ABSTRACT
The cell membrane of a Giardia lamblia trophozoite is covered with a single species of variant-specific surface protein (VSP) that is replaced by another VSP every 6 to 13 generations of cell growth, possibly for an evasion of host immunity. Experimentally, only six VSP species have been verified to localize to the cell membrane thus far. By assuming that VSP contains multiple CXXC motifs, 219 vsp genes were annotated in GiardiaDB of the WB isolate. By further assuming that VSP possesses both CXXC motifs and a CRGKA tail at the C terminus, Adam et al. (BMC Genomics 11:424, 2010) identified a total of 303 potential vsp genes in Giardia WB. The discrepancies between these two assumed VSP identities have caused some confusion. Here, we used experimental approaches to further verify what is required of the structures of a VSP to localize to the surface of cell membrane. The data led to the following conclusions. (i) The C-terminal CRGKA sequence is not essential for localizing VSPs to the cell membrane. (ii) A “motif 1” of 45 residues, consisting of two CXXCs separated by 12 to 15 amino acid residues, located close to the C terminus and a hydrophobic “motif 2” of 38 residues at the C terminus are both essential and sufficient for localizing the protein to the cell membrane. (ii) An N-terminal sequence upstream from motif 1 is not required for targeting VSPs to the cell membrane. By these criteria, we are able to identify 73 open reading frames as the putative vsp genes in Giardia.
IMPORTANCE
The intestinal pathogen Giardia lamblia expresses only one variant-specific surface protein (VSP) on the cell membrane surface at a given time, but it changes spontaneously every 6 to 13 generations of growth, presumably for evading the host immunity. Only 6 VSPs have been empirically shown to localize to the cell membrane surface thus far. Here, we used mutations of VSPs and methods of identifying their locations in Giardia cells and found that a “motif 1” of 45 residues, consisting of two CXXCs separated by 12 to 15 amino acid residues, located close to the C terminus and a hydrophobic “motif 2” of 38 residues at the C terminus are the only essential and sufficient structural requirements for localizing a protein to the cell membrane. By these criteria, 73 genes are identified in the Giardia WB strain genome database as the putative repertoire of VSPs.
Introduction
Giardia lamblia is a deeply branched zoonotic intestinal protozoan pathogen. It infects humans, causing the diarrheal disease giardiasis, throughout the world (1, 2). Prolonged infections in humans often cause malabsorption of nutrients with weight loss in the absence of treatment, despite an immune response that would be expected to control the infection (3). Both innate and adaptive immune responses are apparently involved in giardiasis (4, 5). In a likely defense to the host immunity, antigenic variation, a mechanism known for many pathogens to evade the humoral immune response of their vertebrate hosts (5), was shown to occur on the membrane surface of Giardia trophozoites both in vitro and in vivo (6, 7). Individual Giardia trophozoites express only a single species of variant-specific surface protein (VSP) on the surface of the cell membrane at any given time (8) and subjects to replacement by another VSP every 6 to 13 generations of cell growth (9).
The first VSP was identified in the trophozoites of a Giardia WB isolate from a partial mRNA sequence encoding a 170-kDa protein (initially named CRP170 but later designated VSPA6), which was recognized on the cell membrane surface by a 6E7 monoclonal antibody (mAb) (6). Subsequently, the first complete sequence of another VSP (TSA417) was identified in the WB isolate (10), and a third VSP, VSPH7, was found in the GS isolate (11). Both TSA417 and VSPH7 were found localized to the trophozoite membrane surface by immunofluorescence assays using their respective antibodies, anti-rTSA417 and G10/4 (10, 11). A 17- and a 14-amino-acid N-terminal peptide were found missing from TSA417 and VSPH7, respectively, when they were expressed on the membrane surface, suggesting that the peptides could function as targeting signals during their translocalization and were removed during the process, though direct evidence for this conclusion is still missing (10, 11). Three additional VSPs, VSPA6-S1(G3M), VSP1267, and VSP9B10A, have since been recognized by their specific monoclonal antibodies, 6E7, 5C1, and 9B10, respectively, on the trophozoite membrane surface of the WB isolate by immunofluorescence assays (8, 12, 13), which expanded the total number of experimentally verified VSPs in Giardia trophozoites to 6. Surface labeling and electron microscopic analysis demonstrated that the VSPs are the major surface proteins of individual Giardia trophozoites and form a dense coat on the membrane surface (10, 14).
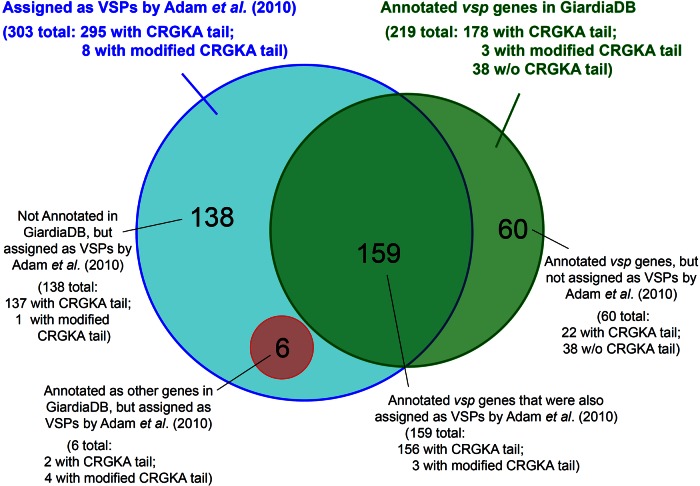
At a close examination of the sequences of the 6 experimentally verified VSPs, one common feature is that they were all rich in cysteine content (12%) and that most of the cysteines are present in CXXC motifs (1, 2). The N-terminal portions of the 6 VSPs present a high degree of sequence variations, whereas the 38 C-terminal residues of the 6 VSPs are 90% conserved. Each of the 6 VSPs carries the CRGKA motif at the C terminus (2). By these observations, the VSPs of Giardia were assumed to have the common features of carrying multiple CXXC motifs and a CRGKA motif at the C terminus (3, 15). Morrison et al. searched for the open reading frames (ORF) that contained multiple CXXC motifs in the GiardiaDB of the WB isolate and were able to predict and annotate a total of 219 potential vsp genes (Fig. 1, green circle) (1). Recently, Adam et al. performed another search of ORFs in the GiardiaDB that have at least two CXXC motifs and at least 3 of the 5 residues in the C-terminal CRGKA motif (3) and resulted in identifying 303 potential vsp genes, with only 159 of them overlapping with those previously annotated by Morrison et al. (Fig. 1) (1).
FIG 1 .
The distribution of the combined 363 putative vsp genes originally annotated in GiardiaDB and subsequently assigned by Adam et al. (3).
We took a careful examination of this seemingly confusing situation and found that 60 of the originally annotated vsp genes by Morrison et al. (1) were not included by Adam et al. (3), even though 22 of them do have the C-terminal CRGKA motif (Fig. 1). We assigned the 60 vsp genes excluded by Adam et al., vsp-292 to vsp-352, to facilitate their further analysis and published this assignment in a previous publication (16). There were also 138 vsp genes identified by Adam et al. (3) that were somehow not found by the previous GiardiaDB annotation even though they all possess the multiple CXXC motifs.
These unexplained contradictions have failed to present a clear profile of the potential VSP repertoire in Giardia. Since the two criteria used in identifying the VSPs, the CXXC motifs and the C-terminal CRGKA motif, were derived from the sequences of only 6 experimentally proven VSPs, these criteria may not be sufficient or essential or even constituting the real structural requirements for a VSP to localize on the membrane surface of Giardia. More experimental data will be needed to better define the essential structural aspects of a VSP.
In the present investigation, we employed the methods of Western blot and immunofluorescence assays of permeabilized and nonpermeabilized Giardia trophozoites expressing various mutants of putative VSPs. We were able to verify that (i) the C-terminal CRGKA motif is unnecessary for localizing a VSP to the cell membrane, (ii) a motif 1 consisting of two CXXCs separated by 12 to 15 amino acid residues and localizing at a specified position near the C terminus is essential, (iii) a hydrophobic motif 2 at the C terminus is necessary, and (iv) an N-terminal sequence upstream from motif 1 is not required for targeting the protein to the cell membrane. By these newly established criteria, the total number of putative VSPs in Giardia trophozoites is now refined to 73, which are presented in the report.
RESULTS
Monitoring VSP expression and localization with Western blot and immunofluorescence assays.
In our previous study (16), crude lysates of Giardia trophozoites were separated, using a specific kit (see Materials and Methods), into organic phase and aqueous phase to roughly classify the proteins into the membrane fraction and the cytoplasmic or nuclear fraction for comparisons on a Western blot. The same population of trophozoites was also stained with an antibody in an immunofluorescence assay to show the expression of a specific protein. Two different methods of fixing the cells were used to make the latter either nonpermeable to the antibody, which is capable of staining only the cell membrane antigens, or totally permeable to it, resulting in staining essentially all the antigens associated with the cell (17). Thus, a comparison between the outcomes from immunofluorescence assays of nonpermeable and permeable cells would demonstrate whether the antigen localizes to the cell membrane surface. Transfected Giardia WB trophozoites expressing N-terminal hemagglutinin (HA)-tagged histone H2A were tested with the two methods using an anti-HA antibody. The result from the Western blot assay showed that HA-histone H2A was primarily in the aqueous phase (see Fig. S1A in the supplemental material), whereas immunofluorescence assays showed no detectable anti-HA stain of the nonpermeable cells but specific staining of the nuclei in the permeable cells (see Fig. S1B). Giardia WB trophozoites expressing a previously experimentally verified VSP9B10A were also examined in both Western and immunofluorescence assays using mAb 9B10 kindly provided to us by Theodore Nash of NIH. The outcome indicated that VSP9B10A was primarily in the organic phase (see Fig. S1C). By immunofluorescence, it was detectable both on the membrane surface and in some internal portions of permeable cells but only on the membrane surface of nonpermeable cells (see Fig. S1D), which agrees with the previous result using the same monoclonal antibody and a somewhat different procedure (13). These experimental data thus verified the usefulness and validity of the two experimental approaches used by us in monitoring VSP expression on the trophozoite membrane surface of Giardia.
The C-terminal hallmark CRGKA is nonessential for localizing VSP to the cell membrane surface.
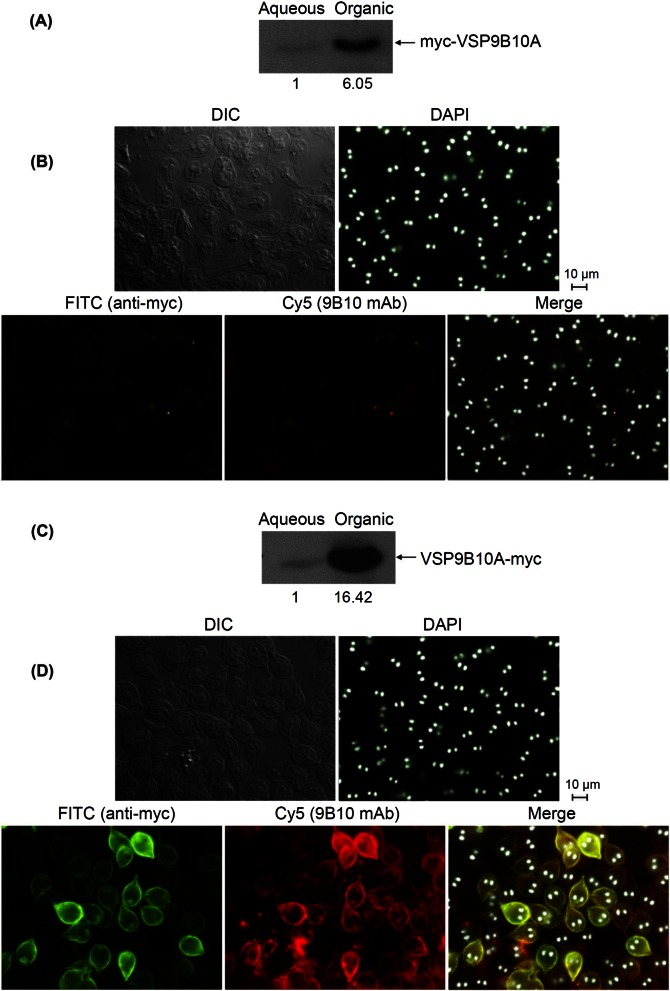
In our planning of examining the expression of a large number of potential VSPs of various structures, the limited number of monoclonal antibodies currently available to us was inadequate for the purpose. We thus tagged a previously experimentally verified VSP, VSP9B10A, with a 3× myc antigen at either the N terminus or the C terminus of the protein (see Materials and Methods) and followed its expression in transfected cells with an anti-myc antibody and the monoclonal antibody 9B10 to see if the tagging would affect the translocalization of the VSP to the membrane surface. The results indicated that myc-VSP9B10A is primarily expressed in the organic phase of the transfected cells (Fig. 2A) but not detectable with either anti-myc antibody or mAb 9B10 on the membrane surface of nonpermeable cells (Fig. 2B). The N-terminal-tagged VSP is thus apparently incapable of translocalizing to the cell membrane surface. When the expression of VSP9B10A-myc was examined, it was also identified largely in the organic phase of transfected cells. But both anti-myc antibody and mAb 9B10 could readily show it on the membrane surface of nonpermeable cells with the same apparent pattern of localization (Fig. 2D). VSP9B10A-myc is thus apparently translocalized to the membrane surface like VSP9B10A, suggesting that the addition of a 42-amino-acid 3× myc tag at the C terminus of a VSP does not affect the translocalization of the VSP. It suggests also that CRGKA does not have to be at the C terminus to enable a VSP to localize to the membrane surface. All the subsequent studies had thus the proteins tagged at their C terminus with 3× myc for studies of localizations.
FIG 2 .
Expression of 3× myc-tagged VSPs on the membrane surface of Giardia trophozoites. (A) Detection of the expression of N-myc-tagged VSP9B10A on a Western blot using myc-Ab. Most of the tagged VSP was present in the organic phase. (B) Immunofluorescence assay of the nonpermeabilized cells with double staining of N-myc-VSP9B10A using myc-Ab (FITC) and mAb 9B10 (Cy5). No cell surface expression of VSP was detected. (C) Detection of the expression of C-myc-tagged VSP9B10A on a Western blot using myc-Ab. Most of the myc-VSP9B10A was present in the organic phase, too. (D) Immunofluorescence assay of the nonpermeabilized cells with double staining of C-myc-VSP9B10A using myc-Ab (FITC) and mAb 9B10 (Cy5). The cell surface was stained by both antibodies and demonstrated the same pattern of staining.
Since only 6 out of the combined total of 363 designated VSPs have been experimentally identified on the trophozoite membrane surface of Giardia thus far, the possibility exists that many of the VSPs designated by the two previously assumed criteria could turn out not to be VSP at all. In order to avoid further confusion, we removed in our present study the prefix “VSP” from all the putative VSPs not yet experimentally verified. Only the number in each case was saved for identification, until it became experimentally verified as a genuine cell membrane protein. At that point, the prefix “VSP” was added back.
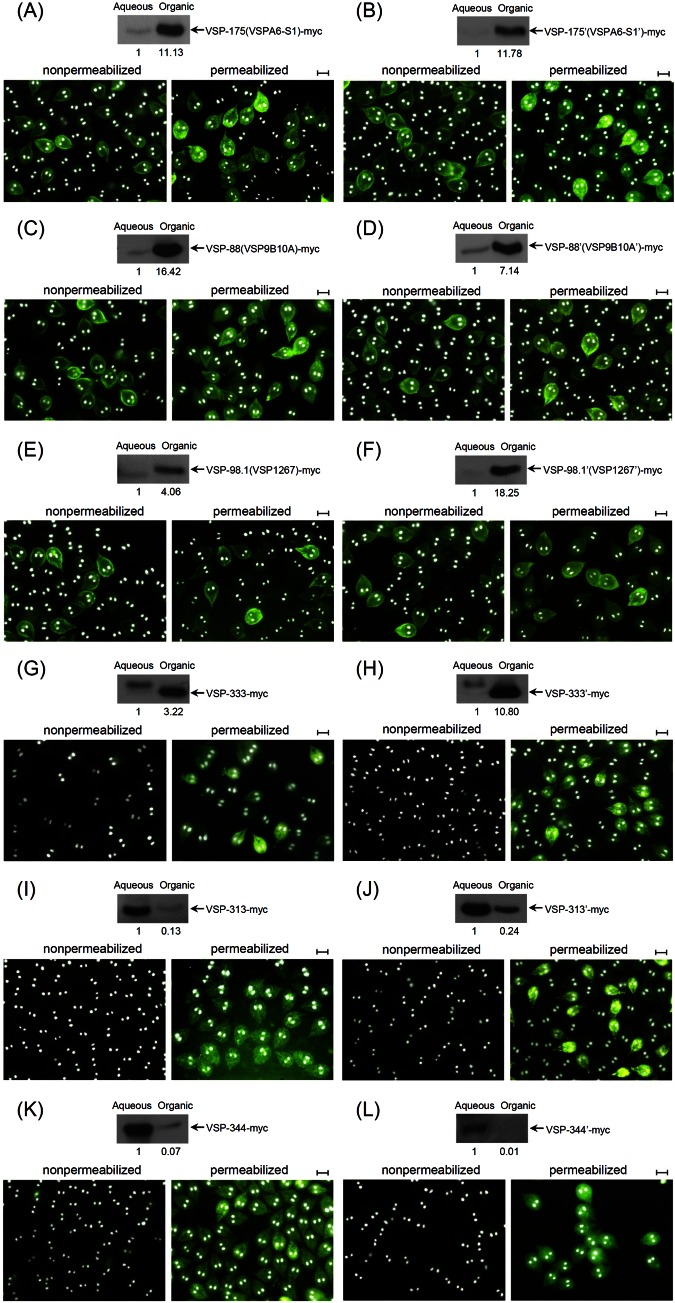
In order to verify further whether the C-terminal hallmark CRGKA is essential for translocalizing a VSP to the trophozoite membrane surface, this common C-terminal pentapeptide in the three experimentally verified VSPs, VSP-175(VSPA6-S1), VSP-88(VSP9B10A), and VSP-98.1(VSP1267), was mutated to CRRET, QSSKE, and WPGSL, respectively, to generate 3 mutants: 175′(A6-S1′), 88′(9B10A′), and 98.1′(1267′). CRRET is the C-terminal pentapeptide of protein 333, QSSKE is that of 344, and WPGSL is that of 313. To verify if these three modified VSPs with changed C-terminal pentapeptides can still localize to the trophozoite membrane surface, their localizations were monitored. Localizations of the three previously assigned VSPs, 333, 344, and 313, were also examined. The C-terminal pentapeptide in each of the three proteins was then replaced with CRGKA, and these mutant proteins were tentatively designated 333′, 344′, and 313′, respectively.
The 12 recombinant proteins, each tagged with a 3× myc at their C termini, were expressed in transfected Giardia trophozoites and monitored for their localizations. The results presented in Fig. 3 indicate that VSP-175(VSPA6-S1)-myc (Fig. 3A), VSP-88(VSP9B10A)-myc (Fig. 3C), and VSP-98.1(VSP1267)-myc (Fig. 3E) are all expressed on the membrane surface as anticipated (8, 12, 13). However, 175′(A6-S1′)-myc (Fig. 3B), 88′(9B10A′)-myc (Fig. 3D), and 98.1′(1267′)-myc (Fig. 3F), each carrying an altered C-terminal pentapeptide, are also localized to the trophozoite membrane surface, suggesting that C-terminal CRGKA is not essential for membrane localization of VSP. These three mutants can be thus also assigned as VSPs. Data in Fig. 3 show that the three putatively assigned VSPs, 333-myc (Fig. 3G), 344-myc (Fig. 3I), and 313-myc (Fig. 3K), which do not possess C-terminal CRGKA, do not locate to the membrane surface and thus should not be classified as VSPs. When their C-terminal pentapeptides were converted to CRGKA, however, none of the three mutants, 333′-myc (Fig. 3H), 344′-myc (Fig. 3J), and 313′-myc (Fig. 3L), were translocalized to the membrane surface either. The C-terminal CRGKA is thus not required for a VSP to localize to the trophozoite membrane surface.
FIG 3 .
C-terminal CRGKA is not required for VSP expression on the Giardia trophozoite membrane surface. The three C-terminal myc-tagged VSPs and their three mutants, which have the C-terminal CRGKA replaced with CRRET, QSSKE, and WPGSL, were each expressed in transfected Giardia trophozoites: VSP-175(VSPA6-S1)-myc (A), VSP-175′(VSPA6-S1′)-myc (B), VSP-88(VSP9B10A)-myc (C), VSP-88′(VSP9B10A′)-myc (D), VSP-98.1(VSP1267)-myc (E), and VSP-98.1′(VSP1267′)-myc (F). Three other C-terminal myc-tagged proteins, previously designated VSPs, 333, 344, and 313, were also expressed in transfected Giardia trophozoites. Their respective C-terminal CRRET, QSSKE, and WPGSL were each replaced with CRGKA, designated 333′, 344′, and 313′, and expressed: 333-myc (G), 333′-myc (H), 344-myc (I), 344′-myc (J), 313-myc (K), and 313′-myc (L). Extracts from the 12 transfectants were analyzed by a Western blot stained with anti-myc-Ab. All 3 previously verified VSPs and their C-terminal pentapeptide mutants were present primarily in the organic phase, whereas protein 333 and its mutant were also largely in the organic phase, but 313 and 344 and their mutants were largely in the aqueous phase. Immunofluorescence assays of the 12 transfectants in both nonpermeabilized and permeabilized forms showed that all 3 VSPs and their mutants were expressed on the cell membrane surface, whereas proteins 333, 313, and 344 and their mutants were all confined in the cytoplasm. C-terminal CRGKA thus does not dictate membrane expression of proteins. Bars = 10 µm.
Data from the Western analysis indicate that for all the proteins that localize to the membrane surface, they are also extracted primarily in the organic phase (Fig. 3A to F). For those non-VSPs that are not on the membrane surface, they are mostly present in the aqueous phase, except for 333-myc and its mutant 333′-myc, which are largely in the organic phase (Fig. 3G to L). Thus, though the VSPs are hydrophobic in nature, hydrophobicity of a protein cannot be regarded as a sufficient feature of a VSP for cell membrane localization.
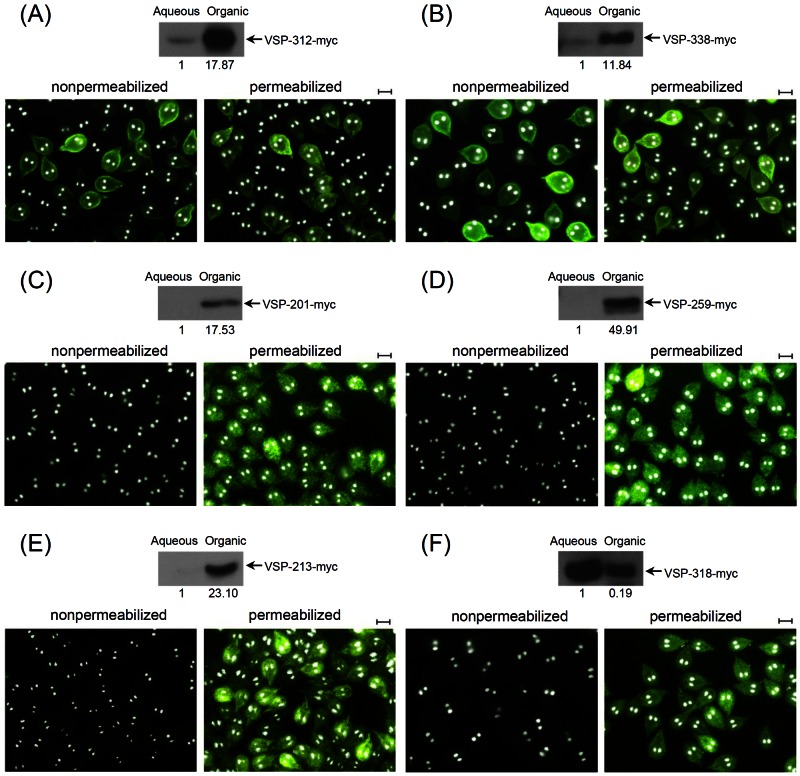
Six additional putative VSPs were randomly chosen for further scrutiny. Among them, 201, 213, and 259 possess the C-terminal CRGKA, whereas 312, 318, and 338 do not. The results show that 312-myc (Fig. 4A) and 338-myc (Fig. 4B), both without a C-terminal CRGKA, localize to the trophozoite membrane surface and thus qualify as VSPs. The other four proteins, 201-myc (Fig. 4C), 259-myc (Fig. 4D), 213-myc (Fig. 4E), and 318-myc (Fig. 4F), are distributed only to the cytoplasm regardless of whether they possess a CRGKA motif or not and thus cannot be classified as VSPs. These results indicate, once again, a lack of correlation between the presence of a C-terminal CRGKA motif and the membrane localization of a protein.
FIG 4 .
Additional evidence that C-terminal CRGKA does not localize a protein to the membrane surface of Giardia trophozoite. Six more previously designated VSPs were randomly chosen for examinations, as described for Fig. 3. 312-myc (A), 338-myc (B), 201-myc (C), 259-myc (D), 213-myc (E), and 318-myc (F). Proteins 201, 259, and 213 carry C-terminal CRGKA, whereas 318, 312, and 338 do not. The results showed that only proteins 312-myc and 338-myc are localized to the membrane surface. Neither protein has a C-terminal CRGKA. Bars = 10 µm.
Conserved motif 1 and motif 2 may be involved in cell membrane localization of VSPs.
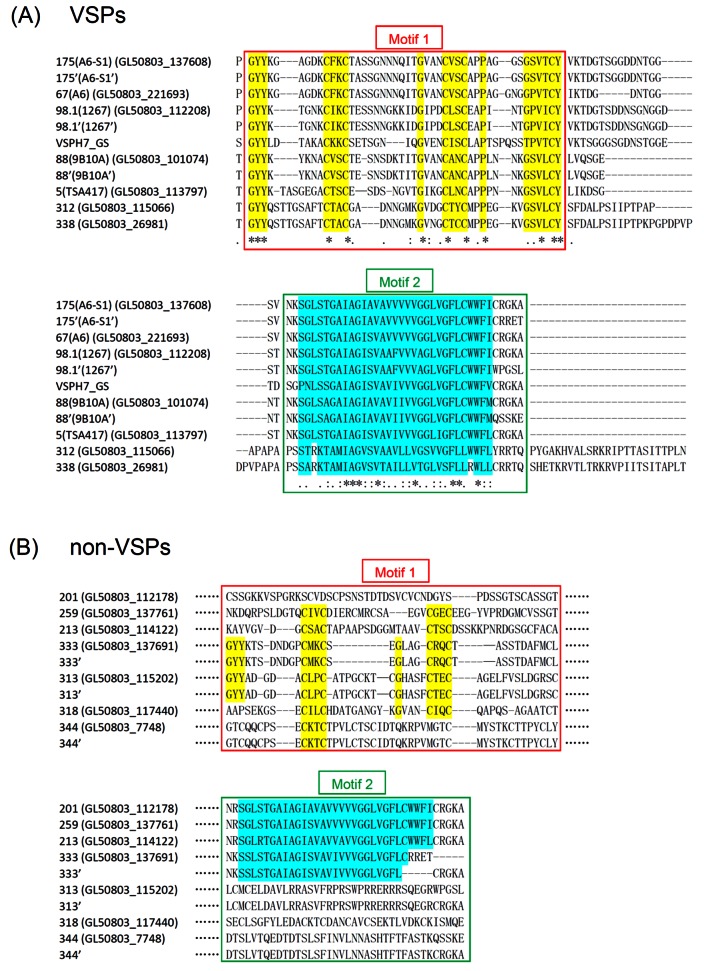
Sequences of the 11 experimentally verified VSPs, 6 from the previous studies (8, 10–13) and 5 from the current experiments, were aligned for potential common features. The results showed high sequence divergence in the N-terminal portion of these proteins (data not shown). However, among the C-terminal 120-amino-acid residues, there is high sequence homology, which could be further divided into two motifs: motif 1 and motif 2 (Fig. 5A). Motif 1 is 45 residues long and about 17 residues upstream from the 38-residue motif 2 located at the C terminus (Fig. 5A). Motif 1 starts with a conserved GYY and ends with a conserved (G/T) (S/P) VXCY with two CXXC moieties separated by 12 to 15 amino acid residues located in the middle of motif 1. Motif 2 consists of a conserved 38 residues, including a stretch of about 25 highly conserved hydrophobic residues, which could be the transmembrane domain of VSP. There is no common sequence among the C-terminal pentapeptides of the 11 VSPs. The two newly identified VSP-312 and VSP-338 both have 25 additional residues to the C terminus of motif 2 and contain no CRGKA sequence. This explains why the membrane localization of a VSP is unaffected by tagging the C terminus with a 3× myc peptide.
FIG 5 .
Motifs 1 and 2 may be involved in localizing VSP to Giardia trophozoite membrane. (A) Sequence alignment of the C-terminal portions of all the 11 VSP proteins shown to localize to the cell membrane surface in Fig. 3 and 4. (B) Sequence alignment of the C-terminal portions of all the 10 non-VSP proteins shown to confine to the cytoplasm in Fig. 3 and 4. Motif 1 is a stretch of 45 amino acids, whereas motif 2 is at the C terminus with a length of 38 residues. The conserved regions in motifs 1 and 2 were highlighted. None of the non-VSPs has the conserved regions in motif 1, but the non-VSPs 201, 259, 213, and 333 have the conserved motif 2.
When the two motifs are outlined in the 10 experimentally proven non-VSPs and compared with those in the VSPs (Fig. 5B), the extent of sequence identity in motif 1 is very low. Only proteins 333 and 313 have the conserved GYY and the CXXC doublet, but motif 2 in 333 is missing part of conserved hydrophobic residues and motif 2 in 313 has poor sequence similarity to that in the VSPs. Proteins 201, 259, and 213 have motif 2s matching that in the VSPs very well, but their motif 1s show little similarity to that in the verified VSPs, suggesting that the presence of motif 2 alone cannot turn a protein into a VSP. Since none of the 10 non-VSPs has a motif 1 with high sequence similarity to that in the verified VSPs, it is not known if motif 1 alone can satisfy the qualifications of a VSP.
Both motif 1 and motif 2 are necessary for VSP membrane surface localization.
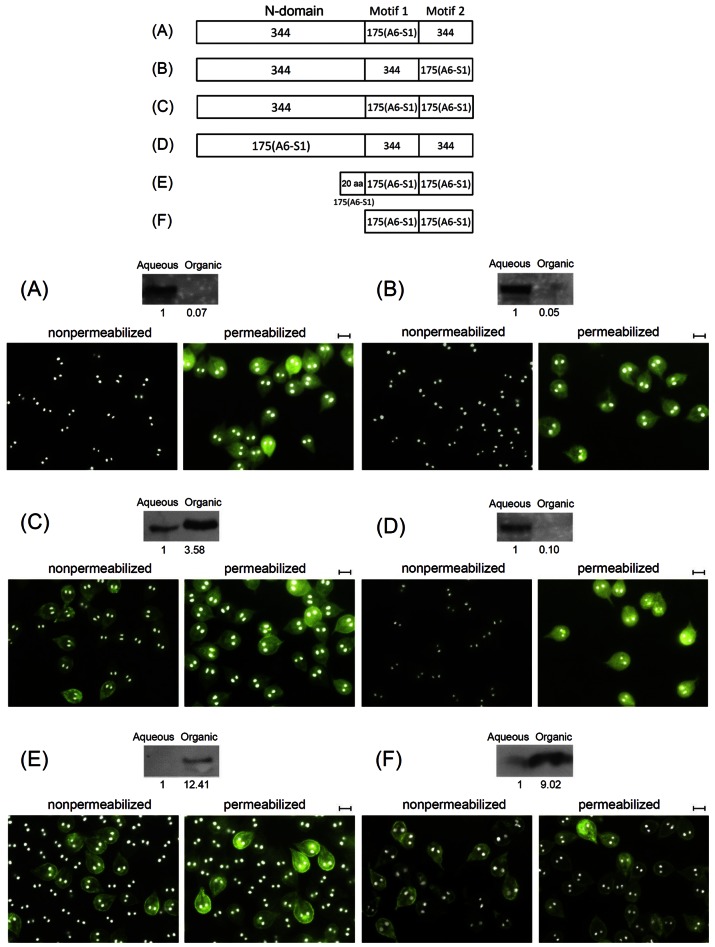
In order to verify if motif 1, motif 2, and the rest of the protein sequence plays an essential role in expressing a VSP on the membrane surface of Giardia trophozoite, we divided a bona fide VSP-175(VSPA6-S1) into the N domain, motif 1, and motif 2 (Fig. 6). We then divided the non-VSP-344, which bears little sequence similarity with VSP-175(VSPA6-S1) (Fig. 5), into the three corresponding domains (Fig. 6). These domains were then exchanged between the two proteins to generate chimera proteins tagged with 3× myc at the C termini and expressed in transfected Giardia trophozoites for their localizations. The results showed that when either the corresponding motif 1 or motif 2 in non-VSP-344 is replaced by that in VSP-175(VSPA6-S1), the chimera protein remains in the cytoplasm (Fig. 6A and B). They suggest that the presence of either motif 1 or motif 2 alone is inadequate in localizing the protein to the membrane surface. But when both motifs in non-VSP-344 are replaced with those from the VSP, the chimera protein becomes expressed on the membrane surface regardless of the presence of the N domain from non-VSP-344 (Fig. 6C). This evidence provides a strong indication that motif 1 and motif 2 are indeed the only essential structural elements in a VSP for its membrane localization. To further examine whether the N domain in a VSP also plays some role in its localization to the membrane surface, motif 1 and motif 2 in VSP-175(VSPA6-S1) were replaced with those from non-VSP-344 (Fig. 6D), while the original N domain remained unchanged. The resulting chimera protein was found staying in the cytoplasm (Fig. 6D), suggesting that the N domain alone in a VSP does not have the capability of localizing it to the cell membrane without the presence of motifs 1 and 2. To further address this hypothesis, two N-domain-truncated mutants of VSP-175(VSPA6-S1) tagged with 3× myc at C termini were generated. When the N domain was truncated, leaving only a 20-amino-acid sequence upstream from motif 1, to resemble a potential targeting signal (Fig. 6E), the mutant was expressed on the cell membrane surface (Fig. 6E). When the N domain was completely deleted, leaving only the motif 1-link-motif 2 portion of VSP-175, the mutant was also expressed on the membrane surface (Fig. 6F). These observations provide a clear indication that motif 1 and motif 2 are the only essential as well as sufficient structural requirement for localizing a VSP to the membrane surface.
FIG 6 .
Motif 1 and 2 are essential and sufficient in localizing VSP to the cell membrane surface. VSP-175(VSPA6-S1) and non-VSP-344 were each divided into N domain, motif 1, and motif 2. Chimera proteins generated from exchanging the three domains between the two proteins were expressed in transfected Giardia trophozoites. (A) Motif 1 in non-VSP-344 was replaced by motif 1 from VSP-175(VSPA6-S1); (B) motif 2 in non-VSP-344 was replaced by motif 2 from VSP-175(VSPA6-S1); (C) both motif 1 and motif 2 in non-VSP-344 were replaced by the corresponding motifs from VSP-175(VSPA6-S1); (D) both motifs in VSP-175(VSPA6-S1) were replaced by the two corresponding motifs from non-VSP-344. Results from immunofluorescence assays showed that only the chimera between the N domain from non-VSP-344 and the two motifs from VSP-175(VSPA6-S1) localizes to the membrane surface (see panel C). This is also the only chimera that was primarily distributed to the organic phase in Western blot analysis. (E) The N domain of VSP-175(VSPA6-S1) was truncated to a 20-amino-acid stretch upstream from motif 1. The truncated mutant was expressed on the membrane surface. (F) The entire N domain of VSP-175(VSPA6-S1) was removed to leave only motif 1 and motif 2. The mutant protein was expressed on the cell membrane. Bars = 10 µm.
The N terminus of VSP cannot be blocked without affecting its cell membrane localization.
Although the N domain of VSP-175(VSPA6-S1) does not participate in localizing the latter to the membrane surface (Fig. 6D), our previous data in Fig. 2A and B indicated that tagging the N terminus of VSP9B10A with 3× myc resulted in a failure of its expression on the membrane surface. Other previous observations that an N-terminal ~14- to 17-amino-acid sequence was missing from TSA417 and VSPH7 when they were isolated from cell membranes suggested that the N-terminal sequence could function as a targeting signal for translocalization and become removed after completion of the process (10, 11). In view of the highly varied N-terminal sequences among the verified VSPs and the localization of motif 1-link-motif 2 of VSP175 to the membrane surface without an N domain (Fig. 6F), there is the possibility that a specific N-terminal targeting signal may not be needed for VSP translocalization. To clarify this possibility further, VSP-175(VSPA6-S1), VSP-98.1(VSP1267), VSP-312, and VSP-338 were each tagged with 3× myc at the N termini, expressed, and monitored for their localizations. The results showed that, like myc-VSP9B10A (Fig. 2A and B), they all failed to be expressed on the membrane surface (see Fig. S2 in the supplemental material). A plausible explanation for it would be that there is no targeting signal at the N terminus of VSP, while the translocalization of VSP is blocked by tagging the N terminus of VSP with 3× myc.
There are 73 vsp genes in the G. lamblia WB isolate.
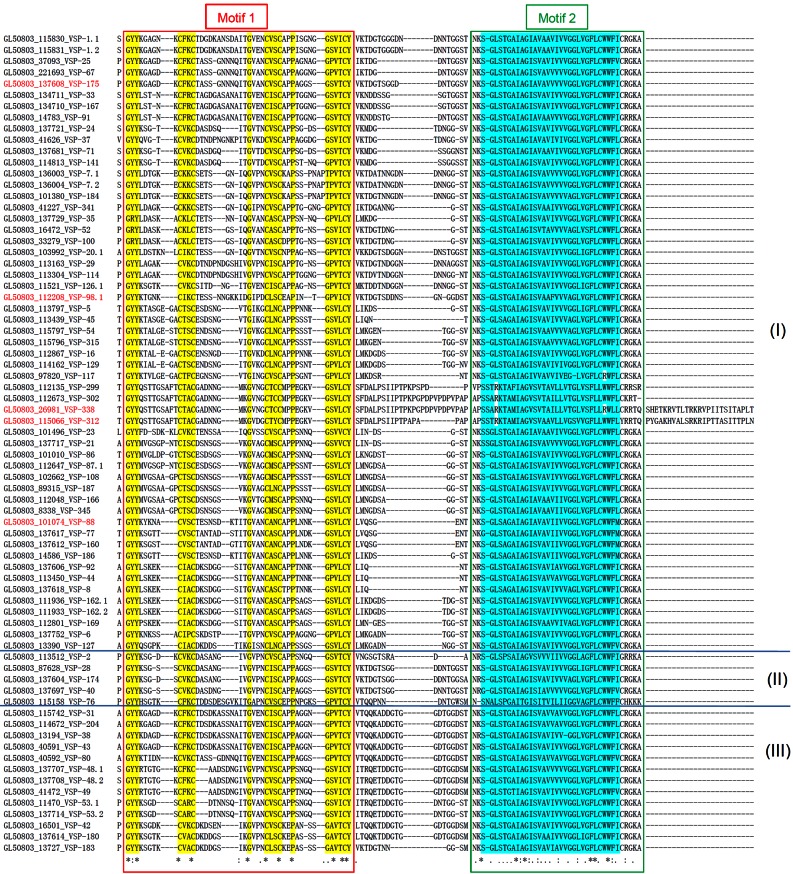
With the establishment of motif 1 and motif 2 as the two essential and sufficient structural elements in a VSP molecule for translocalization, we used them to screen the GiardiaDB and were able to identify a total of 73 vsp genes in the G. lamblia WB isolate. The 73 VSPs all carry motif 1 and motif 2 virtually identical to those presented among the 11 experimentally verified VSPs in Fig. 4A and are thus most likely the bona fide VSPs in Giardia. An examination of the sequences in the N domains of the 73 proteins indicated vast diversities, suggesting that they are 73 distinctive proteins without a common N-terminal targeting signal. The molecular masses of the 73 VSPs range from 228.40 kDa for VSP-67 to 10.49 kDa for VSP-117, which consists of primarily only motifs 1 and 2 (see Table S1 in the supplemental material). Among the 73 VSPs, only 5 do not have the C-terminal CRGKA and 3 have modified CRGKA. The amino acid sequences from the beginning of motif 1 to the C-terminal end of motif 2 were aligned among the 73 VSPs (Fig. 7). A dendrogram of the 73 sequences was drawn according to the pairwise similarity of the sequences using Clustal W (see Fig. S3 in the supplemental material). Based on the dendrogram, the 73 VSP protein sequences could fall into three subgroups I, II, and III (Fig. 7; see also Fig. S3). Most of the VSPs are classified to subgroup I, which includes all the VSPs experimentally verified thus far. Although those in subgroups II and III remain to be experimentally verified, chances are that they are also bona fide VSPs due to the extreme sequence similarities among all 73 VSPs (Fig. 7). These 73 VSPs may thus represent the core VSPs in Giardia. Further studies will be required to address whether minor structural modifications of motifs 1 and 2 will be possible without affecting the membrane localization of a VSP.
FIG 7 .
Sequence alignment of the C-terminal portions among the 73 newly identified VSPs in Giardia. Motif 1 is shown in the red box. Motif 2 is shown in the green box. The conserved sequences were highlighted in both motifs. The gene identifier from GiardiaDB and VSP designations for each VSP are listed. The VSPs experimentally tested and confirmed in this study are in red. The three subgroups labeled I, II, and III are derived from a distance tree analysis among the 73 C-terminal sequences using Clustal W (see Fig. S3 in the supplemental material).
DISCUSSION
In the present investigation, we demonstrated experimentally that two motifs, motif 1 and 2, are the essential as well as the sufficient structural elements in a Giardia VSP for its translocalization to the cell membrane. The two CXXCs within motif 1, separated by 12 to 15 amino acid residues, represent the typical structure of a zinc finger, which is known to bind to Zn2+ and other divalent metal ions (18). Membrane proteins of Giardia trophozoites have been previously found capable of binding to Zn2+ (19, 20), which could be attributed to the presence of zinc fingers in motif 1 of VSP. It is also known that Giardia can be cultivated axenically only in the presence of reducing agents such as l-cysteine (21), which is essential for survival, growth, attachment, and motility of the trophozoites (22, 23). When the trophozoites were metabolically labeled with radioactive l-cysteine, most of the label was incorporated into the VSP (24, 25), suggesting that the zinc finger in motif 1 of VSP may be derived from the l-cysteine in the living environment and is essential for parasite viability (26). Since no free thiols could be detected on the surface of Giardia WB clone C6 trophozoites expressing VSP-TSA417 and the l-cysteines were either chelated with metal ions or in disulfide bonds (27), it is likely that the zinc fingers in VSPs need to chelate with metal ions to perform the vital function for the trophozoites. Though the precise function of zinc finger in VSP remains unknown, it has been postulated that colonization of Giardia trophozoites in the highly proteolytic and lipolytic milieu of the upper small intestine may require chelation of metal ions by the VSP zinc fingers. It may render the VSP resistant to proteolytic attack and maintaining the integrity of the cell surface (11).
Another possible explanation of the essential presence of CXXC in VSP is that the latter might be a member of the thioredoxin family. CXXC is at the active site of the enzyme protein that reduces the disulfide bond in its substrate and catalyzes formation of another disulfide bond between the substrate and the recipient. The activity of thioredoxin is critical for protein-folding reactions and plays an important role in defense against oxidative stress (28). The two cysteine residues in CXXC must, however, remain in the reduced form in order to exhibit enzymatic activity. The fact that no free thiol is detectable on the membrane surface of Giardia trophozoites indicates that the VSPs do not possess thioredoxin activity.
In addition to the zinc finger, there are also other conserved regions in motif 1, whose potential significance remains unknown at the present time. There are 3 other proteins identified in the GiardiaDB that contain only motif 1 but not motif 2. They have all been annotated as high cysteine membrane proteins, even though the membrane location has not yet been experimentally verified. The highly conserved hydrophobic region of 23 amino acid residues in the middle of motif 2 constitutes most likely the transmembrane domain anchoring the VSP to cell membrane. The GiardiaDB also shows 21 additional proteins that carry motif 2 at their C termini without motif 1. They include several previously annotated VSPs, one hypothetical protein, and 3 other proteins designated high cysteine membrane proteins. By our current experimental proof that both motif 1 and motif 2 are required for localizing VSPs to the cell membrane surface, none of the above-mentioned proteins could be classified as VSPs.
Our experimental results indicated also that the C-terminal CRGKA is not required for VSP expression on the Giardia cell membrane. This is in contrast to a previous report (29), which showed that, during encystation of Giardia isolate GS, removal of C-terminal CRGKA from a chimera reporter protein containing the 43-amino-acid C terminus of VSPH7 resulted in failure of translocalizing the protein to the plasma membrane. The chimera protein apparently contains only motif 2 but not motif 1 identified in VSPs by us. It is thus not clear how the reporter protein gets translocalized to the plasma membrane at all. Since the chimera contains also the N-terminal leader sequence of a cell wall protein 1 and a Toxoplasma gondii SAG1 exodomain, its membrane expression could be by a different mechanism from that of VSP expression. A subsequent investigation indicating that a CRGKA tail-deleted mutation and a point mutation of the Cys residue or the Arg residue in the CRGKA tail of VSPH7 could still localize the latter to the cell membrane of the trophozoites of Giardia isolate GS appears to be in good agreement with our current finding (30, 31). However, since most of the newly identified 73 VSPs carry CRGKA at their C termini (Fig. 7), this pentapeptide should remain a hallmark of VSPs, performing a certain function not yet identified.
Our study also indicates that the N termini of VSPs cannot be blocked by c-myc, or the translocalization of VSP will be inhibited. But there does not appear to be an N-terminal-specific sequence requirement for the signaling function.
Among the other anaerobic protozoan parasites, Entamoeba histolytica has numerous receptor serine/threonine kinase proteins carrying motifs with >50% sequence similarities to motif 1 near their N termini. But they do not possess motif 2. These proteins were postulated to localize to the cell membrane surface and function in cell signaling (32). About 40 proteins in Trichomonas vaginalis have been found to have motifs at their C termini, with sequences similar to that of motif 2, though motif 1 is absent. Among these proteins, 4 are the surface immunogen P270-related proteins, whereas 18 were detected in the proteomic analysis of the membrane surface proteins from T. vaginalis (33). Neither E. histolytica nor T. vaginalis possesses proteins containing both motif 1 and 2. The presence of these two motifs in Giardia VSPs may thus constitute a specific structural feature.
Another protozoan parasite known to possess a single species of variant surface glycoprotein (VSG) covering the cell membrane surface is the African trypanosome Trypanosoma brucei. The bloodstream form of the T. brucei cell expresses a single species of VSG on the membrane surface at any given time but is regularly replaced by another VSG with an estimated frequency of up to 10−3 per cell per generation in an apparent effort of evading the host immune response (34, 35). The VSGs do not contain either motif 1 or motif 2 and are anchored to the cell membrane by a specific glycolipid anchor (35). The expression of a vsg gene in T. brucei requires a duplication of the gene, which is followed by a translocalization of the duplicated gene to the expression site at subtelomeric locations (36). Most of the vsp genes in Giardia are, however, located in chromosome-internal positions, and a subtelomeric localization is apparently not required for their expression (3). The mechanism of genetic regulation of VSP expression in Giardia has been recently investigated by us. A functional microRNA (miRNA) machinery is apparently present in this organism (37). Among the 6 miRNAs we have identified thus far, 5 of them, miR2, miR4, miR5, miR6, and miR10, have found their corresponding target sites at the 3′ ends of many tentative vsp genes (16, 37–39). Translational repression by these miRNAs on the expression of the encoded proteins carrying the corresponding target sites on the mRNAs was verified by extensive experiments (16, 37–39). With the newly verified spectrum of 73 VSPs made available in our present study, we repeated the search for the target sites of the 5 known miRNAs among the 73 vsp genes (16, 37–39). The results, presented in Table S1 in the supplemental material, indicate that most of the genes (88%) possess at least one target site for one of the 5 known miRNAs. Since we have not yet exhausted the pool of miRNAs in Giardia, we anticipate that more miRNAs will be found involved in regulating the expression of the 73 vsp genes. Each of the genes may thus turn out possessing multiple targeting sites for multiple miRNAs. The expression of most of the vsp genes is thus most likely repressed. The mechanism involved in allowing the expression of only a single VSP species among the 73 at a given time may thus include regulation of the expression of various miRNAs, which could be also the basis for periodic variation of the expression of VSP. Further studies will be required for exploring this fascinating biological phenomenon.
MATERIALS AND METHODS
Cloning of 3× myc-tagged vsp genes.
The entire coding region of each chosen putative vsp gene was PCR amplified from Giardia genomic DNA. The product was cloned into pGEM-T Easy (Promega), sequenced, and subcloned into the pNlop4 vector, a derivative of the pNlop3-GTetR vector kindly provided by Zac Cande of UC Berkeley (40). For expressing N-terminal 3× myc-tagged VSPs, the pNlop4 vector was modified to introduce each vsp gene downstream from a 3× myc epitope. For expressing C-terminal 3× myc-tagged VSPs, the pNlop4 vector was modified to introduce the vsp gene and the 3× myc epitope upstream from the TAA stop codon. Figure S4 in the supplemental material shows the schematic representation of the N-terminal and C-terminal 3× myc-tagged vsp constructs used in this study. The pNlop4 vector that carries no tag was used for cloning individual vsp genes without any tag.
Cell culture, transfection, and selection.
Giardia lamblia (WB clone C6, ATCC 50803) trophozoites were grown anaerobically in plastic culture tubes at 37°C in the modified TYI-S-33 medium supplemented with antibiotics as described (41). Transfections of Giardia trophozoites were carried out using electroporation. Cells at mid- to late-logarithmic phase were harvested by chilling the culture tubes on ice for 10 min and collected by a brief centrifugation (1,000 × g at 4°C for 10 min). The cells were washed twice in phosphate-buffered saline (PBS) and once in electroporation buffer (cytomix buffer; 10 mM K2HPO4-KH2PO4 [pH 7.6], 25 mM HEPES free acid, 120 mM KCl, 0.15 mM CaCl2, 2 mM EGTA, 5 mM MgCl2, 2 mM ATP, 4 mM glutathione) and then suspended to a final concentration of 2.5 × 107 cells/ml. An aliquot of the concentrated cell suspension (400 µl, containing 107 cells) was transferred to a 0.2-cm-gap electroporation cuvette (Bio-Rad) and placed on ice. A sample of 50 µg plasmid DNA was added to the cell suspension. The cells were immediately subjected to electroporation using a Bio-Rad Gene Pulser Xcell (voltage, 450 V; capacitance, 500 mF; resistance, ∞). The electroporated cells were incubated on ice for 10 min, added to prewarmed culture medium, and incubated at 37°C. For selection, 200 µg/ml G418 was added to the medium 16 h after transfection. The selected cells were incubated with 5 µg/ml of tetracycline at 37°C for 16 h to induce expression of the cloned VSPs.
Western blotting.
The Mem-PER eukaryotic membrane protein extraction reagent kit (Thermo Scientific) was used to extract proteins from the cell lysate. The separated aqueous and organic phases were each purified and concentrated using the Pierce SDS-PAGE sample prep kit (Thermo Scientific). The concentration of protein in each sample was quantified using the Bradford method (Bio-Rad). For SDS-PAGE separation, 25 µg of protein from each sample was used. The fractionated proteins were then transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad) using the Trans-Blot SD semidry transfer cell (Bio-Rad). The blot was used for detection with the anti-c-myc--horseradish peroxidase (HRP) antibody (Invitrogen) or the monoclonal antibodies for specific VSPs. The relative intensities of the stained bands were monitored by densitometer tracing for a quantitative estimation of the distribution of the protein between aqueous and organic phases.
Immunofluorescence assay.
For detecting the 3× myc-tagged VSP expression, the harvested Giardia cells were resuspended in 200 µl of modified TYI-S-33 culture medium, placed on a poly-l-lysine-coated coverslip (BD Biosciences), and incubated at 37°C for 30 min to allow the cells to adhere.
For the staining of nonpermeabilized cells (17), cells were fixed with 3% paraformaldehyde in NaPi (100 mM NaPi, pH 7.4) at room temperature for 30 min, washed with NaPi three times for 5 min each, and blocked with 0.2% gelatin in NaPi at room temperature for 30 min. The anti-myc-fluorescein isothiocyanate (FITC) antibody (Invitrogen) was diluted 1:500 with 0.2% gelatin in NaPi and incubated with the fixed cells at room temperature for 30 min in a dark box. The cells were then washed three times with NaPi for 5 min each.
For the staining of permeabilized cells, cells were fixed with 4% paraformaldehyde in PBS at room temperature for 30 min, washed with PBS three times for 5 min each, permeabilized with 0.1% Triton X-100 in PBS at room temperature for 10 min, and then rewashed with PBS, as described before. The cells were blocked with 1% bovine serum albumin (BSA) in PBS at room temperature for 30 min. The anti-myc-FITC antibody (Invitrogen) was diluted 1:500 with 1% BSA in PBS and incubated with the fixed cells at room temperature for 30 min in a dark box. The cells were then washed three times with PBS for 5 min each.
The double staining was done by following the procedure of the staining of nonpermeabilized cells. The fixed and blocked cells were incubated with diluted anti-myc-FITC antibody (1:500; Invitrogen) and Cy5-conjugated 9B10 mAb (1:1,500, specific for VSP9B10) (the monoclonal antibody was kindly provided to us by Theodore Nash of NIH) and then washed three times with NaPi for 5 min each.
After the final wash, the coverslip was placed facedown on clean glass slides with 1 drop of Vectashield (Vector Labs) mounting medium with DAPI (6-diamidino-2-phenylindole) and sealed with paraffin wax. The immunostained cells were examined using a Nikon TE2000E motorized inverted microscope equipped with 60× bright-field and epifluorescence optics. Images were acquired with the NIS-Elements Advanced Research software (Nikon).
SUPPLEMENTAL MATERIAL
The 73 identified VSPs carrying a putative target site(s) for the 5 previously characterized miRNAs in Giardia. The miscellaneous ranges of molecular weights of the VSPs, derived from GiardiaDB, are presented in the right-hand column.
Immunofluorescence assays of nonpermeabilized and permeabilized Giardia trophozoites. (A) Expression of N-terminal HA-tagged histone H2A in transfected Giardia trophozoites showed that, by Western analysis, HA-histone H2A was primarily in the aqueous phase. (B) By immunofluorescence assay, HA-histone H2A was detectable only in the permeabilized cells and confined in the nuclei. (C) Expression of a previously experimentally verified VSP, VSP9B10A, showed that by using MAb 9B10 in Western blot analysis, most of the VSP was present in the organic phase. (D) Immunostaining of VSP9B10A using mAb 9B10 in both nonpermeabilized and permeabilized cells showed expression of VSP9B10A on the membrane surface of Giardia trophozoites. Bars = 10 µm. Download
N-terminal tagging of VSP blocks its expression on the membrane surface of Giardia trophozoite. Expression of myc-VSP-175(VSPA6-S1) (A), myc-VSP-98.1(VSP1267) (B), myc-VSP-312 (C), and myc-VSP-338 (D) in transfected Giardia trophozoites was monitored with Western blot and immunofluorescence assays. All the N-myc-tagged VSPs were in the organic phase, but none was expressed on the cell membrane surface. Bars = 10 µm. Download
The dendrogram from an alignment analysis of the sequences of motif 1 and motif 2 from 73 VSPs generated by Clustal W. The sequences of the newly identified 73 VSPs, which contain motif 1 and motif 2, were aligned, and a rooted tree was created with Clustal W. According to the tree, the VSPs can be classified into three subgroups (I, II, and III), with most of the VSPs belonging to subgroup I. Scale bar, 0.1 amino acid substitutions per residue. Download
Schematic presentation of the N-terminal and C-terminal 3× myc-tagged vsp constructs. Download
ACKNOWLEDGMENTS
We thank Zacheus Cande of UC Berkeley for the pNlop4 vector and Theodore Nash of NIH for providing us the MAbs for VSPs.
This work was supported by a research grant from the National Institutes of Health (R01 AI-30475).
Footnotes
Citation Li W, Saraiya AA, Wang CC. 2013. Experimental verification of the identity of variant-specific surface proteins in Giardia lamblia trophozoites. mBio 4(3):e00321-13. doi:10.1128/mBio.00321-13.
REFERENCES
- 1. Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, Davids BJ, Dawson SC, Elmendorf HG, Hehl AB, Holder ME, Huse SM, Kim UU, Lasek-Nesselquist E, Manning G, Nigam A, Nixon JE, Palm D, Passamaneck NE, Prabhu A, Reich CI, Reiner DS, Samuelson J, Svard SG, Sogin ML. 2007. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317:1921–1926 [DOI] [PubMed] [Google Scholar]
- 2. Adam RD. 2001. Biology of Giardia lamblia. Clin. Microbiol. Rev. 14:447–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adam RD, Nigam A, Seshadri V, Martens CA, Farneth GA, Morrison HG, Nash TE, Porcella SF, Patel R. 2010. The Giardia lamblia vsp gene repertoire: characteristics, genomic organization, and evolution. BMC Genomics 11:424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roxström-Lindquist K, Palm D, Reiner D, Ringqvist E, Svärd SG. 2006. Giardia immunity—an update. Trends Parasitol. 22:26–31 [DOI] [PubMed] [Google Scholar]
- 5. Ankarklev J, Jerlström-Hultqvist J, Ringqvist E, Troell K, Svärd SG. 2010. Behind the smile: cell biology and disease mechanisms of Giardia species. Nat. Rev. Microbiol. 8:413–422 [DOI] [PubMed] [Google Scholar]
- 6. Adam RD, Aggarwal A, Lal AA, de la Cruz VF, McCutchan T, Nash TE. 1988. Antigenic variation of a cysteine-rich protein in Giardia lamblia. J. Exp. Med. 167:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aggarwal A, Nash TE. 1988. Antigenic variation of Giardia lamblia in vivo. Infect. Immun. 56:1420–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nash TE, Conrad JT, Merritt JW., Jr. 1990. Variant specific epitopes of Giardia lamblia. Mol. Biochem. Parasitol. 42:125–132 [DOI] [PubMed] [Google Scholar]
- 9. Nash TE. 1997. Antigenic variation in Giardia lamblia and the host’s immune response. Philos. Trans. R. Soc. Lond. B Biol. Sci. 352:1369–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gillin FD, Hagblom P, Harwood J, Aley SB, Reiner DS, McCaffery M, So M, Guiney DG. 1990. Isolation and expression of the gene for a major surface protein of Giardia lamblia. Proc. Natl. Acad. Sci. U. S. A. 87:4463–4467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luján HD, Mowatt MR, Wu JJ, Lu Y, Lees A, Chance MR, Nash TE. 1995. Purification of a variant specific surface protein of Giardia lamblia and characterization of its metal-binding properties. J. Biol. Chem. 270:13807–13813 [DOI] [PubMed] [Google Scholar]
- 12. Yang YM, Adam RD. 1995. Analysis of a repeat containing family of Giardia lamblia variant-specific surface protein genes: diversity through gene duplication and divergence. J. Eukaryot. Microbiol. 42:439–444 [DOI] [PubMed] [Google Scholar]
- 13. Nash TE, Luján HT, Mowatt MR, Conrad JT. 2001. Variant-specific surface protein switching in Giardia lamblia. Infect. Immun. 69:1922–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pimenta PF, da Silva PP, Nash T. 1991. Variant surface antigens of Giardia lamblia are associated with the presence of a thick cell coat: thin section and label fracture immunocytochemistry survey. Infect. Immun. 59:3989–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mowatt MR, Aggarwal A, Nash TE. 1991. Carboxy-terminal sequence conservation among variant-specific surface proteins of Giardia lamblia. Mol. Biochem. Parasitol. 49:215–227 [DOI] [PubMed] [Google Scholar]
- 16. Li W, Saraiya AA, Wang CC. 2012. The profile of snoRNA-derived microRNAs that regulate expression of variant surface proteins in Giardia lamblia. Cell. Microbiol. 14:1455–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hinners I, Moschner J, Nolte N, Hille-Rehfeld A. 1999. The orientation of membrane proteins determined in situ by immunofluorescence staining. Anal. Biochem. 276:1–7 [DOI] [PubMed] [Google Scholar]
- 18. Laity JH, Lee BM, Wright PE. 2001. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11:39–46 [DOI] [PubMed] [Google Scholar]
- 19. Nash TE, Mowatt MR. 1993. Variant-specific surface proteins of Giardia lamblia are zinc-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 90:5489–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang YY, Aley SB, Stanley SL, Jr, Gillin FD. 1993. Cysteine-dependent zinc binding by membrane proteins of Giardia lamblia. Infect. Immun. 61:520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Visvesvara GS. 1980. Axenic growth of Giardia lamblia in Diamond’s TPS-1 medium. Trans. R. Soc. Trop. Med. Hyg. 74:213–215 [DOI] [PubMed] [Google Scholar]
- 22. Gillin FD, Diamond LS. 1981. Entamoeba histolytica and Giardia lamblia: growth responses to reducing agents. Exp. Parasitol. 51:382–391 [DOI] [PubMed] [Google Scholar]
- 23. Gillin FD, Diamond LS. 1981. Entamoeba histolytica and Giardia lamblia: effects of cysteine and oxygen tension upon trophozoite attachment to glass and survival in culture media. Exp. Parasitol. 52:9–17 [DOI] [PubMed] [Google Scholar]
- 24. Aggarwal A, Merritt JW, Nash TE. 1989. Cysteine-rich variant surface proteins of Giardia lamblia. Mol. Biochem. Parasitol. 32:39–48 [DOI] [PubMed] [Google Scholar]
- 25. Tekwani BL, Mehlotra RK. 1999. Molecular basis of defence against oxidative stress in Entamoeba histolytica and Giardia lamblia. Microbes Infect. 1:385–394 [DOI] [PubMed] [Google Scholar]
- 26. Gillin FD, Reiner DS, Levy RB, Henkart PA. 1984. Thiol groups on the surface of anaerobic parasitic protozoa. Mol. Biochem. Parasitol. 13:1–12 [DOI] [PubMed] [Google Scholar]
- 27. Aley SB, Gillin FD. 1993. Giardia lamblia: posttranslational processing and status of exposed cysteine residues in TSA 417, a variable surface antigen. Exp. Parasitol. 77:295–305 [DOI] [PubMed] [Google Scholar]
- 28. Carvalho AP, Fernandes PA, Ramos MJ. 2006. Similarities and differences in the thioredoxin superfamily. Prog. Biophys. Mol. Biol. 91:229–248 [DOI] [PubMed] [Google Scholar]
- 29. Marti M, Li Y, Schraner EM, Wild P, Köhler P, Hehl AB. 2003. The secretory apparatus of an ancient eukaryote: protein sorting to separate export pathways occurs before formation of transient Golgi-like compartments. Mol. Biol. Cell 14:1433–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Touz MC, Conrad JT, Nash TE. 2005. A novel palmitoyl acyl transferase controls surface protein palmitoylation and cytotoxicity in Giardia lamblia. Mol. Microbiol. 58:999–1011 [DOI] [PubMed] [Google Scholar]
- 31. Touz MC, Rópolo AS, Rivero MR, Vranych CV, Conrad JT, Svard SG, Nash TE. 2008. Arginine deiminase has multiple regulatory roles in the biology of Giardia lamblia. J. Cell Sci. 121:2930–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, Roncaglia P, Berriman M, Hirt RP, Mann BJ, Nozaki T, Suh B, Pop M, Duchene M, Ackers J, Tannich E, Leippe M, Hofer M, Bruchhaus I, Willhoeft U, Bhattacharya A, Chillingworth T, Churcher C, Hance Z, Harris B, Harris D, Jagels K, Moule S, Mungall K, Ormond D, Squares R, Whitehead S, Quail MA, Rabbinowitsch E, Norbertczak H, Price C, Wang Z, Guillén N, Gilchrist C, Stroup SE, Bhattacharya S, Lohia A, Foster PG, Sicheritz-Ponten T, Weber C, Singh U, Mukherjee C, El-Sayed NM, Petri WA, Jr, Clark CG, Embley TM, Barrell B, Fraser CM, Hall N. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433:865–868 [DOI] [PubMed] [Google Scholar]
- 33. de Miguel N, Lustig G, Twu O, Chattopadhyay A, Wohlschlegel JA, Johnson PJ. 2010. Proteome analysis of the surface of Trichomonas vaginalis reveals novel proteins and strain-dependent differential expression. Mol. Cell. Proteomics 9:1554–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Turner CM, Barry JD. 1989. High frequency of antigenic variation in Trypanosoma brucei rhodesiense infections. Parasitology 99:67–75 [DOI] [PubMed] [Google Scholar]
- 35. Pays E, Nolan DP. 1998. Expression and function of surface proteins in Trypanosoma brucei. Mol. Biochem. Parasitol. 91:3–36 [DOI] [PubMed] [Google Scholar]
- 36. Horn D, McCulloch R. 2010. Molecular mechanisms underlying the control of antigenic variation in African trypanosomes. Curr. Opin. Microbiol. 13:700–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saraiya AA, Wang CC. 2008. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 4 e1000224 http://dx.doi.org/10.1371/journal.ppat.1000224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saraiya AA, Li W, Wang CC. 2011. A microRNA derived from an apparent canonical biogenesis pathway regulates variant surface protein gene expression in Giardia lamblia. RNA 17:2152–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li W, Saraiya AA, Wang CC. 2011. Gene regulation in Giardia lamblia involves a putative microRNA derived from a small nucleolar RNA. PLoS Negl. Trop. Dis. 5:e1338 http://dx.doi.org/10.1371/journal.pntd.0001338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun CH, Su LH, Gillin FD. 2005. Influence of 5′ sequences on expression of the Tet repressor in Giardia lamblia. Mol. Biochem. Parasitol. 142:1–11 [DOI] [PubMed] [Google Scholar]
- 41. Keister DB. 1983. Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans. R. Soc. Trop. Med. Hyg. 77:487–488 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The 73 identified VSPs carrying a putative target site(s) for the 5 previously characterized miRNAs in Giardia. The miscellaneous ranges of molecular weights of the VSPs, derived from GiardiaDB, are presented in the right-hand column.
Immunofluorescence assays of nonpermeabilized and permeabilized Giardia trophozoites. (A) Expression of N-terminal HA-tagged histone H2A in transfected Giardia trophozoites showed that, by Western analysis, HA-histone H2A was primarily in the aqueous phase. (B) By immunofluorescence assay, HA-histone H2A was detectable only in the permeabilized cells and confined in the nuclei. (C) Expression of a previously experimentally verified VSP, VSP9B10A, showed that by using MAb 9B10 in Western blot analysis, most of the VSP was present in the organic phase. (D) Immunostaining of VSP9B10A using mAb 9B10 in both nonpermeabilized and permeabilized cells showed expression of VSP9B10A on the membrane surface of Giardia trophozoites. Bars = 10 µm. Download
N-terminal tagging of VSP blocks its expression on the membrane surface of Giardia trophozoite. Expression of myc-VSP-175(VSPA6-S1) (A), myc-VSP-98.1(VSP1267) (B), myc-VSP-312 (C), and myc-VSP-338 (D) in transfected Giardia trophozoites was monitored with Western blot and immunofluorescence assays. All the N-myc-tagged VSPs were in the organic phase, but none was expressed on the cell membrane surface. Bars = 10 µm. Download
The dendrogram from an alignment analysis of the sequences of motif 1 and motif 2 from 73 VSPs generated by Clustal W. The sequences of the newly identified 73 VSPs, which contain motif 1 and motif 2, were aligned, and a rooted tree was created with Clustal W. According to the tree, the VSPs can be classified into three subgroups (I, II, and III), with most of the VSPs belonging to subgroup I. Scale bar, 0.1 amino acid substitutions per residue. Download
Schematic presentation of the N-terminal and C-terminal 3× myc-tagged vsp constructs. Download