht MH, Kounaves SP, Quinn RC, West SJ, Young SM, Ming DW, Catling DC, Clark BC, Boynton WV, Hoffman J, DeFlores LP, Gospodinova K, Kapit J, Smith PH. 2009. Detection of perchlorate and the soluble chemistry of Martian soil at the Phoenix lander site. Science 325:64–67 [DOI] [PubMed] [Google Scholar]
- 13. Böhlke JK, Sturchio NC, Gu B, Horita J, Brown GM, Jackson WA, Batista J, Hatzinger PB. 2005. Perchlorate isotope forensics. Anal. Chem. 77:7838–7842 [DOI] [PubMed] [Google Scholar]
- 14. Rikken GB, Kroon AGM, vanGinkel CG. 1996. Transformation of (per)chlorate into chloride by a newly isolated bacterium: reduction and dismutation. Appl. Microbiol. Biotechnol. 45:420–426 [Google Scholar]
- 15. Wallace W, Ward T, Breen A, Attaway H. 1996. Identification of an anaerobic bacterium which reduces perchlorate and chlorate as Wolinella succinogenes. J. Ind. Microbiol. 16:68–72 [Google Scholar]
- 16. Bender KS, Shang C, Chakraborty R, Belchik SM, Coates JD, Achenbach LA. 2005. Identification, characterization, and classification of genes encoding perchlorate reductase. J. Bacteriol. 187:5090–5096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Melnyk RA, Engelbrektson A, Clark IC, Carlson HK, Byrne-Bailey K, Coates JD. 2011. Identification of a perchlorate reduction genomic island with novel regulatory and metabolic genes. Appl. Environ. Microbiol. 77:7401–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liebensteiner MG, Pinkse MW, Schaap PJ, Stams AJM, Lomans BP. 2013. Archaeal (per)chlorate reduction at high temperature: an interplay of biotic and abiotic reactions. Science 340:85–87 [DOI] [PubMed] [Google Scholar]
- 19. Achenbach LA, Michaelidou U, Bruce RA, Fryman J, Coates JD. 2001. Dechloromonas agitata gen. nov., sp. nov. and Dechlorosoma suillum gen. nov., sp. nov., two novel environmentally dominant (per)chlorate-reducing bacteria and their phylogenetic position. Int. J. Syst. Evol. Microbiol. 51:527–533 [DOI] [PubMed] [Google Scholar]
- 20. Bardiya N, Bae JH. 2011. Dissimilatory perchlorate reduction: a review. Microbiol. Res. 166:237–254 [DOI] [PubMed] [Google Scholar]
- 21. Bruce RA, Achenbach LA, Coates JD. 1999. Reduction of (per)chlorate by a novel organism isolated from paper mill waste. Environ. Microbiol. 1:319–329 [DOI] [PubMed] [Google Scholar]
- 22. Coates JD, Michaelidou U, Bruce RA, O’Connor SM, Crespi JN, Achenbach LA. 1999. Ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 65:5234–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thrash JC, Ahmadi S, Torok T, Coates JD. 2010. Magnetospirillum bellicus sp. nov., a novel dissimilatory perchlorate-reducing alphaproteobacterium isolated from a bioelectrical reactor. Appl. Environ. Microbiol. 76:4730–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thrash JC, Pollock J, Torok T, Coates JD. 2010. Description of the novel perchlorate-reducing bacteria Dechlorobacter hydrogenophilus gen. nov., sp. nov. and Propionivibrio militaris, sp. nov. Appl. Microbiol. Biotechnol. 86:335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vanlaere E, van der Meer JR, Falsen E, Salles JF, de Brandt E, Vandamme P. 2008. Burkholderia sartisoli sp. nov., isolated from a polycyclic aromatic hydrocarbon-contaminated soil. Int. J. Syst. Evol. Microbiol. 58:420–423 [DOI] [PubMed] [Google Scholar]
- 26. Balk M, Altinbaş M, Rijpstra WI, Sinninghe Damsté JS, Stams AJ. 2008. Desulfatirhabdium butyrativorans gen. nov., sp. nov., a butyrate-oxidizing, sulfate-reducing bacterium isolated from an anaerobic bioreactor. Int. J. Syst. Evol. Microbiol. 58:110–115 DOI: 10.1099/ijs.0.65396-0. PubMed [DOI] [PubMed] [Google Scholar]
- 27. Balk M, Mehboob F, van Gelder AH, Rijpstra WI, Damsté JS, Stams AJ. 2010. (Per)chlorate reduction by an acetogenic bacterium, Sporomusa sp., isolated from an underground gas storage. Appl. Microbiol. Biotechnol. 88:595–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Collado L, Levican A, Perez J, Figueras MJ. 2011. Arcobacter defluvii sp. nov., isolated from sewage samples. Int. J. Syst. Evol. Microbiol. 61:2155–2161 [DOI] [PubMed] [Google Scholar]
- 29. Figueras MJ, Levican A, Collado L, Inza MI, Yustes C. 2011. Arcobacter ellisii sp. nov., isolated from mussels. Syst. Appl. Microbiol. 34:414–418 [DOI] [PubMed] [Google Scholar]
- 30. Debruyne L, Gevers D, Vandamme P. 2008. Taxonomy of the family Campylobacteraceae. American Society for Microbiology, Washington, DC [Google Scholar]
- 31. Collado L, Figueras MJ. 2011. Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin. Microbiol. Rev. 24:174–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Snelling WJ, Matsuda M, Moore JE, Dooley JS. 2006. Under the microscope: Arcobacter. Lett. Appl. Microbiol. 42:7–14 [DOI] [PubMed] [Google Scholar]
- 33. Kim HM, Hwang CY, Cho BC. 2010. Arcobacter marinus sp nov. Int. J. Syst. Evol. Microbiol. 60:531–536 [DOI] [PubMed] [Google Scholar]
- 34. Kern M, Simon J. 2009. Electron transport chains and bioenergetics of respiratory nitrogen metabolism in Wolinella succinogenes and other Epsilonproteobacteria. Biochim. Biophys. Acta 1787:646–656 [DOI] [PubMed] [Google Scholar]
- 35. McClung CR, Patriquin DG. 1980. Isolation of a nitrogen-fixing Campylobacter species from the roots of Spartina-alterniflora Loisel. Can. J. Microbiol. 26:881–886 [DOI] [PubMed] [Google Scholar]
- 36. Léonard D, Youssef CB, Destruhaut C, Lindley ND, Queinnec I. 1999. Phenol degradation by Ralstonia eutropha: colorimetric determination of 2-hydroxymuconate semialdehyde accumulation to control feed strategy in fed-batch fermentations. Biotechnol. Bioeng. 65:407–415 [DOI] [PubMed] [Google Scholar]
- 37. Kojima Y, Itada N, Hayaishi O. 1961. Metapyrocatechase—new catechol-cleaving enzyme. J. Biol. Chem. 236:2223–2228 [PubMed] [Google Scholar]
- 38. Arai H, Ohishi T, Chang MY, Kudo T. 2000. Arrangement and regulation of the genes for meta-pathway enzymes required for degradation of phenol in Comamonas testosteroni TA441. Microbiology 146:1707–1715 [DOI] [PubMed] [Google Scholar]
- 39. Grote J, Schott T, Bruckner CG, Glöckner FO, Jost G, Teeling H, Labrenz M, Jürgens K. 2012. Genome and physiology of a model epsilonproteobacterium responsible for sulfide detoxification in marine oxygen depletion zones. Proc. Natl. Acad. Sci. U. S. A. 109:506–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pati A, Gronow S, Lapidus A, Copeland A, Del Rio TG, Nolan M, Lucas S, Tice H, Cheng JF, Han C, Chertkov O, Bruce D, Tapia R, Goodwin L, Pitluck S, Liolios K, Ivanova N, Mavromatis K, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Detter JC, Rohde M, Goker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Klenk HP, Kyrpides NC. 2010. Complete genome sequence of Arcobacter nitrofigilis type strain (CI(T)). Stand. Genomic Sci. 2:300–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 42. Miller WG, Parker CT, Rubenfield M, Mendz GL, Wösten M, Ussery DW, Stolz JF, Binnewies TT, Hallin PF, Wang G, Malek JA, Rogosin A, Stanker LH, Mandrell RE. 2007. The complete genome sequence and analysis of the epsilonproteobacterium Arcobacter butzleri. PLoS One 2:e1358 http://dx.doi.org/10.1371/journal.pone.0001358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Toh H, Sharma VK, Oshima K, Kondo S, Hattori M, Ward FB, Free A, Taylor TD. 2011. Complete genome sequences of Arcobacter butzleri ED-1 and Arcobacter sp strain L, both isolated from a microbial fuel cell. J. Bacteriol. 193:6411–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wirsen CO, Sievert SM, Cavanaugh CM, Molyneaux SJ, Ahmad A, Taylor LT, DeLong EF, Taylor CD. 2002. Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl. Environ. Microbiol. 68:316–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Coates JD, Jackson A. 2008. Principles of perchlorate treatment. In situ bioremediation of perchlorate in groundwater. Springer Verlag, Norwell, MA [Google Scholar]
- 46. Chakraborty R, O’Connor SM, Chan E, Coates JD. 2005. Anaerobic degradation of benzene, toluene, ethylbenzene, and xylene compounds by Dechloromonas strain RCB. Appl. Environ. Microbiol. 71:8649–8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Coates JD, Chakraborty R, Lack JG, O’Connor SM, Cole KA, Bender KS, Achenbach LA. 2001. Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature 411:1039–1043 [DOI] [PubMed] [Google Scholar]
- 48. Weelink SA, Tan NC, ten Broeke H, van den Kieboom C, van Doesburg W, Langenhoff AA, Gerritse J, Junca H, Stams AJ. 2008. Isolation and characterization of Alicycliphilus denitrificans strain BC, which grows on benzene with chlorate as the electron acceptor. Appl. Environ. Microbiol. 74:6672–6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Coates JD, Bruce RA, Patrick J, Achenbach LA. 1999. Hydrocarbon bioremediative potential of (per)chlorate-reducing bacteria. Bioremediat. J. 3:323–334 [Google Scholar]
- 50. Wrighton KC, Agbo P, Warnecke F, Weber KA, Brodie EL, DeSantis TZ, Hugenholtz P, Andersen GL, Coates JD. 2008. A novel ecological role of the Firmicutes identified in thermophilic microbial fuel cells. ISME J. 2:1146–1156 [DOI] [PubMed] [Google Scholar]
- 51. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. Silva: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Accession numbers for microorganisms used in phylogenetic tree and RT-PCR primers used. Download
Effect of pH on the growth rate of strain CAB grown in medium containing 10 mM (each) acetate and perchlorate. Error bars represent standard deviations of the average of triplicate samples. Download
Effect of temperature on the growth rate of strain CAB grown in medium containing 10 mM (each) acetate and perchlorate. Error bars represent standard deviations of the average of triplicate samples. Download
Effect of salinity on the growth rate of strain CAB grown in medium containing 10 mM (each) acetate and perchlorate. Error bars represent standard deviations of the average of triplicate samples. Download
Growth curve of Arcobacter sp. strain CAB on 10 mM acetate and 8 mM nitrate. Cell number increase was monitored by optical density at 600 nm. Diamonds, cell density; squares, nitrite concentration (mM); circles, nitrate concentration (mM); triangles, cell density for no-acceptor control (mM). Error bars represent standard deviations of the average of triplicate samples. Download
Spectrophotometric scans (300 to 800 nm) of filtrates from triplicate cultures of strain CAB grown with 1 mM catechol and 5 mM perchlorate. Download
2-HMS and perchlorate concentration of Arcobacter sp. strain CAB grown on 1 mM catechol and 5 mM perchlorate. Squares, perchlorate concentration (mM); diamonds, 2-HMS concentration (μM). Error bars represent standard deviations of the average of triplicate samples. Download
LC/QTOF-MS of Arcobacter sp. strain CAB culture grown on 1 mM catechol and 5 mM perchlorate. Calculated 2-HMS concentration is 35.95 µM (based on absorbance at 375 nm). An ion of m/z 141.0192 was observed, consistent (accuracy, 0.71 ppm) with (M-H)− of 2-HMS (theoretical m/z 141.0193). Download
(A) RT-PCR of catechol-grown (lanes 3 to 5) and acetate-grown (lane 7) cultures of strain CAB. Lane 1 is a PCR negative control, and lane 2 is a genomic DNA positive control. Lane 6 is an acetate-grown culture of strain CAB that was transferred from a stock grown on catechol and perchlorate. (B) Proposed pathway of catechol oxidation in strain CAB. Red text, enzymes identified in the genome; black text, intermediates of the pathway. Download