Abstract
Cancer cells exhibit metabolic dependencies that distinguish them from their normal counterparts1. Among these addictions is an increased utilization of the amino acid glutamine (Gln) to fuel anabolic processes2. Indeed, the spectrum of Gln-dependent tumors and the mechanisms whereby Gln supports cancer metabolism remain areas of active investigation. Here we report the identification of a non-canonical pathway of Gln utilization in human pancreatic ductal adenocarcinoma (PDAC) cells that is required for tumor growth. While most cells utilize glutamate dehydrogenase (GLUD1) to convert Gln-derived glutamate (Glu) into α-ketoglutarate in the mitochondria to fuel the tricarboxylic acid (TCA) cycle, PDAC relies on a distinct pathway to fuel the TCA cycle such that Gln-derived aspartate is transported into the cytoplasm where it can be converted into oxaloacetate (OAA) by aspartate transaminase (GOT1). Subsequently, this OAA is converted into malate and then pyruvate, ostensibly increasing the NADPH/NADP+ ratio which can potentially maintain the cellular redox state. Importantly, PDAC cells are strongly dependent on this series of reactions, as Gln deprivation or genetic inhibition of any enzyme in this pathway leads to an increase in reactive oxygen species and a reduction in reduced glutathione. Moreover, knockdown of any component enzyme in this series of reactions also results in a pronounced suppression of PDAC growth in vitro and in vivo. Furthermore, we establish that the reprogramming of Gln metabolism is mediated by oncogenic Kras, the signature genetic alteration in PDAC, via the transcriptional upregulation and repression of key metabolic enzymes in this pathway. The essentiality of this pathway in PDAC and the fact that it is dispensable in normal cells may provide novel therapeutic approaches to treat these refractory tumors.
The prognosis of patients with PDAC remains dismal. The disease is extremely aggressive and is profoundly resistant to all forms of therapy3. Thus, there is a strong impetus to identify new therapeutic targets for this cancer. In recent years, there has been renewed interest in understanding the altered metabolism in cancer, and how such dependencies can be targeted for therapeutic gain. However, achieving a successful therapeutic index remains a major challenge to the development of effective cancer therapies that target metabolic pathways.
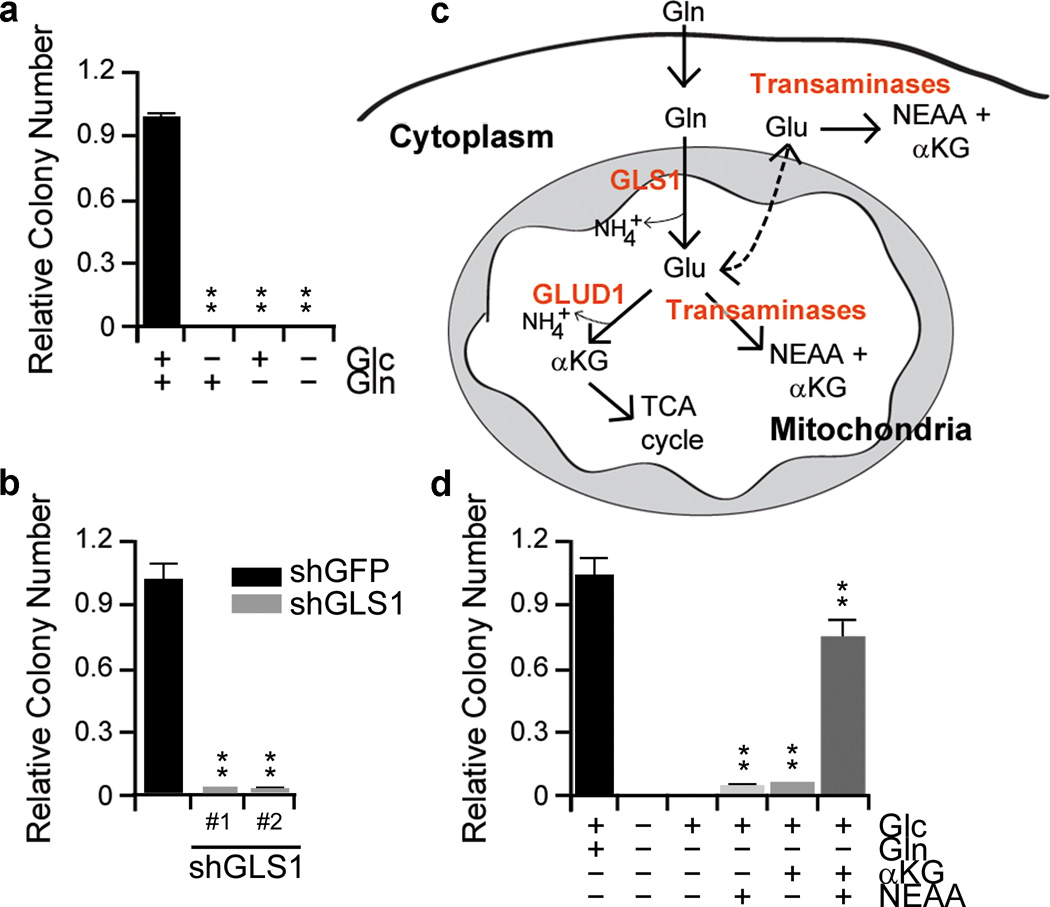
Recent evidence demonstrates that some cancer cells utilize glutamine (Gln) to support anabolic processes that fuel proliferation2. However, the importance of Gln metabolism in pancreatic tumor maintenance is not known. Thus, we sought to explore the dependence of PDAC on Gln, and to examine the functional role of Gln in PDAC metabolism. As expected from our previous work4, glucose was required for PDAC growth. Additionally, PDAC cells were also profoundly sensitive to Gln deprivation, indicating that Gln is also critical for PDAC growth (Fig. 1a and Supplementary Fig. 1).
Figure 1. PDAC utilize a non-canonical glutamine metabolism pathway.
a, PDAC proliferation requires both glucose and glutamine. Cells were plated in the complete media (10mM glucose and 2mM Gln) which was replaced the following day with glucose or Gln-free medium supplemented with 10% dialyzed FBS. b, Relative clonogenic growth of 8988T cells expressing a control shRNA (shGFP) or two independent shRNAs to GLS1. c, Schematic overview of GLUD1- or transaminase-mediated Glu metabolism. d, Relative clonogenic growth of 8988T cells. αKG (4mM), NEAA mixture (0.1 mM glycine, alanine, aspartate, asparagine, proline and serine) or the combination was added to media following Gln-withdrawal. αKG, α-ketoglutarate; Glc, glucose; Gln, glutamine; NEAA, non-essential amino acid. Error bars represent s.d. of triplicate wells from a representative experiment. **, p< 0.01.
Gln provides a carbon source to fuel the TCA cycle and nitrogen for nucleotide, nonessential amino acid (NEAA) and hexosamine biosynthesis5,6. To assess the role of Gln metabolism in PDAC growth, we first impaired glutaminase (GLS) activity using RNA interference (RNAi). Notably, GLS knockdown markedly reduced PDAC growth (Fig. 1b and Supplementary Fig. 2a, b). Consistent with this observation, Glutamate (Glu) was able to support growth in Gln-free conditions (Supplementary Fig. 2c).
Glu can be converted into α-ketoglutarate (αKG) to replenish the TCA cycle metabolites through two mechanisms1; either by glutamate dehydrogenase (GLUD1) or transaminases (Fig. 1c). Indeed, many cancer cells rely on GLUD1-mediated Glu deamination to fuel the TCA cycle7, and αKG has been shown to be an essential metabolite in Gln metabolism8. Surprisingly, dimethyl αKG9 did not restore growth upon Gln deprivation (Fig. 1d), whereas the combination of αKG and an NEAA mixture (the output of transaminase-mediated Glu metabolism) dramatically rescued proliferation in multiple PDAC lines (Fig. 1d and Supplementary Fig. 2d, e). Together, this data suggests that PDAC cells metabolize Gln in a manner that is different from canonical models10 and that this class of enzymes may be critical for Gln metabolism in PDAC.
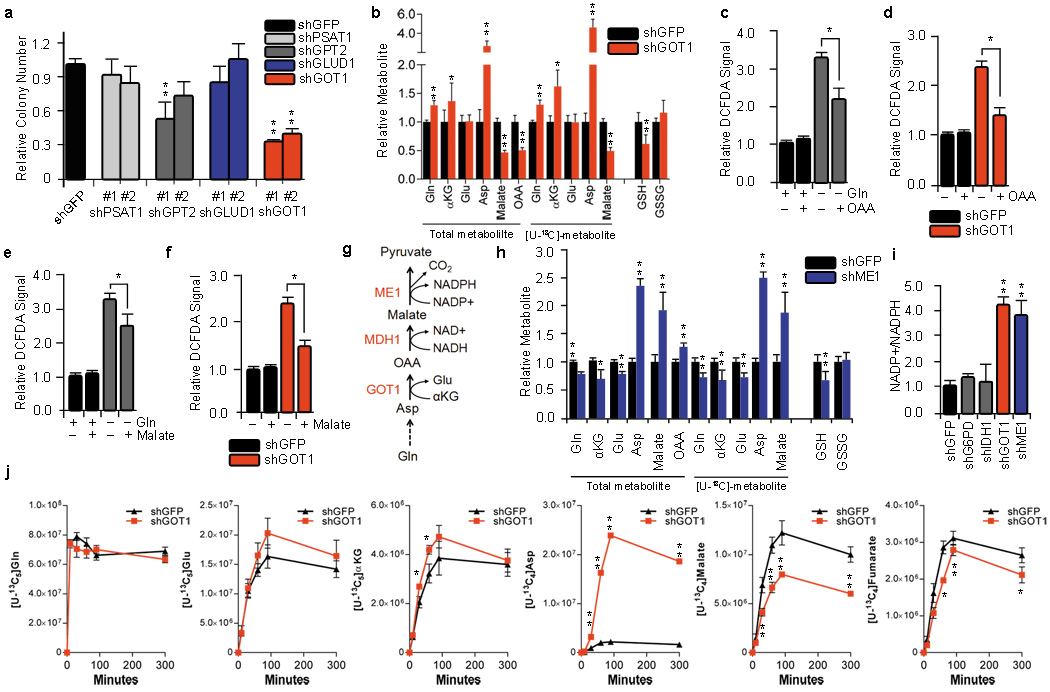
To confirm the importance of transaminases in PDAC Gln metabolism, we treated PDAC cells with either aminooxyacetate (AOA), a pan-inhibitor of transaminases11, or epigallocatechin gallate (EGCG), an inhibitor of GLUD112. While EGCG had no effect on PDAC growth, AOA treatment robustly inhibited the growth of multiple PDAC cell lines (Supplementary Fig. 3). Consistent with these results, GLUD1 knockdown also had no effect on PDAC growth (Fig. 2a). To identify the specific transaminase(s) involved in PDAC Gln metabolism, we inhibited a panel of Glu-dependent transaminases (aspartate, alanine and phosphoserine transaminase) individually using RNAi and examined the effect on PDAC growth. Interestingly, knockdown of the aspartate transaminase GOT1 significantly impaired PDAC growth in multiple PDAC cell lines and primary PDAC cells (Fig. 2a and Supplementary Fig. 4, 5).
Figure 2. GOT1 is essential for redox balance and growth in PDAC.
a, Relative clonogenic growth of 8988T cells expressing a control shRNA (shGFP) or two independent shRNAs targeting GLUD1, GOT1, GPT2 or PSAT1. Error bars represent s.d. of triplicate wells from a representative experiment. b and h, Relative metabolite abundance in 8988T cells grown in [U-13C5]-Gln upon GOT1 or ME1 knockdown. Data are presented as the total metabolite pool (encompassing both metabolite derived from Gln and that not Gln-derived, left) and the [U-13C]-labeled and Gln-derived metabolite pool (right). Error bars represent the s.d. of three independently prepared samples. c, d, e and f, Relative ROS levels in 8988T cells under conditions indicated as determined by DCFDA staining. DCFDA assay was performed 24hr after supplementing Gln-free media with either OAA (4mM) or dimethyl malate (4mM). Each bar represents the mean of three independent experiments with error bars representing the s.d. g, Schematic depiction of the cytoplasmic reactions that convert Asp into pyruvate. i, NADP+/NADPH ratio in 8988T cells expressing a control shRNA (shGFP), or an shRNA to G6PD, IDH1, GOT1 or ME1. Error bars represent s.d. of five replicate wells from a representative experiment. j, Flux of the Gln carbon skeleton into downstream metabolites as a function of time. The reads for uniformly 13C-labeled metabolites, presented in ion current, are plotted for cells expressing the shGFP control or shRNA to GOT1. Asp, aspartate; OAA, oxaloacetate; Glu, glutamate; GSH, reduced glutathione; GSSH, oxidized glutathione. *, p< 0.05; **, p< 0.01.
We next explored the direct effects of GOT1 on Gln metabolism by performing targeted metabolomic analysis in GOT1 knockdown PDAC cells using uniformly 13C-labeled Gln ([U-13C5]-Gln) as a tracer4,13. GOT1 catalyzes the conversion of aspartate (Asp) and αKG into OAA and Glu in the cytoplasm. Indeed, GOT1 knockdown led to increased Gln-derived Asp (and total Asp) and decreased OAA (Fig. 2b and Supplementary Fig. 6a). Interestingly, we also observed a significant decrease in the ratio of reduced-to-oxidized glutathione (GSH:GSSG; Fig. 2b and Supplementary Fig. 6b), suggesting that GOT1 may play a role in the maintenance of cellular redox homeostasis. It should be noted that the changes in metabolite abundance described in this experiment are representative of the total cellular metabolite pool, due to technical limitations associated with organelle-specific metabolite isolation. Importantly however, the results we obtained are consistent with what one would expect if flux through GOT1 was impaired.
In PDAC, we recently demonstrated that oncogenic Kras enhances the flux of glycolytic intermediates specifically through the non-oxidative arm of the pentose phosphate pathway (PPP) (for DNA/RNA biosynthesis) without effecting the NADPH-producing oxidative arm4. This decoupling of ribose biogenesis from NADPH production by Kras suggests that PDAC cells may rely on an alternative mechanism to maintain cellular redox balance. Indeed, inhibition of the oxidative PPP in PDAC by knockdown of the rate limiting enzyme G6PD had minimal effect on reactive oxygen species (ROS) levels, and, consistent with this, glucose deprivation had only a modest impact on ROS (Supplementary Fig. 6c). Therefore, we speculated that the GOT1-mediated conversion of Gln-derived Asp into OAA functions in a pathway that is used to generate the NADPH which could be used to maintain redox balance. To test this hypothesis, we assessed ROS levels upon Gln deprivation in the absence or presence of OAA. Indeed, Gln deprivation induced ROS and OAA could partially rescue the elevated ROS levels (Fig. 2c). GOT1 knockdown also increased ROS levels, which again were significantly restored upon supplementation with OAA (Fig. 2d).
Given that Gln-derived malate (and total malate) was significantly reduced upon GOT1 knockdown (Fig. 2b), we suspected that Gln-derived OAA is metabolized into malate, which is utilized by malic enzyme (ME1) to create NADPH, providing the reducing power to maintain reduced glutathione pools14. Indeed, malate was able to partially rescue the oxidative stress imposed by Gln-deprivation (Fig. 2e) or GOT1 knockdown (Fig. 2f). These data are consistent with a model whereby Gln-derived Asp is converted by GOT1 into OAA, then converted into malate by malate dehydrogenase (MDH1) and subsequently oxidized by ME1 into pyruvate and reducing power in the form of NADPH (Fig. 2g). Consistent with this pathway, metabolomic analysis of [U-13C5]-Gln tracing in ME1 knockdown cells revealed a significant increase in Asp, malate and OAA and decreased GSH (Fig. 2h and Supplementary Fig. 6d). Furthermore, knockdown of GOT1 and ME1 markedly increased the cellular NADP+/NADPH ratio (Fig. 2i), whereas inhibition of other cytosolic sources of NADPH (G6PD or isocitrate dehydrogenase, IDH1) had no effect on NADP+/NADPH ratios or ROS (Fig. 2i and Supplementary Fig. 6c). Together the data suggest that PDAC utilize Gln through the pathway depicted in Fig. 2g to increase the NADPH/NADP+ ratio for maintenance of redox homeostasis. Lastly, Gln tracing kinetic flux experiments in GOT1 knockdown cells clearly demonstrate decreased flux through this pathway (Fig. 2j and Supplementary Fig. 7). Interestingly, lactate labeling in the 13C-labeling experiments was typically at very low levels, indicating that the pyruvate produced by ME1 is not utilized to make lactate by lactate dehydrogenase and has a yet undetermined fate.
The majority of Asp in PDAC cells (50–75%) is derived from Gln, as evidenced by 13C-labeling (Supplementary Fig. 6a, d and 8a). In principle, uniformly 13C-labeled Asp can be derived from Gln following either (i) the GLUD1-mediated conversion of Glu to αKG (and its subsequent traversing through the TCA cycle) or (ii) the mitochondrial aspartate transaminase (GOT2)-mediated conversion of Glu and OAA to αKG and Asp. Of these two enzymes, only GOT2 knockdown significantly impacted PDAC growth (Fig. 2a and Supplementary Fig. 8b). Consistent with this observation, GLUD1 knockdown did not affect Asp biosynthesis from Gln, whereas GOT2 knockdown resulted in a significant decrease in Gln-derived Asp in PDAC cells (Supplementary Fig. 8a, c).
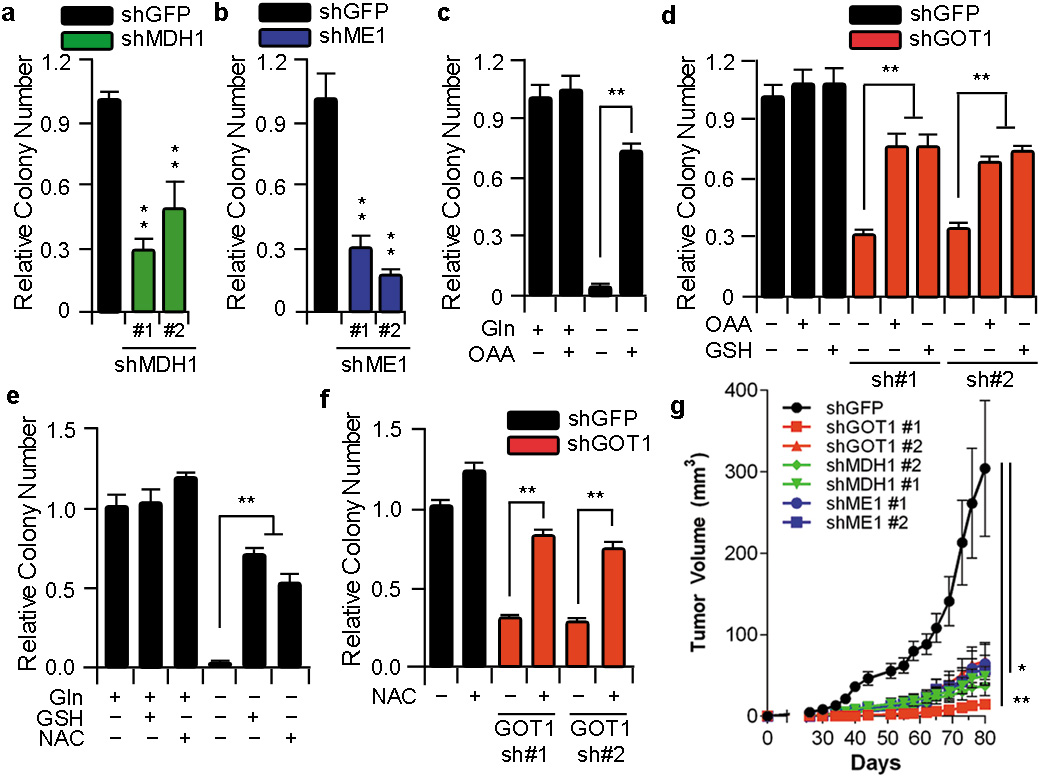
We next tested whether other components of this pathway are also necessary to support PDAC growth. Indeed, knockdown of either MDH1 or ME1 also dramatically inhibited clonogenic survival of PDAC cells (Fig. 3a, b and Supplementary Fig. 9) in a manner similar to GOT1 knockdown. As a further test of enzyme knockdown specificity, we investigated the ability of central metabolites in this pathway to rescue PDAC growth upon Gln deprivation. First, we confirmed that exogenously added metabolites permeated the cell and populated metabolite pools (Supplementary Fig. 10). Next, we demonstrated that the combination of GOT1 substrates, Asp and αKG, could rescue cell growth in Gln-free conditions (Supplementary Fig. 11a). Additionally, OAA permitted PDAC growth under Gln-free conditions in multiple PDAC cell lines (Fig. 3c and Supplementary Fig. 11a–d) as well as upon both GLS (Supplementary Fig. 12a) and GOT1 knockdown (Fig. 3d and Supplementary Fig. 13a, b). Lastly, the addition of dimethyl-malate15 was able to partially rescue PDAC cell growth upon Gln deprivation (Supplementary Fig. 13c) or GOT1 knockdown (Supplementary Fig. 13d).
Figure 3. Metabolism of the Gln carbon skeleton through GOT1, MDH1 and ME1 supports PDAC growth by maintaining redox balance.
a and b, Relative clonogenic growth of 8988T cells expressing a control shRNA (shGFP) or two independent shRNAs to MDH1 or ME1. c and e, Relative clonogenic growth of 8988T cells under conditions indicated. Cells were plated in complete culture media (10mM glucose and 2mM Gln), which was replaced the following day with Gln-free medium supplemented with OAA (4mM), GSH (4mM) or N-acetylcysteine (NAC) (4mM). d and f, Relative clonogenic growth of 8988T cells expressing a control shRNA (shGFP) or two independent shRNAs to GOT1 with or without OAA (4mM), GSH (4mM) or NAC (4mM). Error bars represent the s.d. of triplicate wells from a representative experiment. g, Xenograft growth of 8988T cells expressing a control shRNA (shGFP), shRNAs to GOT1 (#1 and #2), MDH1 (#1 and #2) or ME1 (#1 and #2) in mice. Error bars represent s.e.m (n=10). *, p< 0.05; **, p< 0.01.
Next, to confirm that the OAA- or malate rescue of PDAC growth upon Gln deprivation was through maintenance of redox homeostasis, we treated cells grown in Gln-free conditions with a cell permeable GSH analog. Remarkably, GSH dramatically rescued clonogenic growth following Gln-deprivation (Fig. 3e and Supplementary Fig. 11c) or GOT1 knockdown (Fig. 3d and Supplementary Fig. 13a, b). GSH was also able to rescue either MDH1 or ME1 knockdown (Supplementary Fig. 14a). Near identical results were also seen with the antioxidant N-acetylcysteine (NAC) (Fig. 3e and f and Supplementary Fig. 14b, c). Together, these data support the idea that Gln is utilized by PDAC cells to maintain redox homeostasis, which is required to support continued tumor growth.
As further confirmation of the importance of this pathway in PDAC, we suppressed GOT1, MDH1, and ME1 expression using two lentiviral shRNAs in PDAC cells and assessed their ability to grow as xenografts. Consistent with our in vitro results, both GLUD1 shRNAs had no effect on tumor growth (Supplementary Fig. 15). In contrast, GOT1, MDH1, and ME1 knockdown each robustly diminished tumor growth (Fig. 3g). These data provide further support for the critical role of this pathway in Gln metabolism and PDAC tumor growth.
In contrast to PDAC, this pathway appears to be dispensable in normal cells. Indeed, treatment of non-transformed human pancreatic ductal cells (HPDE) and human diploid fibroblasts (IMR90) with AOA had only modest effects on growth (Supplementary Fig. 16a). HPDE cells, unlike PDAC cells, were significantly sensitive to EGCG, suggesting a greater reliance on the activity of GLUD1 (Supplementary Fig. 16b). Consistent with these results, GOT1 knockdown did not impair the growth of HPDE and IMR90 (Supplementary Fig. 16c). We also obtained similar results in mouse ductal epithelial cells (mPDE) and mouse PDAC cell lines, with the mPDEs being highly insensitive to AOA and GOT1 knockdown and two independently derived mouse PDAC lines exhibiting significant sensitivity to AOA and GOT1 knockdown (Supplementary Fig. 16d, e). Furthermore, using an inducible shGOT1 construct, we demonstrated that mouse PDAC rely on GOT1 to sustain tumor growth in vivo (Supplementary Fig. 17). Collectively, these data demonstrate that the GOT1-mediated utilization of the Gln carbon skeleton is a metabolic adaptation that PDAC, and not normal cells, have uniquely acquired to support growth.
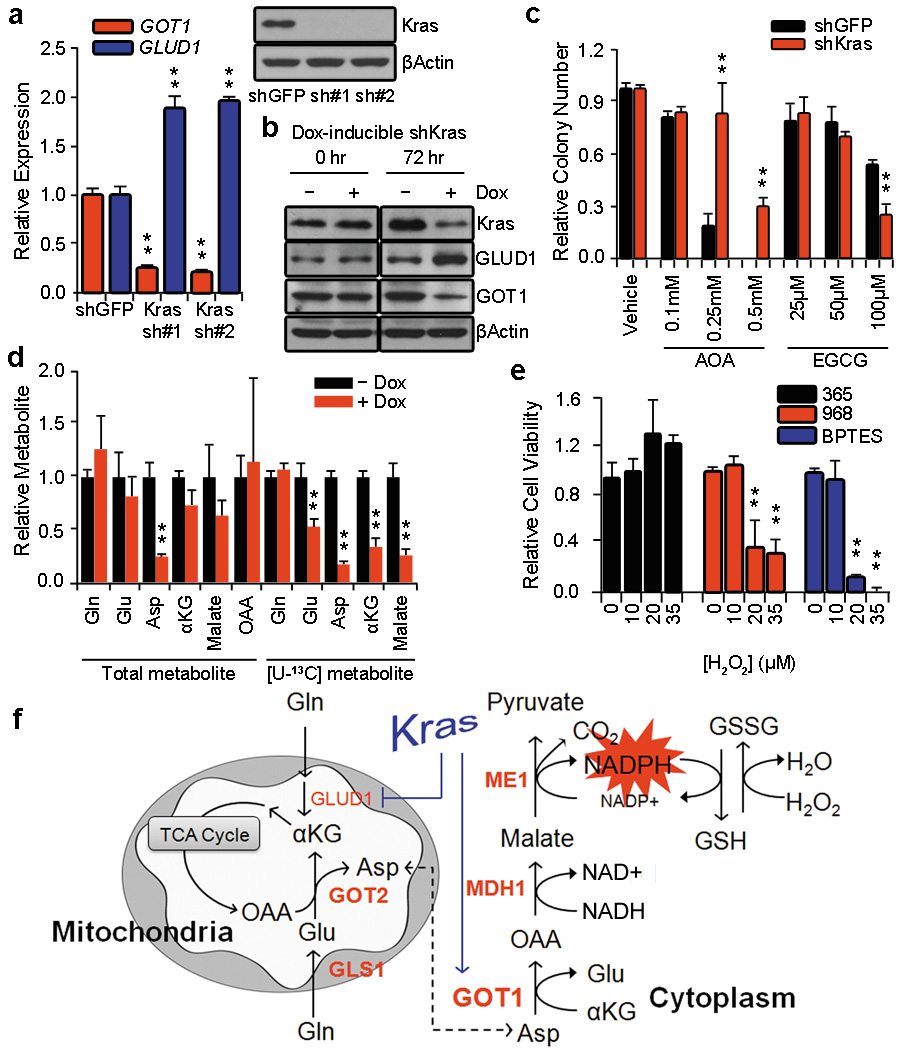
Our previous work demonstrated that anabolic glucose metabolism in PDAC is controlled by oncogenic Kras, which leads to altered expression of a number of rate-limiting metabolic enzymes4. To investigate the role of Kras in the reprogramming of Gln metabolism, we assessed the expression of GOT1 and GLUD1 upon knockdown of Kras in PDAC cells. Interestingly, Kras knockdown resulted in a marked increase in GLUD1 and a decrease in GOT1 expression at the transcriptional level (Fig. 4a), as well as the protein level (Fig. 4b) in multiple PDAC lines (Supplementary Fig. 18a). Additionally, using five independent orthotopic tumors derived from our inducible Kras PDAC model4, we show that expression of GOT1 increased and GLUD1 decreased in an oncogenic Kras-dependent manner in vivo (Supplementary Fig. 18b). These findings demonstrate that, in PDAC, oncogenic Kras plays a critical role in coordinating the shift in Gln metabolism to maintain tumor growth and survival.
Figure 4. Oncogenic Kras mediates Gln reprogramming in PDAC.
a, Expression of GLUD1 and GOT1 was determined by quantitative RT-PCR in 8988T cells expressing a control shRNA (shGFP) or two independent shRNAs targeting Kras. Western blot confirmed knockdown of Kras expression. b, The effect of Kras knockdown on GLUD1 or GOT1 protein levels in Panc1 cells expressing a doxycycline-inducible Kras shRNA. c, Relative clonogenic growth of 8988T cells expressing a control shRNA (shGFP) or shRNA to Kras following treatment with AOA or EGCG. Error bars represent the s.d. of triplicate wells from a representative experiment. d, Relative metabolite abundance in Miapaca2 cells grown in [U-13C5]-Gln upon doxycycline-inducible Kras knockdown. Data are presented as the total metabolite pool (encompassing both metabolite derived from Gln and that not Gln-derived, left) and the [U-13C]-labeled and Gln-derived metabolite pool (right). Error bars represent the s.d. of three independently prepared samples. e, Relative cell viability of 8988T cells treated with GLS inhibitors 968 (active) (10µM), 365 (inactive analog) (50µM), or BPTES (100nM) with increasing concentrations of H2O2. . Error bars represent the s.d. of triplicate wells from a representative experiment. f, Model depicting the Kras-regulated Gln metabolic reprogramming in PDAC used to maintain redox and support growth. AOA, aminooxyacetate; EGCG, epigallocatechin gallate. *, p< 0.05; **, p< 0.01.
We next assessed the sensitivity of PDAC cells to either AOA or EGCG upon Kras knockdown using 8988T cells, a cell line that is not dependent on Kras for survival16. Consistent with our previous results, AOA significantly inhibited clonogenic growth, whereas EGCG had minimal effects. Interestingly, Kras knockdown made the cells significantly more resistant to AOA and sensitive to EGCG (Fig. 4c and Supplementary Fig. 18c). To confirm the role of oncogenic Kras in the reprogramming of Gln metabolism, a targeted metabolomic analysis using [U-13C5]-Gln was performed upon Kras knockdown. Indeed, the changes observed were consistent with Kras-supporting the anabolic metabolism of Gln, where multiple metabolites in the GOT1-dependent pathway were significantly deregulated (Fig. 4d and Supplementary Fig. 19).
Given the importance of Gln metabolism in maintaining the redox state of PDAC, we speculated that inhibition of anabolic Gln metabolism may sensitize PDAC to oxidative stress. To test this concept, we inhibited Gln metabolism in PDAC cells using a GLS inhibitor and examined whether this would synergize with hydrogen peroxide (H2O2) treatment. Indeed, two chemically distinct GLS inhibitors8,17 had a growth suppressive effect on both human and mouse PDAC cells (Supplementary Fig. 20a, b), consistent with the GLS knockdown data (Fig. 1b). Furthermore, when combined with H2O2, this effect was dramatically augmented, indicating that PDAC cells are markedly more sensitive to ROS when Gln metabolism is impaired (Fig. 4e and Supplementary Fig. 20c). This finding may have significant therapeutic implications, given that clinical grade GLS inhibitors are being developed18 and that standard PDAC therapies (such as radiation) lead to the generation of ROS. Moreover, since this aspect of Gln metabolism does not appear as critical in normal cells, these data suggest an accessible therapeutic window.
Collectively, our data reveal a novel dependence on the transaminases GOT2 and GOT1 for metabolism of the Gln carbon skeleton in PDAC. These reactions lead to the cytosolic conversion of Asp into OAA, malate and then pyruvate and are required sustain PDAC growth, likely through maintaining redox balance. Importantly, our work also demonstrates that oncogenic Kras mediates this reprogramming of Gln metabolism (Fig. 4f). While this pathway is critical for redox balance and cell growth in PDAC, it does not preclude the involvement of other pathways that may contribute to redox balance such as glutathione synthesis9,19,20 or an NRF2-dependent mechanism21. Furthermore, there may be cell-type specific differences, as Kras-transformed fibroblasts require both GLUD1 and transaminases for cell growth22. Lastly, these findings may have implications for future therapeutic approaches as inhibition of Gln metabolism in PDAC can potentially synergize with therapies that increase intracellular ROS such as chemotherapy and radiation.
Methods Summary
Proliferation and clonogenic assays were performed as previously described23. To characterize Gln metabolism, targeted liquid chromatography-tandem mass spectrometry was performed4. Briefly, cells were grown in complete media and transferred into Gln-free media supplemented with [U-13C5]-Gln overnight (steady state) or for the indicated timepoints (flux analyses). For subcutaneous xenografts, PDAC cells infected with lentivrial shRNAs to suppress target gene expression were suspended in 100 µl HBSS and injected subcutaneously into the lower flank of NCr nude mice. For mouse xenografts, murine PDAC cells stably infected with a doxycycline-inducible GOT1 shRNA construct were injected. Animals were fed with doxycycline water starting on the day of injection or when tumor volume reached ~50mm3. Complete images of western blots are presented in Supplementary Fig. 21.
Methods
Cell culture
Cell lines were obtained from the American Type Culture Collection or the German Collection of Microorganisms and Cell Cultures. All cell lines were tested routinely, and prior to all metabolomic analyses, for mycoplasma contamination. RPMI-1640, fetal bovine serum and dialyzed fetal bovine serum (dFBS) were purchased from Invitrogen. Glucose free DMEM (containing 2 mM Gln), dimethyl αKG, Asp, GSH reduced ethyl ester, OAA, dimethyl malate and DMEM power (without glucose and Gln) were obtained from Sigma, and Gln-free RPMI 1640 was purchased from Cellgro. Cosmic calf serum (CCS) was obtained from Thermo Scientific. Cells were cultured in the following media: 8988T, Panc1, MPanc96, Miapaca2 and PL45 in DMEM supplemented with 10 mM glucose and 10% CCS; 8902 in RPMI with 10% CCS; IMR90 in MEM with 10% FBS; HPDE cells were cultured as described previously24. Primary human PDAC lines were generated from ascites fluid under IRB approved protocols 02–240 and 2007P001918. Lines were sequenced and confirmed to have Kras mutations.
Cell proliferation assay
Cells were plated in 24-well plates at 2,000 cells per well in 0.5 mL of media. To deprive Gln, cells were plated in complete culture media (10mM glucose and 2mM Gln) which was exchanged with Gln-free medium supplemented with 10% dFBS the following day. Media was not changed throughout the course of the experiment. At the indicated time points, cells were fixed in 10% formalin and stained with 0.1% crystal violet. Dye was extracted with 10% acetic acid and the relative proliferation was determined by OD at 595 nm.
Clonogenic assay
Cells were plated in 6-well plates at 300 cells per well in 2 mL of media. Media was not changed throughout the course of the experiment. After 7–10 days, colonies were fixed in 80% methanol and stained with 0.2% crystal violet.
Quantitative RT-PCR
Total RNA was extracted using TRIzol (Invitrogen) and reverse transcription was performed from 2 µg of total RNA using oligo-dT and MMLV HP reverse transcriptase (Epicentre), according to the manufacturer’s instructions. Quantitative RT-PCR was performed with SYBR Green dye using an Mx3000PTM (Stratagene). PCR reactions were performed in triplicate and the relative amount of cDNA was calculated by the comparative CT method using the 18S ribosomal RNA sequences as a control. Primer sequences available upon request.
Xenograft studies
For subcutaneous xenografts, 8988T cells were infected with lentiviral shRNAs targeting GLUD1 (n=2), GOT1 (n=2), MDH1 (n=2), ME1 (n=2) and GFP (control hairpin, n=1) and subjected to a short puromycin selection (2 µg/mL); shRNA sequences below. 1.5×106 cells, suspended in 100 µL Hanks Buffered Saline Solution (HBSS), were injected subcutaneously into the lower flank of NCr nude mice (Taconic). Tumor length and width were measured twice weekly and the volume was calculated according to the formula (length × width2)/2. All xenograft experiments with human PDAC lines were approved by the HMS Institutional Animal Care and Use Committee (IACUC) under protocol number 04–605. For mouse xenografts, a doxycycline-inducible GOT1 shRNA construct was first generated. For generation of the construct, oligonucleotides to mouse GOT1 shRNA (forward : CCGGCCACATGAGAAGACGTTTCTTCTCGAGAAGAAACGTCTTCTCATGTGGTTTTTG; reverse : AATTCAAAAACCACATGAGAAGACGTTTCTTCTCGAGAAGAAACGTCTTCTCATGTGG) were digested to generate sticky ends (AgeI and EcoRI) and immediately subcloned into the AgeI-EcoRI sites of the pLKO-Tet-on vector. For subcutaneous xenograft, 106 stably infected murine PDAC cells were suspended in 100 µl Hanks Buffered Saline Solution and injected subcutaneously into the lower flank of NCr nude mice (Taconic). Animals were fed with doxycycline water (doxycycline 2g/L, sucrose 20g/L) starting on the day of injection or when tumor diameter reached 50mm. Tumor volumes were measured every third day starting from day 4 post-injection and calculated as above. These xenograft experiments were approved under MDACC IACUC protocol 111113931.
Western blot analysis
After SDS-PAGE, proteins were transferred to Hybond-N Nitrocellulose (Amersham Biosciences). Membranes were blocked in Tris-buffered saline (TBS) containing 5% non-fat dry milk and 0.1% Tween 20 (TBS-T), prior to incubation with the primary antibody overnight at 4°. The membranes were then washed with TBS-T followed by exposure to the appropriate horseradish peroxidase-conjugated secondary antibody for 1h and visualized on Kodak X-ray film using the enhanced chemiluminescence (ECL) detection system (Thermo Scientific). The following antibodies were used: Kras (F234, Santa Cruz), GOT1 (NBP1-54778, Novus), GLUD1 (ab55061, Abcam) and β-Actin (A2066, Sigma).
ROS Quantification
DCFDA assay was performed 24hr after supplementing Gln-free media with either OAA (4mM) or dimethyl malate (4mM). Cells were incubated with 5µM 2’,7’-dichlorodihydrofluorescein diacetate (DCFDA, Invitrogen) for 30 min. Excess DCFDA was removed by washing the cells twice with PBS, and labeled cells were then trypsinized, rinsed, and resuspended in PBS. Oxidation of DCFDA to the highly fluorescent 2’,7’-dichloro-fluorescein (DCF) is proportionate to ROS generation and was analyzed by flow cytometry.
Metabolomics
For steady state metabolomic analysis, PDAC cell lines were grown to ~50% confluence in growth media (DMEM, 2 mM Gln, 10 mM glucose, 10% CCS) on 10 cm dishes in biological quadruplicate. A complete media change was performed two hours prior to metabolite collection. To trace Gln metabolism, PDAC cell lines were grown as above and then transferred into Gln-free DMEM (with 10 mM glucose) containing 10% dialyzed FBS and 2 mM [U-13C5]-Gln (Cambridge Isotope Labs) overnight (for steady state labeling) or for the indicated timepoints in the flux analyses. Additionally, fresh media containing [U-13C5]-Gln was exchanged 2 hr prior to metabolite extraction for steady state analyses. The quantity of the metabolite fraction analyzed was adjusted to the corresponding protein concentration calculated upon processing a parallel 10cm dish. Metabolite fractions were collected and analyzed by targeted LC-MS/MS via selected reaction monitoring (SRM), as described5,14. Processed data was analyzed in Cluster 3.0 and TreeViewer.
Measurement of Sensitization of PDAC cells to ROS
PDAC cell lines were plated into 96-well plates at 103 cells/well in 200 µL of growth media. The following day, growth media was replaced with that containing GLS inhibitors and/or H2O2. Parallel plates were analyzed at 3, 6 and 9 days by Cell Titer Glo analysis (Promega), per the manufacturer’s instruction. The GLS inhibitors 968 (active) and 365 (structurally similar, inactive) were provided as a kind gift from the Cerione laboratory9. BPTES was a kind gift from Jaime Escobedo (Forma Therapeutics).
Lentiviral-mediated shRNA Targets
All shRNA vectors were obtained from the RNA Interference Screening Facility of Dana Farber Cancer Institute. The sequences and RNAi Consortium clone IDs for each shRNA are as follows. shGLS-1 : GCACAGACATGGTTGGTATAT (TRCN0000051135), shGLS-2 : GCCCTGAAGCAGTTCGAAATA (TRCN0000051136), shMDH1-1 : CCCTGTTGTAATCAAGAATAA (TRCN0000221892), shMDH1-2 : GCAACAGATAAAGAAGACGTT (TRCN0000221893), shME1-1 : GCCTTCAATGAACGGCCTATT (TRCN0000064728), shME1-2 : CCAACAATATAGTTTGGTGTT (TRCN0000064729), shGLUD1-1, CCCAAGAACTATACTGATAAT (TRCN0000220878), shGLUD1-2 : GCAGAGTTCCAAGACAGGATA (TRCN0000220880), shGOT1-1 : GCGTTGGTACAATGGAACAAA (TRCN0000034784), shGOT1-2 : GCTAATGACAATAGCCTAAAT (TRCN0000034785), shGPT2-1 : CGGCATTTCTACGATCCTGAA (TRCN0000035024), shGPT2-2 : CCATCAAATGGCTCCAGACAT (TRCN0000035025), shPSAT1-1 : GCCAAGAAGTTTGGGACTATA (TRCN0000035264), shPSAT1-2 : CCAGACAACTATAAGGTGATT (TRCN0000035265), shKras-1 : CCTCGTTTCTACACAGAGAAA (TRCN0000040148), shKras-2 : GAGGGCTTTCTTTGTGTATTT (TRCN0000033260).
Regents
NADP+/NADPH ratios were determined using the NADP/NADPH assay kit (Abcam; ab65349) according to the manufacturer’s instructions. Briefly, 105 cells (n=6 wells of a 6-well dish) were collected on ice in extraction buffer and subject to two rounds of freeze-thaw at −80°C. NADP+ and NADPH values were determined in biological sextuplet and concentration was obtained by comparison to standard curves. OAA was not analyzed by targeted LC/MS-MS due to its limited stability in aqueous solvents at room temperature25. As such, the abundance of this metabolite was determined using a quantification kit (Biovision), according the manufacturer’s instruction. Briefly, 2×106 cells (n=4 10cm dishes) were collected during log-phase growth by trypsinization, re-suspended immediately in the buffers provided (on ice), analyzed and compared to standard curves. The signals obtained were normalized to the protein concentration calculated upon processing a parallel 10cm dish. NEAA mixture consisted of a mixture of 0.1 mM glycine, alanine, aspartate, asparagine, proline and serine.
Supplementary Material
Acknowledgements
We would like to thank Dr. Dimitrios Anastasiou for advice and helpful comments on the manuscript; Min Yuan and Susanne Breitkopf for technical help with mass spectrometry experiments; Drs. Jon Erickson, Richard Cerione (968 and 365) and Jaime Escobedo (BPTES) for the GLS inhibitors. Grant support derives from the Dana Farber Cancer Institute (A.C.K.), National Cancer Institute Grant R01 CA157490 (A.C.K), Kimmel Scholar Award (A.C.K.), AACR-PanCAN Career Development Award (A.C.K.), NIH grants T32 CA009382-26 (H.Y.) and P01 CA117969 (L.C.C. and R.A.D.). C.A.L. is the Amgen Fellow of the Damon Runyon Cancer Research Foundation (DRG-2056-10). NIH grants 5P01CA120964-05 and Dana–Farber/Harvard Cancer Center Support Grant 5P30CA006516-46 (J.M.A.). A.C.K. is a Consultant for Forma Therapeutics. L.C.C. owns equity in, receives compensation from Agios Pharmaceuticals, and serves on the Board of Directors and Scientific Advisory Board of Agios Pharmaceuticals. Agios Pharmaceuticals is identifying metabolic pathways of cancer cells and developing drugs to inhibit such enzymes in order to disrupt tumor cell growth and survival.
Footnotes
Author Contributions:
J.S., C.A.L., L.C.C., and A.C.K. designed the study, interpreted the data and wrote the manuscript. J.S., C.A.L., H.Y., and X.W. performed the experiments. J.M.A., E.M., and N.S. helped with the metabolomic studies and with S.H., M.C.H and R.A.D. assisted in data interpretation. M.L., R.M.P., C.R.F., Y.K., N.B. and J.B.F. developed essential reagents and resources.
References
- 1.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 4.Ying H, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26:877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeBerardinis RJ, Cheng T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shanware NP, Mullen AR, DeBerardinis RJ, Abraham RT. Glutamine: pleiotropic roles in tumor growth and stress resistance. J Mol Med (Berl) 2011;89:229–236. doi: 10.1007/s00109-011-0731-9. [DOI] [PubMed] [Google Scholar]
- 8.Wang JB, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wise DR, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choo AY, et al. Glucose addiction of TSC null cells is caused by failed mTORC1-dependent balancing of metabolic demand with supply. Mol Cell. 2010;38:487–499. doi: 10.1016/j.molcel.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7:872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 15.Huypens P, et al. The dicarboxylate carrier plays a role in mitochondrial malate transport and in the regulation of glucose-stimulated insulin secretion from rat pancreatic beta cells. Diabetologia. 2011;54:135–145. doi: 10.1007/s00125-010-1923-5. [DOI] [PubMed] [Google Scholar]
- 16.Singh A, et al. A gene expression signature associated with "K-Ras addiction" reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLaBarre B, et al. Full-length human glutaminase in complex with an allosteric inhibitor. Biochemistry. 2011;50:10764–10770. doi: 10.1021/bi201613d. [DOI] [PubMed] [Google Scholar]
- 18.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 19.Trachootham D, et al. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell. 2006;10:241–252. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 20.DeBerardinis RJ, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeNicola GM, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaglio D, et al. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol Syst Biol. 2011;7:523. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang S, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ouyang H, et al. Immortal human pancreatic duct epithelial cell lines with near normal genotype and phenotype. Am J Pathol. 2000;157:1623–1631. doi: 10.1016/S0002-9440(10)64800-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bajad SU, et al. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.