SUMMARY
Eukaryotic proteins entering the secretory pathway are translocated into the ER by signal sequences that vary widely in primary structure. We now provide a functional rationale for this long-observed sequence diversity by demonstrating that differences among signals facilitate substrate-selective modulation of protein translocation. We find that during acute ER stress, translocation of secretory and membrane proteins is rapidly and transiently attenuated in a signal sequence-selective manner. Their cotranslational rerouting to the cytosol for degradation reduces the burden of misfolded substrates entering the ER and represents a pathway for pre-emptive quality control (pQC). Bypassing the pQC pathway for the prion protein increases its rate of aggregation in the ER lumen during prolonged stress and renders cells less capable of viable recovery. Conversely, pharmacologically augmenting pQC during ER stress proved protective. Thus, protein translocation is a physiologically regulated process that is utilized for pQC as part of the ER stress response.
INTRODUCTION
Eukaryotic proteins destined for the extracellular environment, cell surface, or compartments of the secretory pathway are first translocated across or integrated into the endoplasmic reticulum (ER) membrane (Wickner and Schekman, 2005). Their initial segregation to the ER requires a signal sequence, often encoded at the N terminus, that is cotranslationally recognized by the signal recognition particle (SRP) (Shan and Walter, 2005). The SRP-ribosome-nascent chain complex is subsequently targeted, via an interaction with the SRP receptor, to an ER protein translocon whose central channel is composed of the Sec61 complex (Osborne et al., 2005). The signal sequence is recognized again, this time by the Sec61 complex, to facilitate insertion of the nascent chain into the translocation channel and tight docking of the ribosome at the translocon (Jungnickel and Rapoport, 1995). Further protein synthesis is accompanied by Sec61-mediated translocation of the nascent chain across the ER membrane, or in the case of membrane proteins, integration into the lipid bilayer.
These basic steps of substrate recognition, targeting, engagement of the translocon, and translocation are thought to be universally applicable to essentially all secretory and membrane proteins. Whether any step in this process can be modulated under certain cellular conditions to selectively regulate protein translocation remains unknown. However, a strictly constitutive system of translocation would seem unlikely since essentially every other basic cellular process (from transcription to protein synthesis to degradation) is regulated for at least some substrates at one time or another. How then might protein translocation be regulated?
Given the essential role of the signal sequence in mediating both targeting and initiation of translocation, any regulatory process would presumably involve modulation of signal sequence function. Although such modulation has yet to be demonstrated, a growing number of studies are beginning to question the widely held view that signal sequences are functionally equivalent and largely interchangeable. For example, analyses of signal sequence-translocon interactions suggest an unexpectedly broad range of efficiencies in initiating translocation (Kim et al., 2002). Surprisingly, only a minority of signal sequences, such as the one from the well-studied model secretory hormone Prolactin (Prl), are highly efficient in vitro and in vivo (Kim et al., 2002; Levine et al., 2005). These same efficient signal sequences also seem to be the only signals that do not depend substantially on accessory translocon components like TRAM (Voigt et al., 1996) and the TRAP complex (Fons et al., 2003) for translocation in vitro. If the only role for a signal sequence was to guarantee translocation across the ER, it is difficult to rationalize the existence of such diversity in sequence, efficiency, or complexity in their requirements for additional factors. Yet, these functional differences in efficiency are often evolutionarily conserved (Kim et al., 2001, 2002), even in instances in which inefficiencies in translocation would appear disadvantageous.
A particularly notable example is the signal sequence from the mammalian Prion protein (PrP), which is detectably less efficient in its interaction with the translocon than the signal from Prl (Rutkowski et al., 2001; Kim et al., 2002; Levine et al., 2005). In vivo, slight inefficiency of the PrP signal constantly generates a nontranslocated cytosolic form of PrP (cyPrP) that is degraded by the proteasome (Drisaldi et al., 2003; Rane et al., 2004). Although cyPrP represents a very low abundance form of PrP under normal conditions, it rapidly accumulates and aggregates upon inhibition of the proteasome (Ma and Lindquist, 2002; Drisaldi et al., 2003; Rane et al., 2004). CyPrP and the aggregates formed from it can be cytotoxic in cultured cells (Ma et al., 2002; Rane et al., 2004; Grenier et al., 2006) and cause neurodegeneration when generated in transgenic mice (Ma et al., 2002).
Similarly, slight inefficiency of the PrP signal sequence also permits the generation of CtmPrP (Kim and Hegde, 2002), a transmembrane form of PrP whose slight overrepresentation can lead to neurodegeneration in mice and humans (Hegde et al., 1998a, 1999). Remarkably, the generation of both cyPrP and CtmPrP can be markedly reduced or even eliminated simply by replacing the PrP signal sequence with the more efficient signal from Prl (Rutkowski et al., 2001; Kim et al., 2002; Rane et al., 2004). Even CtmPrP-favoring mutations in the mature domain of PrP that ordinarily cause neurodegeneration can be reversed by increasing signal sequence efficiency (Kim and Hegde, 2002). Based on these findings, it is puzzling that the signal sequence of PrP has not evolved a few amino acid changes that increase its hydrophobicity to improve its functional efficiency. Yet, comparisons across multiple species have revealed that, although several polymorphic changes have occurred in the PrP signal (Schatzl et al., 1995), its slight but measurable inefficiency is precisely maintained for unknown reasons (Kim et al., 2001).
To resolve this apparent paradox, we hypothesized that differences between signal sequences among substrates might allow translocation to be modulated selectively under certain cellular conditions. Hence, there may exist situations when a seemingly imperfect signal sequence (such as from PrP), although potentially detrimental under some conditions, has additional (and beneficial) functionality that is not available with a “constitutive” and maximally efficient signal sequence (such as from Prl). In exploring this concept for PrP, we have now discovered a stress-induced pathway of translocational attenuation that acts to minimize PrP entry into and misfolding within the ER lumen. Remarkably, this pathway appears to be broadly utilized by the cell for many substrates in a signal sequence-dependent manner. Thus, the long-observed diversity in signal sequences (von Heijne, 1985) appears to encode regulatory information that permits the cell to selectively modulate translocation in a substrate-specific manner. These findings not only reveal protein translocation as a regulated rather than constitutive process but also identify a previously unappreciated protective response to ER stress.
RESULTS AND DISCUSSION
Reduced Translocation of PrP into the ER during ER Stress Defines a pQC Pathway
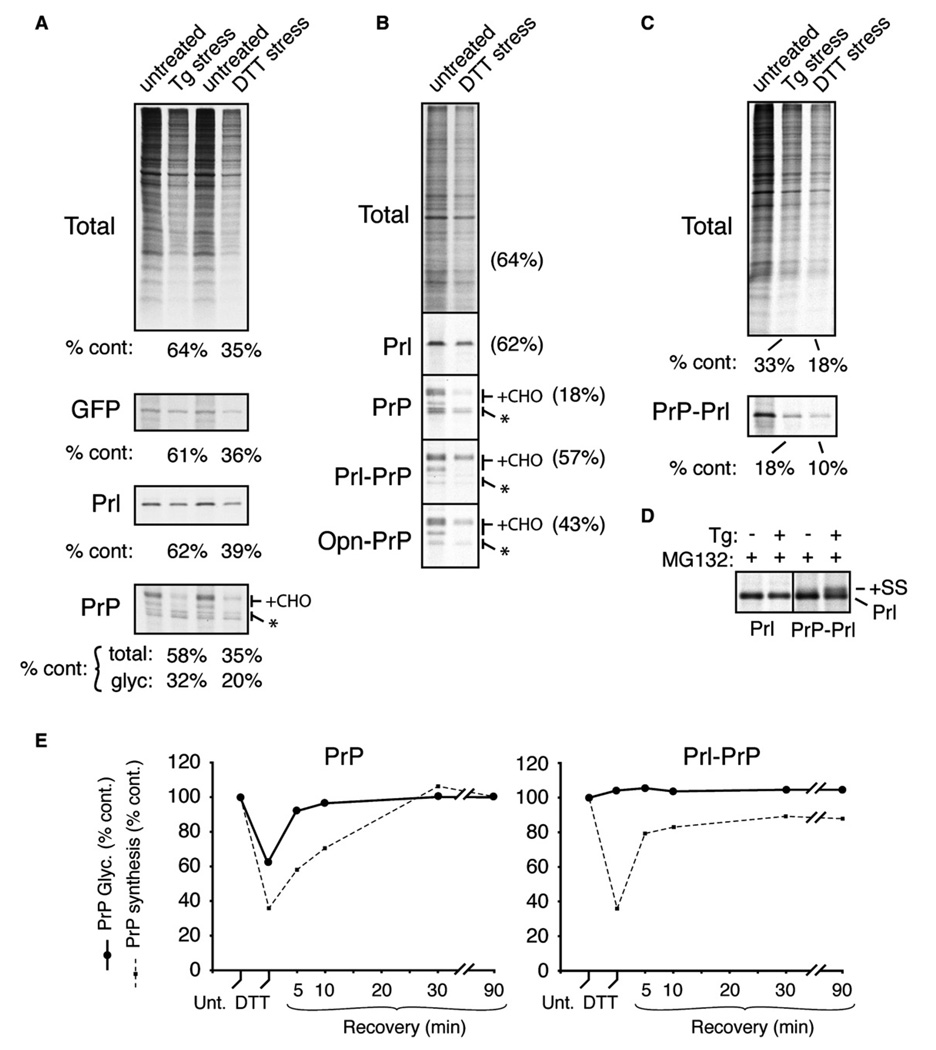
To look for potential examples of translocational regulation, we sought to identify conditions when the translocation of PrP is differentially modulated relative to Prl. A logical situation is during ER stress (Rutkowski and Kaufman, 2004), when entry into the ER of certain misfolding-prone proteins such as PrP may be disadvantageous to the cell. We therefore analyzed biosynthesis of PrP, Prl, and GFP (a cytosolic control) in pulse-labeled cultured cells acutely treated with two qualitatively different ER stressors: DTT, a reducing agent that induces ER stress by preventing productive folding of many secretory and membrane proteins, and thapsigargin (Tg), which depletes ER Ca2+ and influences chaperone function. Relative to untreated cells, Tg- and DTT-treated cells showed translational attenuation (see Figure S1 in the Supplemental Data available with this article online). This general translational inhibition was quantitatively mirrored by GFP and Prl (Figure 1A). Furthermore, all of the Prl synthesized under stressed and nonstressed conditions was processed by signal peptidase (Figure 1A; see also Figure 1D) and found by fractionation to be noncytosolic (data not shown). Although total PrP synthesis during Tg and DTT stress was attenuated to approximately the same levels as Prl and GFP, the amount of fully glycosylated PrP was preferentially reduced. The loss of glycosylated PrP was accompanied by a corresponding relative increase in the unglycosylated forms (asterisk, Figure 1A), at least some of which appeared to still contain an uncleaved signal sequence (S.-W.K. and R.S.H., unpublished data; Orsi et al., 2006; see Supplemental Data, Note 1).
Figure 1. Signal Sequence-Specific Translocational Attenuation of PrP during Acute Stress.
(A) Immunoprecipitation of transfected products (GFP, Prl, or PrP) from pulse-labeled (15 min) cultured HeLa cells treated for 30 min with 10 µM Tg or 10 mM DTT. Radiolabeled products recovered from stressed cells were quantified relative to untreated cells. For PrP, the amount of glycosylated species (+CHO) was also quantified separately. Asterisk is unglycosylated PrP.
(B) The indicated constructs were analyzed as in (A), except COS-7 cells were used, labeling was for 10 min, and 100 µM ALLN (a proteasome inhibitor) was included.
(C) PrP-Prl was analyzed and quantified as in (A).
(D) Prl and PrP-Prl were immunoprecipitated from pulse-labeled transfected cells treated with 10 µM Tg in the presence of 10 µM MG132 (a proteasome inhibitor) as indicated. Note that PrP-Prl (but not Prl) generates signal sequence-containing precursor (+SS, indicative of nontranslocated protein) in a stress-dependent manner, illustrating its translocational attenuation.
(E) Time course of recovery from translocational attenuation. COS-7 cells that were either untreated, acutely treated (30 min) with 10 mM DTT, or recovered for between 5 and 90 min were pulse labeled for 10 min and analyzed by immunoprecipitation of PrP. The efficiencies of glycosylation (solid line) and synthesis (dashed line) of either PrP (left graph) or Prl-PrP (right graph) relative to untreated cells are plotted.
Among the various possible reasons for this observation (see Supplemental Data, Note 2), the explanation proved to be a selective decrease in PrP translocation (but not Prl) during ER stress. This could be shown by domain swap experiments in which signal sequences were exchanged between PrP and Prl. Fusion of the Prl signal to PrP (Prl-PrP) now allowed Prl-PrP to be glycosylated with comparable efficiency in both untreated and DTT-stressed cells, with the decrease in fully glycosylated Prl-PrP during the stress paralleling the degree of translational attenuation (Figure 1B). A similar effect was also observed (albeit to a lesser extent) with another efficient signal sequence (from the protein Osteopontin [Opn]). Comparable results were also seen with Tg stress and validated further by cell fractionation experiments (Figure S3). Conversely, PrP-Prl biosynthesis was decreased below that attributable to general translational attenuation during acute ER stress (Figure 1C), with detection of a nontranslocated precursor when the proteasome is inhibited (Figure 1D).
Stress-dependent attenuation of PrP translocation was initiated almost immediately upon addition of the stressor (within ~5 min; data not shown). Furthermore, reversal of translocational attenutation occurred within minutes of removing the stress, even faster than recovery from translational attenuation (Figure 1E). Similar effects were observed in a variety of cell types (albeit to different extents) with both DTT and Tg (data not shown). Notably, other (non-ER) cellular stressors that also cause translational attenuation (such as serum starvation or amino acid deprivation) did not influence PrP translocation (data not shown). Based on these results, we conclude that during acute ER stress, a significant amount of nascent PrP is rerouted in a signal sequence-selective manner from its normal fate of being translocated into the ER to a pathway of proteasome-mediated degradation. We have termed this process “pre-emptive” quality control (pQC) to denote a pathway by which proteins are cotranslationally triaged for degradation at a step in their biosynthesis before they engage the conventional quality control systems in the ER lumen. The machinery utilized for degradation of translocationally aborted proteins during pQC remains to be identified but could potentially involve the recruitment of chaperones to the translocon by p58IPK (Oyadomari et al., 2006).
PrP Is Susceptible to Terminal Misfolding in the ER during Stress
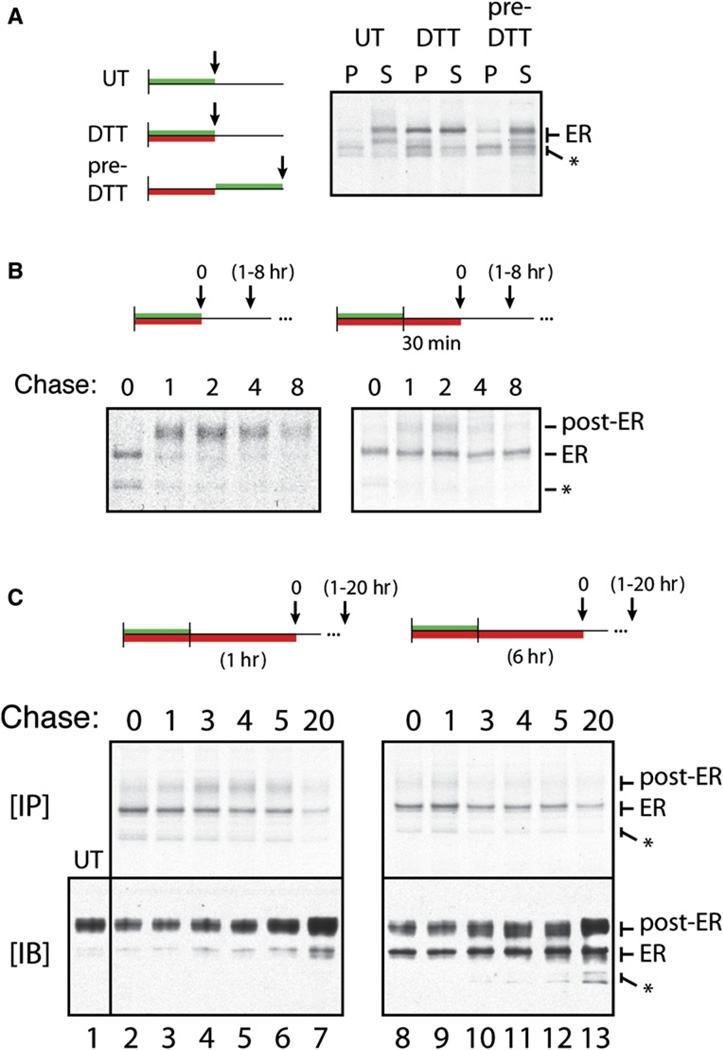
Given that the functional properties of the PrP signal sequence appear to be evolutionarily conserved (Kim et al., 2001, 2002), we hypothesized that stress-dependent translocational attenuation of PrP may provide some benefit to the cell. One possibility is that translocational attenuation serves to avoid an adverse consequence of continued translocation during ongoing ER stress. Indeed, PrP translocated into the ER during DTT-induced stress was more prone to aggregation (as judged by decreased solubility in detergent solution) than newly synthesized PrP made in unstressed cells (Figure 2A). This effect was rapidly reversed (within minutes) upon removal of the stressor, at which point the PrP entering the ER lumen was again made in a soluble form. Pulse-chase experiments showed that PrP translocated into the ER during stress was prevented from subsequent exit to post-ER compartments (Figure S4A), further supporting the conclusion that it was misfolded.
Figure 2. Terminal Misfolding of PrP during Prolonged ER Stress.
(A) The left panel shows treatment protocols using N2a cells for 30 min pulse labeling (green bars) and 30 min treatment with 10 mM DTT (red bars) relative to harvesting (arrows) and analysis by solubility assays. PrP in the detergent-insoluble (P) and -soluble (S) fractions was recovered by immunoprecipitation and visualized by autoradiography. The core-glycosylated ER form and nonglycosylated species (*) of PrP are indicated.
(B) Treatment protocols for labeling, DTT treatment, and chase (for 1–8 hr) prior to harvesting and immunoprecipitation are shown above the respective autoradiographs. Unglycosylated (*), core-glycosylated (ER), and complex glycosylated (post-ER) forms of PrP are indicated.
(C) Cell lysates from the indicated treatment protocols were divided and analyzed for radiolabeled PrP by immunoprecipitation and autoradiography ([IP], top panels) or for total PrP by immunoblots ([IB], bottom panels).
Misfolded PrP in the ER was capable of being refolded and trafficked out of the ER, provided the stressor was removed promptly (within ~15 min; Figure 2B). However, continuing the stress an extra 30–360 min caused progressively larger proportions of the ER-lumenal PrP to become terminally misfolded, being retained in the ER in a largely insoluble state for prolonged times (Figures 2B and 2C and Figure S4B). Western blotting of total lysates from these same cells demonstrated that, after 6 hr of stress, the ER form of PrP had accumulated to ~30%– 50% of total PrP (bottom panels, Figure 2C). This misfolded PrP persisted for the ensuing 20 hr despite ER function returning to normal (as judged by replenishment of fully mature PrP [the “post-ER” form] on the cell surface). Importantly, artificially retaining PrP in the ER lumen using brefeldin A (BFA) did not cause it to become insoluble (N.S.R. and R.S.H., unpublished data) or incapable of subsequent trafficking upon BFA removal (Figures S4C and S4D). Thus, the results in Figure 2 illustrate that PrP entering the ER during acute ER stress is prone to terminal misfolding and aggregation, after which it is neither refolded nor degraded efficiently, even if the stressor is subsequently alleviated. This terminal misfolding occurs over time scales of under an hour (e.g., Figure 2B and Figure S4B), well before the transcriptional responses to ER stress have had an opportunity to upregulate the ER biosynthetic and folding machinery (Yoshida et al., 2003).
The Consequences of Bypassing the pQC Pathway
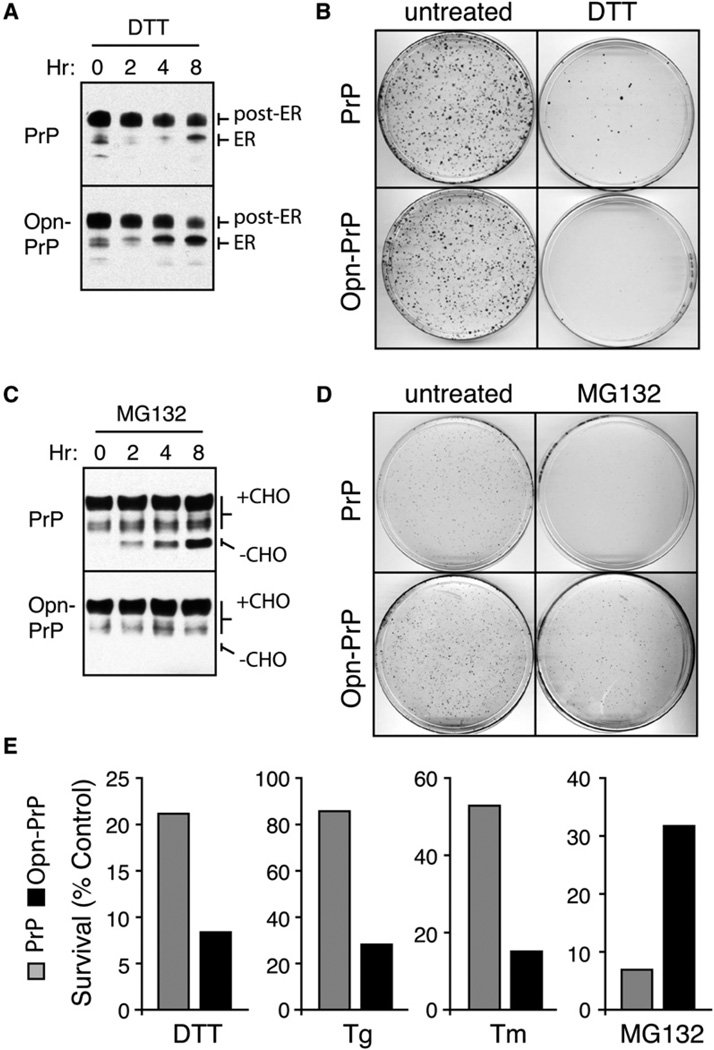
The relative ease with which PrP becomes irrevocably misfolded in the ER lumen combined with its comparatively rapid degradation in the cytosolic environment suggest that stress-dependent translocational attenuation (i.e., access to the pQC pathway) may be a protective response to ER stress. To investigate this hypothesis, we determined the consequences of denying PrP access to the pQC pathway. We therefore generated stable cell lines that overexpress either PrP (which is subject to pQC) or Opn-PrP (which is largely refractory to pQC). The cell lines were analyzed by western blotting, immunofluorescence, and glycosidase digestion to confirm comparable expression, localization, and trafficking of PrP (Figures S5A–S5C).
When these same cells were subjected to DTT stress, Opn-PrP progressively accumulated the glycosylated ER form at a noticeably higher rate than PrP over the course of 8 hr (Figure 3A). Importantly, simply preventing ER-to-Golgi transport with BFA (which does not acutely induce ER stress and does not mediate translocational attenuation; data not shown) caused the accumulation of the ER form at comparable rates for both PrP and Opn-PrP (Figure S5D). Thus, the rate of entry into the ER is very similar for PrP and Opn-PrP in the absence of ER stress but differs sufficiently during stress to influence the accumulation of misfolded ER-lumenal PrP. Remarkably, the increased rate of misfolded ER-lumenal Opn-PrP accumulation led to a diminished capacity to recover from the ER stress, as measured using a cell replating viability assay (Figures 3B and 3E). Similar effects on viability were also seen with other ER stressors (Figure 3E). By contrast, partial inhibition of the proteasome in these same cells caused a nonglycosylated form of PrP (but not Opn-PrP) to accumulate in the cytosol (Figure 3C). Replating viability assays showed the PrP cells to be less viable than Opn-PrP cells after chronic proteasome inhibition (Figures 3D and 3E).
Figure 3. Consequences of Bypassing the pQC Pathway for PrP.
(A) N2a cells stably expressing PrP or Opn-PrP were treated for 0–8 hr with 10 mM DTT and analyzed for total PrP by immunoblot. Note the increased accumulation of the ER form in Opn-PrP cells relative to PrP cells, especially obvious at the 4 hr time point.
(B) Cells stably expressing PrP or Opn-PrP were treated with 10 mM DTT for 24 hr, replated in normal media, and visualized 10 days later by staining with crystal violet.
(C) Cells stably expressing PrP or Opn-PrP were treated for 0–8 hr with 5 µM MG132 and analyzed for total PrP by immunoblot. Note the increased accumulation of unglycosylated species (−CHO) for PrP, but not for Opn-PrP.
(D) Cells treated with 5 µM MG132 for 24 hr were replated in normal media and visualized 8 days later by staining with crystal violet.
(E) Quantification of replating viability assays for survival of cells expressing PrP (gray bars) or Opn-PrP (black bars) after the indicated treatments for 24 hr (5 µM MG132), 6 hr (10 mM DTT), 18 hr (1 µg/ml Tunicamycin; Tm) or 5 min (5 µM Tg).
We conclude from Figure 3 that inefficiencies in PrP translocation necessitate constant proteasomal degradation of nontranslocated material that can be highly aggregation prone and cytotoxic if left undegraded. Although the more-efficient Opn signal sequence minimizes these problems, it becomes a liability when the ER environment is compromised. Under these conditions, constitutively high translocation efficiency of Opn-PrP results in a higher rate of accumulation of misfolded PrP in the ER lumen and decreased recovery from the stressor when compared to PrP. Thus, bypassing stress-mediated translocational attenuation of PrP sensitizes cells to ER stress. This result suggests that, for PrP, the pQC pathway is a physiologically important facet of the cellular response to an altered folding environment in the ER. The basis of this effect correlated directly with the minimization of protein misfolding, aggregation, and accumulation in the ER lumen.
Accentuating the pQC Pathway during ER Stress Is Protective
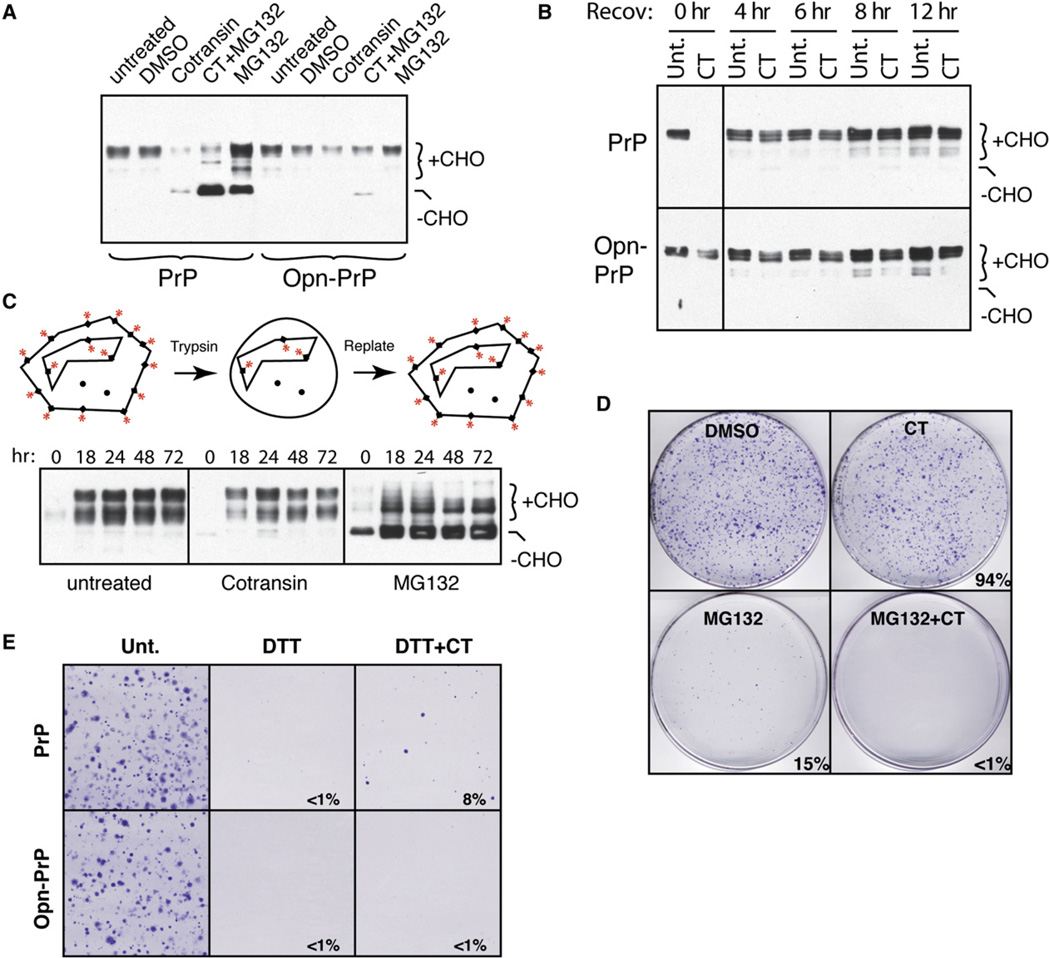
The increased sensitivity of Opn-PrP-expressing cells to ER stress suggests that constitutive translocation under these conditions is more detrimental than rerouting PrP directly to the cytosol. Although a reduction in PrP translocation might have seemed problematic given previous results identifying cytosolic PrP as highly cytotoxic (Ma et al., 2002) and aggregation prone (Ma and Lindquist, 2002; Drisaldi et al., 2003; Rane et al., 2004; Grenier et al., 2006), this proved not to be the case. Pharmacologic inhibition of PrP translocation in vivo with cotransin (CT; see Figures S3B and S5E) for up to 24 hr did not lead to the accumulation of PrP aggregates in the cytosol or obvious cell death (Figures 4A and 4D). Instead, total PrP simply decreased over time due to turnover of preexisting PrP from the cell surface and rapid degradation of nontranslocated PrP (which could be visualized by proteasome inhibition; Figure 4A).
Figure 4. Pharmacologic Induction of pQC during Prolonged ER Stress Is Protective.
(A) PrP and Opn-PrP cells were treated for 6 hr (5 µM CT, 5 µM MG132, and/or 0.1% DMSO) and analyzed by immunoblotting. The glycosylated (+CHO) and unglycosylated (−CHO) forms of PrP are indicated.
(B) PrP and Opn-PrP cells treated for 12 hr with solvent (0.1% DMSO) or 5 µM CT were recovered in regular media for the indicated times and analyzed by immunoblotting for PrP.
(C) PrP-expressing cells were treated for 4 hr with either 5 µM CT or 5 µM MG132, followed by trypsinization and replating in normal media for the indicated times before harvesting and analysis by immunoblotting. Diagram illustrating the experimental design is shown.
(D) PrP-expressing cells treated for 12 hr as indicated were replated in normal media and visualized 10 days later by staining with crystal violet. Quantification of viability relative to control is indicated.
(E) PrP and Opn-PrP cells treated for 24 hr as indicated were replated in normal media and visualized 10 days later by staining with crystal violet. Quantification of viability relative to control is indicated.
Upon removal of CT, cell surface PrP was readily replenished, with no accumulation of cytosolic PrP even over the course of 72 hr (Figures 4B and 4C). This contrasted sharply with proteasome inhibition, where aggregates of nontranslocated PrP appeared within a few hours (Figure 3C) and persisted long after alleviation of proteasome inhibition (Figure 4C). Furthermore, once initiated, PrP aggregates continue to accumulate (by a poorly understood “self-propagation” process that amplifies even trace amounts of PrP aggregates; Ma and Lindquist, 2002), eventually leading to decreased cell viability (Figure 3D). The consequences of proteasome inhibition on PrP accumulation and cell viability were worsened by simultaneous inhibition with CT (Figure 4D) or DTT (Figure S6A), both of which result in increased delivery of PrP to the cytosol.
Since even a complete block in PrP translocation is not inherently cytotoxic, we could ask whether accentuating translocational attenuation of PrP could be protective from the consequences of prolonged ER stress. Indeed, simultaneous treatment with CT during chronic DTT stress was able to partially improve viability for PrP-expressing cells (Figure 4E). This effect was due at least in part to the effect of CT on PrP, since a similar rescue was not effected for cells expressing Opn-PrP (Figure 4E), whose translocation is only partially inhibited by CT (Figure 4A and Figure S5E). With less-severe DTT stress, even Opn-PrP cells or nontransfected cells could be rescued by simultaneous treatment with CT (Figure S6B and data not shown), presumably because CT inhibits several other signal-containing proteins to reduce the overall burden of substrates entering the ER (see Figure S7). Thus, complementary to the adverse consequences of bypassing pQC (Figure 3), accentuating pQC for PrP is protective during ER stress.
The pQC Pathway Is Broadly Utilized
Since highly efficient signals like those from Prl and Opn are found on a relative minority of proteins, we surmised that the pQC pathway might be utilized by many proteins in addition to PrP. To examine this idea, we took two parallel approaches: analysis of global translocation efficiency using N-linked glycosylation as a surrogate marker for entry into the ER, and individual analyses of various secretory and membrane proteins.
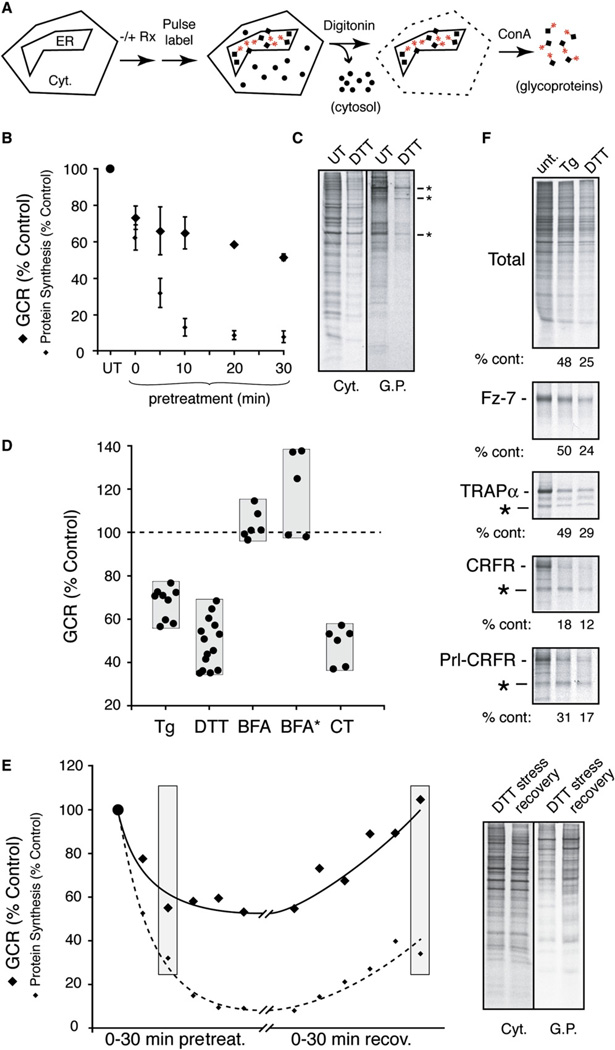
In the first approach (Figure 5A), pulse-labeled adherent cultured cells are first permeabilized with low concentrations of digitonin to selectively extract the cytosolic contents. The remainder of the cell is then solubilized and the glycoproteins isolated by binding to immobilized conconavalin A (ConA). Quantitative analysis of the ConA and digitonin fractions by SDS-PAGE and phosphorimaging is used to derive a “glycoprotein-to-cytosolic protein ratio” (GCR), a parameter that should change with any acute changes to the efficiency of protein sequestration into the ER. This approach was tested and validated using CT to directly influence translocation efficiencies (Figure S7).
Figure 5. Analysis of Global Glycoprotein Biosynthesis during Acute ER Stress.
(A) Experimental design.
(B) HeLa cells treated with 10 mM DTT for the indicated times were pulse labeled (for 15 min), fractionated, and quantified to determine the glycoprotein-to-cytosolic protein ratio (GCR). The GCR at each time point (normalized to untreated cells) is plotted along with the overall level of protein synthesis (mean ± SD for three experiments).
(C) Radiolabeled cytosolic proteins and glycoproteins from untreated and DTT-stressed (30 min) HeLa cells. Asterisks indicate bands that are minimally attenuated relative to other glycoproteins.
(D) GCR (normalized to untreated cells analyzed in parallel) for HeLa cells acutely treated (for 30 min) with 10 µM Tg, 10 mM DTT, 10 µg/ml BFA, or 10 µM CT. Each point represents an individual experiment, with the gray bar showing the range observed. BFA* indicates treatment for 12 hr.
(E) GCR (solid line) and overall translation (dotted line) was measured in cells pretreated for between 0 and 30 min with 10 mM DTT, or treated for 30 min followed by recovery in normal media for 0–30 min. Samples from time points shaded in gray are shown to the right. Note that during acute stress, many (but not all) glycoproteins are attenuated in their biosynthesis relative to the situation during recovery.
(F) Analysis as in Figure 1A of Frizzled-7 (Fz7), TRAPα, CRF1 receptor (CRFR), and Prl-CRFR by pulse labeling and immunoprecipitation from transiently transfected cells treated for 30 min with 10 µM Tg or 10 mM DTT. Asterisks indicate nonglycosylated forms of each glycoprotein (except Frizzled-7, in which a nonglycosylated form was not detectable). The amount of the translocated and glycosylated form generated under each condition is quantified relative to untreated cells. The decrease in Frizzled-7 and TRAPα paralleled the level of translational attenuation caused by the stress.
Upon acute DTT stress, the GCR promptly decreased by ~30% within 10 min (Figure 5B). At very short times after DTT treatment, several glycoproteins (but no cytosolic proteins) were selectively reduced to a much greater level than others (Figure S8). At longer treatment times (Figure 5C), there was both a greater degree of translational attenuation and a larger number of glycoproteins whose levels were yet lower (Supplemental Data, Note 3). Interestingly, some glycoproteins were far less affected than others at all treatment times (asterisks in Figure 5C).
Compilation of numerous experiments examining GCR upon treatment of cells with various agents (Figure 5D) revealed a similar, albeit smaller, effect with the ER stressor Tg. Time course experiments with Tg also showed a rapid onset of decreased GCR, concomitant with or slightly faster than translational attenuation (data not shown). Treatments with BFA, serum starvation, or amino acid starvation did not show obvious changes in GCR (Figure 5D and data not shown). The stress-mediated reduction in GCR was rapidly reversible within minutes after removal of the stressor (Figure 5E), well before translational attenuation was reversed. This argued that the GCR effect was not a direct consequence of decreased translation per se. For example, direct comparison of two samples with essentially equal translational repression (~70%) but markedly different GCR clearly illustrated the selective effect on glycoproteins relative to cytosolic proteins (Figure 5E).
The rapid, reversible, and substrate-selective reduction of glycoprotein biosynthesis (beyond that accounted by translational attenuation alone) was reminiscent of stress-mediated signal sequence-specific translocational attenuation of PrP, suggesting that changes in GCR could potentially be caused by changes in protein translocation efficiencies. The rapid induction (less than 10 min) argued against a transcriptional suppression of glycoproteins, while the rapid reversibility was not compatible with a major contribution from selective degradation of transcripts coding for glycoproteins (Hollien and Weissman, 2006). Furthermore, pulse-chase and inhibitor experiments (Figure S9) argued against a substantial increase in the retrotranslocation (or ERAD) pathway at such short times after initiating stress. Together, these findings pointed toward translocational attenuation as the principal basis for the decreased GCR observed during acute ER stress.
To verify this conclusion further, we examined several individual proteins. Among our still-cursory survey, we found that some membrane glycoproteins such as TRAPα, Frizzled-7, vesicular stomatitis virus glycoprotein (VSVG), and vascular cell adhesion molecule (VCAM) were essentially unaffected in their biosynthesis during stress beyond that caused by translational attenuation (Figure 5F; S.-W.K. and R.S.H., unpublished data). By contrast, angiotensinogen, interferon-γ, and the corticotropin-releasing factor receptor (CRFR) were each attenuated to varying degrees during stress (Figure 5F; S.-W.K. and R.S.H., unpublished data). Replacement of the CRFR signal sequence with that from Prl partially rescued its stress-dependent attenuation (Figure 5F), further validating the fact that the effect on CRFR was at the level of its translocation into the ER. Thus, the pQC pathway is not unique to PrP and appears to be more broadly utilized based on both the glycoprotein profiling experiments and survey of several secretory and membrane proteins.
Reconstitution of Signal Sequence-Specific Translocational Attenuation In Vitro
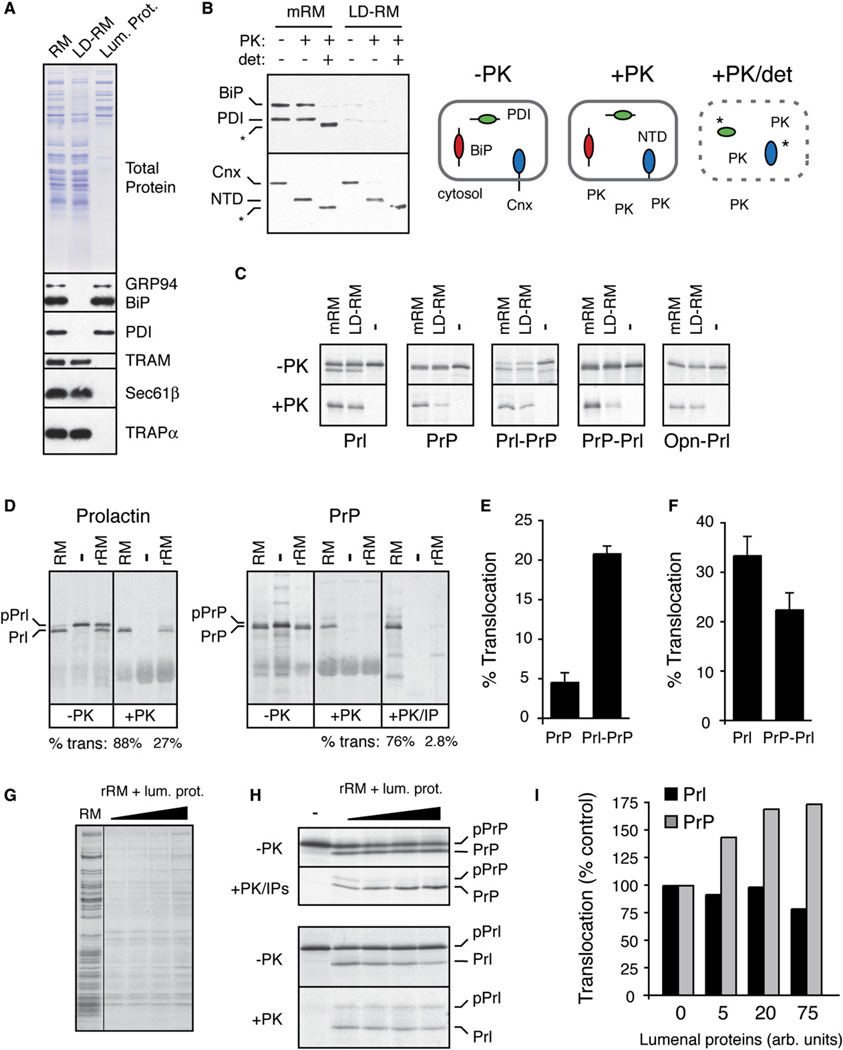
To gain insight into the mechanisms underlying pQC, we sought to reconstitute its salient features in vitro. In initial experiments, we analyzed translocation of endogenous mRNAs using semipermeabilized cells pretreated with ER stressors (Supplemental Data, Note 4 and Figure S10). These experiments not only supported the conclusions from the in vivo studies but suggested that translocation is likely to be attenuated at the ER after nascent polypeptides are targeted to and docked at the translocon.
One of the earliest events in the ER-stress response is thought to be the titration of lumenal chaperones by their association with an increased burden of misfolded proteins (Rutkowski and Kaufman, 2004). To determine whether this acute decrease in functionally available lumenal chaperones might directly influence translocation, we analyzed the in vitro translocation of Prl and PrP in rough microsomes (RM) containing or lacking lumenal contents. Soluble lumenal proteins were selectively extracted from RM with low concentrations of detergent well below those needed to solubilize membrane proteins. The resulting membranes (LD-RM) were substantially depleted (by ~80%–90%) of several lumenal chaperones (and, presumably, nonchaperone proteins) relative to mock-extracted RM (mRM), and both preparations were intact and of the correct orientation (Figures 6A and 6B).
Figure 6. Reconstitution of Signal Sequence-Specific Translocational Attenuation In Vitro.
(A) Selective removal of lumenal proteins from rough microsomes (RM) by treatment with 0.075% deoxyBigCHAP. Equivalent aliquots of RM, the lumenal protein-depleted RM (LD-RM), and lumenal protein fraction were analyzed by Comassie staining (top) and immunoblots for various lumenal and membrane proteins (bottom). Although not seen in this blot, semiquantitative blotting showed ~80%–90% depletion of BiP and PDI (data not shown).
(B) Analysis of LD-RM and mock-extracted RM (mRM) for vesicle integrity and orientation by a protease protection assay. LD-RM and mRM were digested with Proteinase K (PK) in the absence or presence of detergent (det; 0.5% Triton X-100) and analyzed by immunoblotting for the N terminus of calnexin (Cnx), BiP, and PDI. The N-terminal lumenal domain of Cnx is indicated by NTD. Asterisks indicate core domains of Cnx and PDI that are resistant to complete protease digestion. A schematic of the results is shown.
(C) Analysis of protein translocation into mRM and LD-RM. The indicated constructs were translated in vitro without (−) or with membranes (either mRM or LD-RM) and analyzed for translocation by protease protection. Aliquots of the samples before and after PK digestion are shown. The relative translocation efficiency in LD-RM (relative to translocation in mRM) is indicated for each construct. All translation reactions contained a peptide inhibitor of glycosyation to simplify the analysis.
(D) Analysis of protein translocation in RM and rRM (proteoliposomes reconstituted from a total membrane protein extract of RM) as in (C). An aliquot of the PrP samples was also immunoprecipitated with anti-PrP (+PK/IP) to better visualize the translocation products. Translocation efficiencies are given below the lanes. The positions of precursor and processed products for Prl and PrP are indicated.
(E and F) The relative translocation efficiencies of the indicated constructs were analyzed in rRM and quantified from five experiments (mean ± SD). All constructs were translocated into RM with greater than 75% efficiency (data not shown).
(G) Coomassie stain of rRM reconstituted in the presence of increasing concentrations of lumenal proteins. Note that the concentration of lumenal proteins incorporated into the rRM is low and does not approach that found in RM (Supplemental Data, Note 5).
(H) Translocation of PrP and Prl was analyzed as in (D) using rRM containing increasing amounts of lumenal proteins. Note that in rRM, signal sequence cleavage is not complete, resulting in some translocation (and hence protease protection) of unprocessed protein.
(I) The experiment in (H) was quantified, normalized to translocation in rRM (lacking lumenal proteins), and graphed.
While translocation of Prl was modestly decreased (by ~30%) in LD-RM, translocation of PrP was reduced by ~70% (Figure 6C). This decrease was largely reversed upon replacing the PrP signal sequence with that from Prl. Conversely, Prl translocation became more dependent on lumenal proteins when its signal sequence was replaced with that from PrP, but not from Opn. When lumenal proteins were depleted even more thoroughly (>98%) by reconstituting total detergent-solubilized membrane proteins into proteoliposomes (to generate rRM), the differential in translocation efficiencies between Prl and PrP widened: while both were translocated into RM with comparable efficiency (~75%–85%), translocation of PrP into rRM was nearly 10-fold lower than translocation of Prl (Figure 6D).
Translocation into rRM of Prl-PrP was several-fold more efficient that PrP (Figure 6E), while PrP-Prl translocation was modestly less efficient than Prl (Figure 6F). This result, along with the similar observations in LD-RM (Figure 6C), illustrates that the signal sequence contributes significantly to substrate-specific differences in the dependence on lumenal proteins for translocation. And finally, although coreconstitution of lumenal proteins into rRM was rather inefficient (Figure 6G; see Supplemental Data, Note 5), we could nonetheless detect a modest stimulatory effect on PrP translocation, but not Prl translocation (Figures 6H and 6I). Considered together, these in vitro analyses demonstrate that while translocation of PrP and Prl are comparably efficient under normal conditions, modulation of lumenal protein availability has a significantly greater impact on PrP translocation in a signal sequence-selective manner. These findings suggested that changes in the function and/or availability of lumenal proteins (such as chaperones) during acute ER stress in vivo could explain the substrate-specific effects on translocation.
Induction of pQC Correlates with Reduced Lumenal Chaperone Availability
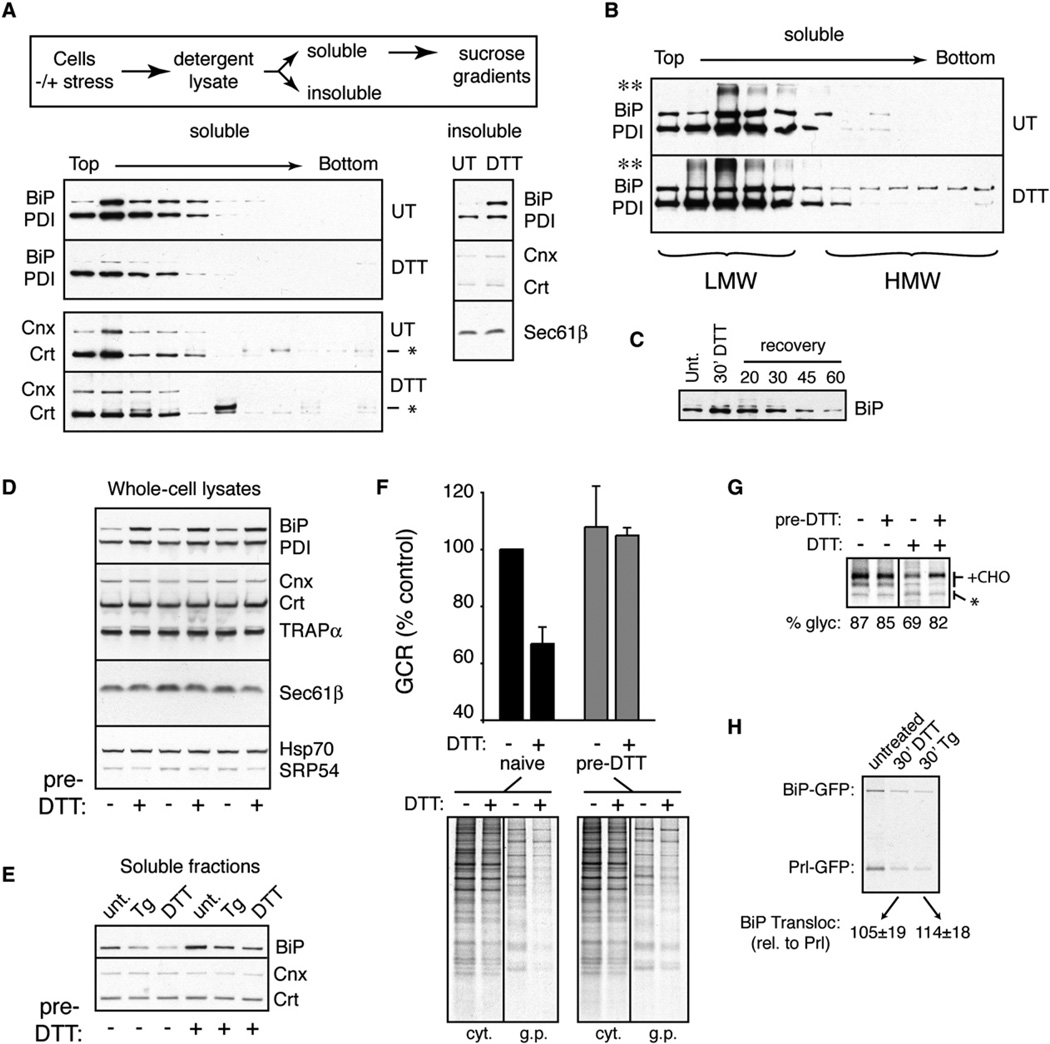
To examine this idea, we assessed the biochemical state of lumenal chaperones in unstressed and acutely stressed cells. Because interactions between BiP and its substrates could be readily stabilized after cell lysis (by ATP depletion), we focused on this lumenal chaperone. During acute DTT stress, the amount of BiP that is unoccupied with substrate (as judged by its solubility and native size on sucrose gradients) decreased noticeably (Figure 7A). The remainder of BiP was engaged in heterogeneous complexes and recovered in a combination of the “insoluble” fraction, high-molecular-weight fractions of the sucrose gradient, and SDS-resistant material at the top of the gel (Figure 7B). Monitoring BiP levels in the insoluble fraction revealed a rapid reversal of this effect upon removal of the stressor (Figure 7C) that paralleled the time course of recovery from translocational attenuation (Figures 1E and 5E). The amount of unengaged PDI was also reduced (but much more modestly), while calnexin (Cnx) and calreticulin (Crt) were unchanged by this assay.
Figure 7. Changes in Lumenal Chaperone Availability Accompany pQC.
(A) The sizes of chaperone-containing complexes were analyzed in unstressed or acutely stressed (10 mM DTT for 30 min) HeLa cells by detergent extraction, fractionation by sucrose gradients, and immunoblotting (outlined on top). Note the decrease in BiP (and to a lesser extent, PDI) in the soluble fractions, with a corresponding increase in the insoluble fraction. The band slightly larger than Crt (*) is a background band that was inconsistently observed in some lanes.
(B) Analysis of overloaded samples prepared as in (A) reveals increased amounts of BiP in both high-molecular-weight fractions (HMW) and previously described SDS-resistant complexes (**; see Marciniak et al. [2004]) in stressed cells relative to control cells.
(C) The level of BiP in the insoluble fraction of HeLa cells acutely treated with 10 mM DTT for 30 min or treated and recovered for between 20 and 60 min.
(D) The levels of various proteins were analyzed by immunoblotting in HeLa cells pretreated with 10 mM DTT for 1 hr followed by recovery for 18 hr. Triplicate samples are shown. Note induction of BiP and, to a lesser extent, PDI by the pretreatment protocol.
(E) Cells were either left untreated or preconditioned with DTT as in (D). Subsequently, the levels of BiP, Cnx, and Crt in the soluble fractions (prepared as in [A]) were analyzed by immunoblotting before or after acute stress for 30 min with 10 µM Tg or 10 mM DTT.
(F) Naive or DTT-preconditioned cells were analyzed for changes in GCR (mean ± SD for three replicates) upon acute ER stress for 15 min with 10mM DTT. The autoradiograph from a representative experiment is shown.
(G) Naive or DTT-preconditioned cells were analyzed for changes in PrP translocation upon acute ER stress for 30 min with 10 mM DTT. Note that while naive cells showed translocational attenuation (as judged by decreased PrP glycosylation), preconditioned cells were largely refractory. Translational attenuation upon DTT stress was equal in both cells (data not shown).
(H) Analysis of BiP-GFP translocation during acute stress. HeLa cells cotransfected with BiP-GFP and Prl-GFP (a constitutively translocated control) were subjected to 10 µM Tg or 10 mM DTT for 15 min prior to pulse labeling for 15 min. The cytosolic fraction was extracted with digitonin, and the noncytosolic (i.e., translocated) protein was recovered by immunoprecipitation with anti-GFP. Note that the level of translocated BiP-GFP during stress closely parallels Prl-GFP, indicating no obvious translocational attenuation. BiP-GFP translocation relative to Prl-GFP in the same cells was tabulated for three experiments and shown below the gel.
Although total chaperone levels were unchanged during the acute stress treatments employed in this study, recovery for ~16–20 hr led to substantial upregulation of BiP (and to a lesser extent PDI) due to induction of the unfolded protein response (Figure 7D). Even in this preconditioned state, treatment with acute ER stress led to decreased BiP levels in the soluble fraction (with corresponding increases in the insoluble fraction; Figure 7E and data not shown). However, the increased reservoir of BiP in preconditioned cells still left enough in the soluble fraction even during stress to maintain levels comparable to unstressed nonpreconditioned cells (compare lane 1 to lanes 5 and 6 in Figure 7E). Remarkably, preconditioned cells showed little or no translocational attenuation during acute stress, as judged by analyses of either the GCR (Figure 7F) or PrP biosynthesis (Figure 7G). Thus, using BiP as a marker, we find that the available (i.e., unengaged) levels of this lumenal chaperone correlate inversely with substrate-specific translocational attenuation. Interestingly, translocation of BiP, whose upregulation is a critical facet of the stress response, is not attenuated even during maximal acute stress (Figure 7H). Together with the biochemical analysis of translocation in vitro (Figure 6), these results point to rapid changes in lumenal chaperone availability and/or function during acute stress as one (but perhaps not the only) basis for substrate-specific translocational attenuation and pQC (see Supplemental Data, Note 6 for possible models).
Conclusions
This study illustrates that protein translocation into the ER lumen is not a constitutive or deterministic process but instead can be regulated in response to changes in cellular conditions. In the context of acute ER stress, changes in translocation efficiency are substrate specific, reversible, and physiologically important (see Supplemental Data, Note 7 and model in Figure S11). Selectivity of translocational attenuation is determined (at least in part) by signal sequences, whose length, hydrophobicity, charge, and amino acid composition vary widely between substrates (von Heijne, 1985). Our discovery that this structural diversity among signals imparts differential functionality during translocation provides a rationale for why signal sequences are often conserved in a substrate-specific manner (Kim et al., 2001; Kim et al., 2002) and appear to evolve more slowly than expected for such a highly variable motif (Williams et al., 2000).
In the case of PrP, a relative weak and modulatable signal sequence may be especially important for minimizing the risk of permanantly producing potentially toxic species in enclosed compartments like the ER lumen. A similar logic may apply to other misfolding-prone secretory and membrane proteins. An analogous (and non-mutually exclusive) explanation for signal sequence diversity is that certain highly overproduced secretory proteins like prolactin may need to contain signal sequences that can escape normal stress-induced attenuation mechanisms that might be induced during rapid changes in secretory activity. This rationale presumably applies to BiP, which sometimes needs to be translocated effectively even at high expression levels during ongoing ER stress. Thus, sequence differences among signal sequences may provide a means to regulate the translocation efficiency of some substrates independently of others for various physiological purposes.
Of note, the most efficient signal sequences (as judged in vitro) may prove to be the least regulatable. This is analogous to many other biological systems, in which optimal efficiency comes at a cost of reduced dynamic range. For example, highly regulatable promoters often have very low basal activity and are dependent on many accessory transcription factors, while extremely strong promoters are less modulatable. A similar concept may apply to translocation, in which signal sequences whose interactions with the translocon are highly efficient and less dependent on accessory factors are less amenable to modulation in trans. Such a view would provide a logical explanation for the otherwise paradoxical observation that most signal sequences appear to be less than maximally efficient (Kim et al., 2002; Levine et al., 2005).
The mechanism by which differences among signal sequences permit substrate-specific and stress-dependent attenuation of translocation remains to be studied. Intrestingly, the posttargeting interaction between the signal sequence and translocon (Jungnickel and Rapoport, 1995; Plath et al., 1998; Mothes et al., 1998) is not only highly variable (Kim et al., 2002) but is differentially influenced by trans-acting factors (Voigt et al., 1996; Fons et al., 2003) and small molecules (Garrison et al., 2005; Besemer et al., 2005). It is therefore tempting to speculate that such differences in the signal-translocon interaction are analogously exploited during ER stress for selective lumenal protein-dependent translocational attenuation to initiate pQC. Analysis of this step in mechanistic detail for multiple signal sequences that are either sensitive or refractory to translocational attenuation will be required to elucidate the molecular basis of substrate-specific regulation of translocation. The reconstitution of signal sequence-selective translocational attenuation in a biochemical system amenable to fractionation (Figure 6) should now facilitate these future studies.
EXPERIMENTAL PROCEDURES
Experiments in this study generally utilized well-characterized procedures described in previous studies as cited below. Time points, treatment conditions, and concentrations of pharmacologic agents specific to individual experiments are provided in the respective figure legends. Additional (previously published) details such as antibody epitope sequences, buffer conditions, and explanations of experimental methods can be found in the Supplemental Data.
Materials
Antibodies were either from commercial sources or described previously, and constructs were made using standard methods (details provided in Supplemental Data). CT was prepared as described (Garrison et al., 2005). DTT was from Roche and dissolved in water. Tg, Tm, BFA, MG132, and ALLN were from Calbiochem, dissolved in DMSO or EtOH (BFA), and used at concentrations indicated in the figure legends. Digitonin was from Calbiochem. Immobilized ConA was from Amersham-Pharmacia.
Cell Culture and Biochemical Analyses
HeLa, COS-7, and N2a cells were cultured in DMEM containing 10% FBS at 5% CO2 and transfected with Effectene (Qiagen) or Lipofectamine 2000 (Invitrogen). Stable cell lines were generated by selection in Zeocin using standard methods (see Supplemental Data). Replating viability assays were performed and quantified by minor modification of published procedures (Marciniak et al., 2004). Pulse-labeling, fractionation, immunoprecipitation, and solubility analyses were performed by minor modifications of published procedures (Fons et al., 2003; Rane et al., 2004; Levine et al., 2005). Exact times and conditions are provided in individual figure legends. Quantification of radiolabeled products utilized a Typhoon Phosphorimager and accompanying software (Molecular Dynamics).
In Vitro Reconstitution and Translation
Translation in reticulocyte lysate and analyses of translocation by protease protection were as before (Fons et al., 2003). Preparation of RM, mRM, LD-RM, rRM, and rRM containing lumenal proteins was as described (Hegde et al., 1998b; Fons et al., 2003; Garrison et al., 2005).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Martina Alken for providing several constructs, Jesse Yonkovich for performing some of the initial pulse-chase experiments with PrP, Ryen Fons for preparing and characterizing the LD-RM, Aarthi Ashok for comments on this text, and members of the Hegde lab for insightful discussions. This work was supported by the Intramural Research Program of NICHD at the National Institutes of Health.
Footnotes
Supplemental Data
Supplemental Data include 11 figures, supplemental notes, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://www.cell.com/cgi/content/full/127/5/999/DC1/.
REFERENCES
- Besemer J, Harant H, Wang S, Oberhauser B, Marquardt K, Foster CA, Schreiner EP, de Vries JE, Dascher-Nadel C, Lindley IJ. Selective inhibition of cotranslational translocation of vascular cell adhesion molecule 1. Nature. 2005;436:290–293. doi: 10.1038/nature03670. [DOI] [PubMed] [Google Scholar]
- Drisaldi B, Stewart RS, Adles C, Stewart LR, Quaglio E, Biasini E, Fioriti L, Chiesa R, Harris DA. Mutant PrP is delayed in its exit from the endoplasmic reticulum, but neither wild-type nor mutant PrP undergoes retrotranslocation prior to proteasomal degradation. J. Biol. Chem. 2003;278:21732–21743. doi: 10.1074/jbc.M213247200. [DOI] [PubMed] [Google Scholar]
- Fons RD, Bogert BA, Hegde RS. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell Biol. 2003;160:529–539. doi: 10.1083/jcb.200210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison JL, Kunkel EJ, Hegde RS, Taunton J. A substrate-specific inhibitor of protein translocation into the endoplasmic reticulum. Nature. 2005;436:285–289. doi: 10.1038/nature03821. [DOI] [PubMed] [Google Scholar]
- Grenier C, Bissonnette C, Volkov L, Roucou X. Molecular morphology and toxicity of cytoplasmic prion protein aggregates in neuronal and non-neuronal cells. J. Neurochem. 2006;97:1456–1466. doi: 10.1111/j.1471-4159.2006.03837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde RS, Mastrianni JA, Scott MR, DeFea KA, Tremblay P, Torchia M, DeArmond SJ, Prusiner SB, Lingappa VR. A transmembrane form of the prion protein in neurodegenerative disease. Science. 1998a;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- Hegde RS, Voigt S, Rapoport TA, Lingappa VR. TRAM regulates the exposure of nascent secretory proteins to the cytosol during translocation into the endoplasmic reticulum. Cell. 1998b;92:621–631. doi: 10.1016/s0092-8674(00)81130-7. [DOI] [PubMed] [Google Scholar]
- Hegde RS, Tremblay P, Groth D, DeArmond SJ, Prusiner SB, Lingappa VR. Transmissible and genetic prion diseases share a common pathway of neurodegeneration. Nature. 1999;402:822–826. doi: 10.1038/45574. [DOI] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Jungnickel B, Rapoport TA. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell. 1995;82:261–270. doi: 10.1016/0092-8674(95)90313-5. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Hegde RS. Cotranslational partitioning of nascent prion protein into multiple populations at the translocation channel. Mol. Biol. Cell. 2002;13:3775–3786. doi: 10.1091/mbc.E02-05-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Rahbar R, Hegde RS. Combinatorial control of prion protein biogenesis by the signal sequence and transmembrane domain. J. Biol. Chem. 2001;276:26132–26140. doi: 10.1074/jbc.M101638200. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Mitra D, Salerno JR, Hegde RS. Signal sequences control gating of the protein translocation channel in a substrate-specific manner. Dev. Cell. 2002;2:207–217. doi: 10.1016/s1534-5807(01)00120-4. [DOI] [PubMed] [Google Scholar]
- Levine CG, Mitra D, Sharma A, Smith CL, Hegde RS. The efficiency of protein compartmentalization into the secretory pathway. Mol. Biol. Cell. 2005;16:279–291. doi: 10.1091/mbc.E04-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Lindquist S. Conversion of PrP to a self-perpetuating PrPSc-like conformation in the cytosol. Science. 2002;298:1785–1788. doi: 10.1126/science.1073619. [DOI] [PubMed] [Google Scholar]
- Ma J, Wollmann R, Lindquist S. Neurotoxicity and neurodegeneration when PrP accumulates in the cytosol. Science. 2002;298:1781–1785. doi: 10.1126/science.1073725. [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothes W, Jungnickel B, Brunner J, Rapoport TA. Signal sequence recognition in cotranslational translocation by protein components of the endoplasmic reticulum membrane. J. Cell Biol. 1998;142:355–364. doi: 10.1083/jcb.142.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi A, Fioriti L, Chiesa R, Sitia R. Conditions of endoplasmic reticulum stress favor the accumulation of cytosolic prion protein. J. Biol. Chem. 2006;13:2067–2078. doi: 10.1074/jbc.M605320200. [DOI] [PubMed] [Google Scholar]
- Osborne AR, Rapoport TA, van den Berg B. Protein translocation by the Sec61/SecY channel. Annu. Rev. Cell Dev. Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- Oyadomari S, Yun C, Fisher EA, Kreglinger N, Kreibich G, Oyadomari M, Harding HP, Goodman AG, Harant H, Garrison JL, et al. Cotranslocational degradation protects the stressed endoplasmic reticulum from protein overload. Cell. 2006;126:727–739. doi: 10.1016/j.cell.2006.06.051. [DOI] [PubMed] [Google Scholar]
- Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- Rane NS, Yonkovich JL, Hegde RS. Protection from cytosolic prion protein toxicity by modulation of protein translocation. EMBO J. 2004;23:4550–4559. doi: 10.1038/sj.emboj.7600462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Lingappa VR, Hegde RS. Substrate-specific regulation of the ribosome- translocon junction by N-terminal signal sequences. Proc. Natl. Acad. Sci. USA. 2001;98:7823–7828. doi: 10.1073/pnas.141125098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatzl HM, Da Costa M, Taylor L, Cohen FE, Prusiner SB. Prion protein gene variation among primates. J. Mol. Biol. 1995;245:362–374. doi: 10.1006/jmbi.1994.0030. [DOI] [PubMed] [Google Scholar]
- Shan SO, Walter P. Co-translational protein targeting by the signal recognition particle. FEBS Lett. 2005;579:921–926. doi: 10.1016/j.febslet.2004.11.049. [DOI] [PubMed] [Google Scholar]
- Voigt S, Jungnickel B, Hartmann E, Rapoport TA. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J. Cell Biol. 1996;134:25–35. doi: 10.1083/jcb.134.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J. Mol. Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- Wickner W, Schekman R. Protein translocation across biological membranes. Science. 2005;310:1452–1456. doi: 10.1126/science.1113752. [DOI] [PubMed] [Google Scholar]
- Williams EJ, Pal C, Hurst LD. The molecular evolution of signal peptides. Gene. 2000;253:313–322. doi: 10.1016/s0378-1119(00)00233-x. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K. A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell. 2003;4:265–271. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.