Abstract
Objective
To identify novel viral determinants in HIV-1 protease, Gag, and envelope V3 that relate to outcomes to initial protease inhibitor-based antiretroviral therapy.
Design
A longitudinal cohort study of protease inhibitor-naive, HIV-infected individuals was designed to identify genetic variables in viral Gag and envelope sequences associated with response to antiretroviral therapy.
Methods
Genetic and statistical models, including amino acid profiles, phylogenetic analyses, receiver operating characteristic analyses, and covariation analyses, were used to evaluate viral sequences and clinical variables from individuals who developed immune reconstitution with or without suppression of viral replication.
Results
Pretherapy chemokine (C–X–C motif) receptor 4-using V3 regions had significant associations with viral failure (P = 0.04). Amino acid residues in protease covaried with Gag residues, particularly in p7NC, independent of cleavage sites. Pretherapy V3 charge combined with p6Pol and p2/p7NC cleavage site genotypes produced the best three-variable model to predict viral suppression in 88% of individuals. Combinations of baseline CD4 cell percentage with genetic determinants in Gag–protease predicted viral fitness in 100% of individuals who failed to suppress viral replication.
Conclusion
Baseline genetic determinants in Gag p6Pol and p2/p7NC, as well as envelope, provide novel combinations of biomarkers for predicting emergence of viral resistance to initial therapy regimens.
Keywords: antiretroviral therapy response, coreceptors, HIV-1 Env V3, HIV-1 Gag, HIV-1 protease inhibitors
Introduction
Selective pressures of drug therapy, target cell type, and host immune mechanisms, combined with a rapid rate of error-prone replication, contribute to the evolution of HIV-1 in its host. Genetic diversity is advantageous for continued viral propagation but can elicit a fitness cost, especially if interactions between proteins are essential for function [1,2]. For example, mutations in Gag cleavage sites compensate for drug-related mutations in protease and modulate protease processing activity and fitness [3–7], whereas Gag determinants outside cleavage sites enhance replicative fitness and sensitivity to inhibitors among viruses with drug-resistant protease alleles [8–10].
In addition to genotype in HIV-1 Gag and protease, host cell targets contribute to viral fitness [11,12]. Cellular tropism of HIV-1 is related to coreceptor preference, which is reflected by amino acid characteristics of the envelope V3 loop [13–15]. Positively charged residues at positions 11 and 25 in V3 are independent predictors of poor immunological response and accelerated mortality [16]. Combination antiretroviral therapies (ARTs), including protease inhibitors, can lead to transient phenotypic shifts from chemokine (C–X–C motif) receptor 4 (CXCR4) (X4) to chemokine receptor 5 (CCR5) (R5) viruses [17] or to suppression of X4 viruses [18]. However, no study has comprehensively related response to combination ART with pretherapy viral genotypes in protease, Gag, and envelope among protease inhibitor-naive individuals.
Therapy response is classified by immune and viral parameters. In our previous study, combination of clinical variables with pretherapy protease, but not reverse transcriptase, genotype predicted response to protease inhibitor combination ART in 75–80% of protease inhibitor-naive individuals who achieved significant CD4 T-cell reconstitution independent of viral suppression [19]. An inability to predict therapy response in all individuals indicated that additional factors contribute to ART responses. In the current study, genetic characteristics in envelope V3 and in Gag (p7NC–p6/p6Pol, including cleavage sites), combined with protease, were evaluated prior to and during treatment with protease inhibitor-containing ART in a cohort of individuals who developed immune reconstitution, with or without complete suppression of virus replication. The goal was to identify regions in HIV-1 Gag and Env that can serve as genotypic markers for viral response to initial ART.
Methods
Study participants
The study involved a retrospective analysis of archived blood samples from 17 HIV-infected children and adolescents selected from a prospectively accrued study population of 50 protease inhibitor therapy-naive individuals enrolled in a treatment trial to examine viral and immune response to combination ART. The study enrolled between August 1995 and September 2002 with approval by Institutional Review Boards of the Universities of Florida and South Florida. Informed consent was obtained from legal guardians for all children; assent was obtained from children over 7 years of age. Patients were between the ages of 1 and 18 years with a range of CD4 T-cell counts and viral loads more than 3.5 log10 copies/ml (Table 1). Therapy included one protease inhibitor [either indinavir (IDV) or ritonavir (RTV)] and two nucleoside reverse transcriptase inhibitors (NRTIs), according to guidelines for treatment of HIV-infected children [20]. Previous exposure to RTI was allowed if the patient was naive to at least one NRTI in the new regimen. Optimal drug dosing was based on pharmacokinetic studies of protease inhibitor in children, and adherence to therapy was confirmed [21,22].
Table 1.
Clinical characteristics of study population.
| Variable | Timepoint | Viral success | Viral failure | Viral failureLow | Viral failureHigh |
|---|---|---|---|---|---|
| No. of patients (n = 17) | 6 | 11 | 5 | 6 | |
| Age (years)a | 8 (4–14) | 7 (2–17) | 9 (3–17) | 7 (2–16) | |
| CD4 T cells (/µl)b | Baseline | 225 (156–425) | 302 (93–480) | 256 (181–540) | 306 (48–503) |
| 24 Weeks | 562 (446–1218) | 495 (454–698) | 486 (399–751) | 509 (462–738) | |
| CD4 cell (%)b | Baseline | 12 (8–32) | 18 (8–22) | 18 (15–24) | 13 (8–22) |
| 24 Weeks | 28 (24–36) | 24 (20–28) | 27 (25–30) | 22 (16–24) | |
| Plasma viral RNA (log10 copies/ml)b | Baseline | 5.0 (4.0–5.4) | 5.1 (4.2–5.7) | 4.3 (4.0–4.7) | 5.5 (5.1–5.9) |
| 24 Weeks | 2.6 (2.6–2.6) | 4.5 (3.7–5.3) | 3.6 (3.4–4.4) | 4.9 (4.5–5.4) | |
| CD4:CD8 ratiob | Baseline | 0.20 (0.12–1.30) | 0.41 (0.24–0.66) | 0.41 (0.37–0.56) | 0.35 (0.11–0.71) |
| 24 Weeks | 0.64 (0.44–2.12) | 0.89 (0.42–1.06) | 1.00 (0.79–1.12) | 0.54 (0.39–0.90) |
Median value (range).
Median value (25th–75th interquartile range).
Response to therapy
Response following 24 weeks of therapy was determined for each individual based on viral and immune parameters. Immune success was defined as increased CD4 T-cell counts (absolute number and/or percentage) by 24 weeks of therapy as previously described [19,21–24]. Viral success was defined by a decline in plasma viral RNA of more than 1.5 log10 copies/ml during the first 4 weeks of therapy, with sustained suppression at undetectable levels for at least 16 weeks [19]. Detectable viral load following 8 weeks of therapy was defined as viral failure. Viruses unsuppressed by therapy were classified on the basis of replicative fitness in vivo as high fit viruses (viral load decreased <1 log10 copies/ml at 4 weeks and was ≥4 log10 copies/ml) at 24 weeks of therapy) or low fit viruses that failed to meet these criteria [24]. Eleven (22%) individuals who developed immune reconstitution, despite incomplete suppression of viral replication, were compared with six individuals who developed similar CD4 T-cell reconstitution but optimally suppressed viral replication.
Amplification, cloning, and sequencing
Plasma and peripheral blood mononuclear cells collected at baseline and during combination ART were used to evaluate viral gag, protease, and envelope clonal genotypes. Pretherapy reverse transcriptase genotypes are not predictive of outcome [19] and were not examined in this study. Briefly, the gag–protease region (HIVHXB2 nucleotides 1817–2611) was amplified using first round primers Gag 7 and Pol 4 and second round primers G100 and Pol I [25]. The env gp120 region (HIVHXB2 nucleotides 6516–7332) was amplified using first round primers D1 and 194G and second round primers Env5 and 195C [26]. All reactions used primers and reagents obtained from Invitrogen (Carlsbad, California, USA) and conditions described previously [8,25,26]. Following preparation of amplicon libraries, sequences were generated with DYEnamic ET dye terminator cycle sequencing kit and analyzed on a MegaBACE 1000 (GE Healthcare, Chalfont St. Giles, UK) in the Genome Sequencing Core at the University of Florida. Approximately, 1500 sequences were edited, verified, and submitted to GenBank (accession numbers GQ145598 – GQ146432).
Genetic analysis
Net positive charge of Env V3 loops (HIVHXB2 amino acid residues 7110–7217) was calculated for every sequence [13]. Each individual was characterized as having all low charge (≤+4), all high charge (≥+5), or a mixture of low and high charge viruses. Pretherapy amino acid residues in Gag (p2 through p6), p6Pol, and protease were classified as polymorphic based on differences from HIVHXB2. Pretherapy protease polymorphisms were categorized as therapy specific or nonspecific, if the amino acid was or was not associated with reduced sensitivity to the administered protease inhibitor [27]. Posttherapy genotypes were compared with baseline within each individual to assess accumulation of mutations.
The best fitting nucleotide substitution model was tested with a hierarchical likelihood ratio test [28] using a neighbor-joining tree with LogDet corrected distances and selected to infer maximum likelihood trees and maximum likelihood-estimated substitution parameters. The heuristic search for the best tree started with a neighbor-joining tree using the tree bisection–reconnection (TBR) branch-swapping algorithm. Calculations were performed with PAUP* 4.0b10 (D. L. Swofford, Sinauer Associates, Sunderland, Massachusetts, USA). Neighbor-joining trees were also inferred using maximum likelihood-estimated distances and the best-fitting nucleotide substitution model for each data set. Statistical support for internal branches in the neighbor-joining tree was obtained by bootstrapping (1000 replicates) and with the maximum likelihood-based zero branch length test for the maximum likelihood tree.
Statistical analysis
Clinical variables among therapy outcome groups were summarized as median and interquartile range. Mutual information (M), calculated for all possible pairwise combinations of variable positions in protease and Gag, was used as a measure of covariation [29]. Because of the sample size, an exact P value, instead of a permutation test, was calculated. Pairs of positions achieving a P value of less than 3 × 10−5 were considered significantly covariant [30].
Comparisons between protease and Gag genotypes with therapy outcome were performed using Fisher’s exact test. Comparisons of CD4 cell percentage and V3 charge between viral failure and viral success responses were performed using Wilcoxon rank sum test. Statistical significance was set at P value of less than 0.05. To develop a model with increased predictive value, multivariate analysis included baseline viral load and CD4 cell percentage, pretherapy genotype for protease, the p2/p7NC cleavage site, p7NC, p6, p6Pol, and envelope V3 charge. Univariate ordinal regression models were used to examine the relationship between each variable and therapy outcome. A receiver operating characteristic (ROC) analysis was performed to determine whether combinations of two or three variables could predict therapy outcome, as described previously [19]. All statistical analyses were performed using Statistical Analysis Software (SAS) software (SAS Institute, Cary, North Carolina, USA).
Results
Immune and viral responses to therapy
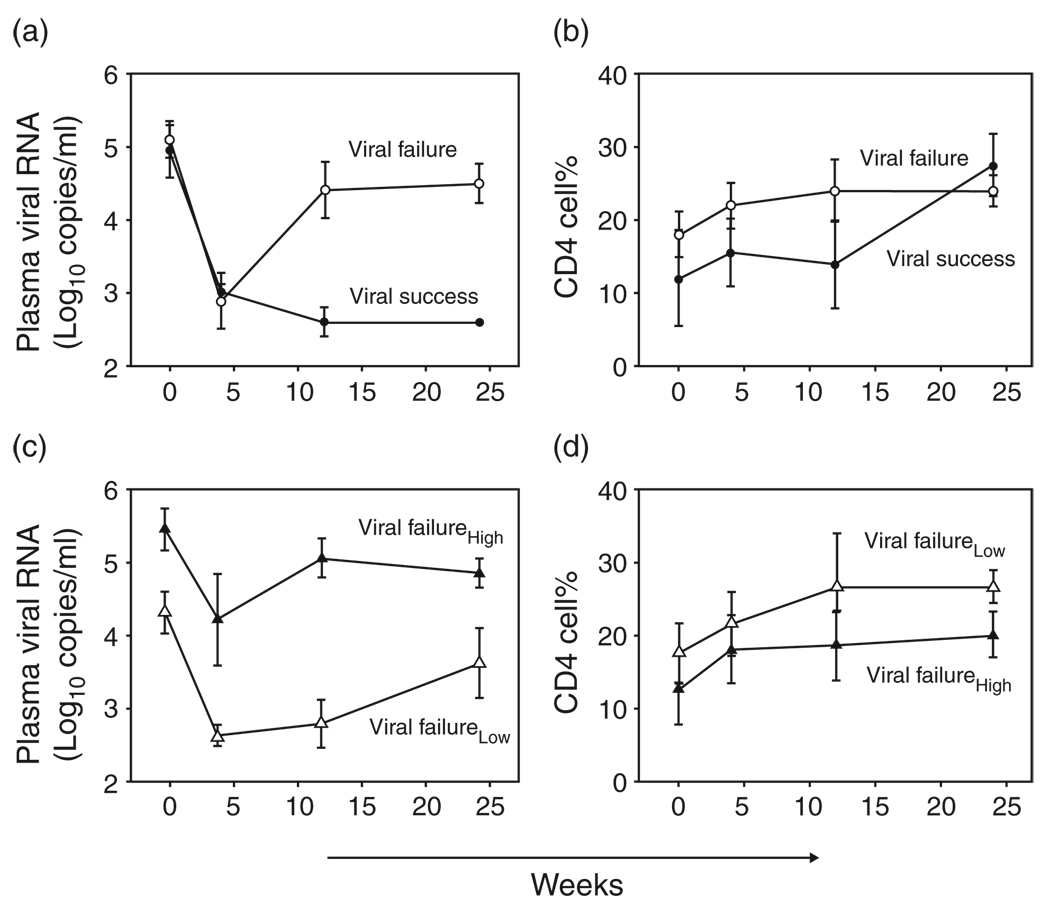
All individuals displayed similar phase I decline in viremia and CD4 cell percentage rebound at 4 weeks of treatment (Fig. 1a and b, respectively). Over 24 weeks of therapy, plasma viral levels of less than 400 log10 copies/ml persisted among viral success individuals (Fig. 1a) but rebounded rapidly by more than 1.4 log10 copies/ml among viral failure individuals. Independent of viral replication, CD4 cell percentage increased in each individual. CD4 cell rebound was sustained over 24 weeks (Fig. 1b), and as long as 96 weeks [19].
Fig. 1. Viral and immune responses over 24 weeks of combination antiretroviral therapy.
(a) Viral response to therapy for viral failures (open circles) and viral successes (filled circles). Median plasma viral RNA (log10 copies/ml) is shown on the y-axis. Weeks on therapy are shown on the x-axis. (b) Immune response to therapy for viral failures (open circles) and viral success (filled circles). Median CD4 cell percentage is shown on the y-axis. Weeks on therapy are shown on the x-axis. (c) Viral response to therapy for viral failure patients with low fit viruses (open triangles) and high fit viruses (filled triangles). Median plasma viral RNA (log10 copies/ml) is shown on the y-axis. Weeks on therapy are shown on the x-axis. (d) Immune response to therapy for viral failure patients with low fit viruses (open triangles) and high fit viruses (filled triangles). Median CD4 cell percentage is shown on the y-axis. Weeks on therapy are shown on the x-axis. Error bars represent standard error of the mean.
Failure to suppress virus replication resulted in two levels of replicative fitness. Viruses that rebounded to pre-therapy levels (high fit) emerged in one group of individuals, whereas viruses with diminished replicative capacity (low fit) developed in a second group [24]. High fit posttherapy viruses had less initial decline and greater rebound compared with low fit viruses (Fig. 1c). Independent of posttherapy viral set point, CD4 T cells increased (Fig. 1d).
Envelope V3 characteristics
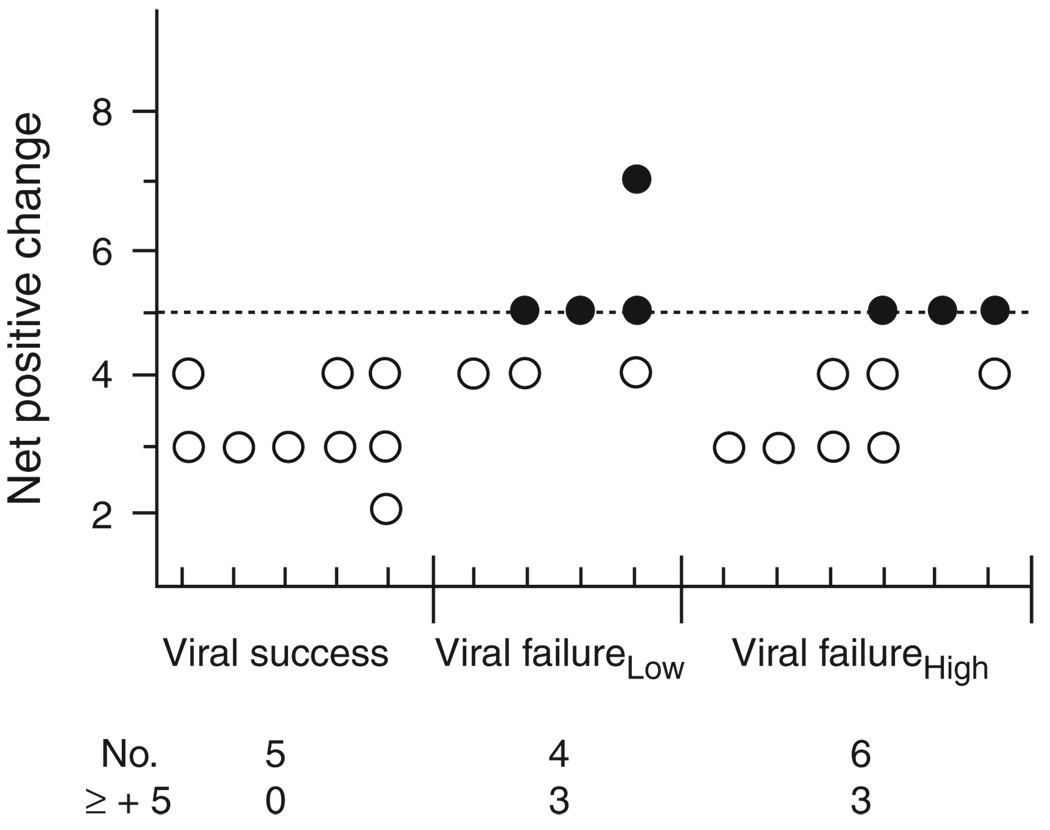
CCR5 or CXCR4 coreceptor use is determined primarily, although not exclusively, by amino acid characteristics of envelope V3 domains. V3 net charge less than 5 predicts CCR5 coreceptor use, whereas net charge at least 5 is associated with use of CXCR4 [13, 31 – 33]. To assess the relationship between pretherapy envelope genotype and viral outcome, V3 charge for 15 individuals was calculated (Fig. 2). Viral success response was related to pretherapy V3 charge less than 5 (5/5) and predicted CCR5 coreceptor use, whereas baseline X4-using envelopes, alone or in combination with R5 envelopes, were associated significantly with failure to suppress viral replication, independent of viral fitness (P = 0.04). After 24 weeks of treatment, baseline coreceptor use persisted among all individuals (data not shown).
Fig. 2. Envelope V3 charge characteristics prior to therapy.
Net V3 charge of viruses present before therapy is shown on the y-axis, with response groups shown on the x-axis. Each tick mark on the x-axis summarizes V3 charge for one patient. Open circles represent low charge sequences and filled circles represent high charge sequences. The total number of patients and the number of patients with high charge (≥+5) viruses are noted below each response group.
Divergence of quasispecies and protease inhibitor-associated changes in protease following 24 weeks of antiretroviral therapy
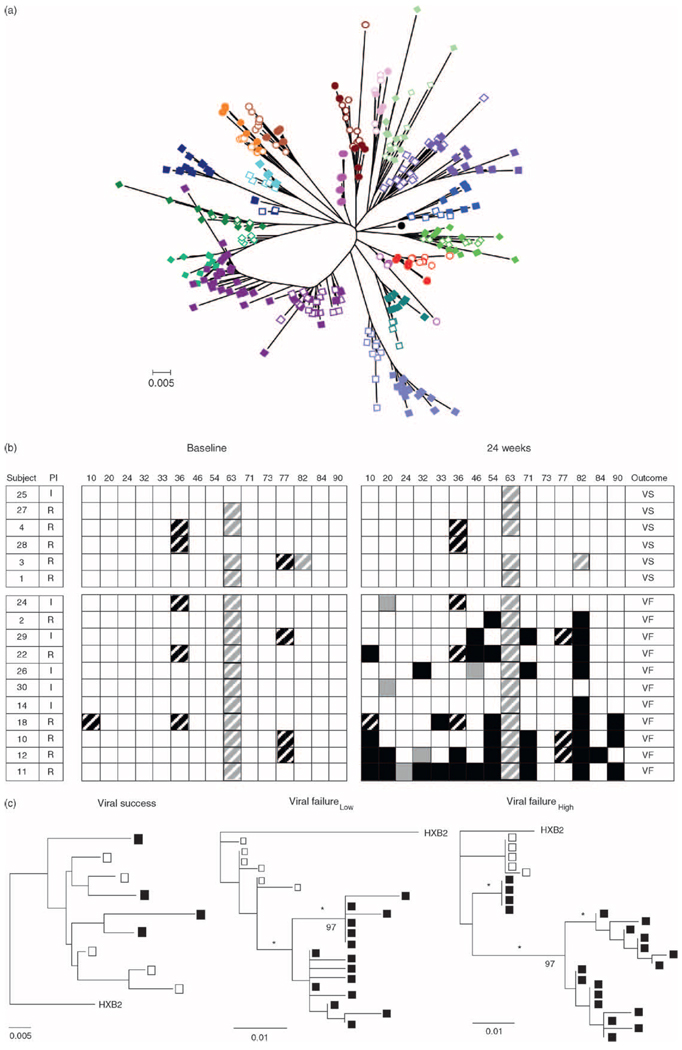
To assess the genetic relationship in protease quasispecies between baseline and 24 weeks of therapy, a phylogenetic tree was constructed from protease nucleotide sequences from all individuals (Fig. 3a). Baseline and 24-week sequences from each individual formed distinct monophyletic branches, thus confirming the integrity of the data set. To investigate the implications of nonsynonymous changes in protease, amino acid polymorphisms among quasispecies in each individual were evaluated. Although a nonspecific V82I polymorphism appeared in one individual, no primary protease resistance mutations were identified prior to therapy (Fig. 3b). In general, baseline polymorphisms were restricted to amino acid positions 10, 36, 63, or 77; polymorphisms at positions 36 and 77 were mutually exclusive. Pretherapy protease genotype defined by amino acid positions associated with reduced sensitivity to protease inhibitor provided no basis to distinguish therapy response in this cohort.
Fig. 3.
(a) Phylogenetic relationship of approximately 500 protease sequences among the cohort of patients. HIVHXB2 (●) was used as the outlier sequence. Each color represents one individual. Open symbols represent pretherapy sequences and closed symbols represent sequences after 24 weeks of therapy. Each branch represents an individual patient. ‘0.005’ indicates genetic distance (substitutions/site). (b) Protease genotypes at baseline and after 24 weeks of combination therapy. Patients are designated by numbers. Black box, new therapy-specific mutation; black-striped box, therapy-specific polymorphism; gray box, new therapy nonspecific mutation; gray-striped box, therapy nonspecific polymorphism; I, indinavir; R, ritonavir; white box, no polymorphism. In response to therapy, all patients were immune successes, and either viral success (VS) or viral failure (VF) with low fit (patients 24, 2, 29, 22, and 26) or high fit viruses (patients 30, 14, 18, 10, 12, and 11). (c) Representative phylogenetic relationship of protease sequences in a viral success subject (left panel), a viral failure low fit patient (center panel), and a viral failure high fit patient (right panel). Open symbols represent pretherapy sequences and closed symbols represent sequences after 24 weeks of therapy. HIVHXB2 was used as the outlier sequence. ‘0.005’ and ‘0.01’ indicate genetic distances. Numbers on the trees represent bootstrap values, and * indicates significant branch lengths.
Following 24 weeks of therapy, baseline protease genotype persisted among viruses in the viral success group, whereas viruses among viral failure individuals accumulated as many as 10 therapy-specific mutations, predicting high level protease inhibitor resistance [27]. Viruses within two individuals (24 and 30) failed to accumulate drug-related changes in protease. High fit viruses developed more new mutations than low fit viruses (median 5 and 3, respectively). Although therapy-related substitutions were dispersed throughout protease independent of replicative fitness, one therapy-specific mutation at position 90 developed in 50% of individuals with high fit viruses, but in none with low fit viruses, providing some basis for distinction between posttherapy levels of virus replication.
Relationship between pretherapy and posttherapy quasispecies was evaluated by construction of phylogenetic trees. Protease sequences from the majority of viral success individuals formed nonstructured trees, reflecting persistence of infected cells, but essentially no evolution in protease (Fig. 3c, left panel). In contrast, protease sequences from viral failure individuals formed structured trees with temporal order. Monophyletic clusters of 24-week sequences indicating a genetic bottleneck developed following rebound by either low or high fit viruses (Fig. 3c, center and right panels, respectively). Trees constructed from Gag sequences had structure similar to their respective protease trees (data not shown).
Cleavage site diversity
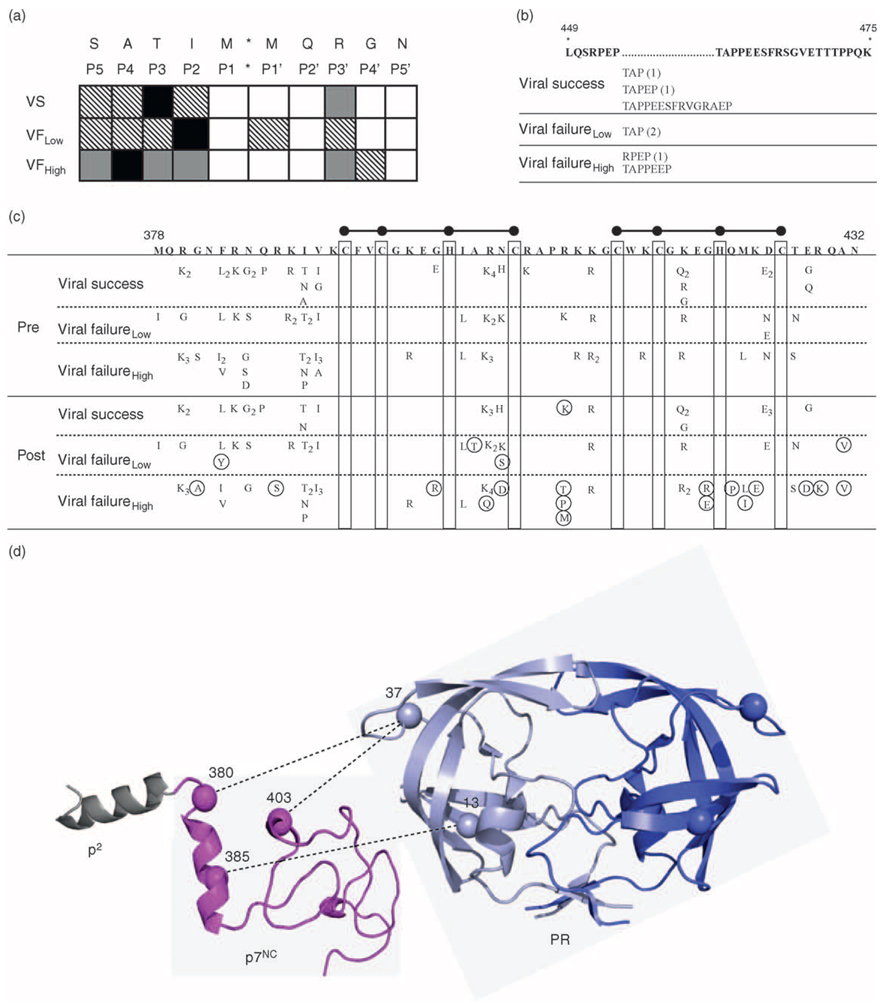
Protease cleavage sites in Gag can display natural polymorphisms, which can be related to therapy response [6,25,34,35]. In our cohort, all cleavage sites between p17MA and protease, with one exception, were virtually identical among individuals, indistinguishable from HIVHXB2, and highly conserved following therapy (data not shown). The p2/p7NC cleavage site was the most heterogeneous prior to therapy, with viruses from every individual showing amino acid differences from HIVHXB2 (Fig. 4a) [25]. Multiple amino acid positions in p2/p7NC cleavage sites were related to viral suppression or levels of posttherapy viral fitness. For example, position P2 was more polymorphic among viruses that continued to replicate (55% or 6/11 viral failure response) than viruses that were subsequently suppressed (17% or 1/6 viral success individuals) by ART. Position P4 was polymorphic in only 20% (1/5) of individuals who developed low fit viruses but 67% (4/6) of individuals who developed high fit viruses. The p2/p7NC cleavage sites remained highly polymorphic with no additional changes accumulating during 24 weeks of therapy (data not shown).
Fig. 4.
(a) Diversity in p2/p7NC cleavage site. Patients are stratified by therapy response: viral success, viral failure with low fit viruses, or viral failure with high fit viruses. Amino acid sequence of HIVHXB2 cleavage site is shown on top line with cleavage site position number shown below each respective amino acid. Black box, polymorphism present in more than 50% of patients; gray box, polymorphism present in 26–50% of patients; hatched box, polymorphism present in 1–25% of patients; white box, no polymorphism. (b) Length variation in p6. Amino acid sequence and position numbers of p6 (HIVHXB2) are shown on top, with regions containing length variation marked with (…). The amino acid compositions of insertions within each response group (number of patients) are shown below each region of length variation. (c) Variability in p7NC. Amino acid sequence and position numbers of p7NC (HIVHXB2) are shown on top. Ball and sticks represent the zinc finger motifs. Polymorphisms present prior to therapy are shown at the top, and polymorphisms after 24 weeks of therapy are shown on the bottom, with subscripts representing the number of patients with a particular polymorphism. Mutations that accumulate in p7NC following 24 weeks of therapy are circled. (d) Model of p2, p7NC, and protease, with specific covarying positions in protease and p7NC noted. Amino acid residues in p7NC are numbered according to HIVHXB2, and amino acid residues in protease are numbered according to position within protease. Dashed lines indicate pretherapy covariation between positions in protease and positions in p7NC.
Gag diversity
Diversity in p6 and the overlapping p6Pol reading frame involved length variation, as well as amino acid polymorphisms. Prior to therapy, length variation ranging from three to 15 amino acids appeared within the p6 Pro–Thr–Ala–Pro (PTAP) domains in nearly half (7/17, 41%) of the cohort, independent of outcome (Fig. 4b). The median number of polymorphisms in baseline p6 was unrelated to viral success or viral failure outcomes (11) but appeared related to posttherapy fitness (high fit, 14; low fit, nine). In contrast, the median number of polymorphisms in p6Pol appeared related to both viral outcome (viral success, 14; viral failure, 11) and posttherapy fitness (high fit, 14; low fit, 10). Protease inhibitor therapy failed to change amino acid length or significantly increase the number of mutations in p6 or p6Pol among response groups (data not shown).
In baseline p7NC, the cysteine and histidine residues of the zinc finger motifs were conserved, but almost half of the remaining amino acid positions, including three positions (384, 398, and 425) at which substitutions can alter protease processing function, were polymorphic (Fig. 4c) [6,35]. Although neither the number nor position of pretherapy polymorphisms provided a basis to distinguish subsequent suppression of virus, posttherapy mutations were associated with level of viral replication, as low fit or high fit viruses accumulated mutations at four or 13 position, respectively. Overall, the number of mutations in both protease and p7NC increased with continued viral replication during therapy.
Covariation between protease and Gag
Covariation, defined as the occurrence of two mutations together at a frequency greater than predicted by chance, can be identified within protease or between protease and Gag cleavage sites without or with therapy [29,34,36–39]. We performed a similar study, but importantly, extended the covariation analysis to assess amino acid residues within protease and regions of Gag outside the cleavage sites. Prior to therapy, position 37 in the hinge region of protease covaried with p7NC residues 380 (P < 0.00001) and 403 (P < 0.00001), whereas protease residue 13 covaried with p7NC residue 385 (P = 0.00001) (Fig. 4d). Following therapy, several drug-associated mutations in protease changed covariation with Gag. For example, protease position 82 and p7NC amino acid 411 covaried significantly after, but not before, therapy (P < 0.00001).
Variables to predict therapy response
To evaluate baseline variables relative to therapy outcome (viral success vs. viral failure), one-variable, two-variable, and three-variable models were constructed. Protease genotype was one of the least sensitive single predictors in this cohort [area under the curve (AUC), 0.54; 95% confidence interval (CI), 0.50–0.84], whereas V3 charge was the best single variable to predict viral success for 65% of individuals (AUC, 0.80; 95% CI, 0.64–0.96). Combining V3 charge with one additional variable only marginally increased predictive value (AUC range 0.78–0.90; prediction rate range 58–71%). In contrast, V3 charge combined with p2/p7NC cleavage site and p6Pol genotype produced a three-variable model (AUC, 1.00; 95% CI, 1.00–1.00) that predicted viral outcome for 88% of individuals.
Outcomes related to posttherapy viral fitness (high vs. low fit) were evaluated as well. Models that combined V3 charge with two additional genetic variables in Gag predicted a high fit viral response among 91% of individuals (AUC, 1.00; 95% CI, 1.00–1.00). However, the best three-variable models, which predicted post-therapy viral fitness among 100% of individuals, included CD4 cell percentage with either protease and p2/p7NC cleavage site genotypes or p7NC and p6 genotypes (AUC, 1.00; 95% CI, 1.00–1.00). These analyses indicate that combinations of baseline Gag and envelope genotypes, in addition to clinical variables, are predictors of response to protease inhibitor therapies.
Discussion
Failure to achieve or maintain viral suppression following ART is not uncommon, although many individuals develop immune reconstitution in spite of viral rebound [19,40–42]. Improving the likelihood of achieving optimal viral suppression to therapy requires expanding the repertoire of predictors of viral response to therapy. Protease genotype has value for selecting switch therapy and is related in part to response to first treatment [19]. Additional regions of the viral genome reflecting interactions between enzymatic function of protease and substrates in Gag and the host cell milieu in which inhibitors function also provide information. The current study included a cohort of therapy-naive children whose first ART uniformly achieved immune reconstitution based on increased numbers of CD4 T cells and improved immune functions, including enhanced thymic output [23,24,43]. The selective pressure of protease inhibitor provides an ideal opportunity to examine HIV-1 evolution because multiple changes are required for development of resistance, and these changes persist over time. Even though the cohort was small, similarity in pretherapy protease genotypes provided increased sensitivity to detect novel determinants outside protease that predicted protease inhibitor therapy outcome and viral fitness. Although the individuals in the cohort were infected by subtype B viruses, naturally occurring polymorphisms appear in Gag and protease regions of non-B HIV-1 subtypes [44–46], suggesting implications for our results for other subtypes of HIV-1. Given that protease has a similar mechanism of action and substrate affinities in different subtypes, it is likely that Gag biomarkers may map to the same regions across subtypes [47]. Overall, a relationship between determinants in Gag, whether cleavage site dependent or independent, and therapy outcome provides a rationale to include extended regions of Gag in evaluations of protease genotype in larger studies.
A recent study [24] from our group identified differential fitness of viruses in vivo among individuals whose treatment failed to suppress virus replication. Differences in Gag and protease genotypes between low fit and high fit viruses raise the question of whether a high fit response is determined by time or a separate resistance pathway. For example, over time, low fit viruses could accumulate a methionine at protease amino acid 90 that was present in 50% of high fit viruses, but none of the low fit viruses, and develop increased fitness. Analysis of sequences at later time points in therapy should provide an answer. Sustained immune reconstitution with persistent high replication in the presence of protease inhibitor could indicate modulation of other functions of the virus related to pathogenesis in vivo. Regardless of pathways to fitness, fewer protease mutations in the low fit group compared with the high fit group bring up the issue of whether amino acid changes function to reduce drug susceptibility or to alter fitness [8].
Amino acid substitutions in Gag accumulate in response to therapy and persist over time, similar to characteristics of drug-related mutations in protease. New insights into the importance of Gag were provided in the covariation analysis. Covariation between amino acid residues could indicate functional interactions. For example, specific amino acids in p7NC and residues in protease covaried prior to therapy, potentially reflecting interactions that optimize processing at the p2/p7NC cleavage site. Covariation between protease and p6/p6Pol (data not shown) suggests that interactions between protease and targets in Gag occur in both cis and trans. It is unlikely that the functional consequences of all covariations between Gag and protease occur simultaneously but more likely occur sequentially during the ordered, stepwise processing of Gag by protease. Importantly, unique covariation developed during therapy, indicating that changes in protease as a result of therapy are strongly associated with changes in Gag.
An association of pretherapy Gag to therapy outcome by multivariate analysis likely has mechanistic explanations. p7NC serves multiple functions in virus replication, including RNA binding and virion assembly. Gag–protease is also a functional domain, and variants of p7NC with limited numbers of amino acid polymorphisms exert a dominant effect on protease processing activity, drug sensitivity, and replicative fitness [6,35]. In addition, two individuals who failed to suppress viral replication without developing drug-resistant protease genotypes did develop posttherapy substitutions in p6/p6Pol, suggesting a functional role for p6/p6Pol in fitness [8].
The majority of viral failure individuals in our cohort had X4 viruses, frequently in combination with R5 viruses that displayed an ability to infect lymphocytes and macrophages (unpublished data). Although genetic linkage between Env and Gag–protease regions was not specifically examined, the prevalence of virus populations with high charge V3 or drug-resistant protease regions suggests that drug resistance developed in X4 viruses. X4 viruses increase genetic diversity of the quasispecies and may exhibit different drug sensitivity than R5 viruses (Ho et al., manuscript in preparation), implicating the complexity of viral-host cell interactions and effectiveness of antiviral drugs in vivo. Furthermore, data from our lab implicate other regions of envelope, such as V1 and V2, in predicting therapy response (Yin et al., manuscript in preparation).
A comprehensive analysis of genetic determinants in regions of Gag and envelope, in combination with protease, revealed that viral failure with immune reconstitution is a complex phenotype that involves the interaction of viral and host factors. Results from this study demonstrate that in vivo, Gag genotype and V3 charge are clinically relevant biomarkers for therapy outcome.
Acknowledgements
The authors are grateful to Dr Marco Salemi for help with the phylogenetic analysis, Dr Brant Burkhart and Dr Dan Tuttle for assistance with the envelope data, and Steven Pomeroy for contributions to data collection and analysis. Research was supported in part by PHS R01 awards HD032259, AI065265, AI028571, and AI047723; Center for Research for Pediatric Immune Deficiency; Laura McClamma Fellowship, Graduate Alumni Fellowship (S.K.H.), and Stephany W. Holloway University Chair for AIDS Research (M.M.G.); Pediatric Clinical Research Center of All Children’s Hospital and the University of South Florida, and the Maternal Child Health Bureau, R60 MC 00003-01, Department of Health and Human Services, Resources and Services Administration.
Footnotes
E.E.P., S.L.R., J.W.S., and M.M.G. were responsible for the study conception and design; S.K.H., E.E.P., S.L.R., and A.C.L. were responsible for data collection, management, and analysis; W.H. and C.M. performed statistical analyses; J.W.S. and R.M.L. managed the clinical cohort; and S.K.H., R.M.C., B.M.D., J.W.S., and M.M.G. participated in writing the manuscript.
There are no conflicts of interest.
References
- 1.Quinones-Mateu ME, Arts EJ. HIV-1 fitness: implications for drug resistance, disease progression, and global epidemic evolution. In: Kuiken C, Foley B, Hahn B, Marx P, McCutchan F, Mellors JW, et al., editors. HIV Sequence Compendium 2001. Los Alamos, New Mexico, USA: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 2001. pp. 134–170. [Google Scholar]
- 2.Martinez-Picado J, Prado JG, Fry EE, Pfafferott K, Leslie A, Chetty S, et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doyon L, Croteau G, Thibeault D, Poulin F, Pilote L, Lamarre D. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J Virol. 1996;70:3763–3769. doi: 10.1128/jvi.70.6.3763-3769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mammano F, Petit C, Clavel F. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J Virol. 1998;72:7632–7637. doi: 10.1128/jvi.72.9.7632-7637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang YM, Imamichi H, Imamichi T, Lane HC, Falloon J, Vasudevachari MB, et al. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J Virol. 1997;71:6662–6670. doi: 10.1128/jvi.71.9.6662-6670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodenow MM, Bloom G, Rose SL, Pomeroy SM, O’Brien PO, Perez EE, et al. Naturally occurring amino acid polymorphisms in human immunodeficiency virus type 1 (HIV-1) Gag p7(NC) and the C-cleavage site impact Gag-Pol processing by HIV-1 protease. Virology. 2002;292:137–149. doi: 10.1006/viro.2001.1184. [DOI] [PubMed] [Google Scholar]
- 7.Whitehurst N, Chappey C, Petropoulos C, Parkin N, Gamarnik A. Polymorphisms in p1-p6/p6* of HIV type 1 can delay protease autoprocessing and increase drug susceptibility. AIDS Res Hum Retroviruses. 2003;19:779–784. doi: 10.1089/088922203769232575. [DOI] [PubMed] [Google Scholar]
- 8.Ho SK, Coman RM, Bunger JC, Rose SL, O’Brien P, Munoz I, et al. Drug-associated changes in amino acid residues in Gag p2, p7(NC), and p6(Gag)/p6(Pol) in human immunodeficiency virus type 1 (HIV-1) display a dominant effect on replicative fitness and drug response. Virology. 2008;378:272–281. doi: 10.1016/j.virol.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nijhuis M, van Maarseveen NM, Lastere S, Schipper P, Coakley E, Glass B, et al. A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism. PLoS Med. 2007;4:e36. doi: 10.1371/journal.pmed.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maguire MF, Guinea R, Griffin P, Macmanus S, Elston RC, Wolfram J, et al. Changes in human immunodeficiency virus type 1 Gag at positions L449 and P453 are linked to I50V protease mutants in vivo and cause reduction of sensitivity to amprenavir and improved viral fitness in vitro. J Virol. 2002;76:7398–7406. doi: 10.1128/JVI.76.15.7398-7406.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aquaro S, Perno CF, Balestra E, Balzarini J, Cenci A, Francesconi M, et al. Inhibition of replication of HIV in primary monocyte/macrophages by different antiviral drugs and comparative efficacy in lymphocytes. J Leukoc Biol. 1997;62:138–143. doi: 10.1002/jlb.62.1.138. [DOI] [PubMed] [Google Scholar]
- 12.Aquaro S, Calio R, Balzarini J, Bellocchi MC, Garaci E, Perno CF. Macrophages and HIV infection: therapeutical approaches toward this strategic virus reservoir. Antivir Res. 2002;55:209–225. doi: 10.1016/s0166-3542(02)00052-9. [DOI] [PubMed] [Google Scholar]
- 13.Briggs DR, Tuttle DL, Sleasman JW, Goodenow MM. Envelope V3 amino acid sequence predicts HIV-1 phenotype (co-receptor usage and tropism for macrophages) AIDS. 2000;14:2937–2939. doi: 10.1097/00002030-200012220-00016. [DOI] [PubMed] [Google Scholar]
- 14.Hwang SS, Boyle TJ, Lyerly HK, Cullen BR. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 15.Goodenow MM, Collman RG. HIV-1 coreceptor preference is distinct from target cell tropism: a dual-parameter nomenclature to define viral phenotypes. J Leukoc Biol. 2006;80:965–972. doi: 10.1189/jlb.0306148. [DOI] [PubMed] [Google Scholar]
- 16.Brumme ZL, Dong WW, Yip B, Wynhoven B, Hoffman NG, Swanstrom R, et al. Clinical and immunological impact of HIV envelope V3 sequence variation after starting initial triple antiretroviral therapy. AIDS. 2004;18:F1–F9. doi: 10.1097/00002030-200403050-00001. [DOI] [PubMed] [Google Scholar]
- 17.Galan I, Jimenez JL, Gonzalez-Rivera M, De Jose MI, Navarro ML, Ramos JT, et al. Virological phenotype switches under salvage therapy with lopinavir-ritonavir in heavily pre-treated HIV-1 vertically infected children. AIDS. 2004;18:247–255. doi: 10.1097/00002030-200401230-00014. [DOI] [PubMed] [Google Scholar]
- 18.Skrabal K, Trouplin V, Labrosse B, Obry V, Damond F, Hance AJ, et al. Impact of antiretroviral treatment on the tropism of HIV-1 plasma virus populations. AIDS. 2003;17:809–814. doi: 10.1097/00002030-200304110-00005. [DOI] [PubMed] [Google Scholar]
- 19.Perez EE, Rose SL, Peyser B, Lamers SL, Burkhardt B, Dunn BM, et al. Human immunodeficiency virus type 1 protease genotype predicts immune and viral responses to combination therapy with protease inhibitors (PIs) in PI-naive patients. J Infect Dis. 2001;183:579–588. doi: 10.1086/318538. [DOI] [PubMed] [Google Scholar]
- 20.Dybul M, Fauci AS, Bartlett JG, Kaplan JE, Pau AK. Guidelines for using antiretroviral agents among HIV-infected adults and adolescents. Recommendations of the Panel on Clinical Practices for Treatment of HIV. MMWR Recomm Rep. 2002;51(RR-7):1–55. [PubMed] [Google Scholar]
- 21.Mueller BU, Sleasman J, Nelson RP, Jr, Smith S, Deutsch PJ, Ju W, et al. A phase I/II study of the protease inhibitor indinavir in children with HIV infection. Pediatrics. 1998;102(1 Pt 1):101–109. doi: 10.1542/peds.102.1.101. [DOI] [PubMed] [Google Scholar]
- 22.Mueller BU, Nelson RP, Jr, Sleasman J, Zuckerman J, Heath-Chiozzi M, Steinberg SM, et al. A phase I/II study of the protease inhibitor ritonavir in children with human immunodeficiency virus infection. Pediatrics. 1998;101(3 Pt 1):335–343. doi: 10.1542/peds.101.3.335. [DOI] [PubMed] [Google Scholar]
- 23.Ghaffari G, Passalacqua DJ, Caicedo JL, Goodenow MM, Sleasman JW. Two-year clinical and immune outcomes in human immunodeficiency virus-infected children who reconstitute CD4 T cells without control of viral replication after combination antiretroviral therapy. Pediatrics. 2004;114:e604–e611. doi: 10.1542/peds.2004-0274. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez CA, Koch S, Goodenow M, Sleasman JW. Clinical implications of discordant viral and immune outcomes following protease inhibitor containing antiretroviral therapy for HIV-infected children. Immunol Res. 2008;40:271–286. doi: 10.1007/s12026-007-0031-1. [DOI] [PubMed] [Google Scholar]
- 25.Barrie KA, Perez EE, Lamers SL, Farmerie WG, Dunn BM, Sleasman JW, et al. Natural variation in HIV-1 protease, Gag p7 and p6, and protease cleavage sites within gag/pol polyproteins: amino acid substitutions in the absence of protease inhibitors in mothers and children infected by human immunodeficiency virus type 1. Virology. 1996;219:407–416. doi: 10.1006/viro.1996.0266. [DOI] [PubMed] [Google Scholar]
- 26.Tuttle DL, Anders CB, Aquino-De Jesus MJ, Poole PP, Lamers SL, Briggs DR, et al. Increased replication of nonsyncytium-inducing HIV type 1 isolates in monocyte-derived macrophages is linked to advanced disease in infected children. AIDS Res Hum Retroviruses. 2002;18:353–362. doi: 10.1089/088922202753519133. [DOI] [PubMed] [Google Scholar]
- 27.Johnson VA, Brun-Vezinet F, Clotet B, Gunthard HF, Kuritzkes DR, Pillay D, et al. Update of the Drug Resistance Mutations in HIV-1: Spring 2008. Top HIV Med. 2008;16:62–68. doi: 10.1007/s11750-007-0034-z. [DOI] [PubMed] [Google Scholar]
- 28.Swofford L, Sullivan JL. The Phylogenetic Handbook: a practical approach to DNA and protein phylogeny. New York: Cambridge University Press; 2003. Phylogeny inference based on parsimony and other methods with PAUP*; pp. 160–206. [Google Scholar]
- 29.Hoffman NG, Schiffer CA, Swanstrom R. Covariation of amino acid positions in HIV-1 protease. Virology. 2003;314:536–548. doi: 10.1016/s0042-6822(03)00484-7. [DOI] [PubMed] [Google Scholar]
- 30.Bickel PJ, Cosman PC, Olshen RA, Spector PC, Rodrigo AG, Mullins JI. Covariability of V3 loop amino acids. AIDS Res Hum Retroviruses. 1996;12:1401–1411. doi: 10.1089/aid.1996.12.1401. [DOI] [PubMed] [Google Scholar]
- 31.Jensen MA, Li FS, ’t Wout AB, Nickle DC, Shriner D, He HX, et al. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J Virol. 2003;77:13376–13388. doi: 10.1128/JVI.77.24.13376-13388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milich L, Margolin B, Swanstrom R. V3 loop of the human immunodeficiency virus type 1 Env protein: interpreting sequence variability. J Virol. 1993;67:5623–5634. doi: 10.1128/jvi.67.9.5623-5634.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Resch W, Hoffman N, Swanstrom R. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology. 2001;288:51–62. doi: 10.1006/viro.2001.1087. [DOI] [PubMed] [Google Scholar]
- 34.Bally F, Martinez R, Peters S, Sudre P, Telenti A. Polymorphism of HIV type 1 gag p7/p1 and p1/p6 cleavage sites: clinical significance and implications for resistance to protease inhibitors. AIDS Res Hum Retroviruses. 2000;16:1209–1213. doi: 10.1089/08892220050116970. [DOI] [PubMed] [Google Scholar]
- 35.Bloom G, Perez E, Parikh S, Kay J, Mills J, Goodenow M, et al. A comparison of gag-pol precursor cleavage in naturally arising HIV variants. Adv Exp Med Biol. 1998;436:53–57. doi: 10.1007/978-1-4615-5373-1_7. [DOI] [PubMed] [Google Scholar]
- 36.Brown AJ, Korber BT, Condra JH. Associations between amino acids in the evolution of HIV type 1 protease sequences under indinavir therapy. AIDS Res Hum Retroviruses. 1999;15:247–253. doi: 10.1089/088922299311420. [DOI] [PubMed] [Google Scholar]
- 37.Koch N, Yahi N, Fantini J, Tamalet C. Mutations in HIV-1 gag cleavage sites and their association with protease mutations. AIDS. 2001;15:526–528. doi: 10.1097/00002030-200103090-00013. [DOI] [PubMed] [Google Scholar]
- 38.Wu TD, Schiffer CA, Gonzales MJ, Taylor J, Kantor R, Chou S, et al. Mutation patterns and structural correlates in human immunodeficiency virus type 1 protease following different protease inhibitor treatments. J Virol. 2003;77:4836–4847. doi: 10.1128/JVI.77.8.4836-4847.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yahi N, Tamalet C, Tourres C, Tivoli N, Ariasi F, Volot F, et al. Mutation patterns of the reverse transcriptase and protease genes in human immunodeficiency virus type 1-infected patients undergoing combination therapy: survey of 787 sequences. J Clin Microbiol. 1999;37:4099–4106. doi: 10.1128/jcm.37.12.4099-4106.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deeks SG, Barbour JD, Martin JN, Swanson MS, Grant RM. Sustained CD4+ T cell response after virologic failure of protease inhibitor-based regimens in patients with human immunodeficiency virus infection. J Infect Dis. 2000;181:946–953. doi: 10.1086/315334. [DOI] [PubMed] [Google Scholar]
- 41.Essajee SM, Kim M, Gonzalez C, Rigaud M, Kaul A, Chandwani S, et al. Immunologic and virologic responses to HAART in severely immunocompromised HIV-1-infected children. AIDS. 1999;13:2523–2532. doi: 10.1097/00002030-199912240-00005. [DOI] [PubMed] [Google Scholar]
- 42.Sleasman JW, Nelson RP, Goodenow MM, Wilfret D, Hutson A, Baseler M, et al. Immunoreconstitution after ritonavir therapy in children with human immunodeficiency virus infection involves multiple lymphocyte lineages. J Pediatr. 1999;134:597–606. doi: 10.1016/s0022-3476(99)70247-7. [DOI] [PubMed] [Google Scholar]
- 43.Yin L, Rodriguez CA, Hou W, Potter O, Caplan MJ, Goodenow MM, et al. Antiretroviral therapy corrects HIV-1-induced expansion of CD8+ CD45RA+ CD2− CD11a(bright) activated T cells. J Allergy Clin Immunol. 2008;122:166–172. doi: 10.1016/j.jaci.2008.04.029. 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abecasis AB, Deforche K, Bacheler LT, McKenna P, Carvalho AP, Gomes P, et al. Investigation of baseline susceptibility to protease inhibitors in HIV-1 subtypes C, F, G and CRF02_AG. Antivir Ther. 2006;11:581–589. [PubMed] [Google Scholar]
- 45.Holguin A, Paxinos E, Hertogs K, Womac C, Soriano V. Impact of frequent natural polymorphisms at the protease gene on the in vitro susceptibility to protease inhibitors in HIV-1 non-B subtypes. J Clin Virol. 2004;31:215–220. doi: 10.1016/j.jcv.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 46.Vergne L, Stuyver L, Van HM, Butel C, Delaporte E, Peeters M. Natural polymorphism in protease and reverse transcriptase genes and in vitro antiretroviral drug susceptibilities of non-B HIV-1 strains from treatment-naive patients. J Clin Virol. 2006;36:43–49. doi: 10.1016/j.jcv.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 47.Coman RM, Robbins AH, Fernandez MA, Gilliland CT, Sochet AA, Goodenow MM, et al. The contribution of naturally occurring polymorphisms in altering the biochemical and structural characteristics of HIV-1 subtype C protease. Biochemistry. 2008;47:731–743. doi: 10.1021/bi7018332. [DOI] [PubMed] [Google Scholar]