Key Points
C/EBPα is needed for transition from stem/progenitor cells to common dendritic cell progenitors.
C/EBPα is dispensable in later stages of dendritic cell maturation.
Abstract
Dendritic cells (DCs) are master regulators of the immune system, but molecular regulation of early DC differentiation has been poorly understood. Here, we report that the transcription factor C/EBPα coordinates the development of progenitor cells required for production of multiple categories of DCs. C/EBPα was needed for differentiation from stem/progenitor cells to common DC progenitors (CDPs), but not for transition of CDP to mature DCs. C/EBPα deletion in mature DCs did not affect their numbers or function, suggesting that this transcription factor is not needed for maintenance of DCs in lymphoid tissues. ChIP-seq and microarrays were used to identify candidate genes regulated by C/EBPα and required for DC formation. Genes previously shown to be critical for DC formation were bound by C/EBPα, and their expression was decreased in the earliest hematopoietic compartments in the absence of C/EBPα. These data indicate that C/EBPα is important for the earliest stages of steady-state DC differentiation.
Introduction
Dendritic cells are bone marrow–derived antigen-presenting cells, and subsets differ with respect to location, phenotype, and function.1-4 Murine conventional CD11c+ B220– dendritic cells (cDC) regulate adaptive immune responses and are distinct from other phagocytic cells involved in innate immunity. CD11c+ B220+ plasmacytoid dendritic cells (pDC) represent another major category of these cells specialized to respond to viral infection and produce substantial amounts of type I interferons (IFN). Because dendritic cells in lymphoid organs have no self-renewing potential, they must be continually replenished from hematopoietic stem and progenitor cells.5-9 However, the routes and transcriptional regulators of differentiation used for production of DCs are not completely understood.
Hematopoiesis is a continuous process in which stem cells differentiate into non–self-renewing progenitors that subsequently give rise to mature blood cells.10 Some studies have considered DCs to be activated or more differentiated monocytes.11 However, several categories of progenitors have been characterized to be committed to DC production, and at least the early stages of these precursors are marked by expression of the Flt3 receptor for Flt3 ligand.12-14 The sequential differentiation from a relatively primitive ckit+ Sca-1+ lineage marker negative (KSL) category may give rise to common myeloid progenitors (CMP), Lin- cKit+ Flt3+ MCSFR+ macrophage/DC progenitors (MDP) and CDPs,5-7,12 and there is substantial evidence that commitment to only a DC fate can occur as early as the KSL fraction.7 These data have clearly shown that steady-state DC populations are not derived from monocytes but rather arise from a newly characterized set of DCPs and subsets, all which have unique and specialized functions.6,7,9,15,16 It has also been suggested that some DC subsets are derived from common precursors, MDPs and CDPs, whose fate depends on cytokine signals or environmental cues. Although this likely represents a major differentiation sequence for producing cDC and pDC, several studies of DC differentiation have shown that DC subsets can be generated from both myeloid and lymphoid progenitors.8,12,17,18 Common lymphoid progenitors (CLPs) can also produce DCs during steady state but have increased potential to do so during infections.12,19,20 In summary, several recent studies indicate that steady-state DCs can be derived from a number of DC-committed progenitors, including KSL.
Production of specialized blood cells depends on the orchestrated actions of transcription factors, and many have been implicated in DC formation.21 IRF4, IRF8, E2-2, Id2, Gfi1, and PU.1 may all contribute, whereas only PU.1 is required for all DC subsets.13,22,23 This may function in part by controlling the expression of Flt3, although rescue of this surface receptor did not overcome the PU.1 deficiency.13 Many of these other transcription factors have broader influence not only on DCs, but also on myeloid lineage cells.1,21
The CCAAT/enhancer binding protein (C/EBP) family of transcription factors has basic region leucine zipper structures with DNA-binding basic regions and leucine zipper dimerization domains.24-26 In particular, C/EBPα is expressed by stem cells, and levels steadily increase with differentiation toward GMPs.27 Mice lacking C/EBPα have cell surface–defined CMPs but lack GMPs and more differentiated granulocytic stages.28,29 However, the importance of C/EBPα in DC formation has not been rigorously assessed. Recent reports suggest that cells in an early progenitor fraction of the thymus that are marked with a C/EBPα–Cre × EYFP reporter displayed enhanced potential to form DCs.19 Here, we demonstrate that C/EBPα is critical for early DC differentiation, especially when these cells are derived from myeloid progenitors. In addition, although C/EBPα is required for the formation of primitive cells with DC potential, it is not needed for the maintenance of DC maturation. These novel findings demonstrate discrete C/EBPα-dependent steps in production of specialized DC subsets. That knowledge is crucial in understanding how these immune response regulators are controlled in health and disease, such as in leukemia.
Materials and methods
Mice
Mice were housed in a sterile barrier facility approved by the IUCAC at the Beth Israel Deaconess Medical Center. MxCre C/EBPα and PU.1 conditional knockout mice have been described previously.28,30 CD11c-Cre mice were purchased from Jackson Laboratory.31
Flow cytometry
Single-cell suspensions were analyzed by flow cytometry using the following conjugated antibodies obtained from BD Pharmingen (BD), BioLegend (San Diego, CA), or eBioscience (San Diego, CA). Cells were analyzed using an LSRII flow cytometer or sorted by FACSAria (BD Biosciences, San Jose, CA). FlowJo (Tree Star) was used for data analysis.
Western blot
75 000 cells were sort-purified into phosphate-buffered saline (PBS). Trichloroacetic acid (TCA) was added to a final concentration of 10% TCA. TCA-treated samples were incubated on ice for 15 minutes and spun down for 10 minutes at 13 200 rpm at 4°C. Pellets were solubilized by adding 4× loading buffer with β-mercaptoethanol and boiled for 10 minutes at 95°C. Proteins were separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels and transferred and blotted on polyvinylidene fluoride membranes (Millipore). Primary antibodies used were rabbit anti-C/EBPα (SC-61), anti-C/EBPβ (SC-150), anti-PU.1 (SC-352), goat anti–Gfi-1 (SC-8558, all from Santa Cruz), or rabbit anti–IRF-8 (#5628), anti-RelB (#4954), or anti-KLF4(#4964; Cell Signaling).
Immunofluorescence C/EBPα staining
Cells were fixed in 4% paraformaldehyde for 10 minutes at room temperature and blocked for 15 minutes with 5% fetal bovine serum in PBS containing 0.1% Triton-X. After incubation with primary antibodies in 1% fetal bovine serum in PBS containing 0.1% Triton-X, cells were washed in PBS and incubated for one additional hour with fluorophore-labeled secondary antibodies. Specimens were analyzed on an Axiovert 200M fluorescence microscope, and images were acquired with a Zeiss Axiocam camera.
T-cell proliferation assay
Sorted CD11c+, CD19–, and CD3– DCs from spleen were mixed with splenic CD4+ T-lineage cells that were purified from the same mice. Increasing numbers of DCs were added to 96-well plates with 105 CD4+ cells to each well. After 72 hours of coculture, tritiated thymidine was added and uptake into proliferating T cells was measured 16 hours later.
Reverse transcriptase–polymerase chain reaction
RNA was purified with an RNeasy Micro Kit (Qiagen). Cells were isolated by flow cytometry and directly resorted into an Eppendorf tube containing 350 uL of RLT buffer provided by the manufacturer (Qiagen). Samples were DNase-treated to remove any trace DNA. RNA was used directly in Taqman reverse transcriptase–polymerase chain reaction (RT-PCR) or was reverse-transcribed and subsequently amplified with a Rotor-Gene 6000 (Corbett). Cytokine expression patterns were produced from splenic control or C/EBPα-deleted DCs. The level of cytokine transcripts was assessed in steady-state DCs or after activation by lipopolysaccharide (LPS) (0.5 μg/mL). Total RNA was extracted from CD11c+ fluorescence-activated cell sorting (FACS)-sorted DCs or those sorted DC incubated with LPS for 10 hours and analyzed for cytokines IL1α, IL1β, IL6, IL12, IL15, IFNγ, TNFα, and TGFβ1.
Cell lines
The stem cell factor (SCF)-dependent EML cell line was maintained in Iscove’s modified Dulbecco medium supplemented with 20% horse serum, and 8% conditioned medium from BHK/MKL cells containing mSCF. EML cells cultured with mSCF (100 ng/mL) were infected with C/EBPα-ER retrovirus and selected with puromycin. Induction of nuclear localization of C/EBPα-ER fusion protein was achieved by the addition of 50 nM 4-hydroxytamoxifen (Sigma-Aldrich, St. Louis, MO) into the culture medium.
Normalization and statistical analysis of gene expression data
Data from wild-type multipotent progenitors (MPPs) and C/EBPαfl/fl MPPs were obtained from the gene expression repository at the Harvard Stem Cell Institutes (bloodprogram.hsci.harvard.edu) or from the Gene Expression Omnibus archive (GSE22432) for CDP. Original .CEL files were obtained and were processed with dChip.32 After normalization using the smoothing spline algorithm, expression values were calculated by applying the perfect match–mismatch difference model algorithm. To identify C/EBPα-related changes, we looked for genes that are commonly expressed between wild-type MPPs and wild-type CDPs, as well as C/EBPα fl/fl MPPs and wild-type MPPs, and at the same time are differentially expressed between C/EBPα fl/fl MPPs and wild-type CDPs.
ChIP-Seq and transcription factor binding site analysis
ChIP was performed as described previously.33 The transcription factors binding site analysis was perform on the C/EPBα peaks using Centdist34 with both Jaspar and Transfac vertebrate databases.35 Identification of potential binding sites was obtained using a custom perl script based on the TFBS library. C/EBPα peaks were scanned by the Jaspar’s C/EBPα (MA0102.2) using a similarity threshold of 80%. If several potential binding sites were found on the same peak with the same matrix, only the ones with the best scores were reported and plotted in the heatmap using the “levelplot” function from the “lattice” R/Bioconductor library (http://bioconductor.org).
Results
C/EBPα is expressed and required for DC differentiation
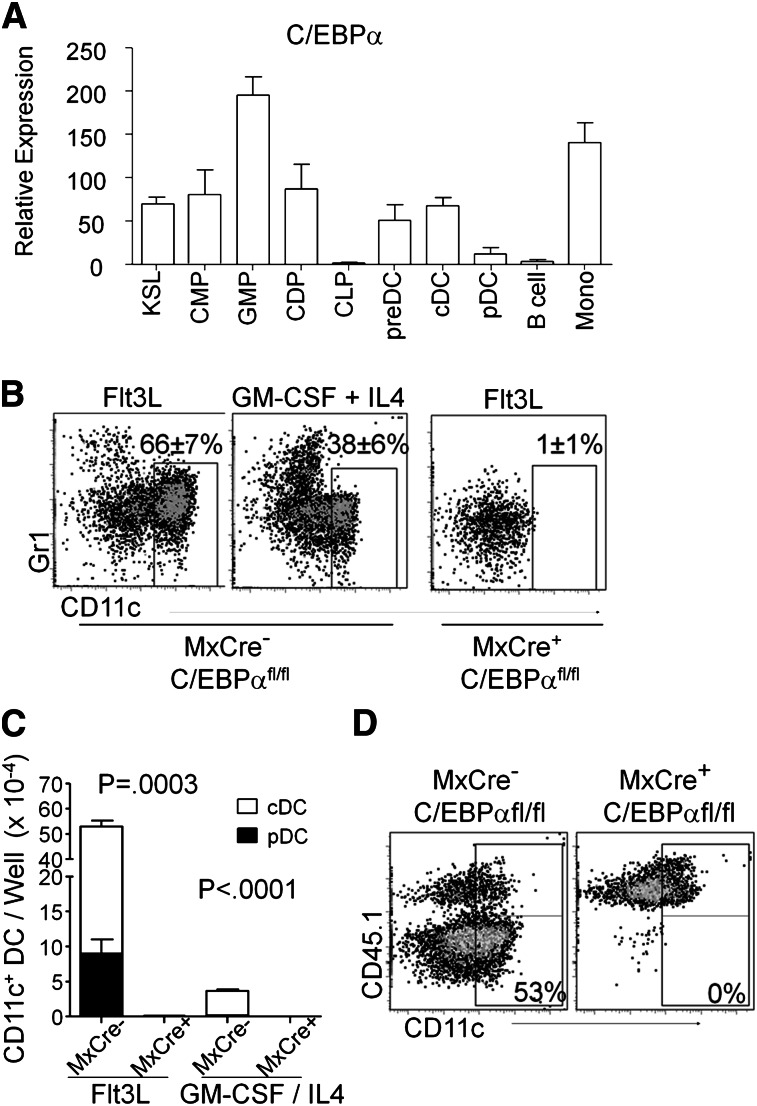
To assess the potential role of C/EBPα in DC development, we measured C/EBPα transcripts by quantitative RT-PCR in mature cDC (CD19/CD3/NK–, CD11c+, MHCII+) and pDC (CD19/CD3/NK–, CD11clo/+, PDCA-1+) that were sorted from the spleens of C57BL/6 mice (Figure 1A). Comparisons were made to pre-DC (Lin–, B220–/lo, CD11clo/+, MHCII–, CD11b/Mac1–), B cells (CD19+, B220+), and monocytes (CD19/CD3/NK–, Gr1+, Mac1+) from the same tissue, as well as the stem/progenitor-rich KSL (cKitHi, Sca1+, Lin–,) fraction and myeloid-related DC progenitors from bone marrow. The latter included CMP (Lin–, Sca1–, cKitHi, CD34+, FcγRII/III–/lo), GMP (Lin–, Sca1–, cKitHi, CD34+, FcγRII/IIIhi), and CDP (Lin–, cKitlo, Flt3+, MCSFR/CD115+, CD11c–) (supplemental Figure 1A). All myeloid-related fractions expressed this gene, whereas C/EBPα transcript levels were undetectable in B cells and low in pDC. Furthermore, C/EBPα protein (p42) could be detected by Western blot and immunostaining in cDC and monocytes, but not in B cells from the spleen (supplemental Figure 1B-C). Thus, this transcription factor is expressed by monocytes, macrophages, myeloid-related progenitors, and DCs.
Figure 1.
Expression of C/EBPα in DC and DC progenitors. (A) Relative quantitative RT-PCR of RNA from sorted KSL (cKitHi, Sca1+, Lin–), CMP (Lin–, Sca1–, cKitHi, CD34+, FcγRII/III–/lo), GMP (Lin–, Sca1–, cKitHi, CD34+, FcγRII/IIIhi), CDP (Lin–, cKitlo, Flt3+, CD115+, CD11c–), preDC (Lin–, B220–/lo, CD11c+, MHCII–, CD11b/Mac1–), cDC (CD19–, CD3–, NK–, B220–, CD11c+, MHCII+), pDC (CD19–, CD3–, NK–, B220+, CD11clo/+, PDCA-1+), B (CD19+, B220+ splenic B cells), and monocytes (CD19–, CD11b/Mac1+, Gr-1+ peritoneal monocytes) to assess expression of C/EBPα. Values are presented relative to that of β2-microglobulin RNA. Data are averages of 4 independent experiments (error bars represent SD). (B) DC loss after deletion of C/EBPα. 106 lysed, WBM cells from poly(IC)-injected MxCre– C/EBPαfl/fl or MxCre+ C/EBPαfl/fl mice were cultured in Flt3L-supplemented media. After 8 days, viable cells were assessed for relative expression of CD11c and Gr1 using flow cytometry. These representative plots show the impact of C/EBPα deletion on the presence of DC (CD11c+). The Gr-1+ myeloid cells can also be seen in these plots. GM-CSF and IL4 cultures with MxCre+ C/EBPαfl/fl WBM cells resulted in no viable cells after 9 days of culture (FACS plot not shown). (C) Shown are the numbers of total live CD11c+ DCs present in the FL or GM-CSF/IL4 cultures for pDC (black, B220+) and cDC (white) from (B). The numbers were generated by hemocytometer counts combined with flow cytometry analysis. These numbers of DCs represent mean ± SD from these 4 experiments where MxCre– and MxCre+ correspond to MxCre– C/EBPαfl/fl and MxCre+ C/EBPαfl/fl cells, respectively. (D) Flow cytometric analysis showing CD45.2+ Mx1-Cre+ C/EBPαfl/fl or C/EBPαfl/fl. WBM cells were cultured together with equal numbers of CD45.1+ BM cells and Flt3L (200 ng/mL) for 8 days. Similarly, cultured cells were stained for cell surface markers CD45.1, CD45.2, CD11c, and MHCII followed by flow cytometry and cell counts. DCs derived from WBM were identified as CD45.2+ CD11c+ Gr1–. The data shown are representative dot plots from 4 experiments with similar results.
Granulocyte–macrophage differentiation has been previously found to be dependent on C/EBP, and the patterns of expression described before suggested the same might be true for dendritic cells. To test this possibility, 12-week-old Mx1-Cre+ C/EBPαfl/fl mice and control (Mx1-Cre– C/EBPαfl/fl) mice were treated with poly(IC). Bone marrow was harvested from these animals 17 to 21 days later, and excision of the C/EBPα gene confirmed by PCR and loss of transcript by quantitative RT-PCR (supplemental Figure 2A). When placed in DC culture conditions containing Flt3L (Flt3 ligand), bone marrow cells from gene-targeted (Mx1-Cre+ C/EBPαfl/fl) bone marrow produced no phenotypic Gr-1Hi myeloid cells or CD11c+ DCs (Figure 1B). An alternate means of DC differentiation is supported by the GM-CSF + IL4 cytokine combination,36 but C/EBPα-deficient stem/progenitors were also defective under those conditions, with no viable cells recovered from cultures. In addition, cells from these cultures were stained by immunofluorescence for the expression of surface markers CD11c and Mac1/CD11b or CD8 (supplemental Figure 1D-E). Because these cultures were initiated with unfractionated marrow, it was conceivable that C/EBPα is required by accessory cells rather than DC progenitors for cDC and pDC production (Figure 1C). However, accompanying CD45.1+ whole bone marrow (WBM) cells generated DCs in 50:50 mixed cultures set up with either control CD45.1 or CD45.2 C/EBPα-deleted marrow (Figure 1D). Thus, marrow progenitors have a cell autonomous C/EBPα requirement rather than a lack of paracrine signaling for formation of DCs in culture.
DC differentiation in vivo is compromised in the absence of C/EBPα
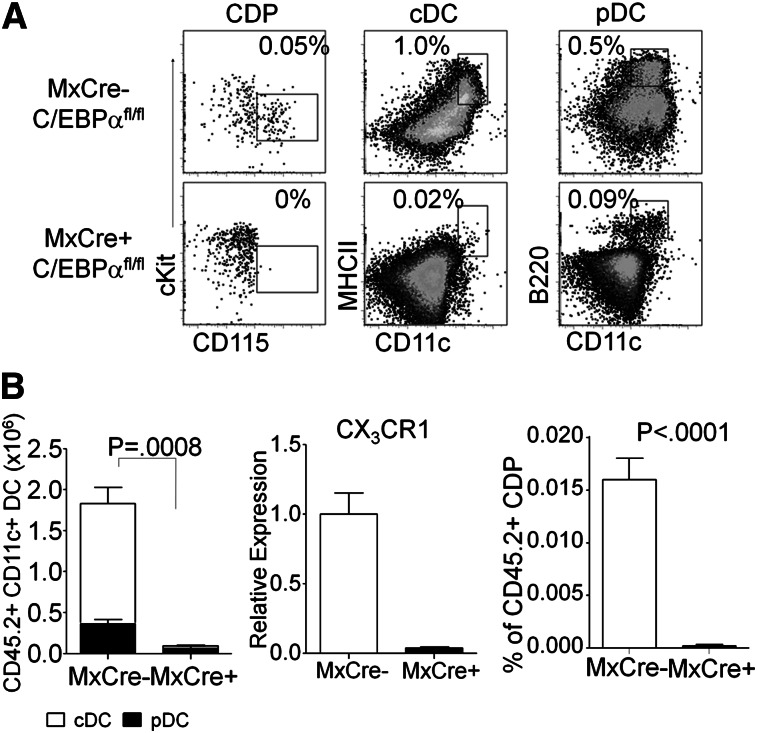
Because cultures do not always replicate in vivo conditions, and multiple routes of DC differentiation have been described,1,21,36 we assessed the importance of C/EBPα in a transplantation model in vivo. The stem/progenitor-rich KSL fraction was sorted from poly(IC) treated Mx1-Cre+ C/EBPαfl/fl mice or Mx1-Cre– C/EBPαfl/fl control mice (CD45.2+) and transferred to sublethally irradiated CD45.1+ recipients. The altered numbers of stem/progenitor cells found in C/EBPα floxed mice compared with controls has been published previously.28,37 From these mice transplanted with KSL, tissues were harvested 1 month later and the differentiation potential of transplanted cells was assessed. Control Mx1-Cre– C/EBPαfl/fl KSL produced lymphocytes, monocytes, granulocytes, and DCs (Figure 2, data not shown). Lymphoid lineages were produced normally from Mx1-Cre+ C/EBPαfl/fl–deleted cells, but no phenotypic monocytes, few cDC (0.02%), and only small numbers of pDC (0.09%) were made (Figure 2B). Macrophage/DC progenitors are characterized by the CX3CR1-GFP reporter; we did not cross these mice to the C/EBPαfl/fl mice but found that expression was reduced in sorted CMP by RT-PCR (Figure 2B). In addition, CDP formation was C/EBPα dependent (Figure 2A-B and supplemental Figure 2B-C). These findings extend those made with our cell culture methods and demonstrate a major requirement for C/EBPα in production of DCs and their progenitors in vivo.
Figure 2.
In vivo assay for DC generation from C/EBPα-targeted progenitors. (A) FACS analysis of CD45.2+ donor–derived cells, from 5000 transplanted KSL of C/EBPαfl/fl or Mx1–Cre+ C/EBPαfl/fl–transplanted mice, for CDP (Lin–, Flt3+, FcγRII/III–, cKit+, CD115+, CD11c–) from bone marrow, or cDC (CD19–, CD3–, NK–, CD11c+, MHCII+) and pDC (CD19–, CD3–, NK–, CD11c+, B220+) from the spleen show contribution of donor cells to each population. The data shown are representative dot plots from 3 mice per group for 3 independent experiments with similar results. (B) Absolute numbers of total CD45.2+ CD19–, CD3–, NK– CD11c+ DCs were calculated from total splenocytes (left). B220+ subset of cells was determined to be pDC (black), and B220– MHCII+ cells were cDC (white) and plotted accordingly. CMPs were sorted from either MxCre– C/EBPαfl/fl or MxCre+ C/EBPαfl/fl mice and analyzed for expression of CX3CR1 by RT-PCR. The percentage of CDP from total CD45.2+ bone marrow are presented from KSL of C/EBPαfl/fl or MxCre+ C/EBPαfl/fl–transplanted mice (right). The numbers represent mean ± SD from 4 similarly transplanted mice. One of 3 experiments giving similar results is presented.
An early requirement for C/EBPα in DC differentiation
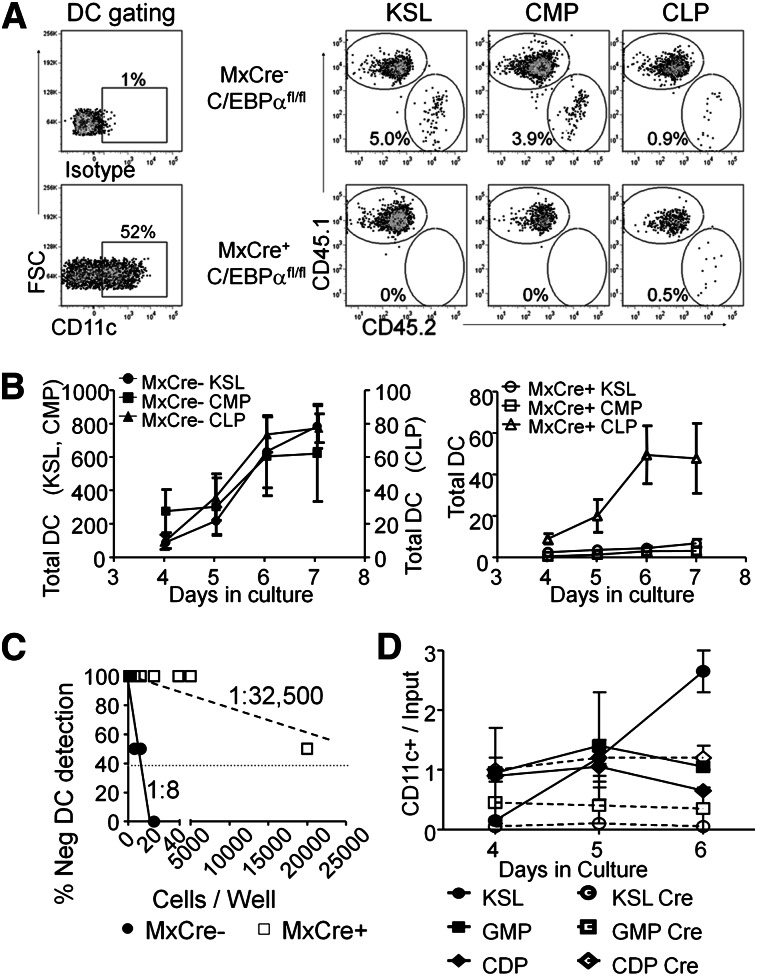
The results shown here suggest that C/EBPα plays a major role in generating DCs and their progenitors. However, we wanted to pinpoint the stages that require this transcription factor. The KSL fraction, as well as phenotypic CMPs and CLPs could be recovered from poly(IC)-treated Mx1-Cre+C/EBPαfl/fl bone marrow, and these 3 subsets of progenitors were tested for Flt3L-mediated DC differentiation in culture. Cultures were again initiated with a mixture of 500 sorted progenitors and 104 WBM cells from normal control CD45.1+ cells (Figure 3). Wells were analyzed for the presence of CD45.2+ CD11c+ DCs, and representative flow cytometry results from day 6 are shown in Figure 3A. Deletion of C/EBPα severely compromised the ability of KSL and CMP subsets of marrow progenitors to generate DCs (Figure 3A-B). Although CLPs are a less efficient source of DCs,12,38 these progenitors maintained the majority of their ability to produce CD45.2+ CD11c+ DCs in the absence of C/EBPα (Figure 3B). A limiting dilution version of this experimental design revealed that ∼1 of 8 Lin– cKit+ cells from control mice gave rise to CD45.2+ CD11c+ cells compared with ∼1 of 32 500 from Mx1-Cre+C/EBPαfl/fl (Figure 3C). To more precisely define C/EBPα dependence of DC formation, Cre-GFP was introduced into sorted KSL, GMP, and CDP from C/EBPαfl/fl mice by retroviral transduction. Double-sorted progenitors were incubated for 24 hours with SCF, Flt3L, and IL6 to allow for Cre-mediated excision before culture with only Flt3L. Although DC production was severely impaired by deletion of C/EBPα in the more primitive progenitors, differentiation from the CDP stage appeared unchanged (Figure 3D). We conclude that C/EBPα is needed for progression to phenotypic CDP and subsequently mature DCs.
Figure 3.
Stage-specific need for C/EBPα in DC progenitors. (A) KSL, CMPs, and CLPs were isolated from the BM of MxCre+ C/EBPαfl/fl or C/EBPαfl/fl mice 2 weeks after 3 poly(IC) injections. 500 progenitor cells were cocultured with 104 Ly5.1+ WBM cells in the presence of Flt3L (200 ng/mL). The resulting cultures were analyzed by flow cytometry for Ly5.1, Ly5.2, MHCII, and CD11c after days 3 to 7. In addition, CD45.1+ cells were subjected to the same culture conditions that were used as controls for efficiency of DC differentiation. Day 5 FACS plots are shown as representative data from triplicate wells of 3 independent experiments. (B) The number of DCs generated in each culture was calculated using cell counts and flow cytometry over days and presented as the number of total DCs (average of 3 wells). On the left are sorted populations from C/EBPα fl/fl, with the DC numbers for KSL and CMPs shown on the left axis and CLP production on the right axis. The graphs on the right are from the same progenitors but sorted from MxCre+ C/EBPαfl/fl mice. Data are presented as mean ± SD of one such experiment that was repeated 3 times. (C) Cloning efficiency was assessed by limiting dilution series of sorted CD45.2+ MxCre+ C/EBPαfl/fl or C/EBPαfl/fl Lin– cKit+ progenitors cocultured with Ly5.1+ Lin– cKit+ BM cells in the presence of Flt3L(200 ng/mL). Cultures were analyzed on day 8 for the presence of CD45.2+ CD11c+ cells. Data are representative of 2 such experiments. (D) The progenitor populations KSL, GMP, and CDP were isolated from the BM of C/EBPαfl/fl mice. These progenitor cells were infected with retrovirus carrying an empty vector with GFP reporter or Cre with GFP reporter. These cells were then cultured with sorted CD45.1+ Lin– cKit+ BM cells in the presence of Flt3L. Cultures were assessed for CD45.2+ CD11c+ GFP+ cells at days 4, 5, and 6. The plot shows a yield of CD11c+ cells per input GFP+ progenitor over time. Data are representative of 2 such experiments.
C/EBPα is not required for mature DCs in lymphoid tissues
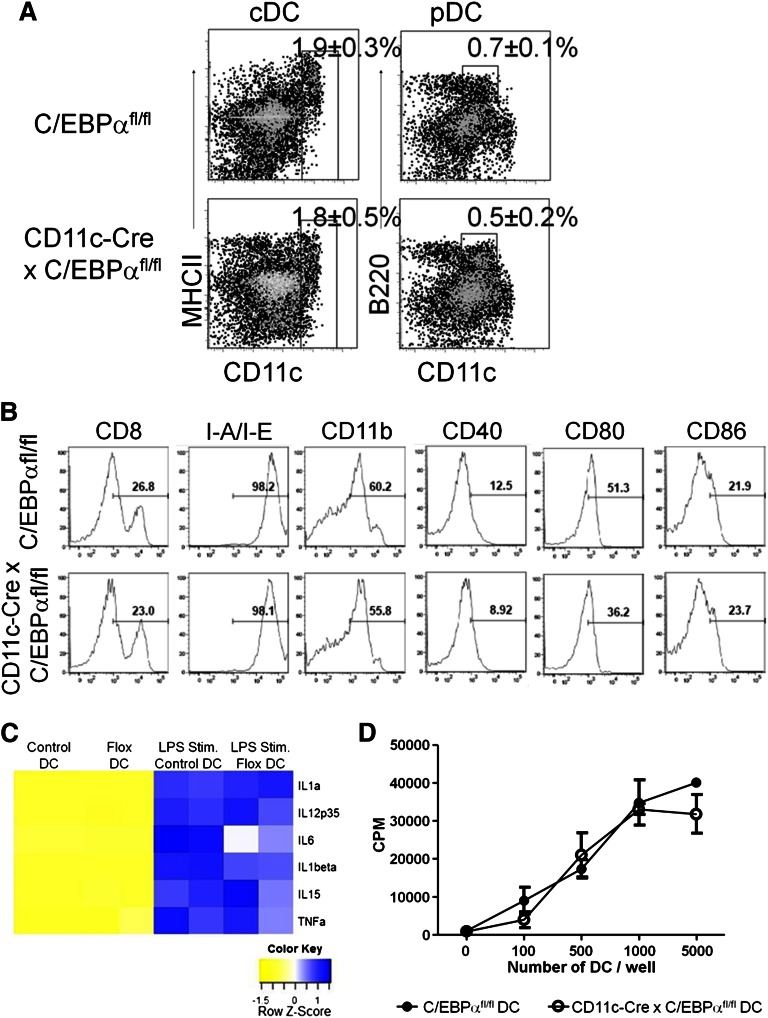
Because some transcription factors have been shown to be required for the maintenance of mature DCs, as with the transcription factor E2-2 in pDC,23 we wanted to know whether C/EBPα was similarly required for maintenance of DCs. Consequently, stage and cell type specificity were achieved with the CD11c-Cre (also known as Itgax-Cre) deleter mouse strain, in which recombination occurs in >90% of splenic pDC and cDC, but not on earlier precursors that are CD11c-negative. Both major DC populations, pDC and cDC, were present in the spleens of 10-week-old mice carrying both C/EBPαfl/fl and CD11c-Cre genes, and were indistinguishable from those in control C/EBPαfl/fl mice (Figure 4A). Further analyses, using known DC cell surface markers resolved subpopulations with no significant differences between control and DC-specific C/EBPα deletions (Figure 4B). PCR analysis of CD11c+ cells sorted from the spleen confirmed C/EBPα gene excision in these cells (supplemental Figure 3A). Absolute numbers of total CD11c+ DCs were measured in lymphoid-specific tissues, and no differences were found between CD11c-Cre+ × C/EBPαfl/fl or control (C/EBPαfl/fl) mice at ∼8 to 10 weeks of age (supplemental Figure 3B). The pattern of cytokine production from C/EBPα-deleted DCs was determined by measuring the mRNA expression in steady-state DCs or after stimulation with LPS. Total RNA was extracted from either CD11c+ CD19– CD3–-sorted splenic DCs or similar cells incubated with LPS (0.5 μg/mL) for 10 hours before RNA isolation. LPS significantly upregulated the expression of IL1α, IL1β, and TNFα transcripts, and, to a lesser extent, the amount of IFNγ and TGFβ1 mRNA (not shown) compared with their steady-state counterpart DCs (Figure 4C). No significant differences in cytokine profiles were observed between control and C/EBPα-deleted DCs with the exception of IL6. In addition, no differences were observed in C/EBPα-deleted splenic DCs in phagocytosis assays (supplemental Figure 3C) or in the ability to activate T-lineage cells to proliferate (Figure 4D). As an assessment of potential DC defects, we double-sorted CD19– CD3– NK– CD11c+ cells from the spleens of C/EBPαfl/fl control or C/EBPα DC–deleted mice and evaluated them for the expression of DC-related transcripts using a commercially available RT-PCR kit (SABiosciences, RT2 Profiler PCR array, PAMM-406A). This confirmed that C/EBPα was deleted and significant expression differences were found in only 6 of 84 DC-related transcripts (supplemental Figure 3D and supplemental Table 1). These data suggest that under these conditions, C/EBPα is not needed for maintenance of mature DCs.
Figure 4.
Targeted C/EBPα deletion has no effect on late-stage DC development. (A) CD11c-Cre+ × C/EBPαfl/fl and C/EBPαfl/fl control mice were analyzed by flow cytometry between 8 and 12 weeks of age. Flow cytometry analysis of DC populations from the spleens of mice of the indicated genotypes are shown as dot plots. cDC are defined as CD19–, CD3–, NK–, CD11c+, and MHCII+, whereas pDC were gated as CD19–, CD3–, NK–, CD11clo, and B220+. Boxes are used to indicate the position of the DC populations in each group with the average percentage per spleen. FACS plots were taken from 3 similar experiments. (B) Histograms of flow cytometric analysis of CD11c+ splenic DC from CD11c-Cre+ C/EBPαfl/fl or C/EBPαfl/fl mice for subpopulations of DCs for CD4, CD8, CD11b, CD40, CD80, CD86, IL3Rα, and M-CSFR. Percentages are representative of similar experiments. (C) CD11c-Cre+ x C/EBPαfl/fl or C/EBPαfl/fl splenic DCs were sorted from 12-week-old mice. These cells were stimulated with LPS for 10 hours and then RT-PCR was performed for expression of cytokine transcripts. RT-PCR analysis displayed in a heatmap for transcripts from steady-state and LPS stimulated DC from both control and flox-deleted DCs. Data are shown from 2 similar experiments. (D) Control or C/EBPα-depleted DCs were sorted as CD11c+ CD19–, and CD3– into 72-hour cultures along with magnetic bead column–enriched CD3+ splenic cells to induce the proliferation of autologous T-lineage cells. The proliferative response of T-lineage cells was measured by tritiated thymidine incorporation after 16 hours. Results are representative of 2 experiments, and each value represents the mean from triplicate wells.
C/EBPα regulates gene expression needed for DC formation
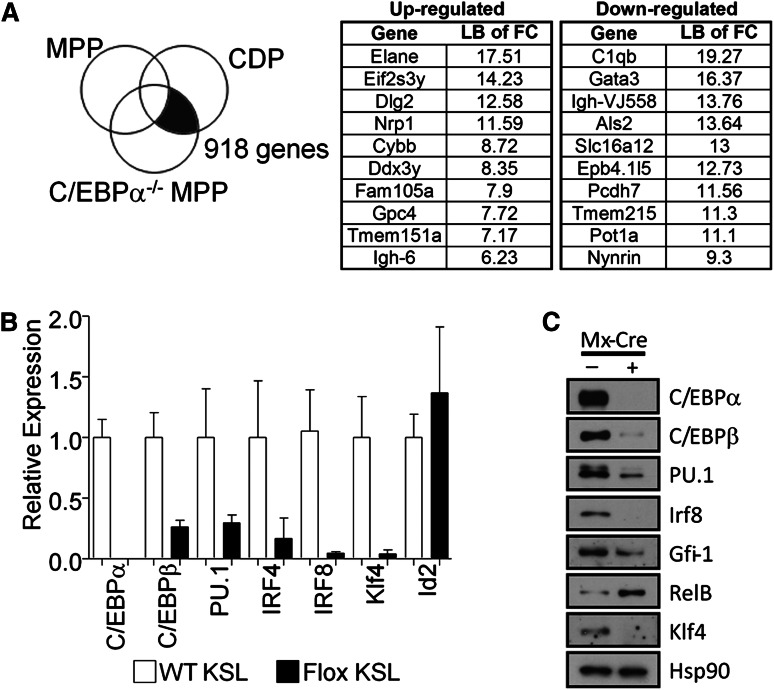
To explore potential mechanisms for C/EBPα regulation of DC formation, we analyzed array data from C/EBPα−/− MPPs and wild-type common CDPs. Genes whose expression differed between these 2 populations were removed if they were also expressed in control MPPs because these were likely not C/EBPα-dependent (Figure 5A). This approach was used to discover C/EBPα-dependent genes that associate with the earliest steps in DC differentiation. These genes are promising candidates for regulation by C/EBPα during the transition from C/EBPα−/− MPPs to CDPs (supplemental Table 2), and a subset of those genes most differentially regulated are shown with the lower bound of fold change values given (Figure 5A).
Figure 5.
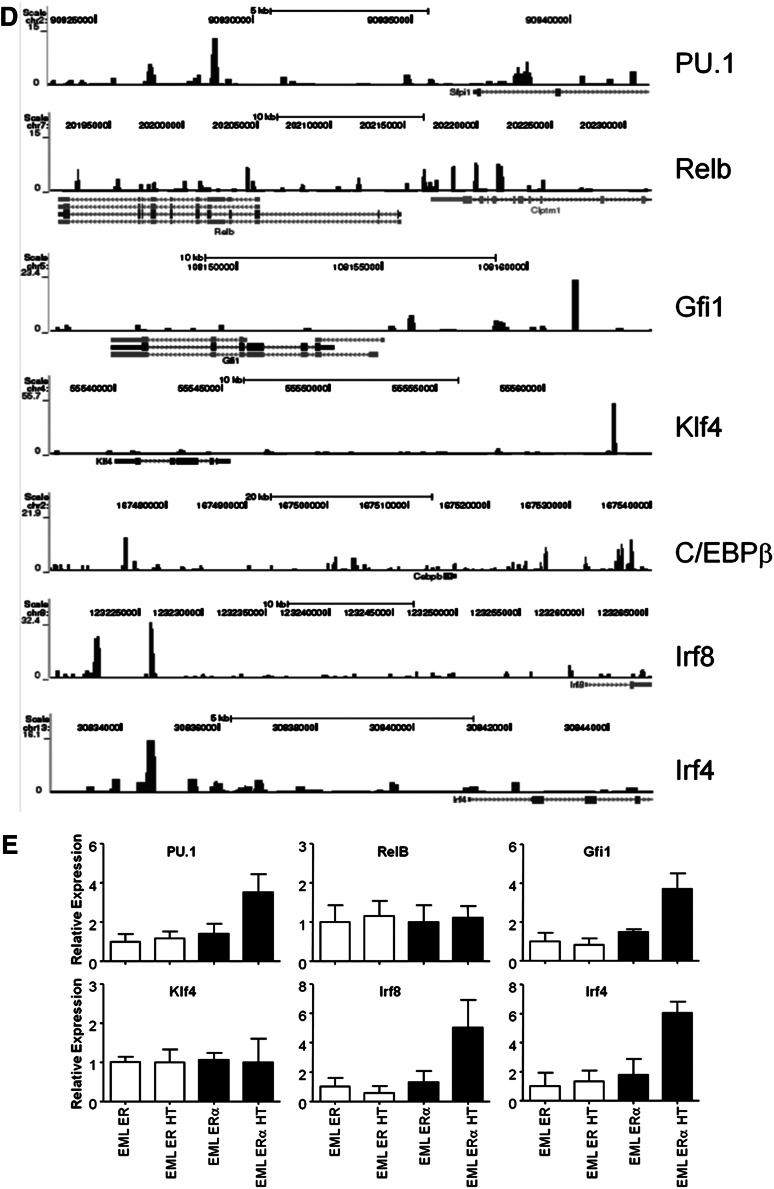
C/EBPα binds the promoter or transcriptional start site of many genes needed for transition from MPP to CDP. (A) Array data from C/EBPα−/− MPPs were compared with that of CDPs, whereas differences between normal MPPs and CDPs were subtracted from this analysis, uncovering genes that are located in the shaded section of the Venn diagram. The most differentially up- and downregulated genes while transitioning to CDP are presented in the right two panels. (B) Expression of C/EBPα, C/EBPβ, PU.1, IRF4, IRF8, Klf4, Id2, and RelB by relative quantitative RT-PCR of RNA from sorted CMPs (cKitHi, Sca1+, Lin– CD34+ FcγRII/IIIlo/–); white bars represent controls and black bars represent floxed progenitors. Values normalized to the control, nonexcised flox CMPs, except with RelB where samples were normalized against the floxed deleted sample. Data are averages of 3 independent experiments (error bars represent SEM). (C) Lin– cKit+ progenitors were sorted from the bone marrow of control or C/EBPα floxed mice. Cells were double-sorted from each group and used for Western blot analysis to determine transcription factor expression compared with HSP-90 (bottom panel). (D) C/EBPα ChIP-seq of CMPs was analyzed for specific genes important during DC differentiation. Plots show ChIP-fragment density at each position in the regions of the genes PU.1, Relb, Gfi1, Klf4, C/EBPβ, Irf8, and Irf4. ChIP-seq profiles are given for genes that were considered altered from (B) and (C). (E) Relative quantitative RT-PCR of transcripts, isolated from EML-ER and EML-C/EBPα-ER, were measured from cell lines cultured for 24 hours with 4-HT to assess expression of PU.1, RelB, Gfi1, Klf4, Irf8, and Irf4. The y-axis indicates the relative expression of the transcription factor relative to that of control treated EML. Data are averages of 2 independent experiments (error bars represent SD).
The differentiation of DCs is controlled by the expression of growth factor receptors that together with transcription factors direct cell fates.39,40 We wanted to test whether C/EBPα was responsible for the induction of well-characterized DC-related transcription factors, so we analyzed the expression patterns of these factors and the interaction of C/EBPα with their promoters, or up- and downstream regulatory elements. To do this, sorted CMPs were used to generate cDNA to determine the expression of DC transcription factors with the loss of C/EBPα (Figure 5B). DC-related transcription factors C/EBPβ, PU.1, IRF4, IRF8, and Klf4 were all decreased with the loss of C/EBPα in CMPs. However, Relb was increased and Id2 was unchanged in C/EBPα-deleted CMPs relative to control cells (not shown); similar analysis for KSL was also conducted (supplemental Figure 4A). In addition, the expression of many of these factors were confirmed through Western blot (Figure 5C), suggesting that C/EBPα regulation of the RNA of many of these transcription factors is reflected in their protein levels. Because PU.1 has been previously shown to be required for DC differentiation,13 our data suggest that C/EBPα could play an important collaborative role with PU.1 during the earliest stages. As evidence for this codependence, overexpression of only PU.1 was not sufficient to rescue DC differentiation from KSL in the absence of C/EBPα, but it did rescue DC differentiation from PU.1 conditionally-deleted KSL (supplemental Figure 4B). Furthermore, to determine whether these changes might be directly regulated by C/EBPα, the interaction of C/EBPα with the regulatory elements of these factors was confirmed by ChIP-seq from sorted progenitors. This analysis was performed on sorted CMP and ChIP-seq profiles for C/EBPα after IgG subtraction was done in the regions of interest. Significant signals were detected in regulatory regions close to these essential transcriptional regulators (Figure 5D). Interestingly, the newly identified Zbtb46, zinc transcription factor specific for cDC, also showed a significant signal upstream of this factor’s start site.41,42 Finally, to determine whether induction of C/EBPα can mediate expression of these DC-related factors, we induced C/EBPα in an EML-inducible system. Estrogen receptor (ER) control clones that contain the ER peptide without C/EBPα, and C/EBPα-ER–expressing clones were cultured in the presence of SCF with or without 4-hydroxytamoxifen (HT). Cells were treated for 24 hours with 4- HT and transcripts were isolated for expression of direct C/EBPα targets. Quantitative RT-PCR showed enrichment of the PU.1, Gfi1, Irf4, and Irf8 upon 4-HT treatment in the C/EBPα-ER–expressing cells, but not in the absence of 4-HT or the ER control cells (Figure 5E). With that data taken together, we have identified a significant number of genes that are associated with DC differentiation and are dependent on C/EBPα. ChIP-seq data suggest that a number of these are likely to be directly regulated by C/EBPα, including those previously defined transcription factors essential for DC differentiation.
Discussion
Limited information was available about expression and function of C/EBPα expressed in several DC subsets, although both are known to be particularly important for early events in hematopoiesis and myeloid differentiation.19 Furthermore, C/EBPα is frequently compromised in hematopoietic diseases and its loss could have consequences for immune system development or function.26 Indeed, we now show that C/EBPα is required for discrete steps in DC production, specifically formation from myeloid progenitors through common DC progenitors. Other genes that could contribute to this process were identified by microarray analyses relative to C/EBPα, and ChIP-seq data showed how multiple factors may cooperate to replenish the innate immune system.
Until now, it was thought that C/EBPα functions as a negative regulator of DC formation.43 This conclusion resulted from artificial overexpression of C/EBPα in primary human cells or cell lines. In addition to the fact that unphysiological levels of C/EBPα were achieved, those approaches would not have been informative about particular steps in the process for which this transcription factor is required. A large body of evidence suggests that C/EBPs regulate lineage choice decisions in a highly stage- and dosage-dependent manner.3,28 Thus, it was essential to explore these questions with lineage- and stage-specific deletion models.
Although macrophage colony-stimulating factor (M-CSF) is not required for DC formation, expression of the M-CSF receptor (M-CSFR) is used to distinguish both MDPs and CDPs.5-7 There is evidence that C/EBPα induces expression of CD115, so it might be expected to find no phenotypic CDPs in C/EBPα knockouts.44 Although that was indeed the case, it is important to stress that downstream stages of DC differentiation were also compromised. Interestingly, recent studies of progenitors and terminally differentiated cells have shown that DCs are more transcriptionally linked to CMPs than GMPs.45 Because CMPs are more likely to be precursors for DC progenitors,46 we found that cells with CMP characteristics were present with C/EBPα deletion. However, they too were unable to generate mature DCs in the absence of C/EBPα.
DC formation is complex, and DCs with similar properties can be made from both myeloid and lymphoid progenitors. The consensus from studying murine models is that the major pathway is through DC progenitors, which arise from the myeloid progenitors and are independent from monocytes.1,38 This process is likely to be altered under normal, disease, and experimental circumstances, making it difficult to determine the relative importance of optional differentiation pathways. We now show that this major route through myeloid progenitors is C/EBPα-dependent. Consistent with the decreased expression of C/EBPα in CLPs, these lymphoid progenitors retained DC potential even in the absence of this transcription factor.
Our unique strategy for identification of genes required for DC formation was highly effective, and many of those upregulated from C/EBPα-deficient MPP to CDP stages were directly bound by C/EBPα in ChIP-seq analysis. Several other transcription factors, including Irf4, Irf8, RelB, Gfi1, Id2, and E2-2, are required for specification and/or differentiation of selected DC subsets, whereas C/EBPα and PU.1 appear to have unique importance.21 Our data suggest that C/EBPα regulates the expression of many of these DC-specific transcription factors through direct binding to the regulatory elements of these genes in progenitors. That some of these factors cooperate rather than complement each other was shown by our finding that PU.1 overexpression did not restore DC potential in C/EBPα-deficient progenitors. As another distinction, PU.1, but not C/EBPα, is required for later stages of DC maturation.
Our new findings show that C/EBPα supports DC formation under normal steady-state conditions. However, previous studies demonstrated that related family members have compensatory functions that are revealed during disease and inflammatory circumstances.27,47 Therefore, alternative mechanisms of DC differentiation may be used in inflammatory environments, such as monocyte activation to DC,11 non–lymphoid tissue DC commitment,48,49 or when C/EBPα is mutated, silenced, or methylated, as in leukemia.26
Overall, our study emphasizes the necessity of C/EBPα to mediate the early events of steady-state DC differentiation. C/EBPα can bind and regulate the expression of many transcription factors previously shown to be necessary for DC differentiation. As one example, PU.1 likely cooperates with C/EBPα during this progression of commitment. These data demonstrate that C/EBPα is needed within a transcriptional network for the earliest differentiation events in normal steady-state DC development.
Supplementary Material
Acknowledgments
The authors thank all members of the Tenen Laboratory for helpful discussions, Junyan Zhang for her help with mice, and the BIDMC flow cytometry facility for their expertise.
This work was supported by the American Italian Cancer Foundation (G.A.), the National Institutes of Health (grants HL56745 and DK080665 [D.G.T.], and HL56745, CA41456, CA66996, CA118316, and DK080665), the José Carreras Leukemia Foundation (FIJC F11/01) (R.S.W.), the German Research Foundation (DFG fellowship BA 4186/1-1) (C.B.) and the Singapore Ministry of Health’s National Medical Research Council under its Singapore Translational Research (STaR) Investigator Award (D.G.T.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: All authors designed research; R.S.W., D.B., G.A., A.C., C.B., K.D.S.A.W., M.Y., H.Z., T.I., and C.J.H. performed research; R.S.W., D.B., G.A., A.C., T.B., C.B., T.I., K.A., and D.G.T. analyzed data; and R.S.W., D.B., G.A., K.A., and D.G.T. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel G. Tenen, Center for Life Sciences, 3 Blackfan Circle, Room 437, Boston, MA 02115; e-mail: daniel.tenen@nus.edu.sg.
References
- 1.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327(5966):656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geissmann F. The origin of dendritic cells. Nat Immunol. 2007;8(6):558–560. doi: 10.1038/ni0607-558. [DOI] [PubMed] [Google Scholar]
- 3.Auffray C, Sieweke MH, Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu Rev Immunol. 2009;27:669–692. doi: 10.1146/annurev.immunol.021908.132557. [DOI] [PubMed] [Google Scholar]
- 4.Liu K, Waskow C, Liu X, Yao K, Hoh J, Nussenzweig M. Origin of dendritic cells in peripheral lymphoid organs of mice. Nat Immunol. 2007;8(6):578–583. doi: 10.1038/ni1462. [DOI] [PubMed] [Google Scholar]
- 5.Fogg DK, Sibon C, Miled C, et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006;311(5757):83–87. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- 6.Onai N, Obata-Onai A, Schmid MA, Ohteki T, Jarrossay D, Manz MG. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat Immunol. 2007;8(11):1207–1216. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- 7.Naik SH, Sathe P, Park HY, et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat Immunol. 2007;8(11):1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- 8.Ishikawa F, Niiro H, Iino T, et al. The developmental program of human dendritic cells is operated independently of conventional myeloid and lymphoid pathways. Blood. 2007;110(10):3591–3660. doi: 10.1182/blood-2007-02-071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu K, Victora GD, Schwickert TA, et al. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324(5925):392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenbauer F, Koschmieder S, Steidl U, Tenen DG. Effect of transcription-factor concentrations on leukemic stem cells. Blood. 2005;106(5):1519–1524. doi: 10.1182/blood-2005-02-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hume DA. Macrophages as APC and the dendritic cell myth. J Immunol. 2008;181(9):5829–5835. doi: 10.4049/jimmunol.181.9.5829. [DOI] [PubMed] [Google Scholar]
- 12.Shigematsu H, Reizis B, Iwasaki H, et al. Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective of their cellular origin. Immunity. 2004;21(1):43–53. doi: 10.1016/j.immuni.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Carotta S, Dakic A, D’Amico A, et al. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity. 2010;32(5):628–641. doi: 10.1016/j.immuni.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Wu L, Liu YJ. Development of dendritic-cell lineages. Immunity. 2007;26(6):741–750. doi: 10.1016/j.immuni.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Ginhoux F, Liu K, Helft J, et al. The origin and development of nonlymphoid tissue CD103+ DCs. J Exp Med. 2009;206(13):3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller JC, Brown BD, Shay T, et al. Immunological Genome Consortium. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13(9):888–899. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Amico A, Wu L. The early progenitors of mouse dendritic cells and plasmacytoid predendritic cells are within the bone marrow hemopoietic precursors expressing Flt3. J Exp Med. 2003;198(2):293–303. doi: 10.1084/jem.20030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karsunky H, Merad M, Cozzio A, Weissman IL, Manz MG. Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J Exp Med. 2003;198(2):305–313. doi: 10.1084/jem.20030323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wölfler A, Danen-van Oorschot AA, Haanstra JR, et al. Lineage-instructive function of C/EBPα in multipotent hematopoietic cells and early thymic progenitors. Blood. 2010;116(20):4116–4125. doi: 10.1182/blood-2010-03-275404. [DOI] [PubMed] [Google Scholar]
- 20.Welner RS, Pelayo R, Nagai Y, et al. Lymphoid precursors are directed to produce dendritic cells as a result of TLR9 ligation during herpes infection. Blood. 2008;112(9):3753–3761. doi: 10.1182/blood-2008-04-151506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merad M, Manz MG. Dendritic cell homeostasis. Blood. 2009;113(15):3418–3427. doi: 10.1182/blood-2008-12-180646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Lee CH, Qi C, et al. IRF8 regulates B-cell lineage specification, commitment, and differentiation. Blood. 2008;112(10):4028–4038. doi: 10.1182/blood-2008-01-129049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh HS, Cisse B, Bunin A, Lewis KL, Reizis B. Continuous expression of the transcription factor e2-2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity. 2010;33(6):905–916. doi: 10.1016/j.immuni.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birkenmeier EH, Gwynn B, Howard S, et al. Tissue-specific expression, developmental regulation and genetic mapping of the gene encoding C/EBP. Genes Dev. 1989;3(8):1146–1156. doi: 10.1101/gad.3.8.1146. [DOI] [PubMed] [Google Scholar]
- 25.Antonson P, Stellan B, Yamanaka R, Xanthopoulos KG. A novel human CCAAT/enhancer binding protein gene, C/EBPepsilon, is expressed in cells of lymphoid and myeloid lineages and is localized on chromosome 14q11.2 close to the T-cell receptor alpha/delta locus. Genomics. 1996;35(1):30–38. doi: 10.1006/geno.1996.0319. [DOI] [PubMed] [Google Scholar]
- 26.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol. 2007;7(2):105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 27.Hirai H, Zhang P, Dayaram T, et al. C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat Immunol. 2006;7(7):732–739. doi: 10.1038/ni1354. [DOI] [PubMed] [Google Scholar]
- 28.Zhang P, Iwasaki-Arai J, Iwasaki H, et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 2004;21(6):853–863. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26(47):6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- 30.Iwasaki H, Somoza C, Shigematsu H, et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106(5):1590–1600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J Exp Med. 2007;204(7):1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98(1):31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan YF, Wansa KD, Liu MH, et al. Regulation of estrogen receptor-mediated long range transcription via evolutionarily conserved distal response elements. J Biol Chem. 2008;283(47):32977–32988. doi: 10.1074/jbc.M802024200. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z, Chang CW, Goh WL, Sung WK, Cheung E. CENTDIST: discovery of co-associated factors by motif distribution. Nucleic Acids Res. 2011;39(Web Server issue):W391-W399. doi: 10.1093/nar/gkr387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Portales-Casamar E, Thongjuea S, Kwon AT, et al. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38(Database issue):D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y, Zhan Y, Lew AM, Naik SH, Kershaw MH. Differential development of murine dendritic cells by GM-CSF versus Flt3 ligand has implications for inflammation and trafficking. J Immunol. 2007;179(11):7577–7584. doi: 10.4049/jimmunol.179.11.7577. [DOI] [PubMed] [Google Scholar]
- 37.Ye M, Zhang H, Amabile G, et al. C/EBPa controls acquisition and maintenance of adult hematopoietic stem cell quiescence. Nat Cell Biol. 2013;15(4):385–394. doi: 10.1038/ncb2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sathe P, Vremec D, Wu L, Corcoran L, Shortman K. Convergent differentiation: myeloid and lymphoid pathways to murine plasmacytoid dendritic cells. Blood. 2012;121(1):11–19. doi: 10.1182/blood-2012-02-413336. [DOI] [PubMed] [Google Scholar]
- 39.Schmid MA, Kingston D, Boddupalli S, Manz MG. Instructive cytokine signals in dendritic cell lineage commitment. Immunol Rev. 2010;234(1):32–44. doi: 10.1111/j.0105-2896.2009.00877.x. [DOI] [PubMed] [Google Scholar]
- 40.Watowich SS, Liu YJ. Mechanisms regulating dendritic cell specification and development. Immunol Rev. 2010;238(1):76–92. doi: 10.1111/j.1600-065X.2010.00949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meredith MM, Liu K, Darrasse-Jeze G, et al. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J Exp Med. 2012;209(6):1153–1165. doi: 10.1084/jem.20112675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meredith MM, Liu K, Kamphorst AO, et al. Zinc finger transcription factor zDC is a negative regulator required to prevent activation of classical dendritic cells in the steady state. J Exp Med. 2012;209(9):1583–1593. doi: 10.1084/jem.20121003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwama A, Osawa M, Hirasawa R, et al. Reciprocal roles for CCAAT/enhancer binding protein (C/EBP) and PU.1 transcription factors in Langerhans cell commitment. J Exp Med. 2002;195(5):547–558. doi: 10.1084/jem.20011465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang DE, Hetherington CJ, Meyers S, et al. CCAAT enhancer-binding protein (C/EBP) and AML1 (CBF alpha2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol Cell Biol. 1996;16(3):1231–1240. doi: 10.1128/mcb.16.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Novershtern N, Subramanian A, Lawton LN, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144(2):296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belz GT, Nutt SL. Transcriptional programming of the dendritic cell network. Nat Rev Immunol. 2012;12(2):101–113. doi: 10.1038/nri3149. [DOI] [PubMed] [Google Scholar]
- 47.Chen SS, Chen JF, Johnson PF, Muppala V, Lee YH. C/EBPbeta, when expressed from the C/ebpalpha gene locus, can functionally replace C/EBPalpha in liver but not in adipose tissue. Mol Cell Biol. 2000;20(19):7292–7299. doi: 10.1128/mcb.20.19.7292-7299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu K, Nussenzweig MC. Development and homeostasis of dendritic cells. Eur J Immunol. 2010;40(8):2099–2102. doi: 10.1002/eji.201040501. [DOI] [PubMed] [Google Scholar]
- 49.Liu K, Nussenzweig MC. Origin and development of dendritic cells. Immunol Rev. 2010;234(1):45–54. doi: 10.1111/j.0105-2896.2009.00879.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.