ABSTRACT
Previous work from our laboratory showed that the Gram-negative aquatic pathogen Vibrio cholerae can take up a much wider repertoire of fatty acids than other Gram-negative organisms. The current work elaborated on the ability of V. cholerae to exploit an even more diverse pool of lipid nutrients from its environment. We have demonstrated that the bacterium can use lysophosphatidylcholine as a metabolite for growth. Using a combination of thin-layer chromatography and mass spectrometry, we also showed that lysophosphatidylcholine-derived fatty acid moieties can be used for remodeling the V. cholerae membrane architecture. Furthermore, we have identified a lysophospholipase, VolA (Vibrio outer membrane lysophospholipase A), required for these activities. The enzyme is well conserved in Vibrio species, is coexpressed with the outer membrane fatty acid transporter FadL, is one of very few surface-exposed lipoprotein enzymes to be identified in Gram-negative bacteria and the first instance of a surface lipoprotein phospholipase. We propose a model whereby the bacterium efficiently couples the liberation of fatty acid from lysophosphatidylcholine to its subsequent metabolic uptake. An expanded ability to scavenge diverse environmental lipids at the bacterial surface increases overall bacterial fitness and promotes homeoviscous adaptation through membrane remodeling.
IMPORTANCE
Our understanding of how bacteria utilize environmental lipid sources has been limited to lipids such as fatty acids and cholesterol. This narrow scope may be attributed to both the intricate nature of lipid uptake mechanisms and the diversity of lipid substrates encountered within an ecological niche. By examining the ability of the pathogen Vibrio cholerae to utilize exogenous lipids, we uncovered a surface-exposed lipoprotein (VolA) that is required for processing the prevalent host lipid lysophosphatidylcholine. VolA functions as a lipase liberating a fatty acid from exogenous lysophospholipids. The freed fatty acid is then transported into the cell, serving as a carbon source, or shunted into phospholipid synthesis for membrane assembly. A limited number of surface-exposed lipoproteins have been found in Gram-negative organisms, and few have enzymatic function. This work highlights the ability of bacteria to exploit exogenous lipids for both maintenance of the membrane and carbon source acquisition.
Introduction
Gram-negative bacteria have developed several mechanisms for scavenging nutrients and other important metabolites from their surroundings. One such mechanism, the fatty acid uptake pathway, allows Gram-negatives to import long-chain fatty acids (LCFAs) through their cell wall via the outer membrane transporter FadL and ligate them to coenzyme A using the fatty acyl coenzyme A ligase FadD. After this ligation event, the activated fatty acids are destined for energy production or membrane construction. Exogenous fatty acids can serve as an energy source through the degradative β-oxidation pathway, which yields numerous activated carrier molecules for metabolic gain. Alternatively, the steep energy requirements for fatty acid biosynthesis can be circumvented by shunting the imported fatty acid into the membrane for phospholipid assembly (1).
Previous research by our laboratory has shown that the Gram-negative human pathogen Vibrio cholerae is capable of utilizing diverse fatty acids from the surrounding environment, including very-long-chain fatty acids with multiple unsaturations (2). This is in contrast to other Gram-negative organisms; Escherichia coli cannot metabolize fatty acids that are longer than 20 carbons nor process those fatty acids with equally high degrees of unsaturation (3–5). This difference in substrate profiles between V. cholerae and other Gram-negatives may be attributed to V. cholerae’s multiple homologs of FadL and FadD. A previous study from our laboratory determined that the bile secreted in the small intestine caused variation in the phospholipid profile of V. cholerae when analyzed via thin-layer chromatography and mass spectrometry (2). All of the major phospholipids—including phosphatidylglycerol, phosphatidylethanolamine, and cardiolipin (see Fig. S1A in the supplemental material)—showed an altered acylation profile. Further experimentation revealed that the fatty acid component of bile specifically caused the altered migratory pattern. The ability of V. cholerae to remodel its membrane phospholipids using exogenous lipid sources is an important advantage. This is further emphasized by the fact that environmental fatty acids found in aquatic sediment were also found to cause migratory shifts (2). Altering the phospholipid profile in this manner can have dramatic effects on fitness via homeoviscous adaptation and reduced energetic requirements for membrane biogenesis.
In their respective niches, Gram-negatives are in contact with diverse lipid chemical species, more than fatty acids alone, which can serve as potential carbon sources. V. cholerae is immersed in lipids when colonizing the human intestine and when living in an aquatic environment (6). For example, copepods, which constitute a common animal reservoir for V. cholerae, are known to synthesize phospholipids with very-long-chain fatty acyl tails, including phosphatidylcholine with attached docosapentaenoic (C22:5) and docosahexaenoic (C22:6) acids (7). The lumen of the human intestine has significant amounts of lysophosphatidylcholine (LPC), derived from lecithinase activity on biliary and dietary sources of phosphatidylcholine (8, 9). Given that V. cholerae has been shown to remodel its membrane architecture using exogenous lipids and that the bacteria have consistent exposure to these alternative lipids, V. cholerae might have evolved mechanisms to utilize these alternate sources of acyl chains for membrane remodeling as well.
Currently, while some steps in the transport of fatty acids into the bacterial cell are well characterized—such as those performed by FadL and FadD—other steps—such as how V. cholerae might utilize alternative lipid sources in conjunction with the fatty acid transport pathway—are enigmatic. This work reports the ability of V. cholerae El Tor O1 to utilize LPC for nutrition and for remodeling of cell wall phospholipids. We have identified a putative surface-exposed lipoprotein, VolA, that is responsible for this activity, and we suggest a mechanism for its function. VolA mediates the unusual lipid utilization characteristics of V. cholerae and represents a newly discovered member of undercharacterized surface-exposed lipoprotein enzymes of Gram-negative bacteria.
RESULTS
Vibrio cholerae can use lysophosphatidylcholine as the sole carbon source.
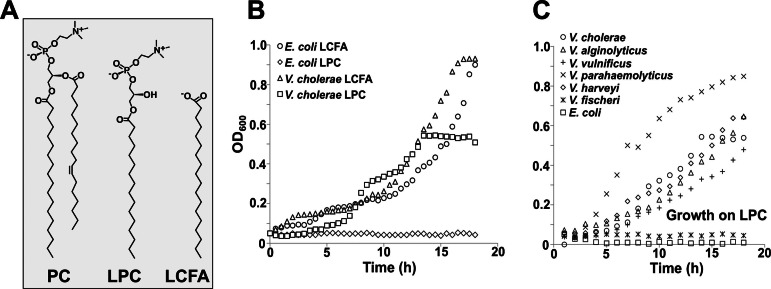
In order to determine if V. cholerae El Tor O1 could utilize lysophospholipids, bacteria (strains are listed in Table S1 in the supplemental material) were grown in defined minimal medium with either 0.2% glucose, 2 mM stearoyl (C18:0)-lysophosphatidylcholine (LPC), or 2 mM stearic acid, serving as the sole carbon source (Fig. 1; see also Fig. S2). When E. coli and V. cholerae were grown with LCFAs, no difference in growth could be detected. However, only V. cholerae was able to utilize LPC. The maximal optical density at 600 nm (OD600) of V. cholerae grown on LPC was approximately half that of the LCFA-grown cultures (Fig. 1B); the slower growth on LPC is likely due to the rate-limiting step of release of the fatty acid from the LPC molecule. Similar results were obtained when the level of growth was measured by determining CFU of viable cells on agar plates (see Fig. S3).
FIG 1 .
Growth of E. coli and Vibrio species on LPC or LCFA. (A) Chemical structures of PC (phosphatidylcholine), LPC (lysophosphatidylcholine), and an LCFA (long-chain fatty acid). (B) Both E. coli and V. cholerae can utilize LCFA as the sole carbon source for growth, while only V. cholerae can utilize LPC. (C) All Vibrio species tested were able to use LPC as the sole carbon source for growth, with the exception of V. fischeri.
After observing this phenotype in V. cholerae El Tor, we expanded our testing to other Vibrio species, including V. alginolyticus, V. fischeri, V. harveyi, V. parahaemolyticus, and V. vulnificus. Bacteria were grown under conditions similar to those of the initial experiments, with either 0.2% glucose (see Fig. S2) or 2 mM LPC (Fig. 1C). A defined salt-rich minimal medium was used for growth of V. fischeri and V. harveyi strains. Interestingly, when grown with 2 mM LPC as the sole source of carbon, all strains were able to grow, with the exception of E. coli and V. fischeri (Fig. 1C).
Bioinformatic analysis of Vibrio strains reveals a putative phospholipase in an operon with a FadL homolog.
Since the ability to use LPC as the sole carbon source was found in multiple species of Vibrio and not in others, a comparative genome search was performed using the String 9.0 database (http://string-db.org). In all species capable of growth on LPC, an additional gene (vca0863 in V. cholerae) was found directly downstream of a FadL homolog (gene vca0862 in V. cholerae) (see Fig. S4 in the supplemental material). A similar genetic organization was conspicuously absent from the V. fischeri genome. Because only those bacterial strains that contained this putative operon were capable of growth on LPC, it suggested that vca0863 might be involved in the LPC growth phenotype. Interestingly, VcA0863 is annotated as a putative lipase (Uniprot Q9KL83 [http://www.uniprot.org]; GenBank accession no. AAF96761.1 [http://ncbi.nlm.nih.gov]) and has an N-terminal lipoprotein signal sequence. VcA0863 shows strong homology to pfam12262, having the known lipase amino acid sequence motif Gly-Xaa-Ser-Xaa-Gly (10). This motif, containing a catalytic serine, has been identified in lipases derived from other aquatic organisms (10) and could potentially act in a similar manner in V. cholerae. A nonaspartate residue is present in the +2 position after the signal peptide cleavage site, and based upon rules governing lipoprotein sorting and localization, the protein should be localized to the outer membrane (11).
The genes vca0862 and vca0863 are cotranscribed in V. cholerae.
The intergenic region between the genes vca0862 and vca0863 in all Vibrio species was between 16 and 23 bp. To confirm that the FadL homolog (encoded by vca0862) and the phospholipase (encoded by vca0863) are encoded in an operon, RNA was purified from wild-type V. cholerae grown in minimal medium supplemented with glucose or glucose and LPC. cDNA was generated and amplified by primers designed for internal regions of vca0862, vca0863, or a region spanning 400 bases centered on the intergenic region (Fig. 2A). Genomic DNA from wild-type V. cholerae was used as a positive control. PCR products were observed for the internal regions of the individual genes under both conditions: a 200-bp band representing vca0862 and a 165-bp band representing vca0863 (Fig. 2B). The presence of LPC in the growth medium was not required for gene transcription compared to the glucose control. A 430-bp band, generated from the intergene-spanning primers, confirms that vca0862 and vca0863 are cotranscribed.
FIG 2 .
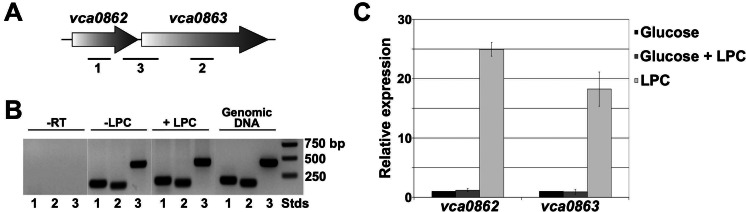
Organization and expression of vca0862 and vca0863. (A) The genetic organization of vca0862 (FadL) and vca0863 (VolA) in V. cholerae, along with primer extension locations used in RT-PCR. (B) RT-PCR of vca0862 and vca0863 show that the genes are cotranscribed and basal levels of expression are independent of the presence of LPC. A genomic DNA template was used to confirm the amplified product sizes, and cDNA without reverse transcriptase (-RT) was used as the negative control verifying DNA-free RNA. (C) qRT-PCR data of the expression of vca0862 and vca0863 grown with glucose and/or LPC. When growth was in minimal medium containing LPC as the sole carbon source, expression of vca0862 and vca0863 increased 25- and 18-fold, respectively, compared to expression for a glucose-grown control.
Quantitative reverse transcription-PCR (qRT-PCR) was performed to determine the expression profiles of vca0862 and vca0863 in wild-type bacteria grown in the presence of glucose, LPC, or glucose and LPC together. The relative expression of both genes was measured as a ratio compared to expression for the glucose-only control. Results showed that there was no statistically significant difference in the expression of vca0862 and vca0863 when LPC was added to the glucose control (Fig. 2C). However, when the strain was grown with LPC as the sole carbon source, expression of the genes increased dramatically, with that of vca0862 increasing ~25-fold and that of vca0863 increasing ~18-fold compared to expression for the glucose control. Analysis of the vca0862 promoter region using a footprinting algorithm (12) revealed binding sites for both cAMP receptor protein (CRP) and FadR. E. coli CRP is a transcriptional activator that binds to DNA in a complex with cyclic AMP (cAMP); in the presence of glucose, levels of cAMP are decreased and CRP does not bind to the DNA (13, 14). FadR is a repressor of many fatty acid metabolism genes, including fadL, and is inhibited by binding coenzyme A (CoA)-activated fatty acids (15). Thus, in the presence of glucose, both genes could be downregulated due to a lack of active CRP; this is consistent with the upregulation that is observed in cultures grown with LPC as the sole carbon source. Additionally, for growth in succinate (see Fig. S5 in the supplemental material), this downregulation is abrogated, and growth with both succinate in the presence of LPC and LPC only shows an upregulation of vca0862 and vca0863 compared to findings for a succinate-only control. This upregulation in the presence of LPC could also be due in part to a lack of downregulation by FadR because of its inhibition by LPC-derived fatty acids.
vca0863 is necessary for utilization of lysophosphatidylcholine.
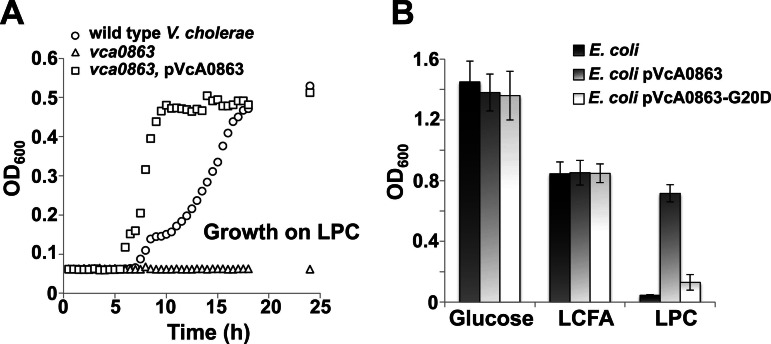
After identification of an operon containing a phospholipase and an outer membrane fatty acid transporter, we investigated whether bacterial growth on LPC was dependent upon expression of vca0863. When a vca0863-deficient V. cholerae strain was incubated with LPC as the sole carbon source, we found that it failed to grow (Fig. 3A). Growth on LPC is restored when vca0863 was expressed in trans from a low-copy-number vector (pVcA0863). However, the doubling time for the complemented strain was decreased ~3-fold compared to that for the wild type and may arise from increased release of LCFA from LPC by the lipase activity.
FIG 3 .
Expression of vca0863 is required for utilization of LPC as the sole carbon source. (A) Growth of wild-type V. cholerae, vca0863 transposon mutant, and the complemented mutant on LPC. (B) Growth of wild-type E. coli and E. coli containing either pVcA0863 or pVcA0863-G20D on glucose, LCFA, or LPC.
Although E. coli K-12 is unable to grow on LPC, heterologous expression of VcA0863 in E. coli conferred growth on LPC (Fig. 3B). However, expression of VcA0863 in which the amino acid glycine at position 20 is replaced with an aspartate residue (VcA0863-G20D) does not support growth. Based upon the rules of lipoprotein sorting, replacement of the glycine residue with an aspartic acid in the +2 position after the lipoprotein signal sequence would result in mislocalization of VcA0863 to the inner membrane (11). These results suggest that expression of VcA0863 is sufficient for the bacterium to utilize lysophospholipids from the environment and that the protein must be localized to the outer membrane.
Strains expressing vca0863 modify their membrane phospholipids using the fatty acyl moiety of lysophosphatidylcholine.
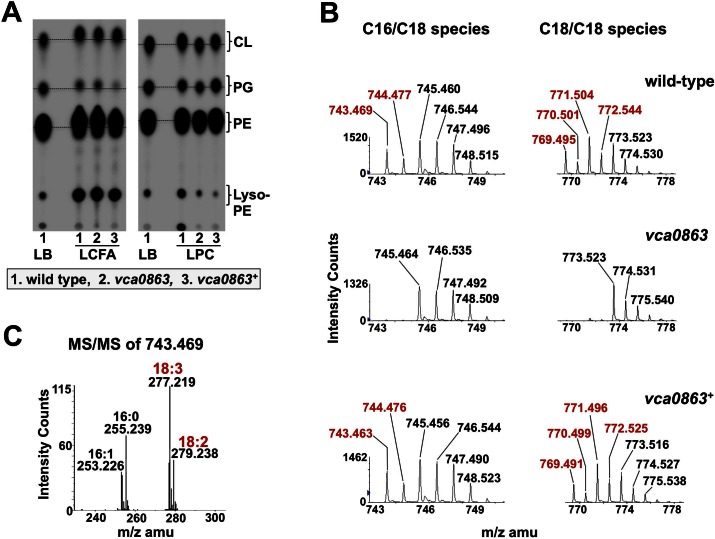
Previously, we reported that V. cholerae underwent substantial membrane remodeling by incorporating long-chain fatty acids from its surrounding environment. Here, we asked if membrane remodeling also occurred in the presence of lysophospholipids. Cultures of wild-type V. cholerae El Tor O1, the vca0863 mutant strain, and the complemented mutant were grown in defined minimal medium containing 2% glucose and 32Pi in the presence of LCFA or LPC. Wild-type E. coli K-12 was included as a negative control (see Fig. S1B in the supplemental material). 32P–labeled phospholipids were extracted and analyzed by thin-layer chromatography (TLC) (Fig. 4A). By using commercially available exogenous lipid sources with acyl chains that cannot be generated by the bacteria de novo, we can monitor the ability of the bacteria to utilize the exogenous lipid in the phospholipid biosynthesis pathway by TLC. Here, we grew cultures in the presence of a mixed sample of either long-chain fatty acids or LPC, predominantly consisting of 18:2 and 18:3 acyl chains; V. cholerae does not synthesize either of these chains de novo (16). When all three strains were grown in LCFA and 2% glucose, an upwards shift in mobility could be observed in all three of the major phospholipids (Fig. 4A), consistent with incorporation of the exogenous LCFAs into phospholipids. When the cultures were grown with glucose and LPC, only strains expressing vca0863 showed the same upward shift in mobility. The lack of a shift in the mobility pattern in wild-type E. coli and the vca0863 transposon mutant is consistent with a requirement for VcA0863 expression to liberate the fatty acid from the LPC molecule.
FIG 4 .
Incorporation of LPC-derived fatty acids into membrane phospholipids of V. cholerae. (A) TLC of LCFA- and LPC-grown cultures of V. cholerae. V. cholerae strains, including the wild type, the vca0863 transposon mutant, and the vca0863-complemented mutant, were grown in the presence of 2 mM LCFA or LPC containing 18:2 and 18:3 unsaturated carbon chains that V. cholerae cannot synthesize de novo. All three strains show a shift in mobility for each of the major phospholipids when treated with LCFA, as expected. When treated with the LPC mix, only strains expressing vca0863 (wild type and complemented mutant) showed a similar shift, indicating that vca0863 is required for generating LPC-derived fatty acid. The dashed line has been included for comparison on mobility shifts. (B) Phosphatidylglycerol (PG) was isolated from wild-type, vca0863 mutant, and vca0863 mutant complement strains of V. cholerae and analyzed by liquid chromatography/ESI-mass spectrometry. Strains that express vca0863 showed a unique set of peaks (shown in red) corresponding to weights of PG that have acyl chains with unsaturations matching those in the LPC mix. These peaks were absent from the vca0863 mutant. (C) Tandem mass spectrometry (MS/MS) of the PG peak, with an m/z of 743.469 found only in vca0863-expressing strains. MS/MS showed that C18:2 and C18:3 acyl chains that originate with exogenous LPC are incorporated into the V. cholerae membrane.
We performed liquid chromatography/electrospray ionization (ESI)-tandem mass spectrometry on lipids extracted from wild-type V. cholerae, vca0863 V. cholerae, and the complemented mutant. Each of the major phospholipids was analyzed. Analyses of spectra revealed distinct differences between the vca0863 transposon mutant and the wild-type and complemented strains. Specifically, results from the Lipid Maps Structure Database prediction algorithm (http://www.lipidmaps.org/) showed several phospholipid species corresponding to phospholipids containing C18:2 and C18:3 that were present only in spectra for wild-type or complemented V. cholerae grown in the presence of C18:2/18:3-LPC (Fig. 4; see also Fig. S6 and S7 in the supplemental material). Masses of phospholipid species specific to LPC growth are highlighted red in Fig. 4B (see also Fig. S6 and S7). A phosphatidylglycerol [M-H]1− ion peak with mass of 743.469 (Fig. 4B) in the wild-type or complemented strain was selected for tandem mass spectrometry. Collision-induced dissociation of the m/z 743.469 ion revealed two phosphatidylglycerol species containing C18:2/C16:1 or C18:3/C16:0, confirmed by the presence of peaks at m/z 277.219 and 279.238, which correspond to C18:3 and C18:2 acyl chains, respectively (Fig. 4C). Similar phenotypes were observed for phosphatidylethanolamine and cardiolipin species, indicating that lipids derived from LPC can be used both for nutrition and for remodeling of all of the membrane phospholipids (see Fig. S6 and S7).
The vca0863 gene product is a surface-exposed lipoprotein.
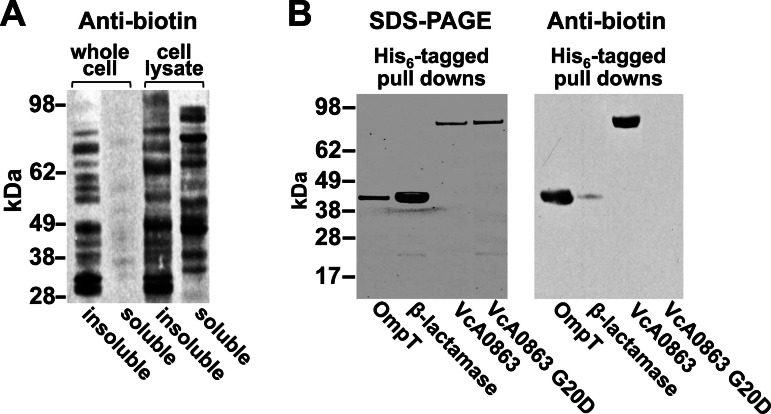
In the course of this study, we observed that a fadL mutant of E. coli failed to grow using LPC as the sole carbon source when heterologously expressing vca0863 (see Fig. S8 in the supplemental material). Because FadL is responsible for the uptake of LCFAs, it is reasonable to assume that the breakdown of LPC occurs prior to entry into the cell. This indicates that VcA0863 may be surface exposed. To confirm localization of VcA0863, whole bacteria were labeled with an amine-reactive biotin (N-hydroxysuccinimide [NHS]-long-chain [LC]-LC biotin), and the labeled proteins were visualized using a streptavidin horseradish peroxidase conjugate (17). The large size and polar nature of the biotin compound prevent passage across the outer membrane, allowing selective labeling of surface-exposed proteins. When whole cells were labeled, only minimal signal could be detected in the soluble fraction, indicating that the biotin compound failed to significantly penetrate the outer membrane (Fig. 5A).
FIG 5 .
Biotin labeling of surface-exposed VcA0863. (A) Antibiotin Western blot of soluble and insoluble protein fractions from either whole V. cholerae cells or cell lysates labeled with NHS-LC-LC-biotin. (B) SDS-PAGE and antibiotin Western blot of His-tagged VcA0863, β-lactamase, OmpT, and mislocalized VcA0863 affinity purified from cell extracts of biotin-labeled whole cells. Protein bands at ~81 kDa (VcA0863-His6 and mislocalized VcA0863-His6) and ~41 kDa (β-lactamase-His6 and OmpT-His6) in size were observed in SDS-PAGE, establishing that all proteins were running according to their molecular masses. The antibiotin Western blot showed two proteins, an 81-kDa band representing wild-type VcA0863 and a 41-kDa band representing OmpT. The fact that VcA0863 was labeled with biotin strongly indicates that VcA0863 is surface exposed. OmpT was also labeled due to its exposure on the surface of the cell. Little or no signal could be detected for either β-lactamase or mislocalized VcA0863 due to lack of surface exposure.
To determine if VcA0863 was surface exposed, a V. cholerae El Tor O1 vca0863-deficient strain was transformed with vectors expressing His-tagged variants of either vca0863 or vca0863 encoding the G20D replacement (vca0863-G20D). Cells were also separately transformed with vectors containing an E. coli-derived β-lactamase (encoded by ampC) (18), serving as a negative control, or V. cholerae ompT, an outer membrane β-barrel acting as a positive control (19). After labeling the V. cholerae strains with NHS-LC-LC biotin, cells were harvested and lysed, and polyhistidine tagged proteins were isolated using cobalt affinity resin and analyzed via Western blotting (Fig. 5B). On the antibiotin blot, controls reflected the surface specificity of the biotin labeling. For example, surface-exposed OmpT was efficiently biotinylated and produced a strong signal. Periplasmic β-lactamase, however, showed minimal labeling even though the protein is overexpressed, suggesting that NHS-LC-LC biotin did not cross the outer membrane to a significant degree. Also detected on the antibiotin blot was a prominent 83-kDa band representing VcA0863, indicating that it is surface exposed. No signal could be detected for the mislocalized VcA0863-G20D mutant.
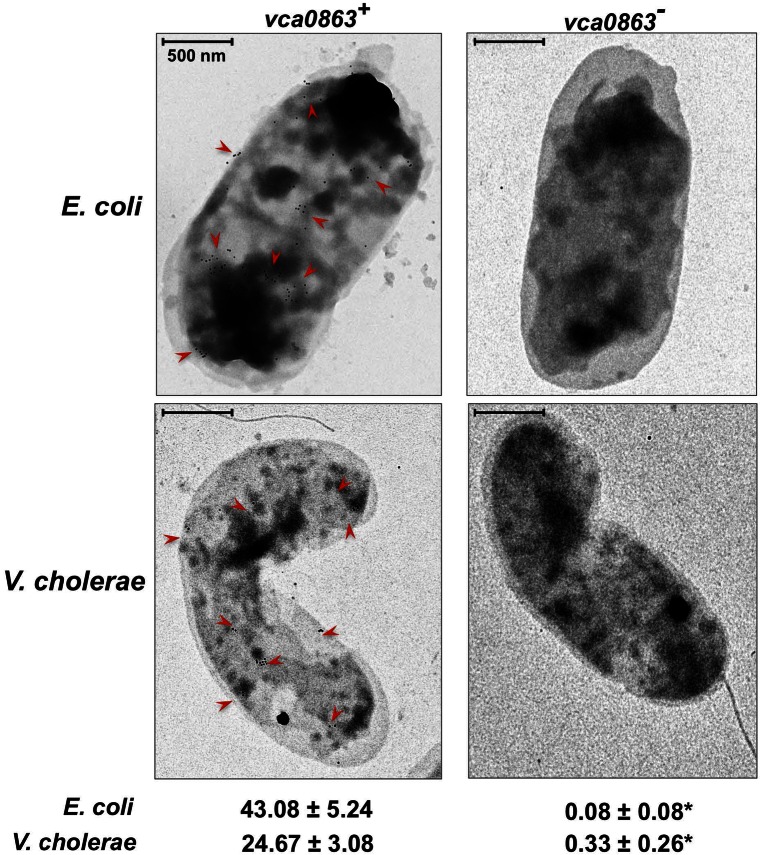
In addition to using surface biotinylation, we independently confirmed that VcA0863 is surface exposed through the use of immunogold electron microscopy. Wild-type E. coli K-12 or wild-type V. cholerae El Tor expressing VcA0863 were incubated with polyclonal anti-VcA0863 antibodies followed by gold-conjugated goat anti-rabbit antibodies. The labeled whole cells were then analyzed via electron microscopy. Both E. coli and V. cholerae cells expressing VcA0863 showed a considerable number of gold particles associated with the cell surface in comparison to strains lacking VcA0863, which showed no surface labeling (Fig. 6). Together with the surface biotinylation of VcA0863, these data indicate a clear surface localization of the lipase. Given that VcA0863 has a lipase domain, is required for growth on lysophospholipids, and is localized to the bacterial surface, we have named the vca0863 gene product VolA for Vibrio outer membrane lysophospholipase A.
FIG 6 .
Immunogold electron micrographs of E. coli and V. cholerae strains expressing vca0863. Gold particles (examples are denoted with a red arrow) indicate the presence of surface-exposed VcA0863. Both E. coli expressing vca0863 from the plasmid and wild-type V. cholerae showed gold particles associated with the surface of the cell, indicating that VcA0863 is exposed to the surface in both strains. Wild-type E. coli or the V. cholerae vcA0863 mutant failed to display any associated gold particles on the bacterial surface. Mean gold particle counts are reported with standard errors (n > 10); statistical significance was observed between results for strains expressing and not expressing vca0863. *, P < 0.0005, Mann-Whitney U test.
DISCUSSION
This study demonstrates an unusual form of lipid utilization in the Gram-negative bacterium Vibrio cholerae. In the course of its life cycle, V. cholerae experiences a variety of environments, from its planktonic existence in the ocean, living as aggregates associated with animal reservoirs in aquatic ecosystems, to infecting a human host. Concentrations of nutrients, ions, and salts vary significantly between these conditions, as does the presence of antimicrobial agents; because of this, V. cholerae has developed specific systems to adapt itself for survival. One such system is the fatty acid uptake pathway, used to incorporate fatty acids from the environment directly into various metabolic pathways. Previous experiments from our laboratory have shown that V. cholerae is capable of using very-long-chain fatty acids from the environment both as a carbon source and to remodel its membrane lipids; these fatty acids have not been shown to be used by other Gram-negatives. This distinct uptake profile could be due to an uncommon evolution of multiple homologs of FadL and FadD found only in V. cholerae and some related species. Elaborating on the utilization of unconventional lipids by V. cholerae, the current work highlights the ability of V. cholerae to process lysophosphatidylcholine (LPC) for use in various metabolic pathways. The ability to use LPC from its surroundings has advantages for V. cholerae fitness in terms of nutrient acquisition and environmental acclimatization; LPC-derived fatty acids can be used as a nutrient source, reducing the energetic needs of the cell, but they can also be used to remodel phospholipids. By using environmental lipids to remodel the membrane architecture of the cell, V. cholerae could respond to membrane stress through homeoviscous adaptation, a mechanism by which bacteria can regulate their membrane fluidity when exposed to a new environment.
LPC and its progenitor phospholipids are commonly found in both aquatic environments and in human host. In the ocean, V. cholerae associates with a variety of organisms, including zooplankton (20, 21), chironomid egg masses (22), flora (23), protozoa (24, 25), mollusks (26), and fish (27). Phospholipids and lysophospholipids are present in and on these various reservoirs (28–31). Secretion of a lecithinase in V. cholerae (32) could free the lyso derivative from phosphatidylcholine in the environment. The ability of V. cholerae to use LPC as a carbon source provides a fitness advantage in nutrient-poor aquatic environments. Furthermore, the ability to scavenge lysophospholipids may reach beyond the usage of LPC. Other environmental lipids present in appreciable amounts could also be processed by VolA or VolA-like proteins.
Once V. cholerae infects a human host, it is consistently exposed to phospholipids and their derivatives. Through human pancreatic phospholipase A2 activity for both dietary and bile-sourced phospholipids, the human intestine has significant levels of lysophospholipids, with LPC as the predominant lipid (33). V. cholerae can modify its membrane lipids using the fatty acid freed from LPC in a manner similar to that of exogenous fatty acid remodeling. By breaking down the LPC that is present and altering its own membrane phospholipids to resemble the host, V. cholerae could protect itself against membrane stress. More importantly, LPC has been shown to be involved in several proinflammatory signaling pathways (34, 35), generation of reactive oxygen species (36), and indirect antimicrobial effects on Gram-negatives (37, 38). A secondary effect of this utilization may be to reduce the local concentration of LPC, thus circumventing host defenses and reducing bacterial clearance.
The importance of lipid utilization in bacteria is not fully understood. While fatty acid uptake has been investigated, few data are available concerning alternate lipid sources. This study is just one example of how other lipids might be used via undiscovered enzymes that work in conjunction with established uptake pathways. Other lipids may play similar roles in nutrition and membrane remodeling in a manner similar to fatty acids and LPC. Cholesterol, for example, has been shown to have significant effects on membrane strength when incorporated into the human pathogen Helicobacter pylori (39); alternatively, it plays a role in nutrition in Mycobacterium tuberculosis (40). It is unclear if lipid uptake plays as substantial a role in other organisms besides Gram-negatives. Uptake of fatty acids has been observed in Gram-positives (41), but it is unknown if the mechanism of uptake is similar to that of Gram-negatives.
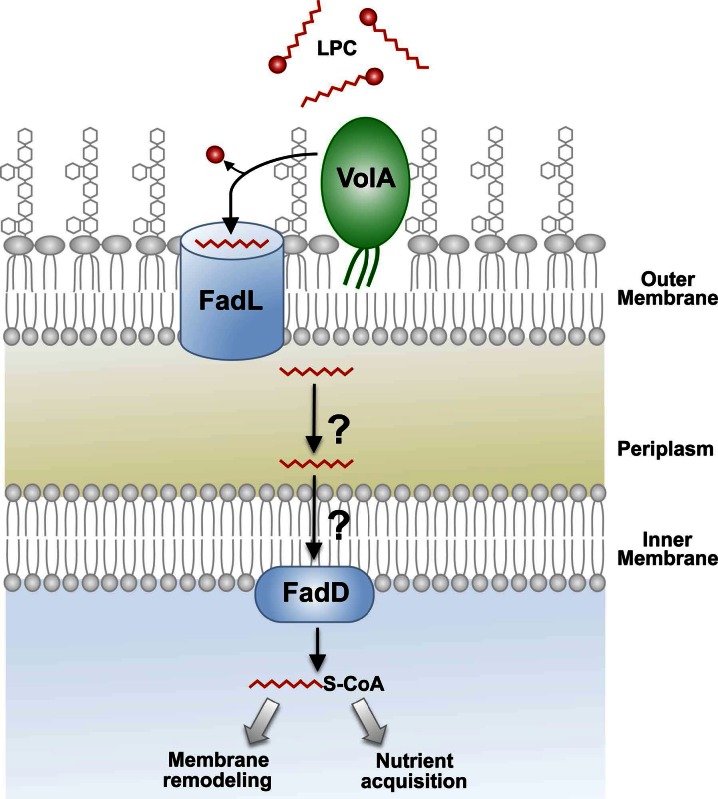
We propose a model for the mechanism of LPC utilization (Fig. 7). In this model, VolA acts on LPC prior to its passage through FadL, supported by the fact that growth on LPC as the sole carbon source is dependent on expression of FadL (see Fig. S8 in the supplemental material). This model is particularly intriguing because there are currently only a few surface-exposed lipoproteins in Gram-negative bacteria: TraTp (42), an F sex factor, WZAK30 (43), a multimeric pore-forming complex involved in K30 capsular polysaccharide expression, and CsgG (44), a member of the secretion apparatus of curli fibers in the Enterobacteriaceae. Additionally, the most abundant protein in E. coli, Lpp, was recently identified by the Silhavy group as being expressed as a surface-exposed lipoprotein (17). Lpp has been shown to have two forms, existing in an ~1:2 ratio: a peptidoglycan-bound form and a “free” form. Cowles et al. (17) demonstrated that nearly all of the free form of Lpp is localized to the surface of the outer membrane of E. coli. While these are several examples of surface-exposed lipoproteins, there is a noticeable lack of discovered lipoprotein enzymes. Pullulanase, a starch-debranching lipoprotein that is exported to the outer surface of Klebsiella species and subsequently released into the growth medium, is one of the only known examples of a surface lipoprotein with enzymatic function (45). The protein described in this work is an instance of a surface-exposed lipase illuminating how lipoproteins can play a role in extracellular enzymatic mechanisms.
FIG 7 .
Proposed model for VolA-dependent utilization of lysophospholipids. Exogenous LPC is initially cleaved by the surface-exposed lipoprotein VolA (VcA0863). One of the V. cholerae homologs of FadL (encoded by vc1042, vc1043, or vca0862) (see Fig. S4 in the supplemental material) can then transport the free fatty acid. Following transport across the periplasm and the inner membrane, the fatty acid is converted to an acyl-CoA by FadD. The activated fatty acid can be utilized for phospholipid biosynthesis or used as a carbon source for the bacterial cell.
It is currently unknown how the protein identified in this study is transported to the surface of V. cholerae. VolA is likely shuttled through the Lol system (46), especially since mutation of the +2 residue after the lipoprotein signal sequence can disrupt localization (Fig. 5). Since the Lol transport machinery ends at the periplasmic face of the outer membrane, how VolA reaches the outer surface of the cell (Fig. 7) is unknown. The fact that VolA can be properly expressed in E. coli, combined with the recent discovery that Lpp is also surface exposed, indicates that both E. coli and V. cholerae express the machinery required to transport lipoproteins to the outer surface. The easily observed growth phenotype associated with the utilization of lysophospholipids presents an opportunity to identify additional machinery required for the transport of VolA to the bacterial surface.
The current study continues to highlight the importance of lipid utilization by Gram-negative bacteria. Vibrio species may have evolved this adaptation as an efficient means for both maintenance of the membrane and carbon source acquisition. The ability to exploit exogenous lipids in different environmental reservoirs would allow the bacterium to adopt a phospholipid profile that reflects the fatty acid composition of its surrounding environment, perhaps as a mechanism of homeoviscous adaptation to survive membrane stress. The roles that VolA and membrane remodeling play in the adaptation of V. cholerae to diverse environments are currently under investigation.
MATERIALS AND METHODS
A description of all bacteria strains, plasmids, and oligonucleotides can be found in Table S1 in the supplemental material. Methods for the purification and characterization of phospholipids, electron microscopy, biotinylation of proteins, and the generation of polyclonal antibodies are all described in the supplemental Materials and Methods. Methods describing recombinant DNA and RNA techniques and the generation of complementation plasmids can also be found in the supplemental Materials and Methods.
SUPPLEMENTAL MATERIAL
Supplemental Materials and Methods. Download
Chemical structures and TLC of the major phospholipids of E. coli K-12. (A) Chemical structures of phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (CL). (B) E. coli was grown in minimal medium containing a 2 mM concentration of a soybean-derived lysophosphatidylcholine (LPC) mix including C18:2 and C18:3 acyl chains or the corresponding LCFA. Phospholipids were extracted and analyzed via TLC. No change in the migration pattern was observed with lipids from cells grown in glucose with and without LPC, confirming that E. coli cannot utilize LPC as a source of LCFA for membrane modification. Download
Growth of different Vibrio species and E. coli K-12 using glucose as the sole carbon source. Cultures of V. cholerae, V. alginolyticus, V. vulnificus, and V. parahaemolyticus were grown along with E. coli in minimal medium containing 0.2% glucose. V. fischeri and V. harveyi were grown in a high-salt defined minimal medium (see supplemental Materials and Methods) with 0.2% glucose. Download
Growth of E. coli and V. cholerae on LPC or LCFA as measured by colony counting. Quantification of bacterial growth was determined by plating assays. Viable colony-forming unit counts show that E. coli fails to utilize LPC as the sole source of carbon. Download
Genomic organization of FadL and VolA (VcA0863) homologues in various Gram-negative species. All Vibrio species possess homologues of vc1042, vc1043, and vca0862 (encoding FadLs), and with the exception of V. fischeri, all also had a homolog of vca0863. E. coli has only a single homolog of FadL and no homolog of vcA0863. Download
qRT-PCR of vca0862 and vca0863. qRT-PCR data of the expression of vca0862 and vca0863 grown with succinate and/or LPC are given. Growth in LPC displayed an upregulation of both vca0862 and vca0863 compared to results for a succinate-grown control. Download
Mass spectrometry of V. cholerae-derived phosphatidylethanolamine. Cultures of wild-type V. cholerae and the vca0863, and vca0863 pVcA0863 strains were grown in a 2 mM mix of soybean-derived LPC. Peaks with masses representing incorporation of C18:2 and C18:3 LPC-derived acyl chains were absent from the vca0863 mutant (masses shown in red). Download
Mass spectrometry of V. cholerae-derived cardiolipin. Cultures of wild-type V. cholerae and the vca0863 and vca0863 pVcA0863 strains were grown in a 2 mM mix of soybean-derived LPC. Peaks with masses representing incorporation of C18:2 and C18:3 LPC-derived acyl chains were absent from the vca0863 mutant (masses are shown in red). Download
FadL is required for use of LPC as a carbon source. Wild-type E. coli and a fadL deletion strain containing pVcA0863 were assayed for growth on 0.2% glucose, 2 mM LCFA, or 2 mM LPC after 24 h. As predicted, when FadL is lost regardless of expression of VcA0863, no growth could be observed on LFCA. Only E. coli expressing both FadL and VcA0863 showed growth above background levels on LPC. Download
Bacterial strains, plasmids and oligos used in this study.
ACKNOWLEDGMENTS
This work was supported by grants AI064184 and AI76322 from the National Institutes of Health (NIH) and by grant 61789-MA-MUR from the Army Research Office to M.S.T. The work was also supported by a LIPID MAPS large-scale collaborative grant GM069338 to Duke University Medical Center.
Footnotes
Citation Pride AC, Herrera CM, Guan Z, Giles DK, Trent MS. 2013. The outer surface lipoprotein VolA mediates utilization of exogenous lipids by Vibrio cholerae. mBio 4(3):e00305-13. doi:10.1128/mBio.00305-13.
REFERENCES
- 1. Campbell JW, Morgan-Kiss RM, Cronan JE., Jr. 2003. A new Escherichia coli metabolic competency: growth on fatty acids by a novel anaerobic beta-oxidation pathway. Mol. Microbiol. 47:793–805 [DOI] [PubMed] [Google Scholar]
- 2. Giles DK, Hankins JV, Guan Z, Trent MS. 2011. Remodelling of the Vibrio cholerae membrane by incorporation of exogenous fatty acids from host and aquatic environments. Mol. Microbiol. 79:716–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nunn WD, Simons RW. 1978. Transport of long-chain fatty acids by Escherichia coli: mapping and characterization of mutants in the fadL gene. Proc. Natl. Acad. Sci. U. S. A. 75:3377–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Overath P, Raufuss EM. 1967. The induction of the enzymes of fatty acid degradation in Escherichia coli. Biochem. Biophys. Res. Commun. 29:28–33 [DOI] [PubMed] [Google Scholar]
- 5. Esfahani M, Barnes EM, Jr, Wakil SJ. 1969. Control of fatty acid composition in phospholipids of Escherichia coli: response to fatty acid supplements in a fatty acid auxotroph. Proc. Natl. Acad. Sci. U. S. A. 64:1057–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lipp EK, Huq A, Colwell RR. 2002. Effects of global climate on infectious disease: the cholera model. Clin. Microbiol. Rev. 15:757–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kattner G, Hagen W. 2009. Lipids in marine copepods: latitudinal characteristics and perspective to global warming, p 257–280 In Arts M, Brett M, Kainz M, Lipids in aquatic ecosystems. Springer, Dordrecht, The Netherlands [Google Scholar]
- 8. Braun A, Treede I, Gotthardt D, Tietje A, Zahn A, Ruhwald R, Schoenfeld U, Welsch T, Kienle P, Erben G, Lehmann WD, Fuellekrug J, Stremmel W, Ehehalt R. 2009. Alterations of phospholipid concentration and species composition of the intestinal mucus barrier in ulcerative colitis: a clue to pathogenesis. Inflamm. Bowel Dis. 15:1705–1720 [DOI] [PubMed] [Google Scholar]
- 9. Ansell GB, Spanner S. 1982. Phosphatidylserine, phosphatidylethanolamine, and phosphatidylcholine, p 1–50 In Neuberger A, Van Deenen LLM, Phospholipids. Elsevier, Philadelphia, PA. [Google Scholar]
- 10. Chuang YC, Chiou SF, Su JH, Wu ML, Chang MC. 1997. Molecular analysis and expression of the extracellular lipase of Aeromonas hydrophila MCC-2. Microbiology 143:803–812 [DOI] [PubMed] [Google Scholar]
- 11. Okuda S, Tokuda H. 2011. Lipoprotein sorting in bacteria. Annu. Rev. Microbiol. 65:239–259 [DOI] [PubMed] [Google Scholar]
- 12. Munch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D. 2005. Virtual footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21:4187–4189 [DOI] [PubMed] [Google Scholar]
- 13. Kolb A, Busby S, Buc H, Garges S, Adhya S. 1993. Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 62:749–795 [DOI] [PubMed] [Google Scholar]
- 14. Zubay G, Schwartz D, Beckwith J. 1970. Mechanism of activation of catabolite-sensitive genes: a positive control system. Proc. Natl. Acad. Sci. U. S. A. 66:104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DiRusso CC, Metzger AK, Heimert TL. 1993. Regulation of transcription of genes required for fatty acid transport and unsaturated fatty acid biosynthesis in Escherichia coli by FadR. Mol. Microbiol. 7:311–322 [DOI] [PubMed] [Google Scholar]
- 16. Raziuddin SR. 1976. Effect of growth temperature and culture age on the lipid composition of Vibrio cholerae 569B (Inaba). J. Gen. Microbiol. 94:367–372 [DOI] [PubMed] [Google Scholar]
- 17. Cowles CE, Li Y, Semmelhack MF, Cristea IM, Silhavy TJ. 2011. The free and bound forms of Lpp occupy distinct subcellular locations in Escherichia coli. Mol. Microbiol. 79:1168–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boman HG, Eriksson-Grennberg KG, Foldes J, Lindstrom EB. 1967. The regulation and possible evolution of a penicillinase-like enzyme in Escherichia coli, p 366–372 In Koningsberger VV, Bosch L, Regulation of nucleic acid and protein biosynthesis, BBA Library, vol. 10 Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 19. Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huq A, Small EB, West PA, Huq MI, Rahman R, Colwell RR. 1983. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl. Environ. Microbiol. 45:275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huq A, Colwell RR, Rahman R, Ali A, Chowdhury MA, Parveen S, Sack DA, Russek-Cohen E. 1990. Detection of Vibrio cholerae O1 in the aquatic environment by fluorescent-monoclonal antibody and culture methods. Appl. Environ. Microbiol. 56:2370–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Broza M, Halpern M. 2001. Pathogen reservoirs. Chironomid egg masses and Vibrio cholerae. Nature 412:40 http://dx.doi.org/10.1038/35083694 [DOI] [PubMed] [Google Scholar]
- 23. Islam MS, Drasar BS, Sack RB. 1994. The aquatic flora and fauna as reservoirs of Vibrio cholerae: a review. J. Diarrhoeal Dis. Res. 12:87–96 [PubMed] [Google Scholar]
- 24. Abd H, Saeed A, Weintraub A, Nair GB, Sandström G. 2007. Vibrio cholerae O1 strains are facultative intracellular bacteria, able to survive and multiply symbiotically inside the aquatic free-living amoeba Acanthamoeba castellanii. FEMS Microbiol. Ecol. 60:33–39 [DOI] [PubMed] [Google Scholar]
- 25. Abd H, Weintraub A, Sandström G. 2005. Intracellular survival and replication of Vibrio cholerae O139 in aquatic free-living amoebae. Environ. Microbiol. 7:1003–1008 [DOI] [PubMed] [Google Scholar]
- 26. Marino A, Crisafi G, Maugeri TL, Nostro A, Alonzo V. 1999. Uptake and retention of Vibrio cholerae non-O1, Salmonella typhi, Escherichia coli and Vibrio harvey by mussels in seawater. New Microbiol. 22:129–138 [PubMed] [Google Scholar]
- 27. Senderovich Y, Izhaki I, Halpern M. 2010. Fish as reservoirs and vectors of Vibrio cholerae. PLoS One 5:e8607 http://dx.doi.org/10.1371/journal.pone.0008607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kariotoglou DM, Mastronicolis SK. 2003. Sphingophosphonolipid molecular species from edible mollusks and a jellyfish. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 136:27–44 [DOI] [PubMed] [Google Scholar]
- 29. Bhaskar N, Kinami T, Miyashita K, Park SB, Endo Y, Fujimoto K. 2004. Occurrence of conjugated polyenoic fatty acids in seaweeds from the Indian Ocean. Z. Naturforsch. C 59:310–314 [DOI] [PubMed] [Google Scholar]
- 30. Ohman MD. 1997. On the determination of zooplankton lipid content and the occurrence of gelatinous copepods. J. Plankton Res. 19:1235–1250 [Google Scholar]
- 31. Ghioni C, Bell JG, Sargent JR. 1996. Polyunsaturated fatty acids in neutral lipids and phospholipids of some freshwater insects. Comp. Biochem. Physiol. B 114:161–170 [Google Scholar]
- 32. Fiore AE, Michalski JM, Russell RG, Sears CL, Kaper JB. 1997. Cloning, characterization, and chromosomal mapping of a phospholipase (lecithinase) produced by Vibrio cholerae. Infect. Immun. 65:3112–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Froehlich F, Gonvers JJ, Fried M. 1995. Role of nutrient fat and cholecystokinin in regulation of gallbladder emptying in man. Dig. Dis. Sci. 40:529–533 [DOI] [PubMed] [Google Scholar]
- 34. Nishi E, Kume N, Ueno Y, Ochi H, Moriwaki H, Kita T. 1998. Lysophosphatidylcholine enhances cytokine-induced interferon gamma expression in human T lymphocytes. Circ. Res. 83:508–515 [DOI] [PubMed] [Google Scholar]
- 35. Stock C, Schilling T, Schwab A, Eder C. 2006. Lysophosphatidylcholine stimulates IL-1beta release from microglia via a P2X7 receptor-independent mechanism. J. Immunol. 177:8560–8568 [DOI] [PubMed] [Google Scholar]
- 36. Schilling T, Eder C. 2010. Importance of lipid rafts for lysophosphatidylcholine-induced caspase-1 activation and reactive oxygen species generation. Cell. Immunol. 265:87–90 [DOI] [PubMed] [Google Scholar]
- 37. Yan JJ, Jung JS, Lee JE, Lee J, Huh SO, Kim HS, Jung KC, Cho JY, Nam JS, Suh HW, Kim YH, Song DK. 2004. Therapeutic effects of lysophosphatidylcholine in experimental sepsis. Nat. Med. 10:161–167 [DOI] [PubMed] [Google Scholar]
- 38. Steel HC, Cockeran R, Anderson R. 2002. Platelet-activating factor and lyso-PAF possess direct antimicrobial properties in vitro. APMIS 110:158–164 [DOI] [PubMed] [Google Scholar]
- 39. Shimomura H, Hosoda K, Hayashi S, Yokota K, Hirai Y. 2012. Phosphatidylethanolamine of Helicobacter pylori functions as a steroid-binding lipid in the assimilation of free cholesterol and 3beta-hydroxl steroids into the bacterial cell membrane. J. Bacteriol. 194:2658–2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Griffin JE, Pandey AK, Gilmore SA, Mizrahi V, McKinney JD, Bertozzi CR, Sassetti CM. 2012. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem. Biol. 19:218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Banchio C, Gramajo HC. 1997. Medium- and long-chain fatty acid uptake and utilization by Streptomyces coelicolor A3(2): first characterization of a gram-positive bacterial system. Microbiology 143:2439–2447 [DOI] [PubMed] [Google Scholar]
- 42. Manning PA, Beutin L, Achtman M. 1980. Outer membrane of Escherichia coli: properties of the F sex factor traT protein which is involved in surface exclusion. J. Bacteriol. 142:285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Drummelsmith J, Whitfield C. 2000. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane. EMBO J. 19:57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. 2006. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 59:870–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pugsley AP, Kornacker MG. 1991. Secretion of the cell surface lipoprotein pullulanase in Escherichia coli. Cooperation or competition between the specific secretion pathway and the lipoprotein sorting pathway. J. Biol. Chem. 266:13640–13645 [PubMed] [Google Scholar]
- 46. Ruiz N, Kahne D, Silhavy TJ. 2006. Advances in understanding bacterial outer-membrane biogenesis. Nat. Rev. Microbiol. 4:57–66 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials and Methods. Download
Chemical structures and TLC of the major phospholipids of E. coli K-12. (A) Chemical structures of phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (CL). (B) E. coli was grown in minimal medium containing a 2 mM concentration of a soybean-derived lysophosphatidylcholine (LPC) mix including C18:2 and C18:3 acyl chains or the corresponding LCFA. Phospholipids were extracted and analyzed via TLC. No change in the migration pattern was observed with lipids from cells grown in glucose with and without LPC, confirming that E. coli cannot utilize LPC as a source of LCFA for membrane modification. Download
Growth of different Vibrio species and E. coli K-12 using glucose as the sole carbon source. Cultures of V. cholerae, V. alginolyticus, V. vulnificus, and V. parahaemolyticus were grown along with E. coli in minimal medium containing 0.2% glucose. V. fischeri and V. harveyi were grown in a high-salt defined minimal medium (see supplemental Materials and Methods) with 0.2% glucose. Download
Growth of E. coli and V. cholerae on LPC or LCFA as measured by colony counting. Quantification of bacterial growth was determined by plating assays. Viable colony-forming unit counts show that E. coli fails to utilize LPC as the sole source of carbon. Download
Genomic organization of FadL and VolA (VcA0863) homologues in various Gram-negative species. All Vibrio species possess homologues of vc1042, vc1043, and vca0862 (encoding FadLs), and with the exception of V. fischeri, all also had a homolog of vca0863. E. coli has only a single homolog of FadL and no homolog of vcA0863. Download
qRT-PCR of vca0862 and vca0863. qRT-PCR data of the expression of vca0862 and vca0863 grown with succinate and/or LPC are given. Growth in LPC displayed an upregulation of both vca0862 and vca0863 compared to results for a succinate-grown control. Download
Mass spectrometry of V. cholerae-derived phosphatidylethanolamine. Cultures of wild-type V. cholerae and the vca0863, and vca0863 pVcA0863 strains were grown in a 2 mM mix of soybean-derived LPC. Peaks with masses representing incorporation of C18:2 and C18:3 LPC-derived acyl chains were absent from the vca0863 mutant (masses shown in red). Download
Mass spectrometry of V. cholerae-derived cardiolipin. Cultures of wild-type V. cholerae and the vca0863 and vca0863 pVcA0863 strains were grown in a 2 mM mix of soybean-derived LPC. Peaks with masses representing incorporation of C18:2 and C18:3 LPC-derived acyl chains were absent from the vca0863 mutant (masses are shown in red). Download
FadL is required for use of LPC as a carbon source. Wild-type E. coli and a fadL deletion strain containing pVcA0863 were assayed for growth on 0.2% glucose, 2 mM LCFA, or 2 mM LPC after 24 h. As predicted, when FadL is lost regardless of expression of VcA0863, no growth could be observed on LFCA. Only E. coli expressing both FadL and VcA0863 showed growth above background levels on LPC. Download
Bacterial strains, plasmids and oligos used in this study.