SUMMARY
The modular SCF ubiquitin ligases feature a large family of substrate receptors that enable recognition of diverse targets. However, how the repertoire of SCF complexes is sustained remains unclear. Real-time measurements of formation and disassembly indicate that SCFFbxw7 is extraordinarily stable but, in the Nedd8-deconjugated state, is rapidly disassembled by the cullin-binding protein Cand1. Binding and ubiquitylation assays show that Cand1 is a protein exchange factor that accelerates the rate at which Cul1–Rbx1 equilibrates with multiple F-box Protein–Skp1 modules. Depletion of Cand1 from cells impedes recruitment of new F-box proteins to pre-existing Cul1 and profoundly alters the cellular landscape of SCF complexes. We suggest that catalyzed protein exchange may be a general feature of dynamic macromolecular machines and propose a hypothesis for how substrates, Nedd8, and Cand1 collaborate to regulate the cellular repertoire of SCF complexes.
INTRODUCTION
Three enzymes work in succession to covalently attach ubiquitin and ubiquitin chains to target proteins: an ubiquitin activating enzyme (E1), an ubiquitin conjugating enzyme (E2), and an ubiquitin ligase (E3) (Dye and Schulman, 2007). The proteasome, a massive multi-subunit protease, recognizes and degrades proteins conjugated to lysine 48- or lysine 11-linked polyubiquitin chains containing at least four ubiquitins (Thrower et al., 2000; Wickliffe et al., 2011). Cullin–RING ubiquitin ligases (CRLs) are the largest family of E3s and are typified by the SCF complexes, which comprise four subunits: the scaffold Cul1, the RING domain protein Rbx1, the adaptor Skp1, and a substrate-binding F-box protein (Petroski and Deshaies, 2005; Dye and Schulman, 2007). Sixty-nine proteins in the human genome have F-box motifs, at least 42 of which form SCF complexes (Lee et al., 2011). Although this modularity of SCF complexes allows for recognition of diverse substrates, how SCF complex formation is regulated remains unclear.
Cullin-associated and neddylation-dissociated protein 1 (Cand1) is a Cul1 associated protein whose binding is mutually exclusive with the F-box Protein–Skp1 sub-complex and is also blocked by attachment of the ubiquitin-like protein Nedd8 to lysine 720 of Cul1 (Liu et al., 2002; Zheng et al., 2002a; Goldenberg et al., 2004; Bornstein et al., 2006; Duda et al., 2008; Siergiejuk et al., 2009). Neddylation of Cul1 induces a major conformational rearrangement in Cul1–Rbx1 that eliminates a Cand1 binding site and stimulates ubiquitin transfer from associated E2s to substrates (Duda et al., 2008; Saha and Deshaies, 2008). In vitro, Cand1 acts as a stoichiometric inhibitor of cullin neddylation, SCF assembly, and SCF ubiquitin ligase activity (Liu et al., 2002; Zheng et al., 2002a; Goldenberg et al., 2004; Bornstein et al., 2006; Duda et al., 2008; Siergiejuk et al., 2009). However, genetic evidence indicates that Cand1 is a positive regulator of SCF and other CRLs in vivo (Zheng et al., 2002a; Chuang et al., 2004; Feng et al., 2004; Lo and Hannink, 2006; Zhang et al., 2008; Bosu et al., 2010; Kim et al., 2010) suggesting that Cand1-mediated recycling of substrate receptor modules is important for proper CRL function (Liu et al., 2002; Cope and Deshaies, 2003; Dye and Schulman, 2007; Zhang et al., 2008; Schmidt et al., 2009). Here, we demonstrate a protein exchange factor activity for Cand1 that resolves the conflicting biochemical and genetic data, and together with other data on the Nedd8 conjugation cycle enables us to propose a specific model for how the cellular repertoire of CRL complexes is controlled.
RESULTS
Dynamics of SCFFbxw7 Assembly and Disassembly
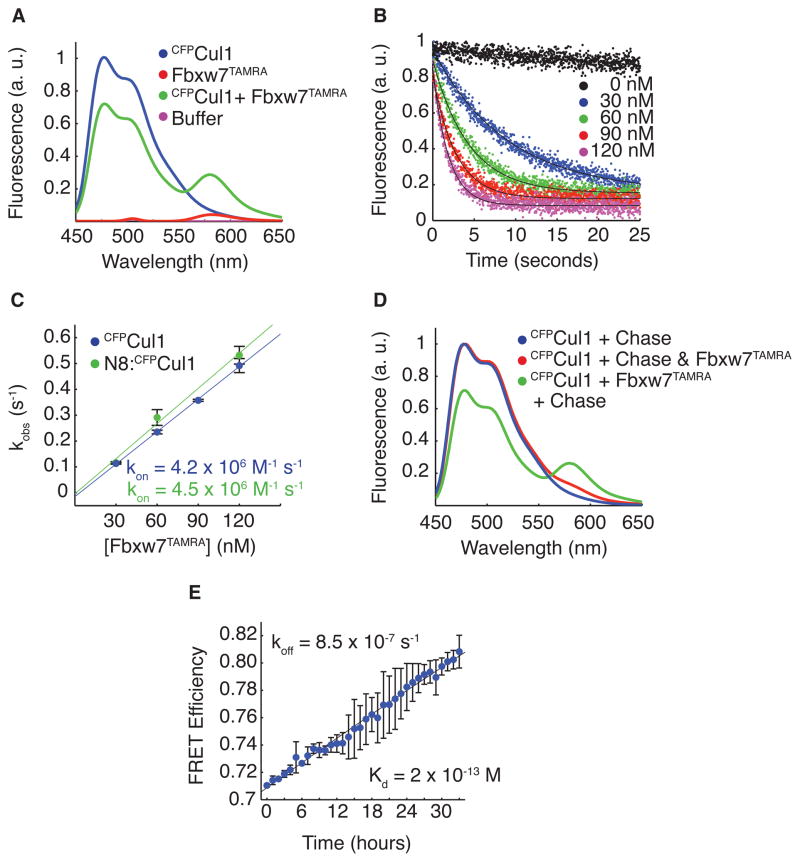
To reconcile the conflicting observations on Cand1, we sought to characterize the assembly properties of SCF complexes by developing a real-time assay based on FRET that monitors the binding dynamics between F-box Protein–Skp1 and Cul1–Rbx1. Fbxw7 co-expressed with Skp1 was tagged at its C-terminus, via the Sortase reaction (Popp et al., 2009; Proft, 2009), with the peptide GGGGK conjugated to the fluorescent dye TAMRA, producing covalently labeled Fbxw7TAMRA. The trans-peptidation reaction was efficient (not shown) and the tag did not compromise ubiquitylation activity (Figure S1A). We observed FRET when Fbxw7TAMRA–Skp1 was mixed with Cul1–Rbx1 in which the N-terminus of Cul1 is fused to cyan fluorescent protein (CFPCul1) (Figure 1A). The association rate constant (kon) for complex assembly, 4 × 106 M−1 s−1, was determined by monitoring donor CFPCul1–Rbx1 fluorescence at varying concentrations of Fbxw7TAMRA – Skp1 in a stopped flow apparatus (Figure 1B and Figure 1C). The FRET observed in our assay could be competed away by excess non-fluorescent Skp2–Skp1 (Figure 1D). Using this chase assay, we measured a dissociation rate constant (koff) for SCFFbxw7 of 9 × 10−7 s−1, or 0.5 week−1 (Figure 1E). These measurements revealed an extraordinarily tight complex with a KD of 200 fM. Neddylation of Cul1 did not affect the maximum FRET efficiency in our assay, nor the rate of association or dissociation of SCFFbxw7, confirming that the neddylation-induced major conformational change in Rbx1 and Cul1’s C-terminal domain does not affect the binding interface of F-box Protein–Skp1 to Cul1’s N-terminal domain (Figure 1C and Figure S1B–S1D) (Zheng et al., 2002b; Petroski and Deshaies, 2005; Duda et al., 2008).
Figure 1. FRET Reveals Properties of SCF Assembly.
(A) Fluorescence emission spectra from excitation at 430 nm of 70 nM CFPCul1–Rbx1, 70 nM Fbxw7TAMRA–Skp1, a mixture of the two, or buffer alone revealed FRET with 30% efficiency upon complex formation. Signals were normalized to peak donor emission at 478 nm.
(B) The change in donor fluorescence versus time in a stopped flow apparatus with 5 nM CFPCul1–Rbx1 and varying concentrations of Fbxw7TAMRA–Skp1. Signal changes were fit to single exponential curves.
(C) The rate of signal change in (B) versus the concentration of Fbxw7TAMRA–Skp1. Fitting the data to (kobs = kon*[Fbxw7] + koff) gave kon of 4 × 106 M−1 s−1 regardless of Cul1’s neddylation status. Error bars: +/− SD, n≥3.
(D) 700 nM Skp2–Skp1 (chase) competed FRET away if pre-incubated with 70 nM Fbxw7TAMRA–Skp1 before, but not after addition of 70 nM CFPCul1 for 5 min.
(E) Fluorescence emission at 478 nm versus time after addition of chase to pre-incubated CFPCul1–Rbx1 and Fbxw7TAMRA–Skp1 normalized to peak donor emission in (D). Single exponential fit with a fixed end point of 1 gave koff of 8.5 × 10−7 s−1. KD is thus 2 × 10−13 M. Error bars: +/− SD, n=3. See also Figure S1.
Cand1 Accelerates SCFFbxw7 Disassembly
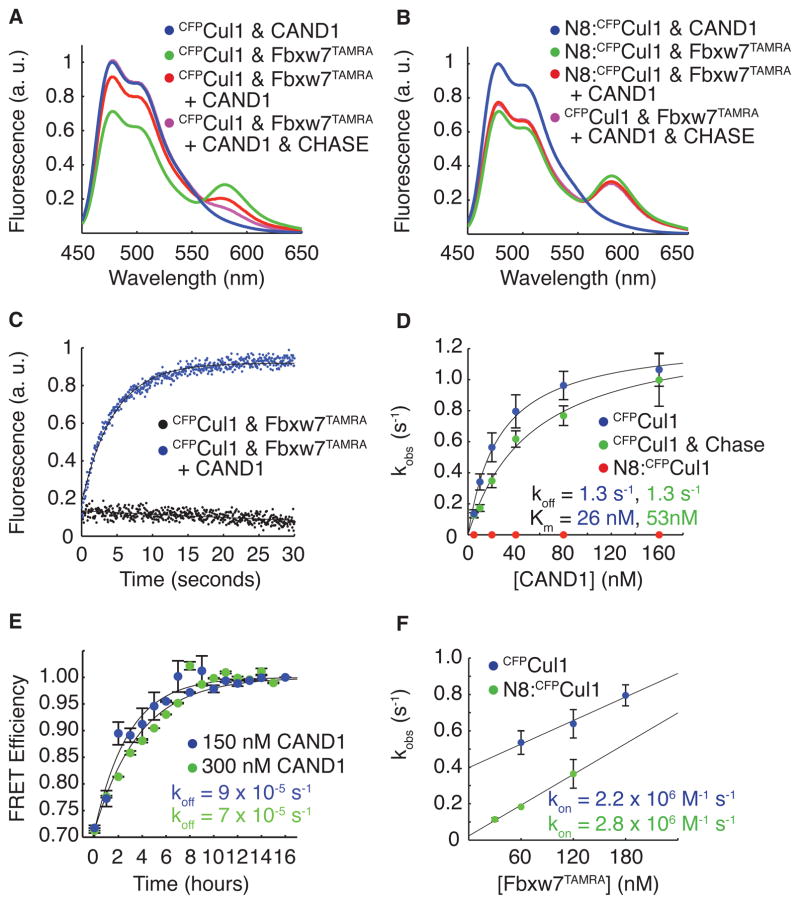
F-box Protein–Skp1 and Cand1 antagonize each other’s binding to Cul1–Rbx1 (Liu et al., 2002; Zheng et al., 2002a; Goldenberg et al., 2004; Bornstein et al., 2006; Duda et al., 2008; Siergiejuk et al., 2009; Lee et al., 2011). To test if this is a general property, we prepared cell lysate from 293 cells that expressed tetracycline-inducible, FLAG-tagged Cul1 and were substantially depleted of Cand1 by stable expression of a lentiviral shRNA construct (Tet-FLAGCul1 Cand1kd cells). SCF complexes should be stable in this lysate due to the near absence of Cand1. This lysate was either mock-treated or supplemented with a large excess of pure Cand1. Addition of Cand1 resulted in reduced recovery of 20 of 21 F-box proteins observed in FLAGCul1 immunoprecipitates, whereas the level of Rbx1 association was not significantly affected (Figure S2; see Supplementary Discussion for a consideration of the one exception). To measure directly Cand1’s effect on SCF assembly in the absence of confounding factors that might be present in cell lysates, we added two-fold molar excess Cand1 to preformed pure SCFFbxw7 assembled from Fbxw7TAMRA–Skp1 and CFPCul1–Rbx1. We observed a significant reduction in FRET after five minutes, indicating that Cand1 interfered with this assemblage (Figure 2A). This observation was not an artifact of our FRET assay, because when we repeated these experiments with neddylated CFPCul1–Rbx1, Cand1’s effect was eliminated (Figure 2B).
Figure 2. Cand1 Actively Removes Fbxw7–Skp1 From Cul1 by Altering Off Rate.
(A) As in Figure 1A and Figure 1D except with the addition of 100 nM Cand1.
(B) As in (A), except using neddylated CFPCul1.
(C) The change in donor fluorescence versus time in a stopped flow apparatus upon addition of 150 nM Cand1 to 50 nM CFPCul1–Rbx1 pre-incubated with 50 nM Fbxw7TAMRA–Skp1.
(D) The single exponential observed rates of SCF disassembly for various Cand1 concentrations mixed with 5 nM CFPCul1–Rbx1 or 5nM neddylated CFPCul1–Rbx1 pre-incubated with 5 nM Fbxw7TAMRA–Skp1. Chase indicates 700 nM Skp2–Skp1. Error bars: +/− SD, n≥3.
(E) As in Figure 1E except with 150 nM or 300 nM Cand1 and 700 nM Skp2–Skp1 chase mixed with 70 nM neddylated CFPCul1 pre-incubated with 70 nM Fbxw7TAMRA–Skp1. Error bars: range of values, n=2.
(F) As in Figure 1C, except with 150 nM Cand1 pre-incubated with 5 nM CFPCul1–Rbx1. Error bars: +/− SD, n≥3. See also Figure S2.
The short time span used for Figure 2A is in direct contrast to the slow dissociation of Fbxw7TAMRA in the presence of excess unlabeled Skp2–Skp1 observed in Figure 1E, suggesting that Cand1 was not a conventional competitive inhibitor that trapped Cul1–Rbx1 as it dissociated from Fbxw7–Skp1, but instead actively promoted the disassembly of the SCFFbxw7 complex. To directly test this hypothesis, we measured the loss of FRET in real time upon addition of Cand1 to a preformed complex in a stopped flow apparatus (Figure 2C). Titration of Cand1 revealed increasingly rapid rates of SCFFbxw7 dissociation that followed saturation kinetics with a maximum rate of 1.3 s−1 and a half maximal concentration (KM) of 26 nM (Figure 2D). To eliminate interference from re-association of Fbxw7TAMRA–Skp1 with CFPCul1–Rbx1, we repeated our measurements with unlabeled Skp2–Skp1 competitor in the reaction (Figure 2D). The maximal rate of Cand1-dependent dissociation remained unchanged while the KM increased to 53 nM (Figure 2D). In agreement with previous results, SCFFbxw7 formed with neddylated Cul1 showed no Cand1-induced dissociation over 30 seconds (Figure 2D). However, when the reactions were allowed to proceed for hours, we observed that saturating Cand1 accelerated disassembly of neddylated SCFFbxw7 by 45-fold (Figure 2E).
Four main points arise from this analysis. First, the KM of 53 nM sets an upper limit on the KD between Cand1 and CFPCul1–Rbx1. Second, the saturation kinetics reveal the existence of a transient complex that contained Cand1, CFPCul1–Rbx1, and Fbxw7TAMRA–Skp1. Third, the maximal observed rate of 1.3 s−1 represents the rate of Fbxw7TAMRA–Skp1 dissociation from the transient complex, which is accelerated by >1 million-fold compared to the spontaneous dissociation of SCFFbxw7. Finally, neddylation of Cul1 attenuated the effect of Cand1 by 30,000-fold.
These observations are reminiscent of a recent analysis that shows that binding of IκBα to NF-κB both slows the association of NF-κB with DNA and causes a significant increase in the rate of dissociation of NF-κB from DNA (Bergqvist et al., 2009). Thus, it seemed likely that Cand1’s dramatic effect on the dissociation rate of SCFFbxw7 would also be accompanied by a decrease in the rate of assembly of SCFFbxw7. However, in the presence of saturating Cand1, we measured an association rate of 2.2 × 106 M−1 s−1 for Fbxw7TAMRA–Skp1 and CFPCul1–Rbx1 (Figure 2F). This is equivalent (within the error of our measurements) to the association rate without Cand1 (Figure 1C). As expected, when CFPCul1 was modified with Nedd8, Cand1 also had no effect on the association rate (Figure 2F).
The above result has profound implications for building a model of Cand1’s function. First, Cand1 is not a competitive inhibitor. Whereas competitive inhibitors decrease kon and have no effect on koff, Cand1 had no effect on kon but increased koff. Second, unlike allosteric inhibitors, such as IκBα, that decrease the association rate of their targets in addition to increasing the dissociation rate, Cand1 only affected the dissociation rate of SCFFbxw7. This specific type of behavior is found in the mechanism of guanine nucleotide exchange factors (GEFs) that actively promote the exchange of GTP for GDP from target GTPases (Klebe et al., 1995; Goody and Hofmann-Goody, 2002; Guo et al., 2005). In these cases, GEF binding to its target GTPase increases the dissociation rate of guanine nucleotides, yet does not inhibit their association rate. This is accomplished by toggling between stable GEF–GTPase and GTPase–nucleotide complexes through a transient GEF–GTPase–nucleotide ternary complex in which the dissociation rates of both GEF and nucleotide are dramatically increased relative to their stable binary complexes (Klebe et al., 1995; Goody and Hofmann-Goody, 2002; Guo et al., 2005). Our results thus far conform to this model. Both Fbxw7–Skp1 and Cand1 formed tight complexes with Cul1–Rbx1 and there exists a transient ternary intermediate that exhibited a greatly increased rate of Fbxw7–Skp1 dissociation from Cul1–Rbx1.
F-box Proteins Remove Cand1 from Cul1
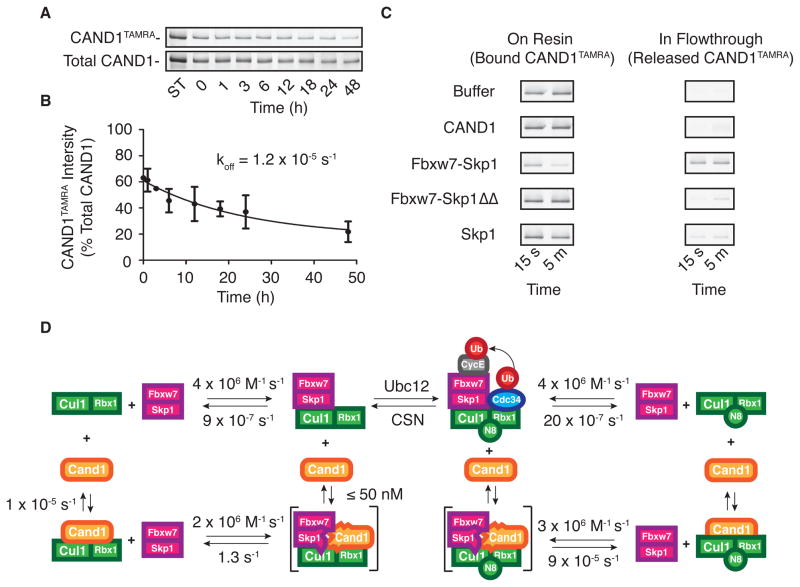
If Cand1 conforms to the behavior of GEFs, Cand1’s dissociation rate from Cul1–Rbx1 should be very slow but should increase dramatically in the presence of F-box Protein–Skp1 complexes. To measure this, we first developed a dissociation assay in which we monitored competitive displacement of Cand1TAMRA from GST-Rbx1–Cul1 in the presence of 10-fold excess of unlabeled Cand1. Displacement was monitored either by native gel electrophoresis (Figure S3A and S3B) or rapid pull-down of GST on glutathione resin (Figure 3A). This protocol revealed that spontaneous dissociation of Cand1TAMRA from Cul1 was extremely slow, with a rate of 1.2 × 10−5 s−1 (Figure 3B). This explains why Ubc12 is unable to conjugate Nedd8 to Cul1 that is bound to Cand1 (Goldenberg et al., 2004; Bornstein et al., 2006; Duda et al., 2008; Siergiejuk et al., 2009).
Figure 3. F-box proteins Rapidly Remove Cand1 from Cul1.
(A) GST-Rbx1–Cul1–Cand1TAMRA (100 nM) was supplemented with 1 μM Cand1. At indicated times, aliquots were removed and incubated with glutathione resin for 15 min. Resin-associated proteins were fractionated by SDS-PAGE and detected by fluorography.
(B) The ratio of released Cand1TAMRA to total Cand1 over time was fit to a single exponential giving koff of 1.2 × 10−5 s−1. Error bars: +/− SD, n=3.
(C) GST-Rbx1–Cul1–Cand1TAMRA (100 nM) pre-incubated with glutathione resin was supplemented with buffer or 1 μM of indicated proteins. Bound and released proteins were collected at indicated times and distribution of Cand1TAMRA was evaluated as in (A).
(D) Summary of the rates measured here. Transient complexes are in brackets. See also Figure S3.
Strikingly, this rate was greatly accelerated in the presence of Fbxw7–Skp1, such that displacement of Cand1TAMRA was largely completed within 5 min (Figure 3C). This key result indicates that Fbxw7–Skp1 is sufficient to displace Cand1 from Cul1, without requiring assistance from the Nedd8 conjugation pathway. This is consistent with reports that loss of Nedd8 conjugation activity has very little effect on the steady-state repertoire of SCF ubiquitin ligases (Bennett et al., 2010; Lee et al., 2011). A similar enhancement of Cand1 displacement by 3 different F-box Protein–Skp1 complexes was observed using an indirect assay wherein removal of Cand1 was evidenced by the ability to conjugate Nedd8 to the liberated Cul1 (Figure S3C and S3D). Displacement of Cand1 by Fbxw7–Skp1 was very specific, because Cand1 was not displaced by either Skp1 alone nor Fbxw7 bound to a mutant of Skp1 (Skp1ΔΔ) used for crystallography that lacks two loops that would be expected to clash with Cand1 (Figure 3C and Figure S3B). Importantly, Skp1ΔΔ was fully able to sustain ubiquitylation, indicating that it was competent to bind Cul1 (Figure S3E).
Combining the results above with the data gathered so far, we constructed a kinetic framework similar to that of GEFs (Klebe et al., 1995; Goody and Hofmann-Goody, 2002; Guo et al., 2005) in which a transient ternary species comprising Fbxw7–Skp1, Cul1–Rbx1, and Cand1 rapidly collapses into sub-complexes that contain Cul1–Rbx1 bound by either Cand1 or Fbxw7–Skp1 (Figure 3D). We can build a similar cycle for neddylated Cul1–Rbx1 in which Cand1 rapidly dissociates from the ternary species, yet Fbxw7–Skp1 slowly dissociates form the ternary species (Figure 3D).
Our observations on Skp1ΔΔ, coupled with the report that deletion of a short β-hairpin from Cand1 allows formation of a stable Cand1–Skp1ΔΔ–Cul1–Rbx1 complex (Goldenberg et al., 2004) leads us to propose that the transient ternary complex we observed in Figure 2D is a high energy intermediate created by clashes involving the flexible acidic loops in Skp1 and the β hairpin in Cand1, enabling rapid dissociation of either Cand1 or F-box Protein–Skp1 from Cul1–Rbx1. In addition, we suggest that the interaction dynamics between Fbxw7–Skp1 and Cand1 should apply to most or all F-box proteins that form SCF complexes given that: (1) the main interaction of F-box proteins with Cul1 occurs through Skp1; (2) three different F- box proteins rapidly evicted Cand1 from Cul1–Rbx1 (Figure S3D); and (3) Cand1 was able to dislodge 20 of 21 F-box proteins from FLAGCul1 in cell lysates (Figure S2).
Cand1 Functions as an F-box protein Exchange Factor
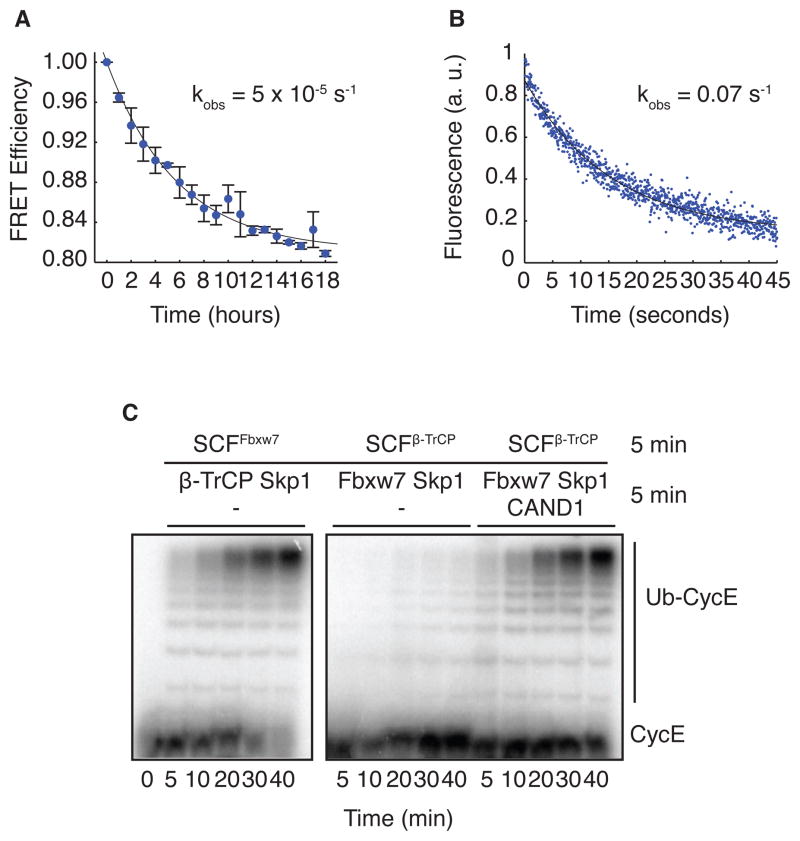
Given that Cand1 can disrupt multiple SCF complexes in a cell lysate, we wondered if Cand1 can promote exchange of one Cul1-bound F-box protein for another. To test this directly, we measured the effect of Cand1 on the ability of Fbxw7TAMRA–Skp1 to gain access to Cul1 sequestered into complexes with β-TRCP–Skp1. As expected, pre-incubation of β-TRCP–Skp1 and CFPCul1–Rbx1 diminished the observed association rate of 210 nM Fbxw7TAMRA–Skp1 with CFPCul1–Rbx1 from 0.9 s−1 to 5 × 10−5 s−1, a reduction of 18,000 fold (Figure 4A). Remarkably, addition of 150 nM Cand1 to this assay increased the observed association rate to 0.07 s−1, a 1,400-fold rescue (Figure 4B). Thus, Cand1 greatly reduced the timescale with which Cul1–Rbx1 equilibrates with the total population of β-TRCP–Skp1 and Fbxw7–Skp1. To determine whether SCFFbxw7 assembled in the presence of Cand1 was active, we measured ubiquitylation of a cyclin E peptide (CycE) that serves as an SCFFbxw7 substrate (Hao et al., 2007; Pierce et al., 2009). Addition of β-TRCP–Skp1 to a preformed SCFFbxw7 did not affect the rate of ubiquitylation of CycE (Figure 4C). However, switching the order in which β-TRCP–Skp1 and Fbxw7–Skp1 were incubated with Cul1–Rbx1 effectively inhibited the ubiquitylation of CycE, because β-TRCP–Skp1 formed stable complexes with Cul1, thereby denying access to CycE–Fbxw7–Skp1. Under these conditions, addition of Cand1 potently stimulated ubiquitylation of CycE (Figure 4C). The same was true if Cul1–Skp1 was preassembled with Skp2–Skp1 (Figure S3F). To our knowledge, all prior in vitro experiments implicated Cand1 as an inhibitor of SCF ubiquitin ligase activity. However, the design of our assay – which more faithfully mimics what happens in vivo as new F-box proteins are being synthesized – highlights the ability of Cand1 to activate an SCF complex by enabling the new F-box protein to gain access to Cul1 assembled into pre-existing SCF complexes.
Figure 4. Cand1 Functions as an F-box protein Exchange Factor.
(A) Fluorescence emission at 478 nm versus time after addition of 210 nM Fbxw7TAMRA–Skp1 to 70 nM CFPCul1 pre-incubated with 70 nM β-TRCP–Skp1. A single exponential fit gave koff of 5 × 10−5 s−1. Error bars: range of values, n=2.
(B) The change in donor fluorescence versus time in a stopped flow apparatus upon addition of 150 nM Cand1 to 70 nM CFPCul1–Rbx1 pre-incubated first with 70 nM β-TRCP–Skp1 and second with 210 nM Fbxw7TAMRA–Skp1.
(C) Cul1–Rbx1 (150 nM) was pre-incubated with 500 nM Fbxw7–Skp1 (lanes 1–6) or 660 nM β-TRCP–Skp1 (lanes 7–18) for 5 min, followed by addition of 600 nM radiolabeled cycE peptide substrate and either 660 nM β-TRCP–Skp1 (lanes 1–6) or 500 nM Fbxw7–Skp1 (lanes 7–18). Either buffer (lanes 1–12) or 200 nM Cand1 (lanes 13–18) were then added, and reactions were incubated an additional 5 min prior to initiation of an ubiquitylation assay (all lanes) by supplementation of all lanes with 60 μM Ubiquitin, 1μM Ubiquitin E1, and 10 μM Cdc34b. See also Figure S4.
Cand1 Modulates the SCF Repertoire in Cells
Consistent with the exchange activity observed with purified components, addition of excess purified β-TRCP–Skp1 to lysates of Tet-FLAGCul1 cells substantially reduced the recovery of 13 out of 15 endogenous F-box proteins detected in anti-FLAG immunoprecipitates (Figure S4). However, this effect was significantly attenuated for 11 of these F-box proteins by shRNA-mediated depletion of Cand1. In contrast, excess β-TRCP–Skp1 had no effect on recovery of Rbx1 regardless of the presence or absence of Cand1. Together with our biochemical data on purified proteins, these results point towards a general ability of Cand1 to act as a protein exchange factor that equilibrates Cul1–Rbx1 with the available pool of F-box Protein–Skp1 complexes.
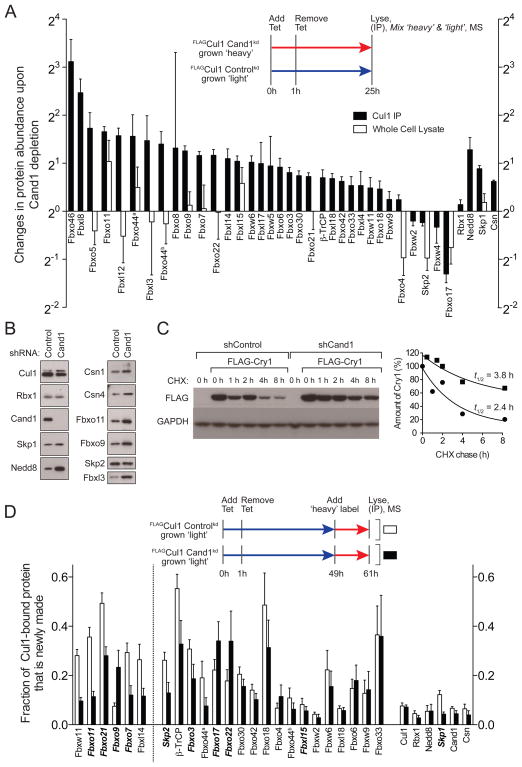
To address whether Cand1’s exchange activity influences SCF complexes in cells, we quantified the steady state and dynamic nature of the Cul1-associated proteome in Tet-FLAGCul1 cells with or without Cand1 depletion. To evaluate the steady state architecture of the SCF proteome, we grew control cells in medium formulated with isotopically light lysine and arginine (‘light’ medium) and Cand1-depleted cells in medium formulated with isotopically heavy lysine and arginine (‘heavy’ medium). Both cultures were pulsed with tetracycline for 1 h at t=0 hours to induce transient synthesis of FLAGCul1. After t=25 hours FLAGCul1 immunoprecipitates were prepared, mixed, and analyzed by quantitative mass spectrometry. Analysis of isotopic ratios revealed that Cand1 depletion had no effect on Cul1 levels but exerted far-reaching effects on the SCF repertoire, with 14 of 34 SCF complexes increasing by ≥2-fold, and 2 decreasing by ≥1.5-fold (Figure 5A, black bars). Meanwhile, Cul1-bound Skp1 was increased 1.7-fold in Cand1-deficient cells. The net assembly of SCF complexes in Cand1-depleted cells is in agreement with the relative abundances of Cul1 (302 nM) and Cand1 (390 nM) in 293 cells (Bennett et al., 2010) and our biophysical data that excess Cand1 causes a net disassembly of SCFFbxw7 (Figure 2A). In addition, the levels of Cul1-associated Nedd8 and CSN were increased by 2.4 and 1.5-fold, respectively. These changes are consistent with the increased overall assembly of SCF complexes in Cand1-depleted cells, and the ability of F-box proteins to stabilize binding of CSN (Enchev et al., 2012) while suppressing deneddylation of Cul1 (Emberley et al., 2012; Enchev et al., 2012). The effect of Cand1 on F-box proteins was specific, because the tightly-bound Cul1 partner Rbx1 exhibited very little change. These and other changes observed in Figure 5A were validated qualitatively by western blotting (Figure 5B). Importantly, mass spectrometry-based quantification of peptides diagnostic for a select group of F-box proteins revealed that the change observed in the steady-state repertoire of SCF complexes in Cand1-depleted cells was not a simple reflection of altered abundance of F-box proteins in total cell lysate (Figure 5A, white bars). To evaluate the relationship between altered abundance of SCF complexes and their role in protein degradation, we examined the degradation of an Fbxl3 substrate, cryptochrome Cry1 (Busino et al., 2007; Siepka et al., 2007). Whereas SCFFbxl3 was more abundant in Cand1-depleted cells (Figure 5A), the rate of degradation of Cry1 was reduced (Figure 5C), as was the association between Cry1 and Cul1 (Figure S5A).
Figure 5. Cellular Cand1 Shapes Steady State and Dynamic SCF Landscape.
(A) Tet-FLAGCul controlkd and Tet-FLAGCul1 Cand1kd cells (kd refers to knockdown) grown in medium with isotopically light or heavy lysine plus arginine, respectively, were induced with 1 μg/mL tetracycline for 1 hour and lysed in 1 μM MLN4924 and 2 mM o-phenanthroline 24 hours later. Two experiments were performed according to this protocol. In the first experiment, we used ‘pseudoMRM’ mass spectrometry to measure the relative amounts of 14 observable F-box proteins in total cell lysate from Cand1-depleted and control cells (white bars). In the second experiment, we retrieved FLAGCul1 and measured the relative amounts of 34 F-box proteins in the immunoprecipitates (black bars). All isotopic ratios were normalized to FLAGCul1’s (0.94), which was set to 1.0. For both experiments, results represent the ratio Cand1kd:controlkd of each protein in anti-FLAG IP measured by mass spectrometry. Each protein had ≥ 2 peptides. Error bars represent standard errors of overall protein group ratios, calculated from bootstrap analysis of two biological replicates (the second replicate was performed as a label-swap). Abundance changes in Cul1 IP for all the proteins listed in Fig5a except for Fbxo8, Fbxw9, and Fbxw4 achieved p-values < 0.05. Fbxo44a and b correspond to IPI00647771 and IPI00414844, respectively. Statistical analysis is provided in Table S1.
(B) Immunoblot validation with indicated antibodies of results in (A).
(C) The same cells used in (A) were transfected with a plasmid that encodes FLAGCry1. Forty-eight hours later, a chase was initiated by addition of 40 μg/mL cycloheximide. Cells were harvested at the indicated times and their content of FLAGCry1 and GAPDH was evaluated by SDS-PAGE and immunoblotting (left panel), and quantified (right panel).
(D) The same cells used in (A) and grown in isotopically light lysine plus arginine were induced with 1 μg/mL tetracycline for 1 hour at t=0 hours, treated with 5 μM epoxomicin at t=48 hours, shifted to isotopically heavy lysine plus arginine at t=49 hours, and lysed at t=61 hours in 1 μM MLN4924 and 2 mM o-phenanthroline. Two experiments were performed according to this protocol. In the first experiment shown here, we used data-dependent mass spectrometry to discover and measure the fraction of F-box proteins in FLAGCul1 IPs that was heavy (i.e. made in the 12 hours prior to lysis). In the second experiment (Figure S5B), we used pseudoMRM to target 9 F-box proteins (italicized) and measure the fraction of heavy-labeled species in total cell lysate from Cand1-depleted and control cells. Each protein had ≥ 2 peptides. Error bars represent standard errors of overall protein group ratios, calculated from bootstrap analysis of two biological replicates. The F-box proteins shown to the left of the dotted line are those for which the different association observed in control and Cand1kd cells achieved a p-value <0.05. See also Figure S5 and Table S2.
To connect our in vitro observations with what happens in cells, we sought to evaluate SCF dynamics in vivo. Control and Cand1-depleted cells growing in ‘light’ medium were pulsed with tetracycline for 1 h at t=0 hours to induce transient synthesis of FLAGCul1. At t=49 hours, the cells were pulsed with ‘heavy’ medium in the presence of the proteasome inhibitor epoxomicin to suppress apparent F-box protein exchange due to degradation. At t=61 hours, FLAGCul1 immunoprecipitates were prepared and analyzed by quantitative mass spectrometry to determine the fraction of each bound protein that was produced during the 12-hour pulse-label. In a parallel experiment, we monitored the heavy:light ratios in total cell lysate of peptides diagnostic for twelve different F-box proteins to evaluate the rate at which newly-synthesized forms of these proteins accumulated. As shown in Figure 5D, several newly-synthesized F-box proteins including Fbxw11, Fbxo11, and Fbxo21 exhibited reduced incorporation into the pool of pre-existing Cul1 in the absence of Cand1, consistent with Cand1 being an F-box protein exchange factor. Importantly, the isotopic ratios for F-box proteins in total cell lysate (including the three mentioned above) were not affected by the presence or absence of Cand1 (Figure S5B), indicating that low penetration of these pulse-labeled F-box proteins into the Cul1-bound pool of Cand1-depleted cells was not due to a reduced rate of synthesis.
DISCUSSION
Cand1 is an F-box Protein Exchange Factor and Modulates the Cellular Repertoire of SCF Complexes
Here we establish the first kinetic framework for the dynamic assembly of SCF complexes. Prior biochemical studies suggested that Cand1 is an inhibitor of SCF complexes, whereas genetic studies indicated that it promotes SCF function. Our results resolve this apparent paradox. We show that Cand1 can unambiguously stimulate SCF activity in vitro by enabling an F-box Protein–Skp1 complex to access Cul1 that was previously occupied by a different F-box Protein–Skp1 complex, and that Cand1 promotes assembly in vivo of new F-box proteins with pre-existing Cul1 molecules. We conclude that Cand1 serves as an exchange factor for F-box Protein–Skp1 complexes and conforms to the scheme originally proposed for GEFs (Klebe et al., 1995; Goody and Hofmann-Goody, 2002; Guo et al., 2005). However, unlike GEFs that exchange GDP for GTP, Cand1 promotes exchange of multiple F-box proteins on the Cul1 scaffold. Because Cand1 can bind other cullins (Lo and Hannink, 2006; Bosu et al., 2010; Chua et al., 2011) and influences ubiquitylation or degradation of multiple CRL substrates in vivo (Zheng et al., 2002a; Chuang et al., 2004; Feng et al., 2004; Lo and Hannink, 2006; Zhang et al., 2008; Bosu et al., 2010; Kim et al., 2010), we suggest that it serves as an exchange factor for CRL adaptors in general. In anticipation of a widespread role for Cand1 exchange activity in CRL biology and by analogy to guanine nucleotide exchange factors (GEFs), we suggest that it be referred to as a ‘substrate receptor exchange factor’ (SREF).
A perplexing feature of prior genetic studies is that Cand1 deficiency causes a partial reduction-of-function for some SCF complexes but not others (Chuang et al., 2004; Feng et al., 2004; Zhang et al., 2008; Bosu et al., 2010; Kim et al., 2010). We suggest that the impact of Cand1 on the steady state distribution of SCF complexes – with some complexes showing large changes in level whereas other complexes exhibit minimal perturbation (Figure 5A) – may partially account for this heretofore unexplained behavior. Another puzzling observation made in both human (Lo and Hannink, 2006) and Arabidopsis (Chuang et al., 2004) Cand1-deficient cells is that accumulation of a given CRL complex paradoxically correlates with stabilization of its substrate – a phenomenon also observed here. Our results suggest the interesting possibility that Cand1 biases the assembly of SCF complexes to favor F-box proteins for which substrates are available.
‘Substrate Sculpting’ of the CRL Repertoire: An Hypothesis
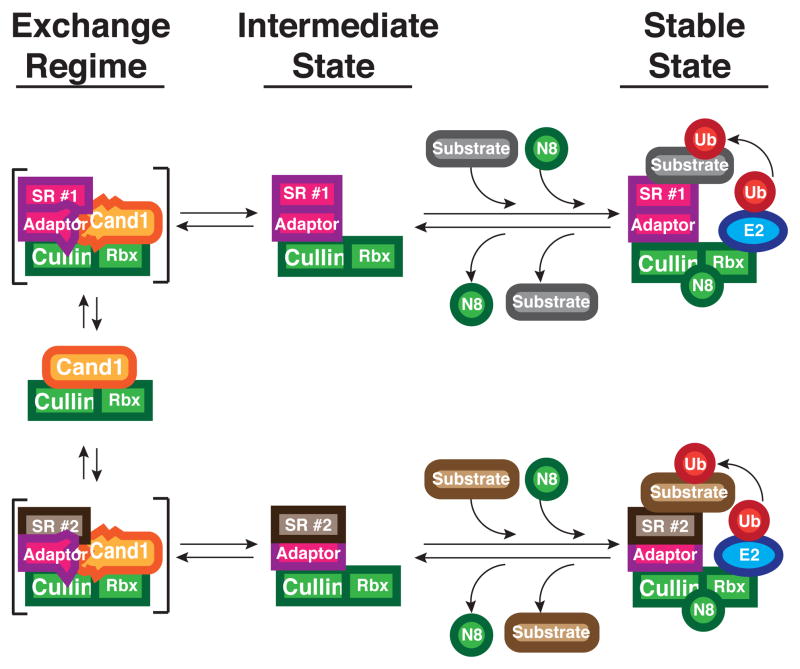
Based on the recent observations that substrates and other ligands can greatly reduce binding and deneddylation of CRLs by CSN (Fischer et al., 2011; Emberley et al., 2012; Enchev et al., 2012) we propose the hypothesis shown in Figure 6, which provides a potential mechanism for how the cellular repertoire of CRL complexes could be optimized to match substrate demand. In our model, which builds upon previous proposals (Cope and Deshaies, 2003; Schmidt et al., 2009), the exchange activity of Cand1 coupled with cycles of Nedd8 conjugation and de-conjugation enables CRL complexes to toggle between two radically different states—the Cand1 ‘exchange’ regime and the Nedd8 ‘stable’ state. When a cullin is conjugated with Nedd8, it can have extraordinary (sub-picomolar) affinity for its adaptor-bound substrate receptor (SR), which binds in a manner that is essentially irreversible (t1/2 ~9 days for Fbxw7–Skp1). Therefore, we envision that a CRL exists in a ‘stable’, active state when it is saturated with substrate, which occludes CSN binding. The Nedd8-conjugated CRL ubiquitylates substrate and can recruit downstream factors involved in substrate degradation (Bandau et al., 2012; den Besten et al., 2012). Once substrate is depleted, the ability of CSN to bind the CRL and remove Nedd8 is enhanced. In this metastable ‘intermediate’ state, the complex can either bind a new substrate and become reactivated by Nedd8 conjugation to return to the ‘stable’ state, or it can bind Cand1 and enter the ‘exchange’ regime, resulting in up to a one million-fold increase in dissociation rate of adaptor–SR. The resulting Cand1–cullin–Rbx complex rapidly assembles with any of the available adaptor–SR complexes to form an unstable ternary intermediate that promptly decays to regenerate Cand1–cullin–Rbx or yield a new CRL complex. Neddylation of the latter species, which is stabilized by substrate, completes the cycle. We envision that substrates, by shielding CRL complexes from the actions of CSN and Cand1, help to sculpt the cellular repertoire of CRL complexes. Other factors are likely to contribute as well, including the rate of synthesis and degradation of substrate receptor proteins, as well as post-translational modifications and localization controls that modulate the access of CRL complexes to the Nedd8–Cand1 cycle.
Figure 6. Hypothesis for Control of CRL Assembly by Substrate, Cand1, and Nedd8.
Rapid exchange of multiple CRL adaptor-bound substrate receptors occurs in the Cand1 exchange regime through the formation and decay of transient ternary complexes shown in brackets. Cand1 and adaptor are drawn as deformed in these complexes, to emphasize the proposal that they clash sterically, yielding an unstable state. In the presence of substrates, CRLs that pass through an intermediate state become neddylated and enter a stable state where ubiquitylation of substrates occurs. Loss of substrates facilitates recruitment of CSN, removal of Nedd8, and a return to the exchange regime effected by Cand1.
An implication of our model is that in Cand1-deficient cells, the assembly state of CRLs would become uncoupled from substrate demand, such that the cullin could potentially become tied up in superfluous and non-productive CRL complexes. This situation may be tolerated in cells with constitutively high turnover of F-box and other substrate receptor proteins, but given the significant phenotypic consequences of Cand1 mutation (Chuang et al., 2004; Feng et al., 2004; Zhang et al., 2008; Bosu et al., 2010; Kim et al., 2010; Helmstaedt et al., 2011), it is apparent that ongoing synthesis and degradation of adaptor–SR modules (Bennett et al., 2010) or other mechanisms of SCF disassembly (Yen et al., 2012) by themselves do not suffice to sustain a fully functional CRL network.
Although other examples of factors that actively promote complex disassembly have been described (Klebe et al., 1995; Goody and Hofmann-Goody, 2002; Guo et al., 2005; Bergqvist et al., 2009; Chan et al., 2009), to our knowledge Cand1 is the first example of a factor that has the potential to promote equilibration of a protein scaffold with a large number of interacting partners. We suggest that ‘protein exchange factors’ that work analogously to Cand1 may play important roles in processes such as DNA replication, transcription, mRNA splicing, and vesicle trafficking that rely on protein machines that engage in transactions notable for their speed and affinity.
EXPERIMENTAL PROCEDURES
FRET Assay
Fluorimeter scans were performed on FluoroLog-3 (Jobin Yvon) in a buffer containing 30 mM Tris pH 7.6, 100 mM NaCl, 0.5 mM DTT, and 1 mg/ml Ovalbumin (Sigma) in a volume of 250 μl. Mixtures were excited at 430 nm and the emissions were scanned from 450 nm to 650 nm. Stopped flow reactions were performed on a Kintek stopped flow machine in the same buffer as the fluorimeter scans.
Ubiquitylation Assay
CycE was incubated with [γ-32P] ATP (132 nM) and Protein Kinase A for 45 min at 30°C to make radiolabeled CycE. Ubiquitylation reactions contained ATP (2 mM), Ubiquitin (60 μM), Ubiquitin E1 (1 μM), Cdc34b (10 μM), Cul1–Rbx1 (150 nM), and Fbxw7–Skp1 (varying concentrations). Additional proteins were included as mentioned in the text. Reactions were performed and quenched in buffers previously described for ubiquitylation assays (Pierce et al., 2009). Reactions were analyzed by running on 16% gels, drying, and quantifying with a phosphor screen (Molecular Devices).
Cul1-Cand1 Dissociation Assay
Cul1 with N-terminal tagged GST-Rbx1 (0.1 μM) was pre-incubated with Cand1TAMRA (0.1 μM) for 10 min and captured on glutathione sepharose 4B resin. Aliquots of resin were transferred to Micro Bio-Spin columns (Bio-Rad), resuspended in 1 μM of the indicated proteins, and incubated for 15 s or 5 min. Reactions were terminated by separation of beads and supernatant by centrifugation, and equivalent portions of each were fractionated by SDS-PAGE. Gels were scanned by a Typhoon™ scanner to quantify Cand1TAMRA. To analyze the dissociation rate of the Cul1-Cand1 complex, at various time points following the addition of 1 μM Cand1 to 0.1 μM GST-Rbx1–Cul1–Cand1TAMRA, an aliquot was withdrawn and incubated with glutathione sepharose 4B resin for 15-min and processed for SDS-PAGE analysis and fluorography. The gels were then stained by SilverQuest™ staining kit (Invitrogen) to detect the total Cand1 bands. The intensity of Cand1TAMRA and total Cand1 band was measured with ImageJ (NIH), and the intensity of Cand1TAMRA was normalized by the intensity of total Cand1 at each time point.
Kinetic Analysis
Regressions were performed in Matlab with the exception of Figure 3B, which was generated in Prism.
Mass spectrometric analyses
All data-dependent liquid chromatography-mass spectrometry and data analyses for Cul1-bound proteins were performed as described previously (Lee et al., 2011) with the following modifications. In experiments where we tested the effect of excess β-TrCP or Cand1 on SCF complex composition, purified β-TrCP or Cand1 was added to the lysate for 2 h at 23 °C, followed by immunoprecipitation for 15 min at the same temperature. To calculate the overall protein group ratio for an experiment, median evidence ratios were first calculated for each biological replicate, and overall ratio was calculated from the mean of the biological replicates. Standard errors of the overall protein group ratios were calculated using bootstrap analysis. PseudoMRM (Greco et al., 2010), also referred to as targeted peptide monitoring (Sandhu et al., 2008; Hewel et al., 2010) or peptide ion monitoring (Kulasingam et al., 2008) was used for the quantification of the selected peptides of F-box proteins in global lysates.
Supplementary Material
Table S1. Changes in the steady-state level of proteins associated with Cul1 in lysate upon Cand1 depletion. Related to Figure 5A.
Table S2. Rate of incorporation of newly synthesized proteins into the pre-existing Cul1 complex in control and Cand1-depleted cells. Related to Figure 5D.
Table S3. Displacement of Cul1-bound proteins upon addition of recombinant Cand1 into Cand1-depleted lysates. Related to Figure S2.
Table S4. Displacement of Cul1-bound proteins upon addition of recombinant β-TrCP into shControl and shCand1 lysates. Related to Figure S4.
Table S5. Rate of protein synthesis in shControl and shCand1 cells. Related to Figure S5.
Figure S1. Neddylation Does Affect Dyamics of SCFFbxw7 Assembly. Related to Figure 1.
(A) 150 nM Cul1–Rbx1 or CFPCul1–Rbx1was pre-incubated with 150 nM Fbxw7–Skp1 or Fbxw7TAMRA–Skp1 and 600 nM radiolabeled CycE and incubated for 5 min. 2 mM ATP, 60 μM Ubiquitin, 1 μM Ubiquitin E1, and 10 μM Cdc34b were pre-incubated for 2 min and added to the mix. Samples were quenched at the indicated times and analyzed by SDS-PAGE followed by phosphorimaging.
(B) As in Figure 1A except with neddylated Cul1.
(C) As in Figure 1B except with neddylated Cul1.
(D) As in Figure 1E except with neddylated Cul1.
Figure S2. Addition of Recombinant CAND1 to CAND1 Depleted Lysates Reduces Fboxes Bound to Cul1. Related to Figure 2.
Tet-FLAGCul1 Cand1kd cell cultures were grown in medium formulated with isotopically ‘heavy’ or ‘light’ lysine and arginine. The cells were simultaneously treated with 1μg/mL tetracycline for 1 h to induce a pulse of FLAGCul1 synthesis, and then returned to tetracycline-free medium for a further 24 hours. Both cultures were then treated with 1 μM MLN4924 for 1 hour. The idea behind this manipulation was to minimize neddylation, which should maximize the sensitivity of SCF complexes to disassembly by Cand1. Cells were then lysed and the lysate from light-labeled cells was supplemented with 9.6 μg Cand1 per mg lysate, whereas the ‘heavy’ lysate was mock-treated. After 2 hours at 23 °C, FLAGCul1 was immunoprecipitated and the ‘light’ and ‘heavy’ samples were combined for analysis by mass spectrometry as described in Methods. The isotopic ratio for Cul1 (1.04) was set to 1.0 and all other proteins were normalized to it. The normalized median values for light/heavy ratios for all sequenced peptides of a given protein are shown. All proteins shown were represented by a minimum of 2 quantified peptides. For CSN, all eight subunits were identified by at least 7 peptides, and the average ratio for all identifications is presented. The ratio for Cand1 was 52, consistent with recruitment of a large amount of recombinant Cand1 relative to the endogenous Cand1 present in FLAGCul1 immunoprecipitates from Cand1-depleted cells. Error bars represent the standard errors of the overall protein group ratios, calculated from the two biological replicates using bootstrap analysis. In the first biological replicate the labeling was done as indicated, and in the second the labeling (and ratio calculation) was reversed.
Figure S3. Gel Shift and Neddylation Assays Reveal that Fboxes can Remove CAND1 from Cul1. Related to Figure 3.
(A) 100 nM Rbx1-Cul1–Cand1TAMRA was supplemented with 1 μM Cand1 for indicated time periods and was fractionated by native PAGE. The first lane contains 100 nM Cand1TAMRA standard.
(B) 100 nM Rbx1-Cul1–Cand1TAMRA was supplemented with indicated proteins at concentrations of 50 nM, 100 nM, 200 nM, 300 nM, 400 nM, respectively, and samples were immediately fractionated by native PAGE. Lane 1: 100 nM Cand1TAMRA. Lane 2: 100 nM Rbx1-Cul1–Cand1TAMRA. Lane 3: 100 nM Rbx1-Cul1–Cand1TAMRA supplemented with buffer.
(C) 100 nM radiolabeled Cul1PKA–Rbx1 was supplemented with a pre-incubated (2 min) mixture containing 2 mM ATP, 15 μM Nedd8, 300 nM F-box–Skp1, 1 μM Nedd8 E1, and 10 μM Ubc12. At the indicated times aliquots were withdrawn and quenched for subsequent evaluation by SDS-PAGE and phosphorimaging.
(D) 100 nM radiolabeled Cul1PKA–Rbx1 was pre-incubated with 300 nM Cand1. Reactions were started with addition of 2 mM ATP, 15 μM Nedd8, 300 nM Fboxes–Skp1, 1μM Nedd8 E1, and 10 μM Ubc12. Reactions were evaluated as in (C).
(E) As in Figure S1A except with either 150 nM Fbxw7–Skp1 or 150 nM Fbxw7–Skp1ΔΔ.
(F) 150 nM Cul1–Rbx1 was pre-incubated with 150 nM Skp2–Skp1 for 5 min then mixed with 150 nM Fbxw7–Skp1, 600 nM radiolabeled CycE and either buffer or 200 nM Cand1. After 5 min, 2 mM ATP, 15 μM Nedd8, 1 μM Nedd8 E1, and 10 μM Ubc12 were added to the mix. After 5 min, pre-incubated 2 mM ATP, 60 μM Ubiquitin, 1 μM Ubiquitin E1, and 10 μM Cdc34b were added to the mix. See also Table S3.
Figure S4. CAND1 Acts as an Fbox Exchange Factor in the Presence of Many SCF Complexes. Related to Figure 4.
Similar to Figure S2, except that four parallel cultures were grown: two cultures of Tet-FLAGCul1 Cand1kd with one labeled heavy and the other light, and two cultures of Tet-FLAGCul1 controlkd cells, one labeled heavy and the other light. The cells were not treated with MLN4924 prior to lysis. Light-labeled lysates from control and Cand1kd cells were supplemented with 5.0 μg β-TRCP–Skp1 per mg lysate, whereas heavy-labeled lysates were mock-supplemented. Following a 2 h incubation at 23 °C, FLAGCul1 was immunoprecipitated from each sample and the Cand1kd samples were mixed together and processed for mass spectrometry and data analysis as in panel (A), and the same was done for the controlkd samples. The pre-normalization isotopic ratios for Cul1 were 1.0 for the Cand1kd samples and 0.98 for controlkd samples. The results for β-TrCP were plotted on a different scale because of the magnitude of signal, most of which is likely to be due to recruitment of ‘light’ recombinant protein. F-boxes are shown to the left of the dotted line and other Cul1 interactors are shown to the right. Error bars and replicates are as described in Figure S2. See also Table S4.
Figure S5. Newly Made Proteins in Whole Cell Lysate. Related to Figure 5.
(A) The lysates from cells infected with lentivirus containing control or Cand1 shRNA that were used for the experiment in Figure 5C were immunoprecipitated with anti-FLAG and then immunoblotted with antibodies against FLAG, Fbxl3, and Cul1. Note that the amount of Cul1 in the IP from Cand1-depleted cells was reduced, even though the amount of Cry1 was slightly greater.
(B) The same cells used in Figure 5D were grown in isotopically light lysine plus arginine, induced with 1 μg/mL tetracycline for 1 hour at t=0 hours, treated with 5 μM epoxomicin at t=48 hours, shifted to isotopically heavy lysine plus arginine at t=49 hours, and lysed at t=61 hours in 1 μM MLN4924 and 2 mM o-phenanthroline. PseudoMRM was used to measure the fraction of heavy-labeled species for 9 observable F-box proteins in total cell lysate from Cand1-depleted (black bars) and control (white bars) cells. To estimate the standard error of the protein ratio, a bootstrap analysis was performed, where a pooled variance was calculated for SILAC pairs of fragment ions within the same MS analysis and each hierarchical level was sampled with replacement. See also Table S5.
Acknowledgments
We thank J. Vielmetter of the Caltech Protein Expression Facility for providing Fbxw7–Skp1, β-TRCP–Skp1, and ubiquitin E1. We thank B. Schulman, L. Busino, M. Pagano, F. Bassermann, O. Schneewind, A. Saha, and W. den Besten for gifts of reagents. We thank G. Smith for assistance with mass spectrometry analyses. We thank all the members of the Deshaies and Shan lab for support and helpful discussions. N.W.P. was supported by the Gordon Ross Fellowship, and an NIH Training Grant. J.E.L. was supported by the Ruth L. Kirschstein NRSA Fellowship (CA138126) from the NIH. R.J.D. is an Investigator of the HHMI. This work was supported in part by NIH GM065997 to R.J.D. The Proteome Exploration lab is supported in part by grants from the Gordon and Betty Moore Foundation and the Beckman Institute and an instrumentation grant from NIH (10565784).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bandau S, Knebel A, Gage ZO, Wood NT, Alexandru G. UBXN7 docks on neddylated cullin complexes using its UIM motif and causes HIF1α accumulation. BMC Biol. 2012;10:36. doi: 10.1186/1741-7007-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of Cullin-RING Ubiquitin Ligase Network Revealed by Systematic Quantitative Proteomics. Cell. 2010;143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergqvist S, Alverdi V, Mengel B, Hoffmann A, Ghosh G, Komives EA. Kinetic enhancement of NF-kappaBxDNA dissociation by IkappaBalpha. Proceedings of the National Academy of Sciences. 2009;106:19328–19333. doi: 10.1073/pnas.0908797106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornstein G, Ganoth D, Hershko A. Regulation of neddylation and deneddylation of cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate. Proc Natl Acad Sci USa. 2006;103:11515–11520. doi: 10.1073/pnas.0603921103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosu DR, Feng H, Min K, Kim Y, Wallenfang MR, Kipreos ET. C. elegans CAND-1 regulates cullin neddylation, cell proliferation and morphogenesis in specific tissues. Developmental Biology. 2010;346:113–126. doi: 10.1016/j.ydbio.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SIH, Draetta GF, Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- Chan C, Beltzner CC, Pollard TD. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr Biol. 2009;19:537–545. doi: 10.1016/j.cub.2009.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua YS, Boh BK, Ponyeam W, Hagen T. Regulation of cullin RING E3 ubiquitin ligases by CAND1 in vivo. PLoS ONE. 2011;6:e16071. doi: 10.1371/journal.pone.0016071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HW, Zhang W, Gray WM. Arabidopsis ETA2, an apparent ortholog of the human cullin-interacting protein CAND1, is required for auxin responses mediated by the SCF(TIR1) ubiquitin ligase. Plant Cell. 2004;16:1883–1897. doi: 10.1105/tpc.021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope G, Deshaies R. COP9 signalosome: A multifunctional regulator of SCF and other cullin-based ubiquitin Ligases. Cell. 2003;114:663–671. doi: 10.1016/s0092-8674(03)00722-0. [DOI] [PubMed] [Google Scholar]
- den Besten W, Verma R, Kleiger G, Oania RS, Deshaies RJ. NEDD8 links cullin-RING ubiquitin ligase function to the p97 pathway. Nat Struct Mol Biol. 2012;19:511–516. doi: 10.1038/nsmb.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural Insights into NEDD8 Activation of Cullin-RING Ligases: Conformational Control of Conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BT, Schulman BA. Structural Mechanisms Underlying Posttranslational Modification by Ubiquitin-Like Proteins. Annu Rev Biophys Biomol Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- Emberley ED, Mosadeghi R, Deshaies RJ. Deconjugation of Nedd8 from Cul1 Is Directly Regulated by Skp1-F-box and Substrate, and the COP9 Signalosome Inhibits Deneddylated SCF by a Noncatalytic Mechanism. Journal of Biological Chemistry. 2012;287:29679–29689. doi: 10.1074/jbc.M112.352484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enchev RI, Scott DC, da Fonseca PCA, Schreiber A, Monda JK, Schulman BA, Peter M, Morris EP. Structural Basis for a Reciprocal Regulation between SCF and CSN. Cell Rep. 2012;2:616–627. doi: 10.1016/j.celrep.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Shen Y, Sullivan JA, Rubio V, Xiong Y, Sun TP, Deng XW. Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein Degradation. Plant Cell. 2004;16:1870–1882. doi: 10.1105/tpc.021949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer ES, Scrima A, Böhm K, Matsumoto S, Lingaraju GM, Faty M, Yasuda T, Cavadini S, Wakasugi M, Hanaoka F, et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell. 2011;147:1024–1039. doi: 10.1016/j.cell.2011.10.035. [DOI] [PubMed] [Google Scholar]
- Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N. Structure of the Cand1-Cul1-Roc1 Complex Reveals Regulatory Mechanisms for the Assembly of the Multisubunit Cullin-Dependent Ubiquitin Ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Goody RS, Hofmann-Goody W. Exchange factors, effectors, GAPs and motor proteins: common thermodynamic and kinetic principles for different functions. European Biophysics Journal. 2002;31:268–274. doi: 10.1007/s00249-002-0225-3. [DOI] [PubMed] [Google Scholar]
- Greco TM, Seeholzer SH, Mak A, Spruce L, Ischiropoulos H. Quantitative mass spectrometry-based proteomics reveals the dynamic range of primary mouse astrocyte protein secretion. J Proteome Res. 2010;9:2764–2774. doi: 10.1021/pr100134n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Ahmadian MR, Goody RS. Guanine nucleotide exchange factors operate by a simple allosteric competitive mechanism. Biochemistry. 2005;44:15423–15429. doi: 10.1021/bi0518601. [DOI] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-Cyclin E Complex: Multisite-Phosphorylated Substrate Recognition by SCF Ubiquitin Ligases. Molecular Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Helmstaedt K, Schwier EU, Christmann M, Nahlik K, Westermann M, Harting R, Grond S, Busch S, Braus GH. Recruitment of the inhibitor Cand1 to the cullin substrate adaptor site mediates interaction to the neddylation site. Molecular Biology of the Cell. 2011;22:153–164. doi: 10.1091/mbc.E10-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewel JA, Liu J, Onishi K, Fong V, Chandran S, Olsen JB, Pogoutse O, Schutkowski M, Wenschuh H, Winkler DFH, et al. Synthetic peptide arrays for pathway-level protein monitoring by liquid chromatography-tandem mass spectrometry. Molecular & Cellular Proteomics. 2010;9:2460–2473. doi: 10.1074/mcp.M900456-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kim HJ, Kim S, Yim J. Drosophila Cand1 regulates Cullin3-dependent E3 ligases by affecting the neddylation of Cullin3 and by controlling the stability of Cullin3 and adaptor protein. Developmental Biology. 2010;346:247–257. doi: 10.1016/j.ydbio.2010.07.031. [DOI] [PubMed] [Google Scholar]
- Klebe C, Prinz H, Wittinghofer A, Goody RS. The kinetic mechanism of Ran--nucleotide exchange catalyzed by RCC1. Biochemistry. 1995;34:12543–12552. doi: 10.1021/bi00039a008. [DOI] [PubMed] [Google Scholar]
- Kulasingam V, Smith CR, Batruch I, Buckler A, Jeffery DA, Diamandis EP. “Product ion monitoring” assay for prostate-specific antigen in serum using a linear ion-trap. J Proteome Res. 2008;7:640–647. doi: 10.1021/pr7005999. [DOI] [PubMed] [Google Scholar]
- Lee JE, Sweredoski MJ, Graham RLJ, Kolawa NJ, Smith GT, Hess S, Deshaies RJ. The Steady-State Repertoire of Human SCF Ubiquitin Ligase Complexes Does Not Require Ongoing Nedd8 Conjugation. Molecular & Cellular Proteomics. 2011;10:M110.006460–M110.006460. doi: 10.1074/mcp.M110.006460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Furukawa M, Matsumoto T, Xiong Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Molecular Cell. 2002;10:1511–1518. doi: 10.1016/s1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- Lo SC, Hannink M. CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1. Molecular and Cellular Biology. 2006;26:1235–1244. doi: 10.1128/MCB.26.4.1235-1244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pierce NW, Kleiger G, Shan SO, Deshaies RJ. Detection of sequential polyubiquitylation on a millisecond timescale. Nature. 2009;462:615–619. doi: 10.1038/nature08595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW-L, Antos JM, Ploegh HL. Site-specific protein labeling via sortase-mediated transpeptidation. Curr Protoc Protein Sci. 2009;Chapter 15(Unit15.3) doi: 10.1002/0471140864.ps1503s56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft T. Sortase-mediated protein ligation: an emerging biotechnology tool for protein modification and immobilisation. Biotechnol Lett. 2009;32:1–10. doi: 10.1007/s10529-009-0116-0. [DOI] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Molecular Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu C, Hewel JA, Badis G, Talukder S, Liu J, Hughes TR, Emili A. Evaluation of data-dependent versus targeted shotgun proteomic approaches for monitoring transcription factor expression in breast cancer. J Proteome Res. 2008;7:1529–1541. doi: 10.1021/pr700836q. [DOI] [PubMed] [Google Scholar]
- Schmidt MW, McQuary PR, Wee S, Hofmann K, Wolf DA. F-Box-Directed CRL Complex Assembly and Regulation by the CSN and CAND1. Molecular Cell. 2009;35:586–597. doi: 10.1016/j.molcel.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siergiejuk E, Scott DC, Schulman BA, Hofmann K, Kurz T, Peter M. Cullin neddylation and substrate-adaptors counteract SCF inhibition by the CAND1-like protein Lag2 in Saccharomyces cerevisiae. The EMBO Journal. 2009;28:3845–3856. doi: 10.1038/emboj.2009.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. The EMBO Journal. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickliffe KE, Williamson A, Meyer H-J, Kelly A, Rape M. K11-linked ubiquitin chains as novel regulators of cell division. Trends in Cell Biology. 2011:1–8. doi: 10.1016/j.tcb.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen JL, Flick K, Papagiannis CV, Mathur R, Tyrrell A, Ouni I, Kaake RM, Huang L, Kaiser P. Signal-Induced Disassembly of the SCF Ubiquitin Ligase Complex by Cdc48/p97. Molecular Cell. 2012 doi: 10.1016/j.molcel.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Ito H, Quint M, Huang H, Noël LD, Gray WM. Genetic analysis of CAND1-CUL1 interactions in Arabidopsis supports a role for CAND1-mediated cycling of the SCFTIR1 complex. Proceedings of the National Academy of Sciences. 2008;105:8470–8475. doi: 10.1073/pnas.0804144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, Wei N, Sun H, Kobayashi R, Zhang H. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Molecular Cell. 2002a;10:1519–1526. doi: 10.1016/s1097-2765(02)00784-0. [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002b;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Changes in the steady-state level of proteins associated with Cul1 in lysate upon Cand1 depletion. Related to Figure 5A.
Table S2. Rate of incorporation of newly synthesized proteins into the pre-existing Cul1 complex in control and Cand1-depleted cells. Related to Figure 5D.
Table S3. Displacement of Cul1-bound proteins upon addition of recombinant Cand1 into Cand1-depleted lysates. Related to Figure S2.
Table S4. Displacement of Cul1-bound proteins upon addition of recombinant β-TrCP into shControl and shCand1 lysates. Related to Figure S4.
Table S5. Rate of protein synthesis in shControl and shCand1 cells. Related to Figure S5.
Figure S1. Neddylation Does Affect Dyamics of SCFFbxw7 Assembly. Related to Figure 1.
(A) 150 nM Cul1–Rbx1 or CFPCul1–Rbx1was pre-incubated with 150 nM Fbxw7–Skp1 or Fbxw7TAMRA–Skp1 and 600 nM radiolabeled CycE and incubated for 5 min. 2 mM ATP, 60 μM Ubiquitin, 1 μM Ubiquitin E1, and 10 μM Cdc34b were pre-incubated for 2 min and added to the mix. Samples were quenched at the indicated times and analyzed by SDS-PAGE followed by phosphorimaging.
(B) As in Figure 1A except with neddylated Cul1.
(C) As in Figure 1B except with neddylated Cul1.
(D) As in Figure 1E except with neddylated Cul1.
Figure S2. Addition of Recombinant CAND1 to CAND1 Depleted Lysates Reduces Fboxes Bound to Cul1. Related to Figure 2.
Tet-FLAGCul1 Cand1kd cell cultures were grown in medium formulated with isotopically ‘heavy’ or ‘light’ lysine and arginine. The cells were simultaneously treated with 1μg/mL tetracycline for 1 h to induce a pulse of FLAGCul1 synthesis, and then returned to tetracycline-free medium for a further 24 hours. Both cultures were then treated with 1 μM MLN4924 for 1 hour. The idea behind this manipulation was to minimize neddylation, which should maximize the sensitivity of SCF complexes to disassembly by Cand1. Cells were then lysed and the lysate from light-labeled cells was supplemented with 9.6 μg Cand1 per mg lysate, whereas the ‘heavy’ lysate was mock-treated. After 2 hours at 23 °C, FLAGCul1 was immunoprecipitated and the ‘light’ and ‘heavy’ samples were combined for analysis by mass spectrometry as described in Methods. The isotopic ratio for Cul1 (1.04) was set to 1.0 and all other proteins were normalized to it. The normalized median values for light/heavy ratios for all sequenced peptides of a given protein are shown. All proteins shown were represented by a minimum of 2 quantified peptides. For CSN, all eight subunits were identified by at least 7 peptides, and the average ratio for all identifications is presented. The ratio for Cand1 was 52, consistent with recruitment of a large amount of recombinant Cand1 relative to the endogenous Cand1 present in FLAGCul1 immunoprecipitates from Cand1-depleted cells. Error bars represent the standard errors of the overall protein group ratios, calculated from the two biological replicates using bootstrap analysis. In the first biological replicate the labeling was done as indicated, and in the second the labeling (and ratio calculation) was reversed.
Figure S3. Gel Shift and Neddylation Assays Reveal that Fboxes can Remove CAND1 from Cul1. Related to Figure 3.
(A) 100 nM Rbx1-Cul1–Cand1TAMRA was supplemented with 1 μM Cand1 for indicated time periods and was fractionated by native PAGE. The first lane contains 100 nM Cand1TAMRA standard.
(B) 100 nM Rbx1-Cul1–Cand1TAMRA was supplemented with indicated proteins at concentrations of 50 nM, 100 nM, 200 nM, 300 nM, 400 nM, respectively, and samples were immediately fractionated by native PAGE. Lane 1: 100 nM Cand1TAMRA. Lane 2: 100 nM Rbx1-Cul1–Cand1TAMRA. Lane 3: 100 nM Rbx1-Cul1–Cand1TAMRA supplemented with buffer.
(C) 100 nM radiolabeled Cul1PKA–Rbx1 was supplemented with a pre-incubated (2 min) mixture containing 2 mM ATP, 15 μM Nedd8, 300 nM F-box–Skp1, 1 μM Nedd8 E1, and 10 μM Ubc12. At the indicated times aliquots were withdrawn and quenched for subsequent evaluation by SDS-PAGE and phosphorimaging.
(D) 100 nM radiolabeled Cul1PKA–Rbx1 was pre-incubated with 300 nM Cand1. Reactions were started with addition of 2 mM ATP, 15 μM Nedd8, 300 nM Fboxes–Skp1, 1μM Nedd8 E1, and 10 μM Ubc12. Reactions were evaluated as in (C).
(E) As in Figure S1A except with either 150 nM Fbxw7–Skp1 or 150 nM Fbxw7–Skp1ΔΔ.
(F) 150 nM Cul1–Rbx1 was pre-incubated with 150 nM Skp2–Skp1 for 5 min then mixed with 150 nM Fbxw7–Skp1, 600 nM radiolabeled CycE and either buffer or 200 nM Cand1. After 5 min, 2 mM ATP, 15 μM Nedd8, 1 μM Nedd8 E1, and 10 μM Ubc12 were added to the mix. After 5 min, pre-incubated 2 mM ATP, 60 μM Ubiquitin, 1 μM Ubiquitin E1, and 10 μM Cdc34b were added to the mix. See also Table S3.
Figure S4. CAND1 Acts as an Fbox Exchange Factor in the Presence of Many SCF Complexes. Related to Figure 4.
Similar to Figure S2, except that four parallel cultures were grown: two cultures of Tet-FLAGCul1 Cand1kd with one labeled heavy and the other light, and two cultures of Tet-FLAGCul1 controlkd cells, one labeled heavy and the other light. The cells were not treated with MLN4924 prior to lysis. Light-labeled lysates from control and Cand1kd cells were supplemented with 5.0 μg β-TRCP–Skp1 per mg lysate, whereas heavy-labeled lysates were mock-supplemented. Following a 2 h incubation at 23 °C, FLAGCul1 was immunoprecipitated from each sample and the Cand1kd samples were mixed together and processed for mass spectrometry and data analysis as in panel (A), and the same was done for the controlkd samples. The pre-normalization isotopic ratios for Cul1 were 1.0 for the Cand1kd samples and 0.98 for controlkd samples. The results for β-TrCP were plotted on a different scale because of the magnitude of signal, most of which is likely to be due to recruitment of ‘light’ recombinant protein. F-boxes are shown to the left of the dotted line and other Cul1 interactors are shown to the right. Error bars and replicates are as described in Figure S2. See also Table S4.
Figure S5. Newly Made Proteins in Whole Cell Lysate. Related to Figure 5.
(A) The lysates from cells infected with lentivirus containing control or Cand1 shRNA that were used for the experiment in Figure 5C were immunoprecipitated with anti-FLAG and then immunoblotted with antibodies against FLAG, Fbxl3, and Cul1. Note that the amount of Cul1 in the IP from Cand1-depleted cells was reduced, even though the amount of Cry1 was slightly greater.
(B) The same cells used in Figure 5D were grown in isotopically light lysine plus arginine, induced with 1 μg/mL tetracycline for 1 hour at t=0 hours, treated with 5 μM epoxomicin at t=48 hours, shifted to isotopically heavy lysine plus arginine at t=49 hours, and lysed at t=61 hours in 1 μM MLN4924 and 2 mM o-phenanthroline. PseudoMRM was used to measure the fraction of heavy-labeled species for 9 observable F-box proteins in total cell lysate from Cand1-depleted (black bars) and control (white bars) cells. To estimate the standard error of the protein ratio, a bootstrap analysis was performed, where a pooled variance was calculated for SILAC pairs of fragment ions within the same MS analysis and each hierarchical level was sampled with replacement. See also Table S5.