Abstract
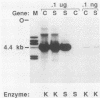
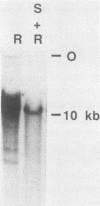
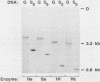
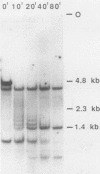
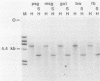
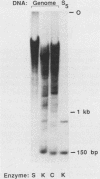
To understand the basis for tissue-specific production and accumulation of alanine tRNA in silkworms, we have examined the organization of the genes that code for silk gland-specific and constitutive alanine tRNAs. We have found that all of the silk gland-specific tRNA(Ala) genes (approximately 20) appear to be tightly clustered at a single locus in the Bombyx genome. These genes are arranged in tandem at intervals of approximately 150 base pairs. In contrast to the arrangement of the silk gland-specific tRNA(Ala) genes, most of the 20 to 30 constitutive tRNA(Ala) genes are dispersed in the genome. Silk gland-specific tRNA(Ala) genes are not amplified or grossly rearranged in the silk gland. Thus it is likely that differential transcription, rather than changes in gene number or structure, accounts for the tissue-specific accumulation of tRNA(Ala).
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borer P. N., Dengler B., Tinoco I., Jr, Uhlenbeck O. C. Stability of ribonucleic acid double-stranded helices. J Mol Biol. 1974 Jul 15;86(4):843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- Conner B. J., Reyes A. A., Morin C., Itakura K., Teplitz R. L., Wallace R. B. Detection of sickle cell beta S-globin allele by hybridization with synthetic oligonucleotides. Proc Natl Acad Sci U S A. 1983 Jan;80(1):278–282. doi: 10.1073/pnas.80.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Ford P. J., Mathieson T. Control of 5S RNA synthesis in Xenopus laevis. Nature. 1976 Jun 3;261(5559):433–435. doi: 10.1038/261433a0. [DOI] [PubMed] [Google Scholar]
- Gage L. P. Polyploidization of the silk gland of Bombyx mori. J Mol Biol. 1974 Jun 15;86(1):97–108. doi: 10.1016/s0022-2836(74)80010-0. [DOI] [PubMed] [Google Scholar]
- Gage L. P. The Bombyx mori genome: analysis by DNA reassociation kinetics. Chromosoma. 1974 Mar 1;45(1):27–42. doi: 10.1007/BF00283828. [DOI] [PubMed] [Google Scholar]
- Garel J. P., Mandel P., Chavancy G., Daillie J. Functional adaptation of tRNAs to fibroin biosynthesis in the silkgland of Bombyx mori L. FEBS Lett. 1970 May 1;7(4):327–329. doi: 10.1016/0014-5793(70)80196-x. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J., Bloomer L. S. Assembly of transcriptionally active 5S RNA gene chromatin in vitro. Cell. 1982 Apr;28(4):781–791. doi: 10.1016/0092-8674(82)90057-5. [DOI] [PubMed] [Google Scholar]
- Gralla J., Crothers D. M. Free energy of imperfect nucleic acid helices. II. Small hairpin loops. J Mol Biol. 1973 Feb 5;73(4):497–511. doi: 10.1016/0022-2836(73)90096-x. [DOI] [PubMed] [Google Scholar]
- Guinta D. R., Korn L. J. Differential order of replication of Xenopus laevis 5S RNA genes. Mol Cell Biol. 1986 Jul;6(7):2536–2542. doi: 10.1128/mcb.6.7.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinta D. R., Tso J. Y., Narayanswami S., Hamkalo B. A., Korn L. J. Early replication and expression of oocyte-type 5S RNA genes in a Xenopus somatic cell line carrying a translocation. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5150–5154. doi: 10.1073/pnas.83.14.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbüchle O., Larson D., Hall G. I., Sprague K. U. The primary transcription product of a silkworm alanine tRNA gene: identification of in vitro sites of initiation, termination and processing. Cell. 1979 Dec;18(4):1217–1229. doi: 10.1016/0092-8674(79)90234-4. [DOI] [PubMed] [Google Scholar]
- Harper M. E., Price J., Korn L. J. Chromosomal mapping of Xenopus 5S genes: somatic-type versus oocyte-type. Nucleic Acids Res. 1983 Apr 25;11(8):2313–2323. doi: 10.1093/nar/11.8.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J., Strathern J. N., Klar A. J. Transposable mating type genes in Saccharomyces cerevisiae. Nature. 1979 Nov 29;282(5738):478–473. doi: 10.1038/282478a0. [DOI] [PubMed] [Google Scholar]
- Karrer K. M., Gall J. G. The macronuclear ribosomal DNA of Tetrahymena pyriformis is a palindrome. J Mol Biol. 1976 Jun 25;104(2):421–453. doi: 10.1016/0022-2836(76)90280-1. [DOI] [PubMed] [Google Scholar]
- LUCAS F., SHAW J. T., SMITH S. G. The silk fibroins. Adv Protein Chem. 1958;13:107–242. doi: 10.1016/s0065-3233(08)60599-9. [DOI] [PubMed] [Google Scholar]
- Larson D., Bradford-Wilcox J., Young L. S., Sprague K. U. A short 5' flanking region containing conserved sequences is required for silkworm alanine tRNA gene activity. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3416–3420. doi: 10.1073/pnas.80.11.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Matsuzaki K. Fractionation of amino acid-specific s-RNA from silkgland by methylated albumin column chromatography. Biochim Biophys Acta. 1966 Feb 21;114(2):222–226. doi: 10.1016/0005-2787(66)90303-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., von Hippel P. H. Formaldehyde as a probe of DNA structure. 3. Equilibrium denaturation of DNA and synthetic polynucleotides. Biochemistry. 1977 Jul 26;16(15):3267–3276. doi: 10.1021/bi00634a001. [DOI] [PubMed] [Google Scholar]
- Meza L., Araya A., Leon G., Krauskopf M. Specific alanine-tRNA species associated with fibroin biosynthesis in the posterior sild-gland of Bombyx mori L. FEBS Lett. 1977 May 15;77(2):255–260. doi: 10.1016/0014-5793(77)80246-9. [DOI] [PubMed] [Google Scholar]
- Morton D. G., Sprague K. U. Silkworm 5S RNA and alanine tRNA genes share highly conserved 5' flanking and coding sequences. Mol Cell Biol. 1982 Dec;2(12):1524–1531. doi: 10.1128/mcb.2.12.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E., Brammar W. J., Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- Narang S. A., Brousseau R., Hsiung H. M., Michniewicz J. J. Chemical synthesis of deoxyoligonucleotides by the modified triester method. Methods Enzymol. 1980;65(1):610–620. doi: 10.1016/s0076-6879(80)65063-0. [DOI] [PubMed] [Google Scholar]
- Pardue M. L., Brown D. D., Birnstiel M. L. Location of the genes for 5S ribosomal RNA in Xenopus laevis. Chromosoma. 1973;42(2):191–203. doi: 10.1007/BF00320940. [DOI] [PubMed] [Google Scholar]
- Rasch E. M. The DNA content of sperm and hemocyte nuclei of the silkworm, Bombyx mori L. Chromosoma. 1974 Mar 1;45(1):1–26. doi: 10.1007/BF00283827. [DOI] [PubMed] [Google Scholar]
- Rose J. K., Mosteller R. D., Yanofsky C. Tryptophan messenger ribonucleic acid elongation rates and steady-state levels of tryptophan operon enzymes under various growth conditions. J Mol Biol. 1970 Aug;51(3):541–550. doi: 10.1016/0022-2836(70)90007-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lund E., Smithies O., Blattner F. R. Hybridization of labeled RNA to DNA in agarose gels. Nucleic Acids Res. 1975 Oct;2(10):1911–1929. doi: 10.1093/nar/2.10.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. Phase variation: genetic analysis of switching mutants. Cell. 1980 Apr;19(4):845–854. doi: 10.1016/0092-8674(80)90075-6. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sprague K. U., Hagenbüchle O., Zuniga M. C. The nucleotide sequence of two silk gland alanine tRNAs: implications for fibroin synthesis and for initiator tRNA structure. Cell. 1977 Jul;11(3):561–570. doi: 10.1016/0092-8674(77)90074-5. [DOI] [PubMed] [Google Scholar]
- Wilson E. T., Larson D., Young L. S., Sprague K. U. A large region controls tRNA gene transcription. J Mol Biol. 1985 May 25;183(2):153–163. doi: 10.1016/0022-2836(85)90209-8. [DOI] [PubMed] [Google Scholar]
- Wormington W. M., Schlissel M., Brown D. D. Developmental regulation of Xenopus 5S RNA genes. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):879–884. doi: 10.1101/sqb.1983.047.01.101. [DOI] [PubMed] [Google Scholar]
- Young L. S., Takahashi N., Sprague K. U. Upstream sequences confer distinctive transcriptional properties on genes encoding silkgland-specific tRNAAla. Proc Natl Acad Sci U S A. 1986 Jan;83(2):374–378. doi: 10.1073/pnas.83.2.374. [DOI] [PMC free article] [PubMed] [Google Scholar]