Abstract
The short half-life of insulin in the human body (4-6 min) prompted the search and discovery of insulin-degrading enzyme (IDE), a 110-kDa metalloprotease that can rapidly degrade insulin into inactive fragments. Genetic and biochemical evidence accumulated in the last sixty years has implicated IDE as an important physiological contributor in the maintenance of insulin levels. Recent structural and biochemical analyses reveal the molecular basis of how IDE uses size and charge distribution of the catalytic chamber and structural flexibility of substrates to selectively recognize and degrade insulin, as well as the regulatory mechanisms of this enzyme. These studies provide a path for potential therapeutics in the control of insulin metabolism by the degradation of insulin.
Keywords: Insulin, Insulin-degrading enzyme, metalloprotease, X-ray crystallography
1. Introduction
The ability of insulin-degrading enzyme (IDE) to degrade insulin was reported nearly sixty years ago (MIRSKY and BROH-KAHN, 1949; MIRSKY et al., 1950). Despite decades of research, the role of IDE in the degradation of insulin, as well as the cellular location of this process, remains controversial (Authier et al., 1996; Hersh, 2006). Yet multiple lines of evidence have accumulated, which support a role for IDE as an important protease involved in insulin's catabolism (Duckworth et al., 1998). IDE has an exceptionally high affinity for insulin (20-85 nM). It also has a thorough non-deterministic cleavage pattern that creates multiple inactive insulin fragments (Duckworth et al., 1998; Chesneau and Rosner, 2000; Grasso et al., 2007). Several examples of in vitro evidence include over-expression studies of IDE in cell lines, which show increased insulin degradation, studies of internalized insulin cross-linking to IDE, as well as injection of monoclonal antibodies to prevent the action of IDE on insulin (Shii and Roth, 1986; Hari et al., 1987; Kuo et al., 1991; Perlman et al., 1993).
Recent evidence has strengthened the physiological relevance of this protease. A decrease in insulin degradation and associated hyperinsulinemia was observed in IDE knockout mice (Farris et al., 2003). In addition, reducing the levels of human IDE in HepG2 cell-line cultured cells using silencing RNA inhibited insulin degradation by up to 76% (Fawcett et al., 2007). The genetic variability of the IDE gene has also been examined in humans and rodents. For example, a large scale human genetic analysis reveals the association of a single nucleotide polymorphism with type 2 diabetes (Sladek et al., 2007). Additional studies in a different population also revealed a single nucleotide polymorphism in the IDE gene with evidence of hyperinsulinemia as compared to the control group (Marlowe et al., 2006). Further genetic evidence of diabetic susceptibility due to polymorphisms of the IDE gene have been found in the Goto-kakizaki (GK) rat, a widely used rodent model of diabetes (Fakhrai-Rad et al., 2000). Yet, clear genetic linkage between IDE dysfunction and type-2 diabetes remains in dispute (Groves et al., 2003; Karamohamed et al., 2003; Gu et al., 2004; Florez et al., 2006). Importantly, IDE is not the sole enzyme responsible for insulin degradation: cathepsin D has also been shown to participate in the lysosomal degradation of insulin (Authier et al., 2002).
IDE (EC 3.4.24.56, insulysin, or insulinase) is an evolutionarily conserved 110-kDa zinc metalloprotease. It has been described principally as a cytosolic enzyme but is also found in multiple cellular compartments including endosomes, peroxisomes, mitochondria, the cell surface and in secreted form (Authier et al., 1995; Qiu et al., 1998; Vekrellis et al., 2000; Sudoh et al., 2002; Leissring et al., 2004; Qiu and Folstein, 2006). Its enzymatic activity is optimal at a physiologically relevant pH range (6.0 – 8.5) and is sensitive to the metalloprotease inhibitor 1,10-phenanthroline but not to other non-metalloprotease inhibitors (Duckworth et al., 1998). Although IDE has greatest affinity for insulin, it has been implicated in the degradation of other amyloidogenic peptides (Duckworth et al., 1998; Kurochkin, 2001; Farris et al., 2003; Shen et al., 2006). Notably, glucagon, a peptide also implicated in glucose metabolism, is degraded by IDE. The recent structural solution of IDE with insulin B chain, glucagon and amylin (Shen et al., 2006) and the substrate-free conformation (Im et al., 2007), has increased understanding of the mechanisms of how IDE interacts with insulin and facilitates its degradation.
These multiple lines of evidence along with recent structural analyses all implicate this highly conserved and ubiquitous metalloprotease in the complex metabolic cycle of insulin. Understanding how IDE itself is regulated has become a vital area of research and improves understanding of how IDE degrades insulin while participating in other cellular processes. We will focus on the molecular structure and regulatory mechanisms of IDE and how this contributes to insulin metabolism.
2. Structure of IDE
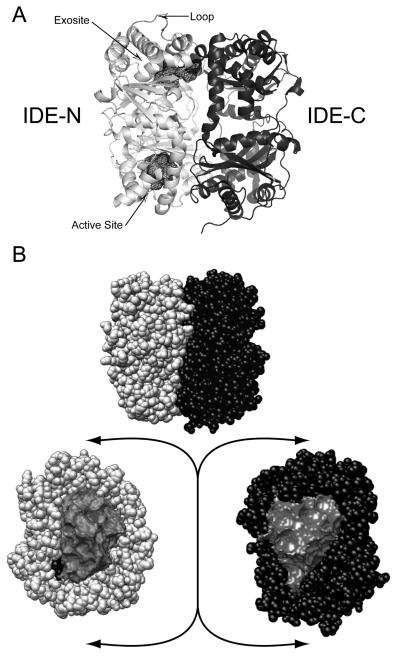
The structural solution of IDE reveals important information about how it associates and degrades substrates. In its monomeric form, IDE is made of two roughly equal sized domains (~55 kDa); IDE-N and IDE-C that are connected by a 28 amino-acid residue loop (Figure 1). When the two halves come together, a crypt is formed (hence crypt forming peptidase) enclosing its substrates and preventing entry or escape. The crypt has a volume of approximately 15,700 Å3, which excludes peptides larger than ~ 70 amino acids (Malito et al., 2008). The two halves, when closed, share a high surface area (11,496 Å2) and possess good shape complementarily (Shen et al., 2006). The catalytic site is located inside the crypt in IDE-N (Figure 1). This site is rich in charged, polar, and hydrophobic patches (Shen et al., 2006), which facilitate interaction with both IDE-C and substrates. IDE-N also possesses an exosite, which is approximately 30 Å away from the catalytic site. Here, the N-termini of ligands are tethered. This site may play a role in the positioning of insulin for cleavage (Shen et al., 2006; Im et al., 2007). This exosite interaction may also contribute to the unique non-deterministic cleavage patterns of IDE's substrates and may be a potential regulatory factor.
Figure 1. Structural conformations of IDE and the presence of a crypt.
A. IDE is a 110,000 Da metalloprotease comprised of two roughly equal-sized domains; IDE-N and IDE-C. IDE-N is the location of several regions that are important in the interaction of substrate to IDE including the exosite and the active site. The active site is also the location of the catalytic zinc ion. The presence of a loop joins the two domains together. B. When closed, IDE forms a crypt with a volume of 15,700 Ǻ3 (Malito et al., 2008)which restricts amyloidogenic peptides to less than 70 amino acids in length. In the closed state, entrance to the catalytic and active site are occluded and substrates may not enter or leave. Structure PDB 2JG4.
Substrate recognition of IDE
The structural solution of IDE in complex with the insulin B-chain, glucagon, and amylin provides insight into how IDE-substrate interaction and cleavage mechanisms occur. All three substrates do not exceed 70 residues and fit entirely within the crypt switching from an α-helix to a β-strand conformation upon binding to IDE (Shen et al., 2006). This conformational change allows the substrate to interact non-covalently with two areas of IDE-N; the catalytic site and the hypothesized exosite. Once trapped, the N-terminus of the substrate is anchored to the exosite of the enzyme by interacting with a β-sheet of IDE-N (Shen et al., 2006). The exosite represents an interesting region of IDE, which is highly conserved and may have a regulatory function contributing to substrate specificity and catalysis (Shen et al., 2006; Im et al., 2007). This interaction serves as a molecular tether, allowing the proper positioning of the C-terminal end of the substrate to the catalytic site where cleavage occurs.
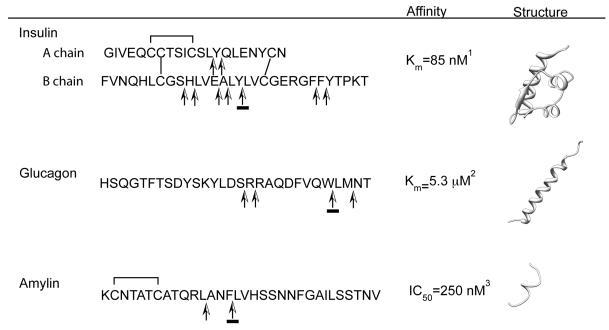
The presence of disulfide bonds are a notable feature of certain substrates of IDEs (Figure 2). For example, insulin has three disulfide bonds. Interestingly, the cleavage of insulin by IDE does not require the reduction and breakdown of these disulfide bonds. Several other substrates of IDE (e.g., amylin, insulin growth factor I and II) also have disulfide bonds, which could influence their interactions with the catalytic chamber of IDE. Yet not all of IDE's substrates possess disulfide bonds (e.g., glucagon, amyloid-beta). How the presence of disulfide bonds influences substrate interaction in the crypt is uncertain and requires further study.
Figure 2. Sequence, substrate cleavage and structure of IDE substrates involved in glucose metabolism.
1 Duckworth et al., 1998, 2 Bennett et al., 2003, 3 Kurochkin, 2000. PBDs; Full chain insulin 1ZNI, glucagon 1GCN, and partial amylin (residues SNNFGAILSS) 1KUW. Arrows indicate sites of cleavage (Grasso et al., 2007, Shen et al., 2006), underlined arrows correspond to P1-P1′ sites of substrate verified in structures 2G56 (Zn2+ free IDE bound to insulin B chain), 2G48 (amylin bound to IDE) and 2G49 (glucagon bound to IDE). Disulfide bonds are indicated by lines joining indicated cysteine residues.
In addition to the exosite and the catalytic site of IDE, other regions of the enzyme are required for its functioning. IDE-N by itself possesses a mere fraction of catalytic activity yet when IDE-C is added (as an individual component with no activity), an ~30% return of activity occurs, suggesting the need for the non-catalytic IDE-C portion (Li et al., 2006). A tyrosine residue (Y831), located in this region, and the primarily positive inner surface of IDE-C may facilitate further positioning of the ligand to the active site by electrostatic tethering. Additional interactions from domain 4 of IDE-C occur with the ligand, including hydrogen bonding from an arginine residue (R824). In both cases, mutation of these residues substantially decreased the specific activity of IDE towards another amyloidogenic peptide, amyloid-beta (Aβ) (Shen et al., 2006).
In order to digest insulin, IDE uses its catalytic chamber (crypt) to engulf insulin entirely. Since the volume of insulin is ~12,000 Å3 it can be fully accommodated (Malito et al., 2008). Since insulin is an acidic peptide hormone, its highly negative charged surface could complement the positive charge surface found on the catalytic chamber of IDE-C (Shen et al., 2006; Im et al., 2007). Only a few selective regions of insulin A and B chains are cleaved by IDE (Figure 2). Future structural study will be needed to elucidate how IDE engulfs insulin and selectively targets these cleavage sites.
Structural evidence for regulatory mechanisms of IDE from the substrate-free conformation
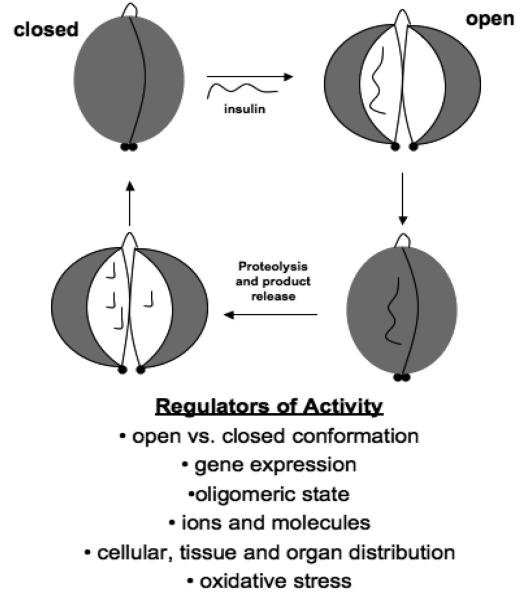
The structural solution of catalytically active IDE in a substrate-free conformation, along with other factors, such as surface and charge complementarity suggest that this enzyme may normally exist in a closed state (Figure 3). When closed, no path to the crypt is seen; thus ligands have no apparent access to the exosite and catalytic site. It has been hypothesized that the closed conformation of IDE is a rate-limiting step for activity since substrates cannot access the catalytic site. Thus, it is important to elucidate factors that stabilize the closed state since they may also regulate activity of the protease. The observation, by mass spectrometry, of multiple cleavage sites on substrates of IDE, including insulin, suggests that during entrapment, multiple cuts are made. It is not clear if one catalytic cycle (i.e., open and close) is sufficient to execute all cuts. However, in the open state, substrates and the by-products of degradation would be available to freely diffuse in and out of the crypt.
Figure 3. Regulators of IDE.
Multiple regulatory mechanisms control the ability of IDE to degrade insulin.
Several factors are thought to play a role in the regulation of the catalytic cycle. Both the high surface complementarity of IDE-N and IDE-C and the charge characteristics of the interior surfaces may facilitate IDE to be in a closed state when inactive (Im et al., 2007). Our previous structures revealed extensive interactions between IDE-N and IDE-C that are predicted to act like a “latch,” which stabilize the closed conformation. Consistent with this model, mutations that destabilize the interactions between IDE-N and IDE-C were found to significantly increase the catalytic rate of IDE.
Consideration of the known structures of IDE and related homologs suggest that the closed state, in the absence of substrate, is unstable. For example, crystal structures of human IDE and a homolog from Arabidopsis thaliana, presequence peptidase (AtPreP), both solved in their closed conformation, also contained substrates bound within their catalytic chambers (Johnson et al., 2006; Im et al., 2007). Furthermore, the structure of the distantly related Escherichia coli pitrilysin, which does not have an associated substrate, is in the open conformation (Maskos, 2004). However, Im et al., 2007, through the structural characterization of catalytically active IDE, revealed that the natural configuration of the active site is in the closed state, suggesting that a ‘molecular switch’ yet to be determined, is responsible for the conformational change between the open and closed state of IDE.
Additional mechanisms of regulation of IDE have been proposed and can be interpreted in the context of the current structural models. A change in oligomeric state, with a dimeric form of IDE showing greater activity versus the tetrameric form has been observed (Song et al., 2003). Such interactions may occur non-covalently. This raises the importance of understanding physiological levels of IDE and how these levels may alter the activity in smaller compartments, such as the peroxisome. Furthermore, the abundance of cysteine residues may play a role in the regulation of IDE via oxidative or possibly nitrosative processes. The relative positions of the side chains of cysteine residues for IDE confirm that several residues show exposed thiol groups on the surface or inside the crypt. Indeed, the ability to regulate IDE and AtPreP, a related cryptidase, has been demonstrated by introducing cysteines to promote disulfide bond formation in oxidizing environments thus locking the cryptidase and prevent substrate access (Johnson et al., 2006; Shen et al., 2006).
3. The regulation of IDE activity
Since IDE regulates insulin levels, mechanisms of the regulation of IDE itself have captured the interest of researchers. This work has resulted in the identification of several regulatory mechanisms of IDE function (Figure 3). This regulation can be observed from the molecular to the organ level. It is important to note that the regulation of IDE may not only affect insulin degradation, but also the degradation of all the other peptide substrates.
At the genetic level, the expression of IDE, insulin, and IGF-1 receptor was demonstrated to be highest in the kidney and liver of rats, indicating an anatomical correlation of IDE in the catabolism of insulin and IGF (Bondy et al., 1994). Additionally, the enzymatic activity of IDE from the homogenates of various rat tissues has been classified in a decreasing order with their insulin degrading activity : liver > pancreas > kidney > testis > adrenal gland > spleen > ovary > lung > heart > muscle > brain > fat (Duckworth and Kitabchi, 1974). The highest enzymatic activity found in the liver and kidneys are consistent with the hypothesis that IDE plays a crucial role in the catabolism of insulin. Interestingly, while IDE is an important contributor to insulin homeostasis, insulin also contributes to the maintenance of IDE levels. Treatment of primary hippocampal neurons with insulin resulted in an increase of IDE protein levels by ~25%, possibly through a feedback mechanism (Zhao et al., 2004). These findings suggest that genetic regulation may account for the differences in expression of IDE among tissues. While the factors that regulate IDE gene expression have not been identified, small molecules like retinoic acid and synthetic retinoic acid analogs have been indirectly implicated in the regulation of IDE expression (Melino et al., 1996). This suggests the possibility of additional small molecules that may also regulate IDE gene expression and thus insulin levels. A thorough genetic analysis of IDE gene expression and its implications on insulin metabolism is necessary for better understanding of insulin-regulated diabetes-related diseases (Jee et al., 2007) and the development of gene therapy as a means of modulating the activity of IDE.
The discovery that IDE is an allosteric enzyme has introduced exciting and new details about its mechanism of regulation. It has been demonstrated that in solution, IDE exists in an equilibrium of dimers and tetramers, with the dimer having higher activity (Song et al., 2003). Upon substrate binding a ‘heterodimer’ of IDE (one wild-type and one mutant subunit) activated the adjacent subunit, restoring the activity of the second subunit (Song et al., 2003). This may occur via conformational changes. This finding was followed by the identification of certain small peptide substrates, such as dynorphin B, that can boost the proteolytic activity of IDE towards the cleavage of Aβ, while at the same time inhibiting the hydrolysis of insulin (Song et al., 2003).
In addition to certain peptide substrates, the activity of IDE is also influenced by other factors. For instance, calcium-depleted muscle tissue has decreased ability to degrade insulin and reduced IDE activity. However, the addition of calcium to muscle returns insulin degradation (Ryan et al., 1985). The catalytic activity of IDE in vitro was also observed to be inhibited by free long chain fatty acids and acyl-CoA (Hamel et al., 2003). This suggests that elevated intracellular long-chain fatty acid concentrations may act directly on IDE to decrease insulin metabolism. This type of regulatory mechanism may explain the correlation between hyperinsulinemia and insulin resistance with elevated fatty acids and obesity (Hamel et al., 2003).
Insulin degradation by IDE is also affected by ATP through the triphosphate moiety (Camberos et al., 2001; Song et al., 2004). A complete inhibition of insulin degradation was observed with the addition of ATP to purified IDE with the inhibitory effect being greater with ATP than with ADP and AMP. ATP also had the ability to shift the oligomeric equilibrium to a monomer suggesting that IDE contains an allosteric site yet to be determined. Thus, the energy status of the cell may serve as feedback inhibition for insulin hydrolysis. In addition to ATP serving as an allosteric regulator by promoting the transition from tetrameric to dimeric forms of IDE, ATP might facilitate the transition from the closed to the open state (Im et al., 2007). Im et a., (2007) studied the activating effect of ATP and found that its effects are reduced in IDE mutants containing mutations that destabilize the closed state. Furthermore, they provided evidence, through biophysical studies, that ATP induces substantial intramolecular conformational changes within IDE. Thus, there is direct evidence that the regulatory mechanism of ATP extends to also facilitating the transition from the closed state to the open state (Im et al., 2007) (Figure 3).
The regulation of IDE extends to the cellular compartmentalization and tissue distribution. The subcellular localization of IDE is dependent on the specific cell type. For the most part, the concentration of IDE is highest in the cytosol, (~95%) and a minor percentage (1% to 5%) is contained in endosomes, peroxisomes, mitochondria, cell surface, and the extracellular milieu (Authier et al., 1995; Duckworth et al., 1998; Qiu et al., 1998; Vekrellis et al., 2000; Leissring et al., 2004). Although IDE has been found in many cellular locations, the exact compartment responsible for insulin hydrolysis is still a controversial issue (Authier et al., 1996; Hersh, 2006). Given the cellular distribution of IDE, insulin hydrolysis may occur intracellularly (either in the cytosol or endosome), at the cell surface, or through the action of secreted IDE. Furthermore, degradation of insulin may occur in different cellular compartments depending on the cell type. For example, primary microglia cells have been shown to secrete IDE while hippocampal neurons possess membrane associated forms (Mentlein et al., 1998; Qiu et al., 1998). Further studies will be necessary to conclusively pinpoint the site of insulin degradation in the cell.
Oxidative stress was recently proposed as another modulator of IDE activity. Enzymes, like IDE, that have metals on or near the active sites can be particularly sensitive to oxidation, which may result in loss of their catalytic function depending on the extent of the oxidative damage (Stocker and Keaney, 2004). A variety of human diseases, such as Alzheimer's disease, diabetes and cardiovascular disease, have been associated with oxidative stress (Stocker and Keaney, 2004) with the modification of several enzymes as a contributing factor. Some examples of proteins that are targeted by oxidative stress include glutamine synthetase, mitochondrial aconitase, adenine nucleotide translocase, carbonic anhydrase III, and calcineurin. IDE has been shown to be vulnerable to treatment with physiologically relevant oxidants in vitro. Yet while insulin degradation was not directly assessed, it is thought that inactivation would extend to insulin cleavage (Shinall et al., 2005). Since it has been demonstrated that oxidative stress can alter IDE activity, it is important to elucidate the molecular and structural basis of IDE oxidation and identify whether this modification can occur in vivo. Together, this may provide the knowledge to spare IDE activity in the presence of oxidative stress.
4. Conclusion
Considerable evidence implicates IDE in the degradation of insulin. While the affinity of this interaction is exceptionally high, the ability to degrade other peptides and an observed gradient of insulin degrading ability based on tissue emphasizes the importance of understanding how this protease is regulated. Structural and biochemical evidence emphasize the importance of several factors including conformational state, presence of ATP, as well as the oligomeric state (Figure 3). These factors, in addition to the presence of a crypt, which engulfs peptides, may all play a role in IDE's ability to trap several different peptides and degrade them thoroughly.
Further structural analysis paired with strategic mutation and biochemical analysis of the regulatory mechanisms of IDE's ability to trap full insulin will extend the knowledge of the nature of these two protein's interactions. Such experiments will increase our ability to understand and influence the degradation of substrates preferentially and so allowing the exploration of the therapeutic potential of IDE. These studies will also determine the most effective strategy for up-or down- regulation and also to determine the physiological consequences of increased or decreased proteolytic activity.
Acknowledgements
This work was supported by NIH R01-GM81539 for W-J T and LAR, and NIH T32-GM07839 for REH
REFERENCES
- Authier F, Cameron PH, Taupin V. Association of insulin-degrading enzyme with a 70 kDa cytosolic protein in hepatoma cells. Biochem J. 1996;319(Pt 1):149–158. doi: 10.1042/bj3190149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Authier F, Metioui M, Fabrega S, Kouach M, Briand G. Endosomal proteolysis of internalized insulin at the C-terminal region of the B chain by cathepsin D. J Biol Chem. 2002;277:9437–9446. doi: 10.1074/jbc.M110188200. [DOI] [PubMed] [Google Scholar]
- Authier F, Bergeron J, Ou W, Rachubinski R, Posner B, Walton P. Degradation of the cleaved leader peptide f thiolase by a peroxisomal proteinase. PNAs. 1995;92:5. doi: 10.1073/pnas.92.9.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy CA, Zhou J, Chin E, Reinhardt RR, Ding L, Roth RA. Cellular distribution of insulin-degrading enzyme gene expression. Comparison with insulin and insulin-like growth factor receptors. J Clin Invest. 1994;93:966–973. doi: 10.1172/JCI117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camberos MC, Perez AA, Udrisar DP, Wanderley MI, Cresto JC. ATP inhibits insulin-degrading enzyme activity. Exp Biol Med (Maywood) 2001;226:334–341. doi: 10.1177/153537020122600411. [DOI] [PubMed] [Google Scholar]
- Chesneau V, Rosner M. Functional Human Insulin-Degrading Enzyme Can Be Expressed in Bacteria. Protein Expression and Purification. 2000;19:91–98. doi: 10.1006/prep.2000.1217. [DOI] [PubMed] [Google Scholar]
- Duckworth W, Bennett R, Hamel F. Insulin degradation: Progess and potential. Endocrine Reviews. 1998:17. doi: 10.1210/edrv.19.5.0349. [DOI] [PubMed] [Google Scholar]
- Duckworth WC, Kitabchi AE. Insulin and glucagon degradation by the same enzyme. Diabetes. 1974;23:536–543. doi: 10.2337/diab.23.6.536. [DOI] [PubMed] [Google Scholar]
- Fakhrai-Rad H, Nikoshkov A, Kamel A, Fernström M, Zierath JR, Norgren S, Luthman H, Galli J. Insulin-degrading enzyme identified as a candidate diabetes susceptibility gene in GK rats. Human Molecular Genetics. 2000;9:2149–2158. doi: 10.1093/hmg/9.14.2149. [DOI] [PubMed] [Google Scholar]
- Farris W, Mansourian S, Chang Y, Lindsley L, Eckman EA, Frosch MP, Eckman CB, Tanzi RE, Selkoe D, Guenette S. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc Natl Acad Sci USA. 2003;100:4162–4167. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett J, Permana PA, Levy JL, Duckworth WC. Regulation of protein degradation by insulin-degrading enzyme: analysis by small interfering RNA-mediated gene silencing. Arch Biochem Biophys. 2007;468:128–133. doi: 10.1016/j.abb.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Florez JC, Wiltshire S, Agapakis CM, Burtt NP, de Bakker PI, Almgren P, Bengtsson Boström K, Tuomi T, Gaudet D, Daly MJ, Hirschhorn JN, McCarthy MI, Altshuler D, Groop L. High-density haplotype structure and association testing of the insulin-degrading enzyme (IDE) gene with type 2 diabetes in 4,206 people. Diabetes. 2006;55:128–135. [PubMed] [Google Scholar]
- Grasso G, Rizzarelli E, Spoto G. AP/MALDI-MS complete characterization of the proteolytic fragments produced by the interaction of insulin degrading enzyme with bovine insulin. J Mass Spectrom. 2007;42:1590–1598. doi: 10.1002/jms.1348. [DOI] [PubMed] [Google Scholar]
- Groves CJ, Wiltshire S, Smedley D, Owen KR, Frayling TM, Walker M, Hitman GA, Levy JC, O'Rahilly S, Menzel S, Hattersley AT, McCarthy MI. Association and haplotype analysis of the insulin-degrading enzyme (IDE) gene, a strong positional and biological candidate for type 2 diabetes susceptibility. Diabetes. 2003;52:1300–1305. doi: 10.2337/diabetes.52.5.1300. [DOI] [PubMed] [Google Scholar]
- Gu HF, Efendic S, Nordman S, Ostenson CG, Brismar K, Brookes AJ, Prince JA. Quantitative trait loci near the insulin-degrading enzyme (IDE) gene contribute to variation in plasma insulin levels. Diabetes. 2004;53:2137–2142. doi: 10.2337/diabetes.53.8.2137. [DOI] [PubMed] [Google Scholar]
- Hamel FG, Upward JL, Bennett RG. In vitro inhibition of insulin-degrading enzyme by long-chain fatty acids and their coenzyme A thioesters. Endocrinology. 2003;144:2404–2408. doi: 10.1210/en.2002-0007. [DOI] [PubMed] [Google Scholar]
- Hari J, Shii K, Roth RA. In vivo association of [125I]-insulin with a cytosolic insulin-degrading enzyme: detection by covalent cross-linking and immunoprecipitation with a monoclonal antibody. Endocrinology. 1987;120:829–831. doi: 10.1210/endo-120-2-829. [DOI] [PubMed] [Google Scholar]
- Hersh L. The insulysin (insulin degrading enzyme) enigma. Cell Mol Life Sci. 2006;63:2432–2434. doi: 10.1007/s00018-006-6238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im H, Manolopoulou M, Malito E, Shen Y, Zhao J, Neant-Fery M, Sun C, Meredith S, Sisodia S, Leissring M, Tang W. Structure of Substrate-free Human Insulin-degrading Enzyme (IDE) and Biophysical Analysis of ATP-induced Conformational Switch of IDE. Journal of Biological Chemistry. 2007;282:25453–25463. doi: 10.1074/jbc.M701590200. [DOI] [PubMed] [Google Scholar]
- Jee S, Hwang D, Seo S, Kim Y, Kim C, Kim B, Shim S, Lee S, Sin J, Bae C, Lee B, Jang M, Kim M, Yim S, Jang I, Cho J, Chae K. Microarray analysis of insulin-regulated gene expression in the liver: the use of transgenic mice co-expressing insulin-siRNA and human IDE as an animal model. Int J Mol Med. 2007;20:829–835. [PubMed] [Google Scholar]
- Johnson K, Bhushan S, Ståhl A, Hallberg B, Frohn A, Glaser E, Eneqvist T. The closed structure of presequence protease PreP forms a unique 10 000 Å3 chamber for proteolysis. EMBO J. 2006;25:1977–1986. doi: 10.1038/sj.emboj.7601080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamohamed S, Demissie S, Volcjak J, Liu C, Heard-Costa N, Liu J, Shoemaker CM, Panhuysen CI, Meigs JB, Wilson P, Atwood LD, Cupples LA, Herbert A. Polymorphisms in the insulin-degrading enzyme gene are associated with type 2 diabetes in men from the NHLBI Framingham Heart Study. Diabetes. 2003;52:1562–1567. doi: 10.2337/diabetes.52.6.1562. [DOI] [PubMed] [Google Scholar]
- Kuo WL, Gehm BD, Rosner MR. Regulation of insulin degradation: expression of an evolutionarily conserved insulin-degrading enzyme increases degradation via an intracellular pathway. Mol Endocrinol. 1991;5:1467–1476. doi: 10.1210/mend-5-10-1467. [DOI] [PubMed] [Google Scholar]
- Kurochkin IV. Insulin-degrading enzyme: embarking on amyloid destruction. Trends Biochem Sci. 2001;26:421–425. doi: 10.1016/s0968-0004(01)01876-x. [DOI] [PubMed] [Google Scholar]
- Leissring MA, Farris W, Wu X, Christodoulou DC, Haigis MC, Guarente L, Selkoe DJ. Alternative translation initiation generates a novel isoform of insulin-degrading enzyme targeted to mitochondria. Biochem J. 2004;383:439–446. doi: 10.1042/BJ20041081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Kuo WL, Yousef M, Rosner MR, Tang W. The C-terminal domain of human insulin degrading enzyme is required for dimerization and substrate recognition. Biochemical and Biophysical Research Communications. 2006;343:1032–1037. doi: 10.1016/j.bbrc.2006.03.083. [DOI] [PubMed] [Google Scholar]
- Malito E, Hulse RE, Tang WJ. Amyloid β-degrading cryptidases: insulin degrading enzyme, presequence peptidase, and neprilysin. Cell Mol Life Sci. 2008 doi: 10.1007/s00018-008-8112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlowe L, Peila R, Benke KS, Hardy J, White LR, Launer LJ, Myers A. Insulin-degrading enzyme haplotypes affect insulin levels but not dementia risk. Neurodegener Dis. 2006;3:320–326. doi: 10.1159/000097300. [DOI] [PubMed] [Google Scholar]
- Maskos K. Handbook of Metalloproteins. John Wiley & Sons, Inc; New York: 2004. [Google Scholar]
- Melino G, Draoui M, Bernardini S, Bellincampi L, Reichert U, Cohen P. Regulation by retinoic acid of insulin-degrading enzyme and of a related endoprotease in human neuroblastoma cell lines. Cell Growth Differ. 1996;7:787–796. [PubMed] [Google Scholar]
- Mentlein R, Ludwig R, Martensen I. Proteolytic degradation of Alzheimer's disease amyloid beta-peptide by a metalloproteinase from microglia cells. J Neurochem. 1998;70:721–726. doi: 10.1046/j.1471-4159.1998.70020721.x. [DOI] [PubMed] [Google Scholar]
- MIRSKY IA, BROH-KAHN RH. The inactivation of insulin by tissue extracts; the distribution and properties of insulin inactivating extracts. Archives of biochemistry. 1949;20:1–9. [PubMed] [Google Scholar]
- MIRSKY IA, SIMKIN B, BROH-KAHN RH. The inactivation of insulin by tissue extracts. VI. The existence, distribution and properties of an insulinase inhibitor. Archives of biochemistry. 1950;28:415–423. [PubMed] [Google Scholar]
- Perlman RK, Gehm BD, Kuo WL, Rosner MR. Functional analysis of conserved residues in the active site of insulin-degrading enzyme. J Biol Chem. 1993;268:21538–21544. [PubMed] [Google Scholar]
- Qiu WQ, Folstein MF. Insulin, insulin-degrading enzyme and amyloid-beta peptide in Alzheimer's disease: review and hypothesis. Neurobiol Aging. 2006;27:190–198. doi: 10.1016/j.neurobiolaging.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Qiu WQ, Walsh DM, Ye Z, Vekrellis K, Zhang J, Podlisny MB, Rosner MR, Safavi A, Hersh LB, Selkoe DJ. Insulin-degrading enzyme regulates extracellular levels of amyloid beta-protein by degradation. The Journal of Biological Chemistry. 1998;273:32730–32738. doi: 10.1074/jbc.273.49.32730. [DOI] [PubMed] [Google Scholar]
- Ryan MP, Gifford JD, Solomon SS, Duckworth WC. The calcium dependence of insulin degradation by rat skeletal muscle. Endocrinology. 1985;117:1693–1698. doi: 10.1210/endo-117-4-1693. [DOI] [PubMed] [Google Scholar]
- Shen Y, Joachimiak A, Rich Rosner M, Tang W. Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature. 2006;443:870–874. doi: 10.1038/nature05143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shii K, Roth RA. Inhibition of insulin degradation by hepatoma cells after microinjection of monoclonal antibodies to a specific cytosolic protease. Proc Natl Acad Sci USA. 1986;83:4147–4151. doi: 10.1073/pnas.83.12.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinall H, Song ES, Hersh LB. Susceptibility of amyloid beta peptide degrading enzymes to oxidative damage: a potential Alzheimer's disease spiral. Biochemistry. 2005;44:15345–15350. doi: 10.1021/bi050650l. [DOI] [PubMed] [Google Scholar]
- Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- Song ES, Juliano MA, Juliano L, Hersh LB. Substrate activation of insulin-degrading enzyme (insulysin). A potential target for drug development. The Journal of Biological Chemistry. 2003;278:49789–49794. doi: 10.1074/jbc.M308983200. [DOI] [PubMed] [Google Scholar]
- Song ES, Juliano MA, Juliano L, Fried MG, Wagner SL, Hersh LB. ATP effects on insulin-degrading enzyme are mediated primarily through its triphosphate moiety. J Biol Chem. 2004;279:54216–54220. doi: 10.1074/jbc.M411177200. [DOI] [PubMed] [Google Scholar]
- Stocker R, Keaney JF., Jr. Role of oxidative modifications in atherosclerosis. Physiol Rev. 2004;84:1381–1478. doi: 10.1152/physrev.00047.2003. [DOI] [PubMed] [Google Scholar]
- Sudoh S, Frosch MP, Wolf B. Differential effects of protease involved in intracellular degradation of amyloid-beta protein between detergent soluble and insolube pools in CHO-695 cells. Biochemistry. 2002;41:9. doi: 10.1021/bi011193l. [DOI] [PubMed] [Google Scholar]
- Vekrellis K, Ye Z, Qiu WQ, Walsh D, Hartley D, Chesneau V, Rosner MR, Selkoe DJ. Neurons regulate extracellular levels of amyloid beta-protein via proteolysis by insulin-degrading enzyme. J Neurosci. 2000;20:1657–1665. doi: 10.1523/JNEUROSCI.20-05-01657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Teter B, Morihara T, Lim GP, Ambegaokar SS, Ubeda OJ, Frautschy SA, Cole GM. Insulin-degrading enzyme as a downstream target of insulin receptor signaling cascade: implications for Alzheimer's disease intervention. J Neurosci. 2004;24:11120–11126. doi: 10.1523/JNEUROSCI.2860-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]