Abstract
Despite ample focus on this endangered species, conservation planning for chimpanzees residing outside Africa has proven a challenge because of the lack of ancestry information. Here, we analysed the largest number of chimpanzee samples to date, examining microsatellites in >100 chimpanzees from the range of the species in Africa, and 20% of the European zoo population. We applied the knowledge about subspecies differentiation throughout equatorial Africa to assign origin to chimpanzees in the largest conservation management programme globally. A total of 63% of the genotyped chimpanzees from the European zoos could be assigned to one of the recognized subspecies. The majority being of West African origin (40%) will help consolidate the current breeding programme for this subspecies and the identification of individuals belonging to the two other subspecies so far found in European zoos can form the basis for breeding programmes for these. Individuals of various degree of mixed ancestry made up 37% of the genotyped European zoo population and thus highlight the need for appropriate management programmes guided by genetic analysis to preserve maximum genetic diversity and reduce hybridization among subspecies.
Keywords: Pan troglodytes, microsatellites, structure, conservation, ancestry
Introduction
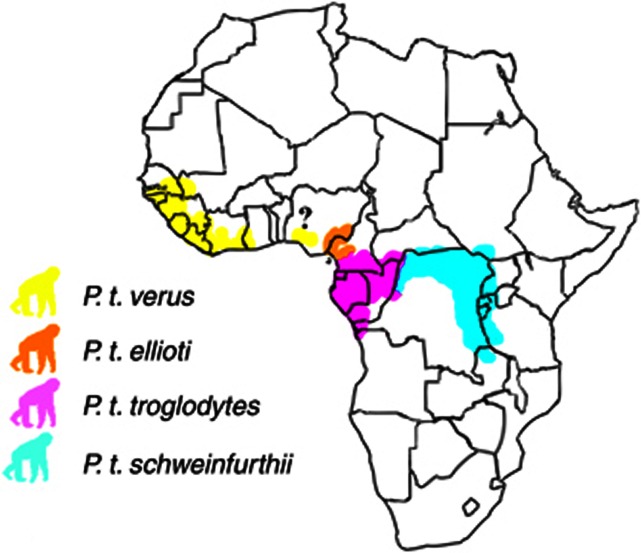
Chimpanzee populations are undergoing a drastic decline in the wild because of threats such as deforestation, hunting and disease (Butynski, 2003; Miles et al., 2005; Oates, 2006; Oates et al., 2008). As a consequence, conservation programmes linking in situ and ex situ efforts are of high importance. Determining conservation priorities requires thorough knowledge of the genetic diversity and structure of the species in focus (Witzenberger and Hochkirch, 2011). The common chimpanzee (Pan troglodytes) is subdivided into four subspecies: the West African chimpanzee (P. t. verus), the Nigeria/Cameroon chimpanzee (P. t. ellioti), the Central African chimpanzee (P. t. troglodytes) and the East African chimpanzee (P. t. schweinfurthii) (Figure 1) (Hill, 1969; Grubb et al., 2003; Oates et al., 2009). The contemporary distribution ranges have been proposed separated by geographical barriers such as large rivers (Hill, 1969; Teleki, 1989; Grubb et al., 2003). However, the location of contact zones between the subspecies is still unclear owing to insufficient sampling. Indeed, disentangling the taxonomy has been surrounded with much attention (Becquet et al., 2007; Caswell et al., 2008; Hey, 2010; Wegmann and Excoffier, 2010; Fischer et al., 2011; Gonder et al., 2011). But while the appropriate taxonomic labelling is still debated, conservation planning has become ever more important as a consequence of threats to the endangered chimpanzee across Equatorial Africa. Globally, the largest existing conservation programme for chimpanzees is the European conservation management programme under the European Association of Zoos and Aquaria (EAZA). This programme targets the lowest recognized taxon, the subspecies level, although at present a breeding programme (European Endangered Species Programme (EEP)) has only been established for P. t. verus because of the taxonomic consensus about the distinctiveness of this subspecies (Becquet et al., 2007; Caswell et al., 2008; Wegmann and Excoffier, 2010; Fischer et al., 2011; Gonder et al., 2011; Bowden et al., 2012).
Figure 1.
Geographical distribution ranges for Pan troglodytes subspecies (Hill, 1969; Grubb et al., 2003; Oates et al., 2008, 2009). The ranges are colour coded: yellow for P. t. verus, orange for P. t. ellioti, pink for P. t. troglodytes and turquoise for P. t. schweinfurthii. The question mark indicates a dispute about the chimpanzees inhabiting the area west of the Niger River in Nigeria and their phylogenetic grouping with either P. t. verus or P. t. ellioti.
Facing possible future re-introduction, it is of high importance that ex situ conservation breeding programmes aim to preserve healthy (physically, behaviourally and genetically) self-sustaining populations that resemble their wild counterparts. Inbreeding and loss of genetic diversity are the major concerns of ex situ conservation efforts. Studbook pedigree information is used to minimize inbreeding by guided breeding. Loss of genetic diversity of the captive population can be reduced by equal representation of genetic lineages (founders) and keeping the effective population size as high as possible. Unfortunately, the origin and genetic relationships of the founders are often not known and studbooks are as a consequence built upon assumptions, which do not accurately describe their true relationship. Many of the founders of the European chimpanzee population are assumed to be of West African origin because of information on disembarkation or place of capture (Peterson and Goodall, 1993; Carlsen, 2009). However, the moderate breeding restrictions before the establishment of a P. t. verus breeding programme and a studbook for the remaining population presumably implies some degree of mixed individuals. Studbook pedigree data, sometimes combined with genetical studies based on mtDNA, were previously the only source upon which breeding programmes were established. For this to be implemented efficiently, knowledge of the genetic structure of the chimpanzee is required. To improve the management of chimpanzee populations in Europe, we have analysed the population structure of the largest number of chimpanzee samples from throughout equatorial Africa to date, coupled with the genotyping of 20% of the European zoo population. Highly polymorphic microsatellites were assessed to answer the following questions: (1) What is the fraction of subspecies in the European conservation management programme? (2) What is the degree of admixture in European zoos? (3) What are the future prospects for chimpanzee management programmes?
Materials and methods
Sample collection
A total of 112 chimpanzee samples of known origin were analysed for genetic variation at 30 selected microsatellite loci (Supplementary Table S1). The samples were collected from 26 P. t. verus, 52 P. t. troglodytes and 34 P. t. schweinfurthii individuals (Supplementary Table S2). Of these, 35 came from European zoos and the remainder from sanctuaries in Africa. In our currently unpublished study, we had information on the geographical region of origin for the majority of the individuals and for the remaining we had a population designation and microsatellite or mtDNA genotypes (Supplementary Table S2). Besides the 35 genotyped zoo specimens of known origin from the European zoo population, we genotyped 128 chimpanzees of unknown origin using our panel of 30 microsatellites (Supplementary Table S3).
Laboratory methods
A panel of 30 microsatellite loci were selected from the study by Becquet et al. (2007). The tetra and tri repeat loci were selected based on the following criteria: (1) distributed throughout all chromosomes; (2) no more than two loci on any chromosome; (3) with two loci on a chromosome, the location should be on the p and q arm and should be located far from each other to maximize the impact of recombination; and (4) the informativeness (Rosenberg et al., 2003) should be as high as possible. A list of the loci selected can be seen in Supplementary Table S1. Forward primers were end-labelled with 6FAM, PET, VIC or NED fluorescent dye (Applied Biosystems, Foster City, CA, USA). The microsatellites were multiplexed in Set1 (4, 10, 13, 15, 20, 26, 28), Set2 (7, 12, 16, 19, 21, 23), Set3 (8, 9, 17, 18, 24, 25) and Set4 (3, 11, 14, 22, 27, 29, 31) (primers are listed in Supplementary Table S1). Each PCR was carried out in a 25 μl reaction volume containing 1 ng genomic DNA (extracted using QIAmp Blood and tissue kit; Qiagen, Valencia, CA, USA), 2 or 4 mℳ MgCl2, 1 × Buffer 10X, balanced amount in μℳ of each primer and 2 U of AmpliTaq Gold DNA polymerase (Applied Biosystems). Negative controls were included in each set of amplification to check for contamination. Amplification included 94 °C for 3 min, followed by 33 cycles of 95 °C for 30 s and annealing temperature range for the four multiplexes between 54 and 66 °C for 30 s, 72 °C for 30 s, followed by 60 °C for 45 min and a hold at 4 °C. A 1.5 μl volume of each sample was mixed with 15 μl formamide and LIZ500 size standard and denatured at 94 °C for 5 min, snap cooled on ice for 2 min and subsequently loaded into an ABI 3130XL Gene Analyser (Applied Biosystems). Data were analysed with Genescan version 3.7 and PCR fragment lengths were scored using Genotyper version 3.7 (Applied Biosystems). All genotyping was carried out in duplicates, amplified on separate dates. No genotypic inconsistencies were observed for any locus among replications. The level of overall missing data was <0.54% across all loci. After evaluation of microsatellite performance, four loci were excluded, which resulted in a final set of 25 autosomal loci and one Y-chromosome locus. The autosomal loci were used in the data analysis. Genotypes are available in Supplementary material and Supplementary Tables S4 and S5.
Data analysis
We calculated common statistics for each population and tested for genotypic linkage disequilibrium using the Markov chain method implemented in Genepop version 4.0.10 (Raymond and Rousset, 1995) on the web (http://genepop.curtin.edu.au/) with 10 000 dememorizations, 1000 batches and 10 000 iterations per batch, using Fisher's method for combining independent test results (Manly, 1985). The statistical significance of these tests was determined by adjusting the probability values for multiple comparisons using sequential Bonferroni correction (Rice, 1989).
We estimated allele frequencies and gene diversity (He) using Arlequin 3.5.1.2 (Excoffier and Lisher 2010), and deviation from Hardy–Weinberg proportions (Fis) (Weir and Cockerham, 1984) using Genepop. Hardy–Weinberg significance values (P) were estimated using the Markov chain method with 10 000 dememorizations, 1000 batches and 10 000 iterations per batch. Probabilities over all loci per population were computed using Fisher's method for combining independent test results (Manly, 1985). Deviation from random mating was calculated for each locus and overall per population.
Population structure
To enable direct comparison with previous studies, analysis of pairwise population differentiation (Fst) was quantified using Arlequin 3.5.1.2 (Excoffier and Lisher, 2010), with 100 permutations and 0.05 as the significance cutoff. It has been highlighted that Fst mistakenly does not capture the unbiased differentiation when estimated based on highly polymorphic markers such as microsatellites (Jost, 2008; Heller and Siegismund, 2009). As a consequence, we applied Jost's unbiased estimator of differentiation Dest (Jost, 2008) to quantify the level of genetic similarity among populations without the confounding effect of diversity. Dest was estimated using SMOGD version 1.2.5 (Crawford, 2010). Mean Dest across loci was calculated as the arithmetic mean, which is considered most reasonable when relatively low numbers of loci and samples are used (Jost, personal communication).
Admixture in in situ chimpanzees
Two complementary methods were applied to characterize population admixture within the chimpanzee data set comprising specimens collected in Africa. We used the program STRUCTURE version 2.3.4 (Pritchard et al., 2000) to estimate the number of genetically distinct clusters in the data set. We applied the admixture model to allow individuals to have ancestry from multiple populations and assumed correlated allele frequencies. We blinded prior population notation and allowed K to be estimated (K=1–10). The individuals were simultaneously assigned probabilistically (90% probability limit) to one population (the population of origin), or more than one cluster (the parental populations) if they were genetically admixed. A model of correlated allele frequencies was used with a burn-in of 100 000 MCMC iterations and 1 000 000 follow-on MCMC iterations. A total of 20 individual repetitions, for each K, were run to check for consistency of the results. Likelihood scores were evaluated using the ad hoc procedure recommended by Evanno et al. (2005) and visualized using Distruct version 1.1 (Rosenberg, 2004). To explore the level of admixture between populations, the same approach was used for the combined data set as well as pairwise analysis of populations. To further asses the population admixture, we analysed the data using principal component analysis (PCA), implemented in the package Adegenet version 1.3-4 (Jombart, 2008). Unlike STRUCTURE, PCA is used exploratory to uncover unknown trends in the data without any user-specified priors. The eigenvectors associated with the principal components are explanatory variables with the first eigenvector separating the samples in a way that explains the largest amount of variability, while the second and subsequent ones explain lesser amounts of variability. The low value of missing data (0.54%) was replaced by the mean of the concerned allele, computed on all available observations without distinction of population (Jombart, 2008).
Admixture in ex situ chimpanzees
To assign origin to the zoo specimens, we used both STRUCTURE and PCA. Geographical origin information from the European studbook for the chimpanzee P. troglodytes (Carlsen, 2009) excluded the likelihood of finding traces of P. t. ellioti in the sampled zoo specimens. Furthermore, preliminary STRUCTURE analysis excluded traces of more than three populations present in the zoo specimens. Thus, our analysis included data from the three African chimpanzee subspecies sampled, P. t. verus, P. t. troglodytes and P. t. schweinfurthii, and the zoo specimens. To empower the PCA analysis, we identified the zoo specimens harbouring >99% ancestry to one subspecies only by the admixture PCA analysis of wild chimpanzees, and included them in the reference populations they were assigned to. In STRUCTURE, we used these reference populations as learning samples to assist ancestry estimation for zoo individuals of unknown origin. For the STRUCTURE analysis, individuals were assumed to be admixed and allele frequencies correlated. To account for asymmetric gene flow between chimpanzee subspecies (Won and Hey, 2005; Wegmann and Excoffier, 2010), we inferred the degree of admixture (alpha) for each separate population with a uniform prior distribution between 0.0 and 0.1. Migration priors were similarly explored in the range 0.01–0.1, as recommended by Pritchard et al. (2000). As preliminary runs showed a trend towards better estimates of likelihood for migration parameters close to 0.1, the final analysis was parameterized with a migration prior of 0.1. Each run was carried out with a burn-in of 100 000 MCMC iterations and 1 000 000 follow-on MCMC iterations.
To test if we could distinguish between zoo specimens of mixed ancestry, we simulated F1 hybrid populations as well as first and second backcrossed populations, each of size n=20 using Adegenet (Jombart, 2008). On the basis of geographical origin information from the European studbook for the chimpanzee P. troglodytes (Carlsen, 2009), the majority of chimpanzees were assumed to be descendants of West African founders and we thus simulated first and second backcrossed populations with unadmixed P. t. verus individuals. As above, the PCA implemented in Adegenet (Jombart, 2008) was used and missing data (0.32%) was replaced by the mean allele frequency. To further explore the zoo population composition, we estimated Ne and mean inbreeding using the PMx population management software version 1.0 (Ballou et al., 2011). The date span for the export, which was set to 1 January 1970–until present, was sufficient to include sensible demographic data associated with modern management. Depending on the geographic span, either a filter for regional zoo association (EAZA) or a geographic filter including all nations with relevant holders was chosen. Living neutered specimens were excluded from the PMx genetic analyses, as these specimens do not actively contribute to future the population. The amount of genetic diversity of wild chimpanzees represented in the European zoo population was estimated using rarefaction by the program HP Rare (Kalinowski, 2005) to account for the expected difference in the number of alleles due to sample sizes. As above, the gene diversity was estimated using Arlequin 3.5.
Results
Hardy–Weinberg and genotypic linkage disequilibrium
The population by locus comparison showed deviations from Hardy–Weinberg after sequential Bonferroni correction at nine loci (Table 1). Significant deviation from Hardy–Weinberg proportions across loci was observed for all three populations (Table 1) likely due to Wahlund effect, caused by population structure in the form of admixture or stratification. We checked the data set for genotypic linkage equilibrium. Across the three populations, we observed genotypic linkage disequilibrium in 8 out of 300 locus pairs after sequential Bonferroni correction.
Table 1. Common statistics.
| P. t. verus | P. t. troglodytes | P. t. schweinfurthii | |
|---|---|---|---|
| GATA104 | 0.74 | 0.93 | 0.87 |
| 0.25 | 0.21 | 0.09 | |
| 0.03 | 0.0005 | 0.49 | |
| GATA129H04 | 0.82 | 0.92 | 0.91 |
| −0.02 | 0.04 | 0.01 | |
| 0.15 | 0.28 | 0.55 | |
| GATA176C01 | 0.79 | 0.91 | 0.86 |
| −0.14 | 0.12 | 0.24 | |
| 0.95 | 0.06 | 0.0004 | |
| GATA71H05 | 0.66 | 0.88 | 0.90 |
| 0.35 | 0.10 | 0.08 | |
| 0.0006 | 0.14 | 0.22 | |
| ATA27A06P | 0.65 | 0.80 | 0.82 |
| −0.03 | 0.16 | 0.04 | |
| 0.58 | 0.02 | 0.38 | |
| GATA43A04 | 0.77 | 0.93 | 0.83 |
| −0.07 | 0.44 | 0.11 | |
| 0.15 | 0 | 0.002 | |
| GATA14E09 | 0.64 | 0.84 | 0.73 |
| −0.14 | −0.02 | 0.20 | |
| 0.85 | 0.57 | 0.47 | |
| GATA116B01N | 0.77 | 0.87 | 0.83 |
| −0.06 | −0.05 | −0.05 | |
| 0.08 | 0.86 | 0.06 | |
| GATA50G06 | 0.76 | 0.89 | 0.88 |
| −0.25 | 0.05 | −0.07 | |
| 0.03 | 0.26 | 0.84 | |
| UT7544 | 0.67 | 0.87 | 0.84 |
| −0.05 | 0.18 | 0.05 | |
| 0.98 | 0.05 | 0.73 | |
| GGAA21G11L | 0.84 | 0.90 | 0.86 |
| −0.03 | 0.05 | 0.02 | |
| 0.85 | 0.15 | 0.995 | |
| GATA28F03 | 0.66 | 0.89 | 0.87 |
| −0.02 | 0.24 | 0.65 | |
| 0.79 | 0.001 | 0 | |
| TCTA017M | 0.79 | 0.79 | 0.79 |
| 0.01 | 0.12 | 0.22 | |
| 0.34 | 0.03 | 0.07 | |
| AGAT120 | 0.80 | 0.86 | 0.74 |
| 0.10 | 0.05 | 0.07 | |
| 0.02 | 0.21 | 0.17 | |
| GATA25A04 | 0.64 | 0.80 | 0.85 |
| −0.05 | 0.18 | 0.12 | |
| 0.40 | 0.01 | 0.06 | |
| GATA125D11N | 0.08 | 0.66 | 0.64 |
| −0.02 | −0.02 | −0.07 | |
| 1 | 0.75 | 0.52 | |
| GATA8C04 | 0.80 | 0.76 | 0.80 |
| 0.02 | 0.05 | −0.007 | |
| 0.51 | 0.0003 | 0.96 | |
| ATA11D10M | 0.64 | 0.83 | 0.72 |
| 0.21 | −0.01 | 0.24 | |
| 0.10 | 0.81 | 0.002 | |
| GATA68A03 | 0.90 | 0.94 | 0.91 |
| 0.05 | 0.02 | 0.15 | |
| 0.60 | 0.05 | 0.003 | |
| GATA24F03 | 0.85 | 0.91 | 0.87 |
| 0.50 | 0.29 | 0.24 | |
| 0 | 0 | 0.002 | |
| GATA29F06z | 0.40 | 0.81 | 0.59 |
| −0.07 | 0.02 | −0.04 | |
| 0.68 | 0.03 | 0.86 | |
| GATA4D07 | 0.84 | 0.87 | 0.83 |
| 0.04 | 0.06 | −0.01 | |
| 0.47 | 0.63 | 0.63 | |
| GATA63D12P | 0.87 | 0.76 | 0.83 |
| 0.28 | 0.04 | 0.02 | |
| 0.47 | 0.63 | 0.30 | |
| GATA88F09 | 0.73 | 0.91 | 0.86 |
| −0.14 | 0.12 | 0.05 | |
| 0.42 | 0.01 | 0.63 | |
| GATA63F09 | 0.73 | 0.78 | 0.75 |
| 0.15 | 0.36 | 0.65 | |
| 0.02 | 0 | 0 | |
| Total average | 0.72 | 0.87 | 0.82 |
| 0.04 | 0.12 | 0.12 | |
| 2.8 × 10−5 | 8.4 × 10−19 | 1.4 × 10−11 | |
Abbreviations: HW, Hardy–Weinberg.
In each cell: gene diversity He (top), deviations from Hardy-Weinberg (HW) proportions Fis (Weir and Cockerham) (middle) and HW(P) (bottom) are estimated using Genepop (http://genepop.curtin.edu.au) and Arlequin 3.5.1.2 (Excoffier and Lisher, 2010). The total HW(P) per population was calculated using Fisher's method for combining independent test results. The P-values of 0 were replaced with 1/number of simulations. Estimates in bold are significant after sequential Bonferroni correction.
Genetic diversity, distance and differentiation
All microsatellite loci, except GATA125D11N for P. t. verus, were highly polymorphic and heterozygosity was generally high (He range 0.40–0.94; Table 1). When looking at deviations from Hardy–Weinberg expectations, the average Fis estimates were due to heterozygote deficits (0.04–0.12). Genetic distance, as measured by pairwise Fst values between populations, increased with geographical distance between subspecies (Table 2). Genetic differentiation followed the same tendency for Dest as observed for Fst, with increasing values with increasing geographical distance between subspecies (Table 2).
Table 2. Population differentiation.
| P. t. verus | P. t. troglodytes | P. t. schweinfurthii | |
|---|---|---|---|
| P. t. verus | 0.54 | 0.59 | |
| P. t. troglodytes | 0.12 | 0.31 | |
| P. t. schweinfurthii | 0.14 | 0.04 |
Abbreviations: ANOVA, analysis of variance; Dest, estimator of differentiation; Fst, fixation index is a measure of pairwise population differentiation.
Lower: Pairwise Fst estimated in Genepop (Raymond and Rousset 1995) on the web (http://genepop.curtin.edu.au/) following standard ANOVA as in Weir and Cockerham (1984). Upper: Pairwise Dest, corrected for small sample sizes (Jost, 2008), estimated in SMOGD (Crawford, 2010). Pairwise population per locus Dest with 1000 bootstrap replicates, summed across loci by taking the arithmetic mean.
Admixture in chimpanzees from the natural distribution range
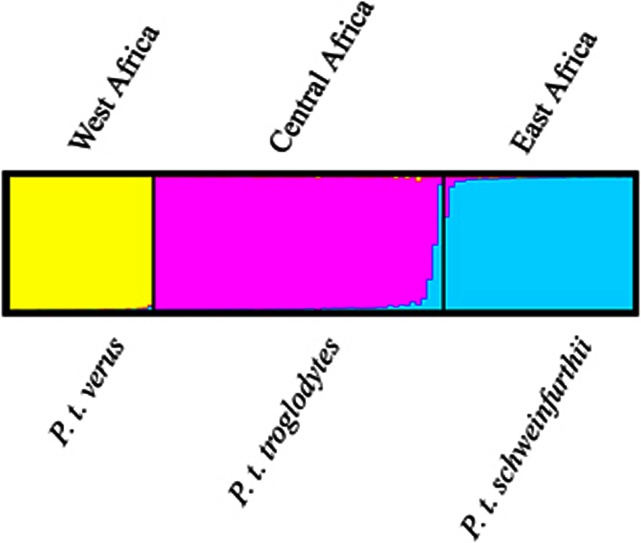
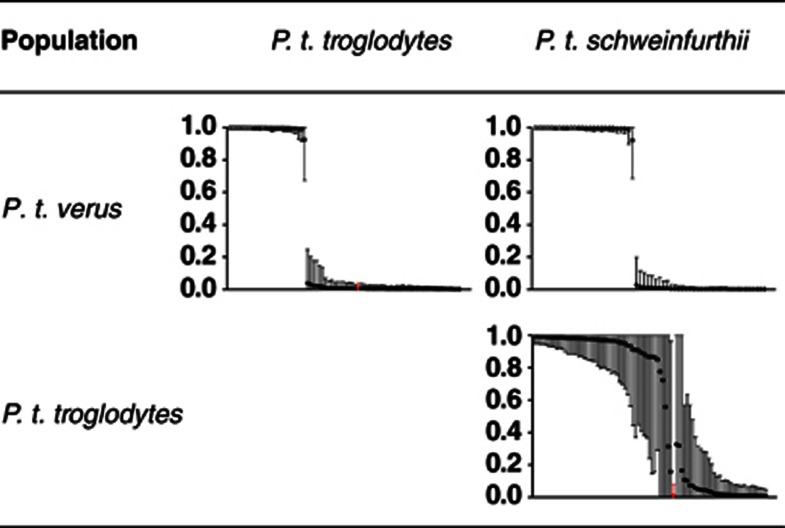
Evidence for subdivision among chimpanzees was explored with STRUCTURE and the analysis supports the division of the samples into either K=2 or K=3 populations according to the ΔK method (Supplementary Figure S1) (Evanno et al., 2005). Dividing the data into two separate clusters, the geographically isolated P. t. verus falls within one separate cluster, while the two geographically adjoined P. t. troglodytes and P. t. schweinfurthii groups within one shared cluster (Supplementary Figure S2). Following the designated number of subspecies, K=3, we observe evidence of admixture between P. t. troglodytes and P. t. schweinfurthii, while all individuals of P. t. verus forms a separate cluster (Figure 2). To further explore the evidence of admixture, we performed pairwise population analysis in STRUCTURE (K=2). When comparing P. t. verus to the other populations, we find estimates of q near 1 and 0, respectively, with non-overlapping probability limits (Figure 3). The pairwise analyses between the populations of P. t. troglodytes and P. t. schweinfurthii (Figure 3) confirmed the admixture observed in the STRUCTURE analysis of the entire data set (n=112) (Figure 2). Using a cutoff level of >5% ancestry from two clusters, we report seven individuals to be of mixed P. t. troglodytes/P. t. schweinfurthii origin (Figure 2).
Figure 2.
Genetic clusters inferred from genotypic data of Pan troglodytes subspecies with known origin. Proportion of membership to each cluster for every sampled individual is indicated by colours in the vertical bars. The bar plots show the genetic structure among all subspecies when three clusters are assumed. Predefined subspecies origin is indicated below and geographical sampling location above. One individual (Choupette) was registered as P. t. troglodytes, but the analysis revealed it to be of P. t. schweinfurthii origin.
Figure 3.
Pairwise comparison of posterior distribution of admixture coefficients (q) between all population pairs based on 25 autosomal loci. The 90% probability intervals are indicated by vertical bars for each individual. Open and full circles indicate population origin for each individual based on its geographical sampling location. Open circles represent individuals from row populations and closed circles denote individuals from column populations. One individual (Choupette) marked with a red asterisk (*) denotes a discrepancy between sampling location and the genotyped subspecies origin (Supplementary Table S2), see also Figure 2 and text.
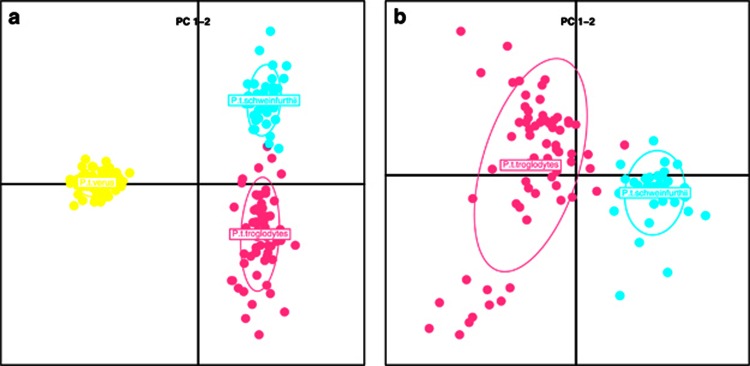
Clustering of chimpanzees based on PCA displayed separated groups. Inspecting the first two eigenvectors confirmed a separation of all three subspecies (Figure 4a). As observed in the STRUCTURE analysis, the groups are separated but with a few individuals falling between when observed pairwise (Figure 4b)—matching the same individuals from the STRUCTURE analysis.
Figure 4.
(a) PCA of chimpanzee individuals from the species distribution range in Africa. Colours: yellow, pink and turquoise for P. t. verus, P. t. troglodytes and P. t. schweinfurthii, respectively. (b) A PCA as above but without P. t. verus is displayed.
Admixture in ex situ chimpanzees
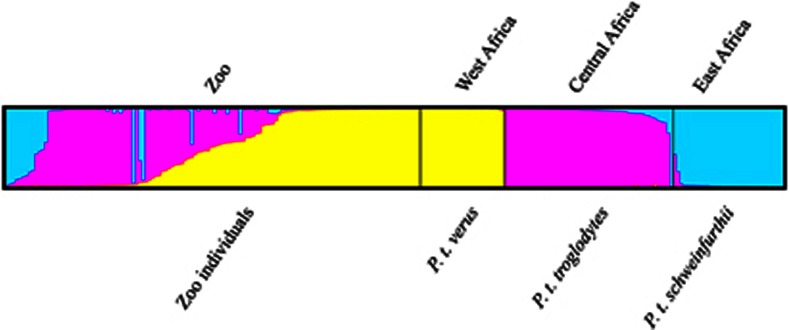
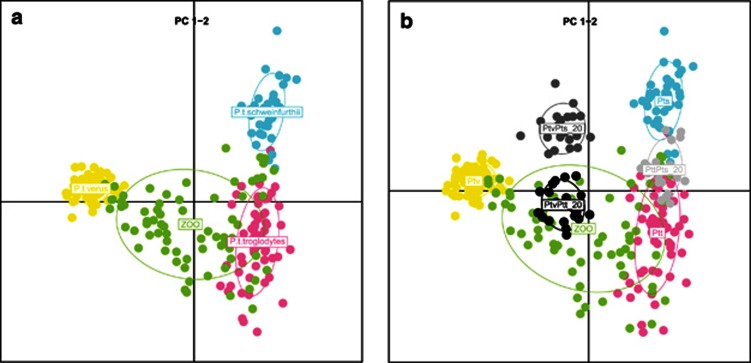
Inspecting the output of STRUCTURE for the individuals of unknown origin revealed 72 individuals with 95% ancestry from one population (Figure 5). The majority, 42, clustered with P. t. verus, 26 with P. t. troglodytes and 4 with P. t. schweinfurthii. Individuals with >5% ancestry (56 individuals) from more than one population were considered as hybrids or backcrosses. We found the mixed individuals to be predominantly with some degree of P. t. verus and P. t. troglodytes ancestry (Figure 5 and Table 3). The same is observed when inspecting the PCA analysis (Figure 6a). When distinguishing between zoo specimens of mixed ancestry, we observed that the simulated P. t. verus/P. t. troglodytes and P. t. troglodytes/P. t. schweinfurthii F1 hybrid populations in Figure 6b fall within the mixed zoo individuals. First backcrossed populations too fall within the mixed zoo cluster and are in addition clearly differentiated from the simulated F1 hybrid populations (Supplementary Figure S3a+Figure 6b). Lastly, for the simulated second backcrossed populations we observed that these, given their high degree of P. t. verus ancestry, do not fall within the mixed zoo cluster, but instead with the reference P. t. verus cluster (Supplementary Figure S3b). Owing to these findings, we compared the results from the STRUCTURE analysis, which identified 72 zoo specimens with 95% ancestry from one population, with the information provided in the common chimpanzee ESB. This uncovered four individuals to be of second, or more, backcrossed origin. As revealed by our simulation analysis, we do not have enough power in our analysis to unambiguously assign these to a subspecies. Excluding these makes a total of 68 individuals with 95% ancestry from one population, 39 clustering with P. t. verus, 25 clustering with P. t. troglodytes and 4 clustering with P. t. schweinfurthii. The total number of genotyped zoo individuals with known and unknown origin belonging to a subspecies is hereby 65 P. t. verus, 30 P. t. troglodytes and 8 P. t. schweinfurthii.
Figure 5.
Inferred ancestry of Pan troglodytes subspecies with unknown origin based on genetic clustering with sampled individuals from the species natural distribution range. Proportion of membership to each cluster for every sampled individual is indicated by colours in the vertical bars Predefined subspecies origin is indicated below and geographical sampling location above.
Table 3. Zoo ancestry.
| P. t. verus | P. t. troglodytes | P. t. schweinfurthii | Mix of all subspecies | |
|---|---|---|---|---|
| P. t. verus | 40% | 22% | 1% | 7% |
| P. t. troglodytes | 18% | 7% | ||
| P. t. schweinfurthii | 5% |
Proportion of individuals genotyped belonging to a subspecies (diagonal). Individuals of mixed ancestry (above diagonal) with more than 5% ancestry to two populations. Mixed individuals of >2 subspecies and individuals of potentially old backcrossed origin are listed in the last right column.
Figure 6.
(a) PCA of chimpanzee individuals from the species distribution range in Africa and zoo specimens of unknown origin. Colours: yellow, pink, turquoise and green for P. t. verus, P. t. troglodytes, P. t. schweinfurthii and zoo specimens, respectively. The pink dot falling within the outer range of the P. t. schweinfurthii cluster represents the, based on the STRUCTURE analysis, misclassified individual Choupette. In (b), three simulated F1 hybrid populations are included, each consisting of 20 individuals. The populations are colour coded light grey, medium grey and dark grey for P. t. troglodytes/P. t. schweinfurthii, P. t. schweinfurthii/P. t. verus and P. t. verus/P. t. troglodytes, respectively.
Status of European ex situ population
In a population, the census size (Nc) may differ from the effective population size (Ne) depending on the contribution of each individual to reproduction. To counteract effectively inbreeding and maintain genetic diversity, Ne should be kept as high as possible. Given that the updated Nc for P. t. verus, including individuals identified in this study, in the European ex situ EEP population counts 201 living individuals, this amounts to an estimated Ne of 58 and a mean inbreeding of 0.0206. However, a large fraction of the European population is still unsampled and it would be of interest to disclose the deduced total Ne for each subspecies. Of the whole genotyped collection, 40% belonged to P. t. verus. The European EAZA chimpanzee population comprises 870 individuals, and if we assume the individuals genotyped in this study to represent a random selection of the population, the 40% belonging to P. t. verus will total an Ne of 100. No breeding programme yet exists for P. t. troglodytes and P. t. schweinfurthii, but if we assume that we can use a similar ratio for Ne and Nc for each subspecies, we get the Ne for them to be 47 and 13, respectively. The census sizes of them have been estimated from the proportions belonging to the two subspecies (Table 3) and a total census size of 870 for chimpanzees in the European EAZA ex situ population. When investigating the overall genetic diversity of wild and zoo populations, we found them to be congruent (Table 4).
Table 4. Diversity estimates.
| Number of individuals |
Genetic diversity based on 46 genes |
Genetic diversity based on four genes |
He | |||
|---|---|---|---|---|---|---|
| Allelic richness | Private allelic richness | Allelic richness | Private allelic richness | |||
| Wild P. t. verus | 26 | 7.59 | 0.67 | 2.74 | 0.66 | 0.72 |
| Zoo P. t. verus | 39 | 7.72 | 0.49 | 2.79 | 0.68 | 0.74 |
| Wild P. t. troglodytes | 52 | 11.95 | 1.41 | 3.29 | 1.03 | 0.87 |
| Zoo P. t. troglodytes | 25 | 11.93 | 1.56 | 3.25 | 0.98 | 0.85 |
| Wild P. t. schweinfurthii | 34 | 9.97 | 1.08 | 3.08 | 0.82 | 0.82 |
| Zoo P. t. schweinfurthii | 4 | — | — | 2.94 | 0.70 | 0.79 |
| Zoo hybrids | 60 | 12.20 | 0.78 | 3.28 | 0.74 | 0.86 |
The number of genes refers to the minimum number of genes sampled at any locus for the given population. Zoo P. t. schweinfurthii was excluded in the first analysis based on 46 genes. Genetic diversity estimates are averaged over loci.
Discussion
Genetic structure of chimpanzees from throughout the species range in Africa was assessed and the knowledge used to assign origin to individuals within the European conservation management programme.
Genetic structure of chimpanzee subspecies
The highest level of admixture was found between P. t. troglodytes and P. t. schweinfurthii, which is in good agreement with these geographically adjoined populations being most closely related as a result of a prolonged period of contact after their divergence, as suggested by previous studies (Becquet and Przeworski, 2007; Hey, 2010; Wegmann and Excoffier, 2010; Fischer et al., 2011; Gonder et al., 2011). The observed admixture between subspecies corroborates with reports of low migration, such as the 1.5 migrants per generation between P. t. troglodytes and P. t. schweinfurthii (Wegmann and Excoffier, 2010). Interestingly, our study reveals one individual (Mgbadolite) being a hybrid between a paternal P. t. troglodytes and maternal P. t. schweinfurthii individual, corresponding with the place of capture (Supplementary Table S2), and is to our knowledge the first genetic evidence of hybridization between P. t. troglodytes and P. t. schweinfurthii in the vicinity of the proposed geographical barrier, the Ubangi River in the Democratic Republic of Congo.
Conservation aspects
Before 2001, no subspecies breeding programme existed for the chimpanzee. On the basis of the geographical information recorded in the European chimpanzee studbook, the majority of the founders in the EAZA programme were expected to be of West African origin. It was thus assumed that a considerable proportion of the genotyped zoo specimens would be wild caught individuals from West Africa, or hybrid individuals with varying degrees of P. t. verus descent. The results from our genotyping corroborate this, with 40% of all genotyped individuals being of West African origin, 18% of Central African origin, 5% East African origin and 23% hybrids with a proportion of P. t. verus ancestry (Table 3). The newly identified P. t. verus individuals will be included in the existing EEP breeding programme. This programme until recently comprised 150 individuals (Carlsen, 2009) and the newly identified living individuals increased the total number of individuals herein with a third to 201 individuals. The potentially new founder pool identified, comprising wild born individuals considered to be unrelated, increases the total number of founders with two-thirds. An effort should thus be taken to bring these into a breeding situation. Interestingly, besides the P. t. verus individuals, we found 30 to be of P. t. troglodytes origin (34 if counting deduced relatives) and 8 of P. t. schweinfurthii origin.
The viability of the subspecies in the European conservation programme can be assessed by looking at their effective population sizes. The Ne of their wild equivalents (Wegmann and Excoffier, 2010) are far from the estimates for the European population of 100, 47 and 13 for P. t. verus, P. t. troglodytes and P. t. schweinfurthii, respectively. Although the total Ne for the two latter might be considered small, they could be managed based on separate studbooks. This requires that the entire ex situ population is genotyped to infer the subspecies status of each individual. It is worth mentioning that the genetic diversity in wild and zoo populations is nearly identical. No diversity has yet been lost, which is not surprising given the long generation time of the species and the fact that the founders of the European population arrived during the 1960–1970s are still members of the current population. Although exchange with wild populations would be desirable (see for example, Lacy, 2013), it is not a solution at present. However, with efficient breeding management ensuring a population growth >1, the populations are viable and self-sustaining for many generations to come, assuming that the expected number of founders for the entire population is correct.
Unfortunately, our assumption regarding a high portion of hybrid individuals within the common chimpanzee ESB was confirmed. As a consequence of the implementation of the European Strategy for Chimpanzees, hybrids will be phased out but taking welfare issues into account, the aim is to establish mixed breeding groups including core breeding individuals that have been assigned to subspecies as well as non-breeding hybrids.
Lastly, the chimpanzee data provide an excellent example of a powerful DNA profiling system which allows us to provide additional data to population management and future efforts should be taken to use molecular data to help validate studbooks.
Conclusion
By analysing the largest number of chimpanzee individuals to date, we have sought to enhance the understanding of a chimpanzee zoo population structure. Our study presents the first analysis of chimpanzees from the natural distribution range coupled with origin assignment of zoo specimens. The results corroborated our hypotheses that the majority of European zoo specimens were of West African origin and a considerable proportion of all individuals genotyped were hybrids. We also found proportions of P. t. troglodytes and P. t. schweinfurthii that seem sufficiently high to consider the proposal of breeding programmes for them as well—contingent that both genders are well represented. These findings hold great importance for future conservation efforts for chimpanzees outside Africa as well as in their home-range countries. The applicability of our results directly helps solve a chimpanzee management problem in EAZA by devising a strategy for future breeding programmes and will hereby contribute sustaining the natural genetic diversity in the European captive population. However, a management problem is not unique for the chimpanzee. Of the large number of taxa kept in zoos, only a limited number are managed by studbooks (ISIS, 2012), and to our knowledge, only a minority of these are guided by genetics. Thus, our findings not only highlights but also suggest the application of extensive genetic surveys of both wild and zoo specimens to help guide future conservation management programmes in other taxa as well.
Data archiving
Data deposited in the Dryad repository: doi:10.5061/dryad.1th15.
Acknowledgments
We thank Werner Schempp, Ronald Bontrop, Benoit Goosens, Ngamba Island Chimpanzee Sanctuary and finally all participating European zoos for generously providing samples. This work was supported financially by the 15. Juni Fund, Torben and Alice Frimodt's Foundation, HR and Grete S Frederiksen's Foundation, Copenhagen Zoo, The Danish Council for Independent Research|Natural Sciences as well as the Industrial PhD programme under the Danish Agency for Science, Technology and Innovation.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Ballou JD, Lacy RC, Pollak JP. PMx: Software for Demographic and Genetic Analysis and Management of Pedigreed Populations (Version 1.0) Chicago Zoological Society: Brookfield, IL, USA; 2011. [Google Scholar]
- Becquet C, Patterson N, Stone AC, Przeworski M, Reich D. Genetic structure of chimpanzee populations. PLoS Genet. 2007;3:617–626. doi: 10.1371/journal.pgen.0030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becquet C, Przeworski M. A new approach to estimate parameters of speciation models with application to apes. Genome Res. 2007;17:1505–1518. doi: 10.1101/gr.6409707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden R, MacFie TS, Myers S, Hellenthal G, Nerrienet E, Bontrop RE, et al. Genomic tools for evolution and conservation in the chimpanzee: Pan troglodytes ellioti is a genetically distinct population. PLoS Genet. 2012;8:e1002504. doi: 10.1371/journal.pgen.1002504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butynski TM.2003The robust chimpanzee Pan troglodytes: taxonomy, distribution, abundance, and conservation statusIn: Kormos R, Boesch C, Bakarr MI, Butynski TM, (eds)West African Chimpanzees Status Survey and Conservation Action Plan IUCN/SSC Primate Specialist Group: Gland, Switzerland; 5–12. [Google Scholar]
- Carlsen F. European Studbook for the Chimpanzee (Pan troglodytes) Copenhagen Zoo: Frederiksberg, Denmark; 2009. [Google Scholar]
- Caswell JL, Mallick S, Richter DJ, Neubauer J, Schirmer C, Gnerre S, et al. Analysis of chimpanzee history based on genome sequence alignments. PLoS Genet. 2008;4:1–14. doi: 10.1371/journal.pgen.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NG. SMOGD: software for the measurement of genetic diversity. Mol Ecol Res. 2010;10:556–557. doi: 10.1111/j.1755-0998.2009.02801.x. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lisher H. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fischer A, Prüfer K, Good JM, Halbwax M, Wiebe V, André C, et al. Bonobos fall within the genomic variation of chimpanzees. PLoS One. 2011;6:e21605. doi: 10.1371/journal.pone.0021605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonder MK, Locatelli S, Ghobrial L, Mitchell MW, Kujawski JT, Lankaster FJ, et al. Evidence from Cameroon reveals differences in the genetic structure and histories of chimpanzee populations. Proc Natl Acad Sci USA. 2011;108:4766–4771. doi: 10.1073/pnas.1015422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens B, Funk SM, Vidal C, Latour S, Jamart A, Ancrenaz M, et al. Measuring genetic diversity in translocation programmes: principles and application to a chimpanzee release project. Anim Conserv. 2002;5:225–236. [Google Scholar]
- Grubb P, Butynski TM, Oates JF, Bearder SK, Disotell TR, Groves C, et al. Assessment of the diversity of African primates. Int J Primatol. 2003;24:1301–1357. [Google Scholar]
- Heller R, Siegismund HR. Relationship between three measures of genetic differentiation G(ST), D(EST) and G'(ST): how wrong have we been. Mol Ecol. 2009;18:2080–2083. doi: 10.1111/j.1365-294x.2009.04185.x. [DOI] [PubMed] [Google Scholar]
- Hey J. The divergence of chimpanzee species and subspecies as revealed in multipopulation isolation–with–migration analyses. Mol Biol Evol. 2010;27:921–933. doi: 10.1093/molbev/msp298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WCO.1969The nomenclature, taxonomy and distribution of chimpanzeesIn: Bourne GH, (ed).The Chimpanzee Karger: Basel; 22–49. [Google Scholar]
- ISIS 2012. International Species Information System. Available at: : http://www.isis.org , last accessed 28 September 2012.
- Jombart T. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- Jost L. G(ST) and its relatives do not measure differentiation. Mol Ecol. 2008;17:4015–4026. doi: 10.1111/j.1365-294x.2008.03887.x. [DOI] [PubMed] [Google Scholar]
- Kalinowski ST. HP-Rare: a computer program for performing rarefaction on measures of allelic diversity. Mol Ecol Notes. 2005;5:187–189. [Google Scholar]
- Lacy R. Achieving true sustainability of zoo populations. Zoo Biol. 2013;32:19–26. doi: 10.1002/zoo.21029. [DOI] [PubMed] [Google Scholar]
- Manly B. The Statistics of Natural Selection on Animal Populations. Chapman & Hall: London, UK; 1985. pp. 432–433. [Google Scholar]
- Miles L, Caldecott J, Nellemann C.2005Challenges to great ape survivalIn: Caldecott J, Miles L, (eds).World Atlas of Great Apes and their Conservation University of California Press: Berkeley, USA; 217–241. [Google Scholar]
- Oates JF. Is the chimpanzee, Pan troglodytes, an endangered species? It depends on what ‘endangered' means. Primates. 2006;47:102–112. doi: 10.1007/s10329-005-0149-5. [DOI] [PubMed] [Google Scholar]
- Oates JF, Groves CP, Jenkins PD. The type locality of Pan troglodytes vellerosus (Gray, 1862), and implications for the nomenclature of West African chimpanzees. Primates. 2009;50:78–80. doi: 10.1007/s10329-008-0116-z. [DOI] [PubMed] [Google Scholar]
- Oates JF, Tutin CEG, Humle T.2008Pan troglodytes. In: IUCN 2012. IUCN Red List of Threatened Species. Version 2012.1 available at: http://www.iucnredlist.org .
- Peterson DJ, Goodall J. Visions of Caliban—On Chimpanzees and People. Houghton Mifflin Company: Boston, MA and New York, NY, USA; 1993. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. Genepop (Version 1.2)—Population–Genetics Software for Exact Tests and Ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. Distruct: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
- Rosenberg NA, Li L, Ward R, Pritchard JK. Informativeness of genetic markers for inference of ancestry. Am J Hum Genet. 2003;73:1402–1422. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleki G. Population Status of Wild Chimpanzees (Pan troglodytes) and Threats To Survival. Harvard University press: Cambridge, MA; 1989. [Google Scholar]
- Wegmann D, Excoffier L. Bayesian inference of the demographic history of chimpanzees. Mol Biol Evol. 2010;27:1425–1435. doi: 10.1093/molbev/msq028. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–137. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Witzenberger K, Hochkirch A. Ex situ conservation genetics: a review of molecular studies on the genetic consequences of captive breeding programmes for endangered animal species. Biodivers Conserv. 2011;20:1843–1861. [Google Scholar]
- Won YJ, Hey J. Divergence population genetics of chimpanzees. Mol Biol Evol. 2005;22:297–307. doi: 10.1093/molbev/msi017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.