Summary
Amphotericin, miconazole and ciclopirox are antifungal agents from three different drug classes that can effectively kill planktonic yeast, yet their complete fungicidal mechanisms are not fully understood. Here we employ a systems biology approach to identify a common oxidative damage cellular death pathway triggered by these representative fungicides in Candida albicans and Saccharomyces cerevisiae. This mechanism utilizes a signaling cascade involving the GTPases Ras1/2 and Protein Kinase A, and culminates in death through the production of toxic ROS in a tricarboxylic acid cycle- and respiratory chain-dependent manner. We also show that the metabolome of C. albicans is altered by antifungal drug treatment, exhibiting a shift from fermentation to respiration, a jump in the AMP/ATP ratio, and elevated production of sugars; this coincides with elevated mitochondrial activity. Lastly, we demonstrate that DNA damage plays a critical role in antifungal-induced cellular death and that blocking DNA repair mechanisms potentiates fungicidal activity.
Introduction
A rapid rise in immunocompromised patients over the past five decades has led to increasing incidence of systemic fungal infections. Despite current treatment options, the morbidity and mortality rates associated with fungal infections, particularly those of Candida species, remain high (Ostrosky-Zeichner et al., 2010).
The polyene amphotericin B (AMB), introduced in the late 1950s, was the first widely used antifungal (AF) drug (Ostrosky-Zeichner et al., 2010). Due to its strong hydrophobicity, AMB penetrates the fungal membrane and binds to ergosterol leading to membrane damage. Azoles, a second class of AFs, became available in the 1980s and act by inhibiting ergosterol biosynthesis to induce the accumulation of a toxic methylated sterol that stops cell growth (Ostrosky-Zeichner et al., 2010). While azoles tend to be fungistatic due to their poor solubility, under certain conditions and formulations, some azoles such as miconazole (MCZ) can be fungicidal (Thevissen et al., 2007). Unlike AMB and MCZ, the primary targets of the synthetic AF ciclopirox olamine (CIC) are not fully understood, though some evidence indicates that CIC acts by affecting DNA repair or directly inducing DNA damage (Leem et al., 2003).
Recent work with select AFs has indicated that AF-induced cellular death may follow apoptotic or necrotic pathways that involve the production of reactive oxygen species (ROS) (Phillips et al., 2003; Thevissen et al., 2007) and the modulation of central metabolism (Gonzalez-Parraga et al., 2011; Yan et al., 2007). However, a comprehensive genetic and metabolic mechanism of AF-induced cellular death remains elusive.
Recent systems biology work from our lab, focused on bacterial cell death, identified a common mechanism of antibiotic action that involves a genetic and metabolic cascade that ultimately results in the formation of toxic ROS (Dwyer et al., 2007; Kohanski et al., 2007; Kohanski et al., 2008). Aspects of this mechanism have been harnessed to potentiate currently available antibiotics and combat the development of resistance (Dwyer et al., 2009; Kohanski et al., 2010; Kohanski et al., 2007; Lu and Collins, 2009). Similar approaches to AF mechanisms could provide important, meaningful insights and identify possible means to improve current treatments.
In this work, we apply a systems biology approach to identify mechanisms by which the aforementioned AFs —AMB, MCZ and CIC— lead to fungal cellular death. We find that, despite their different primary modes of action, all three classes of fungicidal drugs induce a common oxidative damage cellular death pathway in S. cerevisiae and C. albicans that involves alterations to cellular metabolism and respiration, culminating in the formation of lethal ROS.
Results
Fungicide-Dependent ROS Production Leads to Fungal Cell Death
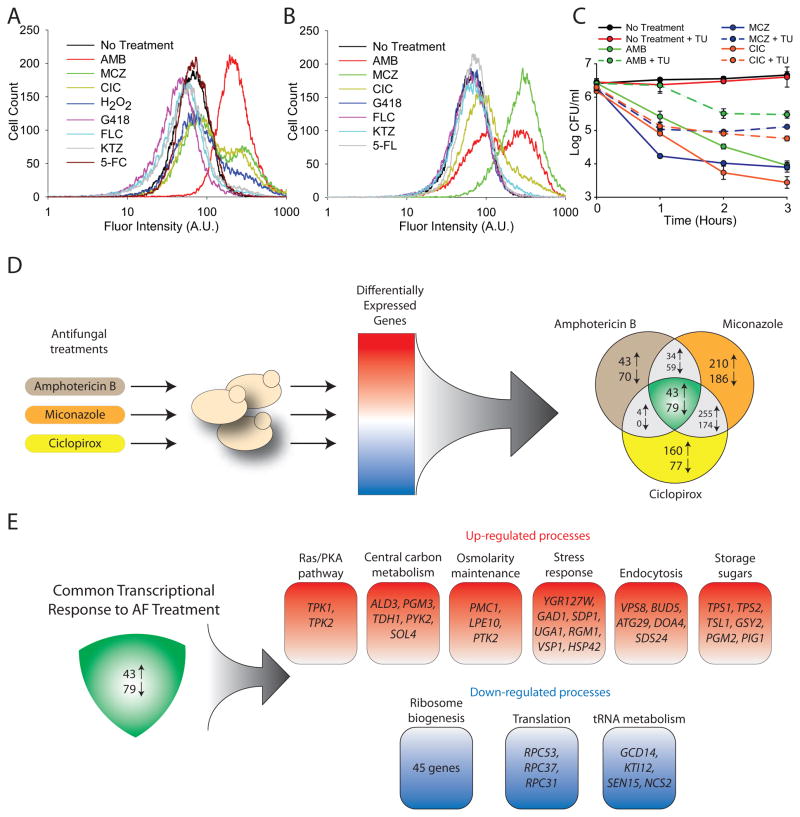
Based on previous work that supports the cidal role of ROS production in many modes of fungal death (Breitenbach et al., 2005; Henriquez et al., 2008), we hypothesized that ROS production is critical to AF-induced cellular death. We measured the formation of ROS following AF treatment in yeast using the dye 3′-(p-hydroxyphenyl) fluorescein (HPF), which is preferentially oxidized by intracellular hydroxyl radicals into a fluorescent product (Kohanski et al., 2007). We treated exponentially growing wildtype S. cerevisiae and C. albicans in synthetic dextrose complete (SDC) medium with the minimum concentration of fungicide required to achieve at least a 90% reduction in colony forming units (CFU) after three hours of exposure. As a positive control, we also treated cells with H2O2, a potent inducer of hydroxyl radical formation (Perrone et al., 2008). After 1.5 hours of treatment, we found that all tested fungicidal drugs and H2O2 lead to dramatic induction of HPF fluorescence (Figures 1A, B and S1), indicating that all tested fungicidal agents induce the formation of ROS. Conversely, fungistatic drugs added at concentrations 10-fold above the minimal inhibitory concentration or at the maximum soluble concentration did not lead to detectable ROS formation (Figures 1A, B and S1).
Figure 1. Fungicide-Dependent ROS Production Leads to Fungal Cell Death and a Common Transcriptional Response.
(A–B) Generation of ROS as measured by a change in HPF fluorescence after 1.5 hours of drug treatment in S. cerevisiae (A) and C. albicans (B). (C) Log of CFU/ml remaining after drug exposure in the presence and absence of 50 mM thiourea (TU). Drug concentrations used: AMB 1 μg/ml, MCZ 50 μg/ml, CIC 75 μg/ml, H2O2 1 mM, geneticin (G418) 100 μg/ml, fluconazol (FLC) 50 μg/ml, 5-flucytosine (5-FC) 15 μg/ml, and ketoconazole (KTZ) 50μg/ml. The reported error is standard deviation (s.d.) with n≥3. (D) A common transcriptional response to antifungal treatment was identified by performing differential expression analysis against a compendium of expression arrays. The common set of differentially expressed genes is represented by the intersection of the three sets of genes. (E) Pathway analysis of the common set of differentially expressed genes identified six major processes that are upregulated in response to antifungals and three processes that are downregulated under the same treatments.
To test whether the observed production of ROS contributes to AF-induced cellular death, we treated cells with thiourea, a potent scavenger of hydroxyl radicals in eukaryotic and prokaryotic cells (Kohanski et al., 2007; Touati et al., 1995). We found that exposing exponentially growing S. cerevisiae to 50 mM thiourea for 30 minutes prior to the addition of AF drugs considerably diminished the toxicity of all three AFs, reducing killing at 3 hours by ~15-fold for AMB and CIC and by ~10-fold for MCZ (Figure 1C). These results indicate that ROS production plays a critical role in cellular death following treatment by AMB, MCZ and CIC.
Identifying a Common Transcriptional Response to AF Treatment
To build a comprehensive model of the yeast response to AFs, we measured changes in global gene expression of S. cerevisiae following treatment with AMB, MCZ and CIC. To assess the effect of each AF treatment on gene expression, we performed a z-test between experiment and control samples, where the average and standard deviation for the expression of each gene was calculated across a compendium of publically available expression datasets (see Supplemental Information). We compared the differential gene expression profile of each treatment at each time point to the no-treatment control to identify the set of genes commonly perturbed by fungicide treatment (Figure 1D). This analysis revealed that 43 genes were commonly upregulated under AF treatment, while 79 genes were downregulated under the same conditions (Figure 1E). To investigate the biological pathways and processes in which these genes are involved, we ran a functional enrichment analysis on the GO terms associated with each one of these genes. The analysis was conducted with the Saccharomyces Genome Database’s GO Term Finder tool using default parameters. The majority of the commonly downregulated genes are involved in protein synthesis, specifically, ribosomal biogenesis and tRNA synthesis. The 43 commonly upregulated genes fall into six general biological processes: the production of storage sugars, endocytosis, general stress response, osmolarity maintenance, central carbon metabolism, and the RAS/PKA signaling pathway (Figure 1E).
Our expression analysis identified the production of storage sugars as a key process upregulated following AF treatment. Specifically, genes involved in glycogen metabolism and the production of the storage sugar trehalose were robustly upregulated after the addition of fungicides (Figure 1E; Table S2). Cellular production of trehalose and glycogen are energy expensive pathways that consume significant amounts of ATP. Consistent with this, the activation of the trehalose pathway has been shown to increase ATP consumption, mitochondrial enzyme content and respiration (Noubhani et al., 2009). Thus, the production of these sugars may contribute to increased mitochondrial activity and elevated ROS production.
TCA Cycle and Electron Transport Chain Play Critical Roles in AF-Induced Cell Death
To identify genes critical to AF action, we tested the AF sensitivity of single-gene knockouts identified through our common transcriptional analysis. In total, we tested the AF sensitivities of 81 single-gene knockouts (Table S1), and found that 12 of them had increased resistance to all three antifungals (AMB, MCZ, CIC).
Actively respiring, energy-producing mitochondria are the major source of ROS in yeast cells. However, S. cerevisiae respiratory activity is usually repressed under normal growth conditions in the presence of glucose (Hardie et al., 1999), though various stress responses have been shown to upregulate respiratory genes even in the presence of glucose (Gasch et al., 2000). We therefore hypothesized that treatment with fungicides induces a switch from normal fermentative growth to mitochondrial respiration.
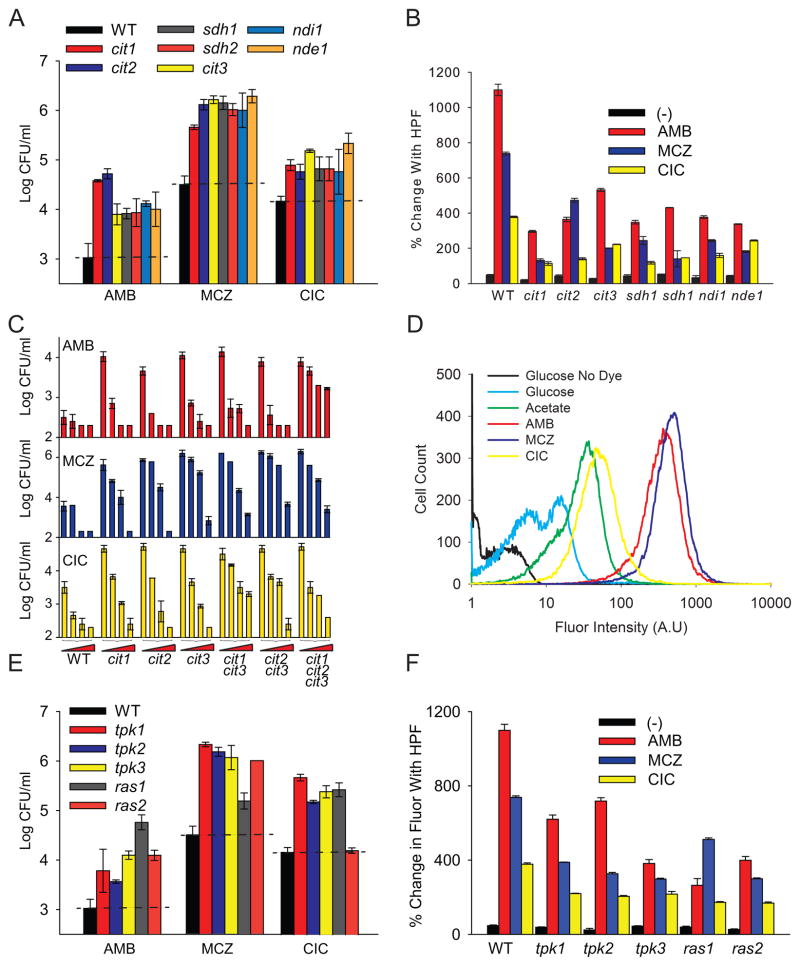
To assess the role of mitochondrial respiration in AF-induced cellular death, we first analyzed the expression of mitochondrial genes in S. cerevisiae in response to AF treatment (Table S2). Of particular interest, we found that the gene encoding the rate-limiting step of the TCA cycle, citrate synthase-1 (CIT1), exhibited a slight increase in expression across all AF treatments. The induction of CIT1 is a hallmark of stress-induced respiration (Gasch et al., 2000). We found that deleting CIT1, CIT2 or CIT3 dramatically reduced yeast sensitivity to all three AFs, compared to the wildtype S. cerevisiae strain (Figures 2A and S2). To parse out the role of the various citrate syntheses in AF-induced cellular death, we created single and double deletions of CIT1 and CIT2 in the cit3 background. We found that deleting the remaining citrate synthases in this background provides additive resistance to AMB, with the triple mutant requiring more than 8-fold higher drug concentrations to achieve the sensitivity of the single mutants (Figures 2C). Interestingly, a similar additive effect was not observed with MCZ or CIC, indicating that AMB toxicity is more sensitive to further changes in citrate metabolism than the other two drugs.
Figure 2. TCA-Dependent Respiration and the Ras/PKA Pathway Play a Critical Role in Antifungal-Induced Cell Death.
(A and E) Log of CFU/ml remaining after three hours of drug exposure of wildtype S. cerevisiae and mutants targeting the TCA cycle, respiration and the Ras/PKA pathway. (B and F) Cellular ROS levels quantified as percent change in fluorescence after the addition of HPF. The indicated strains were treated with antifungal drugs for 1 hour prior to the addition of HPF. Drug concentrations used: AMB 1 μg/ml, MCZ 50 μg/ml, and CIC 75 μg/ml. The reported error is s.d. with n≥3. (C) Log of CFU/ml remaining after three hours of drug exposure. Drug concentrations used, increasing from left to right: AMB (1 μg/ml, 1.5 μg/ml, 2.5 μg/ml and 8 μg/ml), MCZ (50 μg/ml, 75 μg/ml, 100 μg/ml and 150 μg/ml), and CIC (75 μg/ml 100 μg/ml, 125 μg/ml and 150 μg/ml). (D) Yeast mitochondrial content assayed by fluorescent staining with MitoTracker Red probe after one hour of exposure to the indicated drug or metabolite.
We also found that deleting succinate dehydrogenases (SDH1 or SDH2), enzymes that couple the oxidation of succinate to the transfer of electrons to the mitochondrial ETC (Chapman et al., 1992), decreased drug susceptibility. Additionally, we found that blocking the TCA cycle at these two key points reduces the AF-dependent production of ROS (Figure 2B), indicating that this phenomenon involves the TCA cycle.
The TCA cycle produces NADH, which is then fed into the ETC to produce ATP. This activity also leads to the production of ROS as a byproduct of aerobic respiration. We therefore targeted the first committed steps of the ETC, by deleting the intramitochondrial NADH dehydrogenase (NDI1) and the external NADH dehydrogenase (NDE1). The NDI1 and NDE1 deletions exhibited increased resistance to all three AF drugs and a concomitant reduction in AF-dependent ROS production (Figure 2A and B).
We next sought to assess the mitochondrial activity of AF-treated fungal cells. We utilized the MitoTracker Red dye, which enters the mitochondrial matrix by utilizing the proton motive force and thus labels metabolically active mitochondria in viable cells (Arita et al., 2006; Tomas et al., 2011). We incubated exponential phase S. cerevisiae in glucose-free synthetic complete (SC) media for 30 min and then switched them to SC containing 2% glucose or non-fermentable acetate for 1.5 hours. As expected, we detected approximately 5-fold more MitoTracker fluorescence in acetate-incubated cells (Figure 2D), indicating that the dye specifically labels cells with activated mitochondria. Adding CIC, AMB, or MCZ to glucose-incubated cells increased MitoTracker fluorescence by 10-fold, 60-fold and 70-fold, respectively (Figure 2D). All three drugs induced considerably more mitochondrial activity than acetate, indicating that AF treatment has a greater impact on mitochondrial activity than normal respiratory metabolism. These results are consistent with our hypothesis that AF drugs induce a shift from fermentative growth to ROS-producing mitochondrial respiration.
RAS/PKA Pathway is a Key Mediator of Antifungal Toxicity
Having established that AFs induce mitochondrial-dependent ROS production, we next sought to ascertain the signaling events that lead to these metabolic changes. The RAS/PKA pathway, consisting of two GTPases Ras1 and 2 and three Protein Kinase A isoforms (Tpk1-3), has been shown to respond to cellular stress by inducing mitochondrial biogenesis and cellular death through the production of ROS via disordered mitochondrial respiration (Chevtzoff et al., 2010; Leadsham and Gourlay, 2010; Thevelein and de Winde, 1999). We found that aspects of this signaling pathway were upregulated in response to AFs (Table S2). For example, TPK1 and TPK2 were robustly induced in response to AF treatment, and we found an increase in expression of other genes involved in the same pathway (Table S2), namely BMH2, CYR1 and SRV2. Further, we found that PDE2, the cAMP phosphodiesterase that represses the RAS/PKA signaling, was significantly downregulated. Deleting PDE2 in the presence of activated RAS/PKA signaling has been shown to lead to the overproduction of ROS by dysfunctional mitochondria resulting in cellular death (Leadsham and Gourlay, 2010). Together, these results indicate that the RAS/PKA signaling pathway is largely upregulated in response to AF treatment and may contribute to the production of ROS by mitochondria.
We tested the role of the RAS/PKA pathway in AF-induced cellular death by studying single-gene knockouts of the upstream and downstream portions of the pathway. We found that deleting either RAS1 or RAS2 reduced yeast susceptibility to all three AFs (Figures 2E and S2), and deleting the downstream effector kinases, TPK1, TPK2 and TPK3, also reduced or delayed killing. Additionally we found that deleting any one of the key members of the RAS/PKA signaling pathway reduced the drug-dependent buildup of ROS (Figure 2F). This finding suggests that RAS/PKA signaling is critical for the induction of mitochondrial ROS production in response to drug treatment.
Common Metabolic Changes Resulting from Antifungal Treatment
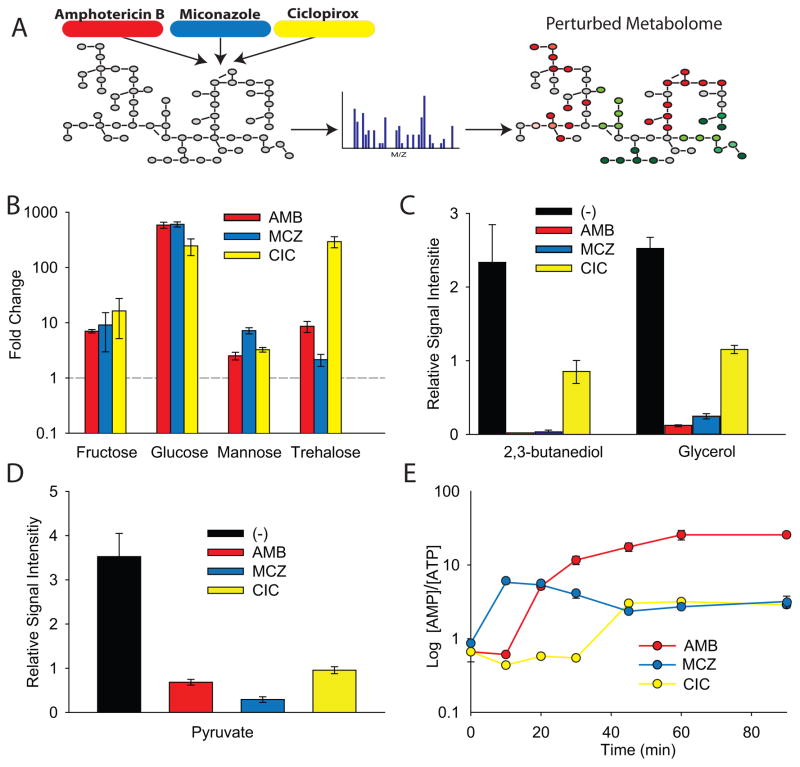
Our results link AF treatment to distinct changes in intracellular metabolic activity as part of an induced common killing mechanism. To further explore the role of metabolism in AF-induced cellular death, we directly measured the effect of AF treatment on the intracellular metabolome of C. albicans. To do so, we utilized a platform for metabolite detection and relative quantification from Metabolon Inc. (Durham, USA) (Evans et al., 2009). We treated exponentially growing C. albicans cells with AFs and collected samples for metabolomic analysis after 1.5 hours of treatment, a time point when we expected at least 90% of cells contributing to the metabolite pool to be irreversibly committed to the death pathway (Figure S3). We found that the three drug treatments (AMB, MCZ, CIC) significantly changed (p ≤ 0.05) the relative abundance of between 155 and 213 metabolites, compared to the no-treatment controls (Supplemental Data File 1).
Glucose was the most highly induced metabolite, found to be greater than 600-fold more abundant in drug-treated cells (Figure 3B). Other carbohydrates, such as fructose and mannose, were also significantly more abundant in all of the treatment groups compared to the control, as was the disaccharide trehalose (Figure 3B). These results are consistent with our microarray data that identified the upregulation of polysaccharide biosynthesis as an important transcriptional response to AF treatment.
Figure 3. Antifungal Treatment Leads to Common Metabolic Changes Resulting in the Production of Sugars and a Dramatic Reduction of ATP Levels.
(A) A schematic of the metabolomics study. Cells were treated with antifungal drugs for 1.5 hours and intracellular metabolites were analyzed using mass spectrometry to identify the AF-perturbed metabolome. (B) Fold change in metabolite levels compared to the no-treatment control. (C and D) Relative signal intensity of select metabolites identified through metabolomic profiling of C. albicans exposed to antifungal drugs for 1.5 hours. Drug concentrations used: AMB 1 μg/ml, MCZ 50 μg/ml, and CIC 75 μg/ml. The reported error is standard error mean with n=6. (E) AMP/ATP ratio in C. albicans treated with drugs over a 90 min period. Drug concentrations used: AMB 0.35 μg/ml, MCZ 50 μg/ml, and CIC 75 μg/ml. The reported error is s.d. with an n≥3.
Our phenotypic analyses in S. cerevisiae suggested that cells may be switching from exclusively fermentative growth to mitochondrial respiration in response to AF treatment. Consistent with this result, we found that 2,3-butanediol and glycerol, two major fermentative waste products (Gonzalez et al., 2000), were substantially reduced after AF treatment (Figure 3C). Furthermore, pyruvate levels were dramatically lower in drug-treated samples (Figure 3D), indicating possible consumption by the TCA cycle; of note, TCA cycle intermediates were also reduced (Supplemental Data File 1) in comparison to the untreated cells. These measurements, combined with our genetic data and measurements of mitochondrial activity and biogenesis, provide support for a common fungicidal mechanism of action that relies on reduced fermentation and induced ROS-producing mitochondrial respiration.
The dramatic elevation of intracellular sugars and the shift from fermentation to respiration suggests that the AF treatments may be increasing ATP consumption. Abundant nucleotides such as ATP are not accurately quantified using the metabolic platform we selected for this study. We therefore sought to measure ATP levels over time using HPLC. We analyzed the AMP/ATP levels in lysates collected from cells treated with the three AFs. We found that the AF treatments elevated the AMP/ATP ratio by between 4- and 24-fold (Figure 3E), through a large drop in ATP levels and a proportional rise in AMP levels. ATP levels are normally static, but can change dramatically under severe stress conditions that induce necrosis (Henriquez et al., 2008; Osorio et al., 2004). Our results suggest that fungicide-stimulated sugar production induces a necrosis-like rapid consumption of ATP.
DNA Repair is a Critical Response to Antifungal-Dependent ROS Production
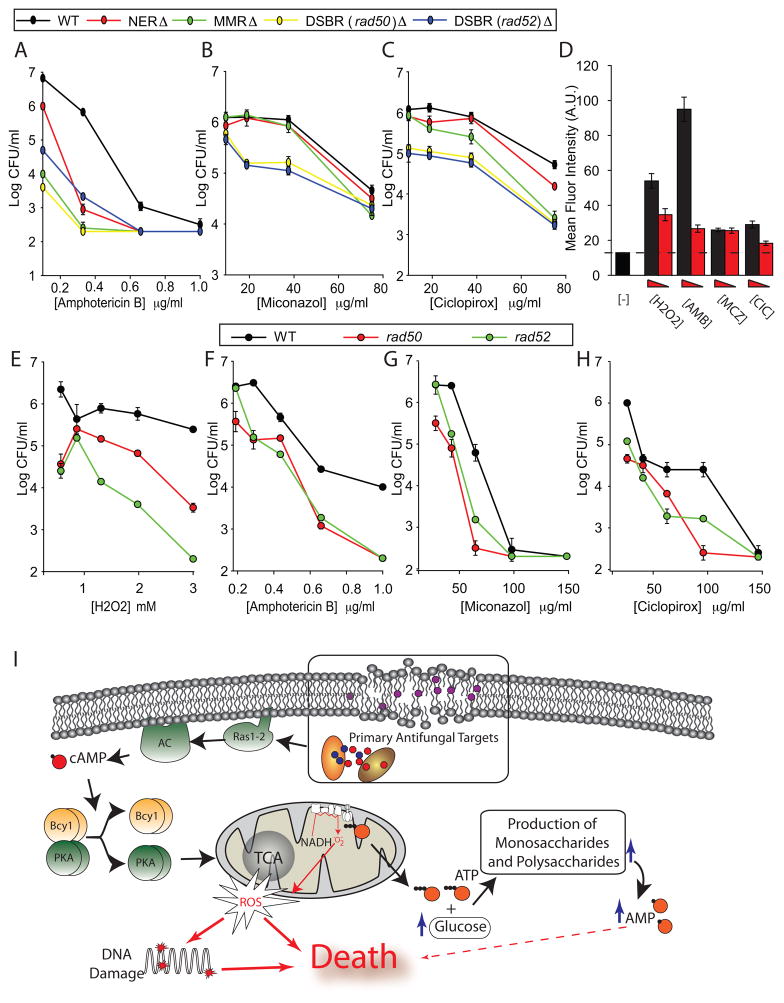
ROS damage multiple cellular targets, including membranes, proteins and DNA (Kohanski et al., 2007; Salmon et al., 2004). Recent work indicates that DNA repair, specifically, double-strand break repair (DSBR), plays a critical role in C. albicans resistance to oxidative damage (Legrand et al., 2007, 2008). We chose to analyze the role of DNA repair in AF-induced cellular death by testing the susceptibility of C. albicans strains with deletions of critical genes in nucleotide excision repair, mismatch repair, and DSBR. We used AMB, MCZ and CIC at a range of concentrations, and found that DSBR mutants were particularly susceptible to all three AFs, compared to wildtype (Figure 4A–C). Specifically, we found that the minimal fungicidal concentrations for the DSBR mutants, rad50/rad50 and rad52/rad52, were reduced by as much as 10-fold compared to wildtype.
Figure 4. DNA Repair is a Critical Response to Antifungal-Dependent ROS Production.
(A–C) Log of CFU/ml remaining after 3 hours of drug exposure of wildtype C. albicans and mutants targeting double-strand break repair (DSBR (rad50/rad50 and rad52/rad52)), nucleotide excision (NER(rad10/rad10)) and mismatch (MMR (msh1/msh1)) DNA repair mechanisms. (D) TUNEL staining of exponentially growing cells assayed by flow cytometry, after 2 hours of treatment. Drug concentrations used, decreasing from left to right: H2O2 (10 mM and 5 mM), AMB (1 μg/ml, and 0.5 μg/ml), MCZ (100 μg/ml, and 50 μg/ml), and CIC (150 μg/ml and 75 μg/ml). (E–H) Log of CFU/ml remaining after 3 hours of drug exposure of wildtype S. cerevisiae and mutants targeting DSBR. The reported error is s.d. with n≥3. (I) The proposed common mechanism of AF action for the tested fungicides: antifungal activity against primary intracellular targets leads to cellular changes sensed by the RAS/PKA signaling pathway. The RAS/PKA signaling cascade induces mitochondrial activity, leading to the production of ROS. Simultaneously, the production of sugars, consistent with the fungal stress response, leads to the rapid consumption of ATP and the production of AMP. Elevated intracellular ROS production leads to cellular death through damage to DNA and other cellular targets.
Next we utilized the terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay to quantify the relative abundance of DSBs in C. albicans cells treated with AFs and H2O2. The TUNEL reagent fluorescently labels both double- and single-strand DNA breaks (Ribeiro et al., 2006). Treating cells with AFs and H2O2 elevated relative fluorescence as measured by flow cytometry (Figure 4D), indicating a considerable induction of DNA breaks.
We also assayed rad50 and rad52 knockouts in S. cerevisiae to test whether the role of DNA damage in AF toxicity is consistent with the results from C. albicans. We found that these mutants were considerably more susceptible to H2O2 and AFs than wildtype S. cerevisiae, although the differences were smaller than those seen with C. albicans. Together these data provide support for the role of DNA damage as a factor contributing to a common mechanism of AF action, and indicates that targeting DNA repair mechanisms could be an effective means for potentiating fungicidal activity.
Discussion
In this work, we utilized a systems biology approach to study the response of fungal cells to AF treatment. We found that despite divergent primary modes of action, fungicides from three distinct classes induce a common signaling and metabolic cascade that leads to ROS-dependent cellular death (Figure 4I). We propose that cellular changes and damage initiated through interaction with primary targets of AFs results in activation of a stress-like response that includes signaling through the RAS/PKA pathway. Through this and possibly other pathways, cells activate mitochondrial activity and shift from fermentation to respiration in response to AFs. This abrupt induction of mitochondrial activity leads to the overproduction of toxic ROS in a manner that depends on the TCA cycle and ETC. Additionally, we found that AF treatment leads to a buildup of monosaccharides and disaccharides, including glucose and trehalose. Both the import and synthesis of these sugars are energetically expensive, requiring the consumption of ATP and the production of AMP. It is likely that these metabolic changes lead to altered respiration and the overproduction of ROS by dysfunctional mitochondria, ultimately resulting in cell death (Figure 4I).
The common AF-dependent responses identified in this work have similarities to many fungal stress response mechanisms (Gasch et al., 2000; Soufi et al., 2009), suggesting that antifungal agents may function by over-activating these pathways to induce cellular death. These similarities include a collapse of ATP levels (Osorio et al., 2004), mitochondrial dysfunction that leads to the production of ROS (Breitenbach et al., 2005), and a central role played by the RAS/PKA pathway (Estruch, 2000). It is likely that AFs cause cellular damage that mirrors the effects of osmotic, heat and alcohol stress, leading to the induction of stress-related pathways. Interestingly, the same responses that protect cells from mild environmental stress may in fact contribute to AF-dependent cellular death. AF insult to fungal cells may be sufficiently powerful such that the over-activation of these general stress responses does not protect the cell, but instead plays a net contributory role in cellular death through the consumption of ATP and the production of ROS.
In addition to similarities with known fungal stress pathways, the identified common mechanism of AF-induced fungal cellular death can also be compared to the previously identified common mechanism of antibiotic-mediated bacterial cellular death (Dwyer et al., 2007; Kohanski et al., 2007). Despite the complex and multilayered eukaryotic mechanism, robust similarities can be found between the two phenomena, indicating possible evolutionary conservation. In both prokaryotes and eukaryotes, cellular damage resulting from the primary drug-target interaction activates key metabolic regulators, such as RAS in yeast and ArcA in bacteria (Kohanski et al., 2008). These universal regulators modulate central metabolism through the induction of the TCA cycle and respiratory activity to produce toxic ROS.
The endpoint of the common antimicrobial mechanisms in both bacteria and yeast is cellular death through the production of toxic ROS. In both prokaryotes and eukaryotes, ROS induces double-strand DNA breaks (Dwyer et al., 2012; Foti et al., 2012) and other forms of cellular damage that lead to cellular death and are potentiated by blocking DNA repair. In yeast, we demonstrated that by blocking DSBR, we could reduce the minimal fungicidal concentration of the tested AF drugs by as much as 10-fold. Similarly, deleting RecA, an E. coli homolog of the yeast RAD genes, substantially enhances bactericidal antibiotic activity in bacteria (Kohanski et al., 2007). These findings suggest that inhibitors of DNA repair mechanisms may be robust potentiators of fungicides and bactericidal antibiotics.
The critical role of ROS in antimicrobial activity indicates that ROS generation may have evolved and been maintained in both prokaryotes and eukaryotes as a central mechanism to overcome environmental stress. Under various stress conditions, low-level ROS production can activate multiple protective responses including the upregulation of antioxidant enzymes and the induction of beneficial mutations (Belenky and Collins, 2011; Gems and Partridge, 2008; Kohanski et al., 2010). However, if ROS production goes above a certain threshold, it no longer serves a protective role and instead induces cellular death. Thus it is likely that many lethal challenges, including fungicides, function by highjacking natural stress response mechanisms to induce ROS production above this threshold.
Experimental Procedures
Fungal Strains and Media
S. cerevisiae strains used in this work were derivatives of BY4742, created as part of the Deletion Consortium (Winzeler et al., 1999). Deletion Consortium strains with reported phenotypes were verified by PCR. Stains PAB202 (BY4742 cit1Δ::kanMX4 cit3Δ::URA3), PAB205 (BY4742 cit3Δ::kanMX4 cit2Δ::LEU2) and PAB208 (PAB202 cit2Δ::LEU2) were created by direct transformation of PCR products as previously described (Brachmann et al., 1998). S. cerevisiae rad50 and rad52 knockouts were derivatives of MKP-0 and generously provided to us by Dr. Simone Moertl (Steininger et al., 2010). Wildtype C. albicans strain, SC5314, was used in metabolomic profiling and HPF measurements. C. albicans DNA repair mutants used in this work were derivatives of DKCa39, generously provided by Dr. David T. Kirkpatrick. A complete list of DKCa strains used in this study is provided in the Supplemental Information. Synthetic dextrose complete (SDC) media and synthetic complete media with 2% acetate (pH 6.5) were prepared as previously described (Burke et al., 2000; Wickerham, 1946).
Fungicidal Killing and Fluorescent Dye Assays
Overnight yeast cultures were diluted into the indicated media and grown to an OD600 of 0.2, at which point the AFs were added. Colony forming units (CFU) were measured by plating six serial dilutions onto YPD agar plates. A more detailed description is included in the supplemental information.
S. cerevisiae Microarrays
Yeast cells were incubated in 25 ml of SDC and grown in 250ml flasks at 30° C and 300 rpm. AFs were added at an OD600 of 0.2. Cells were harvested after treatment, and their RNA was isolated and processed as previously described (Schmitt et al., 1990). The complete microarray analysis procedures are described in the Supplemental Information.
Metabolomic Profiling
C. albicans cells were lysed and assayed by Metabolon Inc. (Durham, USA) as previously described (Shakoury-Elizeh et al., 2010). ATP and AMP were extracted as previously described (Walther et al., 2010) and analyzed by HPLC. This procedure is more fully described in the Supplemental Information
Supplementary Material
Highlights.
Fungicide-dependent ROS production leads to fungal cellular death.
The TCA, ETC and RAS/PKA pathways are involved in fungicide-induced cellular death.
Antifungal agents elevate mitochondrial activity, the AMP/ATP ratio, and sugar production.
DNA damage plays an important role in fungicide-induced cellular death.
Acknowledgments
We thank David T. Kirkpatrick for the set of DKCa strains, Simone Moertl for the MKP-0 strains, Merck & Co., Inc. for providing caspofungin, Katie Vignola and Tim Hyde at Metabolon for their help with metabolomic profiling, and Norman Gerry (Coriell Institute) for processing the microarrays. This work was supported by the Howard Hughes Medical Institute and an NIH Director’s Pioneer Award DP1 OD003644.
Footnotes
The Supplemental Information includes a brief results section, additional experimental procedures, four figures, two tables, and one additional data set.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arita Y, Harkness SH, Kazzaz JA, Koo HC, Joseph A, Melendez JA, Davis JM, Chander A, Li Y. Mitochondrial localization of catalase provides optimal protection from H2O2-induced cell death in lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2006;290:L978–986. doi: 10.1152/ajplung.00296.2005. [DOI] [PubMed] [Google Scholar]
- Belenky P, Collins JJ. Microbiology. Antioxidant strategies to tolerate antibiotics. Science. 2011;334:915–916. doi: 10.1126/science.1214823. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Breitenbach M, Laun P, Gimona M. The actin cytoskeleton, RAS-cAMP signaling and mitochondrial ROS in yeast apoptosis. Trends Cell Biol. 2005;15:637–639. doi: 10.1016/j.tcb.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2000. [Google Scholar]
- Chapman KB, Solomon SD, Boeke JD. SDH1, the gene encoding the succinate dehydrogenase flavoprotein subunit from Saccharomyces cerevisiae. Gene. 1992;118:131–136. doi: 10.1016/0378-1119(92)90260-v. [DOI] [PubMed] [Google Scholar]
- Chevtzoff C, Yoboue ED, Galinier A, Casteilla L, Daignan-Fornier B, Rigoulet M, Devin A. Reactive oxygen species-mediated regulation of mitochondrial biogenesis in the yeast Saccharomyces cerevisiae. J Biol Chem. 2010;285:1733–1742. doi: 10.1074/jbc.M109.019570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer DJ, Camacho DM, Kohanski MA, Callura JM, Collins JJ. Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Molecular cell. 2012;46:561–572. doi: 10.1016/j.molcel.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer DJ, Kohanski MA, Collins JJ. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol. 2009;12:482–489. doi: 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch F. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol Rev. 2000;24:469–486. doi: 10.1111/j.1574-6976.2000.tb00551.x. [DOI] [PubMed] [Google Scholar]
- Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- Foti JJ, Devadoss B, Winkler JA, Collins JJ, Walker GC. Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science. 2012;336:315–319. doi: 10.1126/science.1219192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Partridge L. Stress-response hormesis and aging: “that which does not kill us makes us stronger”. Cell Metab. 2008;7:200–203. doi: 10.1016/j.cmet.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Gonzalez E, Fernandez MR, Larroy C, Sola L, Pericas MA, Pares X, Biosca JA. Characterization of a (2R,3R)-2,3-butanediol dehydrogenase as the Saccharomyces cerevisiae YAL060W gene product. Disruption and induction of the gene. J Biol Chem. 2000;275:35876–35885. doi: 10.1074/jbc.M003035200. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Parraga P, Sanchez-Fresneda R, Zaragoza O, Arguelles JC. Amphotericin B induces trehalose synthesis and simultaneously activates an antioxidant enzymatic response in Candida albicans. Biochimica et biophysica acta. 2011;1810:777–783. doi: 10.1016/j.bbagen.2011.04.012. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Salt IP, Hawley SA, Davies SP. AMP-activated protein kinase: an ultrasensitive system for monitoring cellular energy charge. Biochem J. 1999;338(Pt 3):717–722. [PMC free article] [PubMed] [Google Scholar]
- Henriquez M, Armisen R, Stutzin A, Quest AF. Cell death by necrosis, a regulated way to go. Curr Mol Med. 2008;8:187–206. doi: 10.2174/156652408784221289. [DOI] [PubMed] [Google Scholar]
- Kohanski MA, DePristo MA, Collins JJ. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell. 2010;37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135:679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadsham JE, Gourlay CW. cAMP/PKA signaling balances respiratory activity with mitochondria dependent apoptosis via transcriptional regulation. BMC Cell Biol. 2010;11:92. doi: 10.1186/1471-2121-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leem SH, Park JE, Kim IS, Chae JY, Sugino A, Sunwoo Y. The possible mechanism of action of ciclopirox olamine in the yeast Saccharomyces cerevisiae. Mol Cells. 2003;15:55–61. [PubMed] [Google Scholar]
- Legrand M, Chan CL, Jauert PA, Kirkpatrick DT. Role of DNA mismatch repair and double-strand break repair in genome stability and antifungal drug resistance in Candida albicans. Eukaryot Cell. 2007;6:2194–2205. doi: 10.1128/EC.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand M, Chan CL, Jauert PA, Kirkpatrick DT. Analysis of base excision and nucleotide excision repair in Candida albicans. Microbiology. 2008;154:2446–2456. doi: 10.1099/mic.0.2008/017616-0. [DOI] [PubMed] [Google Scholar]
- Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Natl Acad Sci U S A. 2009;106:4629–4634. doi: 10.1073/pnas.0800442106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noubhani A, Bunoust O, Bonini BM, Thevelein JM, Devin A, Rigoulet M. The trehalose pathway regulates mitochondrial respiratory chain content through hexokinase 2 and cAMP in Saccharomyces cerevisiae. J Biol Chem. 2009;284:27229–27234. doi: 10.1074/jbc.M109.029454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio H, Moradas-Ferreira P, Gunther Sillero MA, Sillero A. In Saccharomyces cerevisiae, the effect of H2O2 on ATP, but not on glyceraldehyde-3-phosphate dehydrogenase, depends on the glucose concentration. Arch Microbiol. 2004;181:231–236. doi: 10.1007/s00203-004-0648-6. [DOI] [PubMed] [Google Scholar]
- Ostrosky-Zeichner L, Casadevall A, Galgiani JN, Odds FC, Rex JH. An insight into the antifungal pipeline: selected new molecules and beyond. Nat Rev Drug Discov. 2010;9:719–727. doi: 10.1038/nrd3074. [DOI] [PubMed] [Google Scholar]
- Perrone GG, Tan SX, Dawes IW. Reactive oxygen species and yeast apoptosis. Biochim Biophys Acta. 2008;1783:1354–1368. doi: 10.1016/j.bbamcr.2008.01.023. [DOI] [PubMed] [Google Scholar]
- Phillips AJ, Sudbery I, Ramsdale M. Apoptosis induced by environmental stresses and amphotericin B in Candida albicans. Proc Natl Acad Sci U S A. 2003;100:14327–14332. doi: 10.1073/pnas.2332326100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro GF, Corte-Real M, Johansson B. Characterization of DNA damage in yeast apoptosis induced by hydrogen peroxide, acetic acid, and hyperosmotic shock. Mol Biol Cell. 2006;17:4584–4591. doi: 10.1091/mbc.E06-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon TB, Evert BA, Song B, Doetsch PW. Biological consequences of oxidative stress-induced DNA damage in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32:3712–3723. doi: 10.1093/nar/gkh696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakoury-Elizeh M, Protchenko O, Berger A, Cox J, Gable K, Dunn TM, Prinz WA, Bard M, Philpott CC. Metabolic Response to Iron Deficiency in Saccharomyces cerevisiae. Journal of Biological Chemistry. 2010;285:14823–14833. doi: 10.1074/jbc.M109.091710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi B, Kelstrup CD, Stoehr G, Frohlich F, Walther TC, Olsen JV. Global analysis of the yeast osmotic stress response by quantitative proteomics. Mol Biosyst. 2009;5:1337–1346. doi: 10.1039/b902256b. [DOI] [PubMed] [Google Scholar]
- Steininger S, Ahne F, Winkler K, Kleinschmidt A, Eckardt-Schupp F, Moertl S. A novel function for the Mre11-Rad50-Xrs2 complex in base excision repair. Nucleic Acids Res. 2010;38:1853–1865. doi: 10.1093/nar/gkp1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein JM, de Winde JH. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- Thevissen K, Ayscough KR, Aerts AM, Du W, De Brucker K, Meert EM, Ausma J, Borgers M, Cammue BP, Francois IE. Miconazole induces changes in actin cytoskeleton prior to reactive oxygen species induction in yeast. J Biol Chem. 2007;282:21592–21597. doi: 10.1074/jbc.M608505200. [DOI] [PubMed] [Google Scholar]
- Tomas E, Stanojevic V, Habener JF. GLP-1-derived nonapeptide GLP-1(28-36)amide targets to mitochondria and suppresses glucose production and oxidative stress in isolated mouse hepatocytes. Regul Pept. 2011;167:177–184. doi: 10.1016/j.regpep.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T, Novo M, Rossger K, Letisse F, Loret MO, Portais JC, Francois JM. Control of ATP homeostasis during the respiro-fermentative transition in yeast. Mol Syst Biol. 2010;6:344. doi: 10.1038/msb.2009.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickerham LJ. A Critical Evaluation of the Nitrogen Assimilation Tests Commonly Used in the Classification of Yeasts. J Bacteriol. 1946;52:293–301. doi: 10.1128/JB.52.3.293-301.1946. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Yan L, Zhang JD, Cao YB, Gao PH, Jiang YY. Proteomic analysis reveals a metabolism shift in a laboratory fluconazole-resistant Candida albicans strain. J Proteome Res. 2007;6:2248–2256. doi: 10.1021/pr060656c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.