Abstract
Enterococcus faecalis is a gram-positive opportunistic pathogen known to form biofilms in vitro. In addition, this organism is often isolated from biofilms on the surfaces of various indwelling medical devices. However, the molecular mechanisms regulating biofilm formation in these clinical isolates are largely unknown. Recent work has suggested that a specific cell surface protein (Esp) of E. faecalis is critical for biofilm formation by this organism. However, in the same study, esp-deficient strains of E. faecalis were found to be capable of biofilm formation. To test the hypothesis that Esp is dispensable for biofilm formation by E. faecalis, we used microtiter plate assays and a chemostat-based biofilm fermentor assay to examine biofilm formation by genetically well-defined, non-Esp-expressing strains. Our results demonstrate that in vitro biofilm formation occurs, not only in the absence of esp, but also in the absence of the entire pathogenicity island that harbors the esp coding sequence. Using scanning electron microscopy to evaluate biofilms of E. faecalis OG1RF grown in the fermentor system, biofilm development was observed to progress through multiple stages, including attachment of individual cells to the substratum, microcolony formation, and maturation into complex multilayered structures apparently containing water channels. Microtiter plate biofilm analyses indicated that biofilm formation or maintenance was modulated by environmental conditions. Furthermore, our results demonstrate that expression of a secreted metalloprotease, GelE, enhances biofilm formation by E. faecalis. In summary, E. faecalis forms complex biofilms by a process that is sensitive to environmental conditions and does not require the Esp surface protein.
Surface-associated bacterial growth is ubiquitous, occurring in any nutrient-sufficient natural, industrial, or medical ecosystem (5). Despite the ubiquity of surface-associated growth, until relatively recently the genetics and physiology of bacteria inhabiting this unique ecological niche remained unknown. It has now become clear that surface-associated bacteria exist in complex microbial communities, known as biofilms, which are typically encased in a self-produced extracellular polymeric matrix (for reviews, see references 5 and 42). Osmolarity, nutrient concentration and composition, and other nascent environmental conditions influence the formation of surface-adherent biofilms by diverse bacterial species (22, 25, 29, 32, 35, 46). In a clinical setting, biofilms have been implicated as etiological agents of chronic infection (6, 11, 26). Biofilm bacteria are differentiated from their free-swimming (planktonic) counterparts, exhibiting biofilm-specific phenotypes such as enhanced tolerance to antibiotic treatment and increased levels of genetic exchange (reviewed in reference 42). Furthermore, many studies have documented biofilm-specific patterns of gene expression or protein production, leading to the proposal that biofilm formation by planktonic bacteria represents a form of microbial development (reviewed in references 28 and 42).
In support of this hypothesis, mutations that interfere with the function of cell density signaling systems lead to alterations of mature biofilm architecture in diverse bacterial species, suggesting that an orchestrated program of changes in gene expression occurs during the formation of mature biofilms. For example, the Pseudomonas aeruginosa lasI gene product catalyzes the production of an acylhomoserine lactone signaling molecule that is required for the expression of a number of virulence genes and which was also found to be required for development of typical mature biofilm architecture by P. aeruginosa, at least under certain environmental conditions (8). Studies of biofilm formation by the oral bacterium Streptococcus mutans yielded similar results: a mutant unable to produce the competence-stimulating peptide required to signal the development of natural genetic competence in streptococci was found to form biofilms with an altered architecture (24). Thus, cell density-dependent signaling systems appear to trigger changes in gene expression required for proper biofilm development by distantly related bacteria.
Enterococcus faecalis is a gram-positive commensal bacterium normally associated with humans as a member of the intestinal microflora (43). However, E. faecalis is also an opportunistic pathogen of increasing clinical significance, particularly as an etiological agent of nosocomial infections (36). The intrinsic antibiotic resistance of enterococci, coupled with their promiscuity in acquisition and dissemination of genetically mobile antibiotic resistance elements, presents serious challenges to the treatment of enterococcal infections.
Infection-derived isolates of E. faecalis have been shown to form biofilms in vitro (3, 38, 44). Furthermore, E. faecalis is often isolated from biofilms on the surfaces of various indwelling medical devices associated with chronic infection (10, 11). However, because these clinical isolates are usually not characterized at the genetic level, little is known about the genetic determinants or molecular mechanisms regulating biofilm formation by E. faecalis. E. faecalis biofilms grown in vitro have been observed to exhibit phenotypic properties similar to biofilms of other organisms. For example, using an in vitro perfused biofilm system, Foley and Gilbert established that E. faecalis biofilms exhibit enhanced tolerance to vancomycin and teicoplanin (16). The enhanced antibiotic tolerance resulting from growth in a biofilm state, coupled with the intrinsic antibiotic resistance of the organism as well as its propensity to serve as a reservoir for the dissemination of antibiotic resistance genes, suggests that biofilm formation of E. faecalis in a hospital setting may be particularly problematic.
Although the genetic basis of biofilm formation by E. faecalis is largely unknown, a recent study suggested that a specific cell surface protein (Esp) is critical for biofilm formation by this organism (44). Esp exhibits overall sequence similarity to Bap, a surface protein of Staphylococcus aureus previously reported to be involved in biofilm formation (7). Toledo-Arana et al. used an esp-specific PCR assay to determine if the esp gene was present in 152 clinical isolates of E. faecalis, which were evaluated for biofilm formation by use of a polystyrene microtiter plate biofilm assay (44). The authors reported a significant correlation between the presence of esp and the capacity to form biofilms in their microtiter assay, suggesting that Esp is involved in biofilm formation by this organism. However, in the same report, two esp-deficient mutants of E. faecalis were constructed and found to be fully capable of biofilm formation. Furthermore, Sandoe et al. recently evaluated 70 bloodstream isolates of E. faecalis in culture media distinct from those used by Toledo-Arana et al., using a microtiter plate assay, and found no association between the presence of esp and biofilm-forming ability (38). Therefore, we hypothesized that Esp is not critical for biofilm formation but that additional, unknown genetic determinants control biofilm formation by E. faecalis.
To test this hypothesis, we investigated biofilm formation by several genetically well-characterized esp-negative strains of E. faecalis. Using a chemostat-based biofilm fermentor system and a microtiter plate biofilm system, we established that multiple esp-negative strains are capable of biofilm formation and that E. faecalis biofilm development is influenced by the prevailing environmental conditions. A secreted protease, GelE, was found to enhance biofilm formation by E. faecalis. Our results further suggest that biofilm formation by E. faecalis, as for other organisms, is controlled by multiple partially overlapping regulatory pathways.
MATERIALS AND METHODS
Bacterial strains, growth media, plasmids, and genetic methods.
E. faecalis strains OG1RF (13, 17), JH2 (21), V583 (30, 37), MMH594 (19, 40), TX5243 (34), and TX5264 (41) have been described. Their relevant properties are described in the text. Bacteria were stored at −80°C in Todd-Hewitt broth (THB; prepared according to the manufacturer's instructions) supplemented with 30% glycerol. Unless otherwise indicated, all culture media were from Difco and all chemicals were from Sigma. Culture media were as follows: Trypticase soy broth without dextrose (TSB; BBL), prepared according to the manufacturer's instructions, without additional exogenous carbohydrate; M17 broth (M17), prepared according to the manufacturer's instructions, without additional exogenous carbohydrate; brain heart infusion (BHI) medium, prepared according to the manufacturer's instructions; Todd-Hewitt yeast extract broth (THYE), prepared as for THB plus 0.3% yeast extract; and M9YE medium (14), a semidefined M9-based medium supplemented with 0.3% yeast extract and 1% Casamino Acids.
Plasmid pMSP3535 is an expression vector containing a nisin-inducible promoter (4). Plasmid pMSP3614 is a derivative of pMSP3535 carrying cloned gelE from strain OG1RF (45), expressed under the control of the nisin-inducible promoter. Transformation of E. faecalis was carried out as previously described (1). When it was required to select for plasmid-bearing E. faecalis, erythromycin was included in culture media at 10 μg/ml. Nisin was included in culture media at 25 ng/ml when required for induction of gelE expression. Expression of GelE was evaluated by culturing strains on THB agar plates containing 3% gelatin at 37°C, as previously described (45).
Biofilm formation in the chemostat-based biofilm fermentor.
Biofilms of E. faecalis OG1RF were grown in the chemostat-based biofilm fermentor and analyzed as previously described for S. mutans (23). Briefly, THYE diluted fourfold and supplemented with 0.01% hog gastric mucin (type III; Sigma) was used to cultivate OG1RF at a dilution rate D of 0.1 h−1 at 37°C and pH 7.0. When the culture reached a steady state (at least 10 mean generation times or 4 days at D = 0.1 h−1), glass rods were aseptically inserted into the fermentor (approximately 4.4-cm2 surface area per rod immersed in fluid medium) for the initiation of biofilm formation. At intervals thereafter, biofilms were quantified by withdrawal of the glass rods followed by sonication in phosphate-buffered saline to remove bacteria from the rod surface and enumeration of viable CFU in the resulting cell suspension. Scanning electron microscopy (SEM) of intact biofilms on unsonicated glass rods was performed as previously described (23).
Microtiter plate assay for biofilm formation.
Polystyrene microtiter plates were used to evaluate biofilm formation of E. faecalis, as previously described for other gram-positive cocci (2, 18, 24, 25, 31), with slight modifications. A test strain was cultivated overnight in THB at 37°C. A portion of the overnight culture was diluted 100-fold in fresh culture medium, as required for a particular experiment, and 0.1-ml aliquots were dispensed into the wells of a 96-well flat-bottomed polystyrene microtiter plate (Corning 3595). When indicated for a particular experiment, fresh culture medium was either diluted fourfold with sterile water, mixed with cell-free conditioned culture supernatants, or supplemented with glucose or sodium chloride (NaCl) prior to the addition of bacteria. Unless otherwise indicated, microtiter plates were incubated aerobically at 37°C without agitation for the specified amount of time to allow bacterial growth and biofilm formation. At the desired times, planktonic cells were routinely removed for determination of cell density at 600 nm, and the plates were washed with distilled water three times by use of a multichannel handheld microplate washer (Bio-Tek Instruments) to remove any remaining nonadherent cells. After drying of the microtiter plates in air at room temperature, the adherent biofilms were stained for 20 min at room temperature with 0.1% safranin. The plates were then washed five times as described above and dried in air at room temperature. Subsequently, the absorbance of the biofilm on the bottom surface of each well of the dried plates was determined at 490 nm using an enzyme-linked immunosorbent assay microplate reader. All experiments included a blank well (culture medium without any bacteria) and seven replicate experimental wells. All experiments were repeated independently on different days a minimum of three times each, with similar results.
Preparation of conditioned medium supernatants.
For preparation of conditioned medium supernatants, a test strain was cultivated overnight in THB at 37°C. A portion of the overnight culture was diluted 100-fold in fresh M17 and incubated at 37°C without shaking for 24 h. The resulting cell suspension was centrifuged for 2 min at 13,000 × g in a microcentrifuge to pellet the cells. The supernatant was passed through a 0.45-μm-pore-size Tuffryn syringe filter (Gelman) to yield a sterile conditioned culture supernatant that was subsequently added directly to fresh M17 to produce a final concentration of 80% fresh to 20% conditioned M17. This mixture was then used as a culture medium for biofilm formation.
For specific experiments, OG1RF-conditioned M17 was subjected to one of several experimental treatments prior to mixing with fresh M17. These treatments consisted of heat treatment of an aliquot of conditioned M17 in a water bath at either 55 or 80°C for 10 min or, alternatively, fractionation of the OG1RF-conditioned M17 with a Microcon YM10 centrifugal filtration device (Millipore) by centrifugation at 13,000 × g for 30 min at room temperature. This procedure yielded the flowthrough fraction (composed of OG1RF-conditioned M17 components that were smaller than approximately 10 kDa) and the retained fraction, or retentate (composed of OG1RF-conditioned M17 components that were larger than approximately 10 kDa). Because the contents of the retentate fraction had been concentrated approximately 16-fold during centrifugation, sterile water was added to restore the retentate fraction to the original volume prior to mixing with fresh M17.
Determination of the presence of esp.
A PCR assay which was previously used for epidemiology of the esp gene in clinical isolates of E. faecalis (40) was used to assess whether the E. faecalis strains evaluated in this study carried the esp gene. It is important that the presence or absence of esp in these strains has been previously reported by other investigators, and as such, our assays served primarily to confirm these findings. Briefly, template DNA was obtained by use of the DNEasy tissue kit (Qiagen). For isolation of chromosomal DNA, test strains were cultivated overnight in THB at 37°C without shaking and processed according to the manufacturer's instructions for gram-positive organisms. Subsequently, 1 μl of the purified DNA was added to a PCR as the source of template. Three esp-specific primer sets were used to test for the presence of the esp gene: esp11-esp12, esp2-esp5, and esp46-esp47 (40). As a control for the quality of each template preparation, an unrelated 0.8-kb fragment of the E. faecalis chromosome was successfully amplified in parallel. A portion of each reaction was analyzed by electrophoresis on a 1% agarose gel and staining with ethidium bromide.
RESULTS
To evaluate the ability of esp-negative E. faecalis to form biofilms, we chose to analyze the well-characterized plasmid-free strain OG1RF. This strain has been used extensively to study the genetics of pheromone-responsive conjugative plasmid transfer (12, 15) as well as cell density-dependent control of extracellular protease expression (33, 34). OG1RF was previously found to lack the recently identified E. faecalis pathogenicity island harboring the coding sequence for Esp (39). Consistent with that result, we were unable to generate any esp-specific PCR amplification products from OG1RF chromosomal DNA (data not shown) with a PCR assay designed to detect the presence of esp (40).
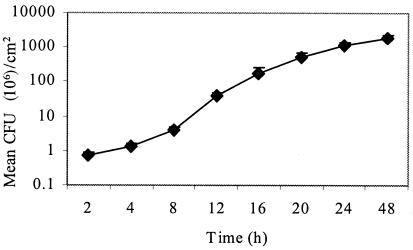
We used a chemostat-based biofilm fermentor to analyze biofilm formation by E. faecalis OG1RF growing under conditions of continuous culture, as previously described for S. mutans (23). Insertion of glass rods into the fermentor provided surfaces for biofilm formation. At specified intervals, rods were withdrawn from the biofilm chamber and surface-adherent bacteria were removed by sonication for enumeration of CFU, revealing that OG1RF formed dense biofilms of >109 CFU/cm2 within 24 h of growth (Fig. 1). This biofilm density is 100-fold higher than the biofilm density achieved by S. mutans under identical conditions (23), suggesting that E. faecalis OG1RF is particularly competent at growth in a biofilm state.
FIG. 1.
Quantitation of biofilm development by E. faecalis OG1RF in the chemostat-based biofilm fermentor. Biofilms were grown as described in Materials and Methods. Surface-adherent CFU were enumerated at intervals during growth of biofilms on glass rods inserted into the fermentor. The results are expressed as the mean CFU ± standard deviations per cm2 of glass surface.
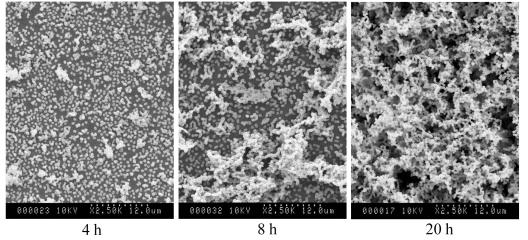
The process of biofilm formation is often characterized as proceeding temporally through a series of defined stages, including initial bacterial attachment to the substratum, microcolony formation, and maturation into complex three-dimensional structures, which typically contain void spaces described as water channels (5, 42). We used SEM to monitor the structure of OG1RF biofilms grown in the biofilm fermentor as a function of time (Fig. 2). The results show that biofilm formation by OG1RF progresses through each of the typical biofilm stages: initial attachment of individual cells (Fig. 2, 4 h), microcolony formation (Fig. 2, 8 h), and development into a mature biofilm with complex architecture containing void spaces, presumably corresponding to water channels (Fig. 2, 20 h). Thus, E. faecalis OG1RF is fully capable of forming dense biofilms that exhibit typical biofilm architecture in the absence of esp.
FIG. 2.
Analysis by SEM of biofilm development by E. faecalis OG1RF in the chemostat-based biofilm fermentor. Glass rods carrying biofilms were removed from the fermentor at the indicated times and processed for SEM. Biofilm growth and processing for microscopy were done as described in Materials and Methods.
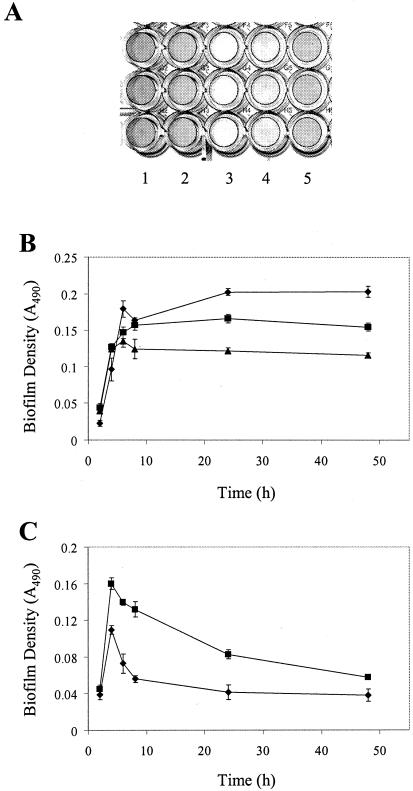
Microtiter plate biofilm assays have been used for many years to evaluate biofilm formation by a wide variety of organisms (18, 24, 25, 29), including clinical isolates of E. faecalis (3, 38, 44). The microtiter plate method utilizes cells grown in batch culture and incubated under static conditions for biofilm growth. In contrast, the biofilm fermentor uses continuously grown cultures subjected to conditions of bulk medium flow for biofilm growth. As a result, bacteria in the microtiter plate assay experience a different chemical and physical environment that may affect their ability to form a biofilm. Therefore, we wanted to evaluate the ability of OG1RF to form biofilms under the distinct conditions found in the microtiter plate system. OG1RF cultures were grown in flat-bottomed 96-well microtiter plates using a variety of laboratory culture media. At intervals after inoculation of the cultures, planktonic cells were removed and the surface-adherent biofilms were washed and stained with safranin. After removal of excess safranin by washing, the density of the stained biofilm on the bottom of the microtiter well (reflecting the biomass present in the biofilm) was determined by measuring the absorbance at 490 nm. By this method, biofilm formation by OG1RF was observed to occur predominantly on the bottom of the microtiter wells (Fig. 3A), as has been observed with biofilms of other gram-positive cocci (18, 24, 25).
FIG. 3.
Biofilm development by E. faecalis OG1RF in the wells of microtiter plates. The experiment was performed as described in Materials and Methods. All quantitative data are presented as the mean absorbance values for seven replicate wells ± standard deviations from a representative experiment. (A) Image of representative stained biofilms of OG1RF formed in the wells of microtiter plates after 24 h of growth. Culture media were as follows: column 1, TSB; column 2, M17; column 3, BHI medium; column 4, THYE; column 5, M9YE. (B) OG1RF biofilm accumulation over time in culture media that support the formation and maintenance of biofilms. Diamonds, TSB; squares, M17; triangles, M9YE. (C) OG1RF biofilm accumulation over time in culture media that support initial biofilm formation but not extended biofilm maintenance. Diamonds, BHI medium; squares, THYE.
Microtiter plate analyses of biofilm accumulation as a function of time revealed that OG1RF exhibited two distinct phenotypes, depending on the growth medium in which biofilms formed (Fig. 3). Biofilm accumulation in TSB, M17, and M9YE media progressed steadily during the first 6 to 8 h of growth. At that time, further accumulation of OG1RF biofilm slowed and gradually achieved a plateau of biofilm density (Fig. 3B). Thereafter, the biofilm density was essentially maintained at a constant level for the duration of the experiment (up to 48 h). On the other hand, OG1RF biofilm accumulation in THYE and BHI media progressed steadily for the first 4 h of growth but then abruptly ceased (Fig. 3C). Thereafter, the density of the nascent biofilm immediately began to decrease, reflecting the loss of preformed biofilm from the polystyrene surface in the microtiter plate wells. Thus, certain environmental conditions promoted long-term biofilm formation and maintenance in the microtiter assay, whereas other conditions led initially to biofilm formation but did not support long-term biofilm maintenance. These results suggest that OG1RF senses its physiological state or the existing environmental conditions and uses this information to modulate biofilm formation and maintenance.
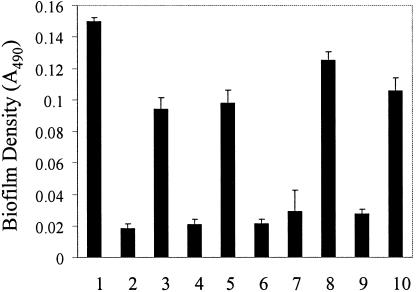
The facile recovery of E. faecalis from the surfaces of various indwelling medical devices in the clinical setting (10, 11) suggests that the ability to form biofilms is common to most isolates of E. faecalis. To determine if characterized strains of E. faecalis other than OG1RF also exhibit biofilm formation, we used the microtiter plate assay to evaluate biofilm formation by three additional, independent isolates of E. faecalis: (i) strain JH2, a plasmid-free isolate, known to be naturally deficient in the production of a secreted metalloprotease (GelE) (21) (JH2, like OG1RF, has been used to study the transfer of pheromone-responsive conjugative plasmids [20]); (ii) strain V583, a vancomycin-resistant clinical isolate whose genome sequence has been determined (30); and (iii) strain MMH594, a virulent clinical isolate responsible for a hospital ward infectious outbreak in the mid-1980s (19), which harbors a complete copy of the only known E. faecalis pathogenicity island (39). Of these three isolates, MMH594 was previously shown to be esp positive (40), V583 was shown to be esp negative (39), and JH2 is thought to be esp negative. Our PCR analysis of chromosomal DNA from these strains yielded amplification products of the expected sizes from the MMH594 template, but no amplification products from the V583 or JH2 template (not shown), confirming the presence of esp in MMH594 and indicating its absence in V583 and JH2.
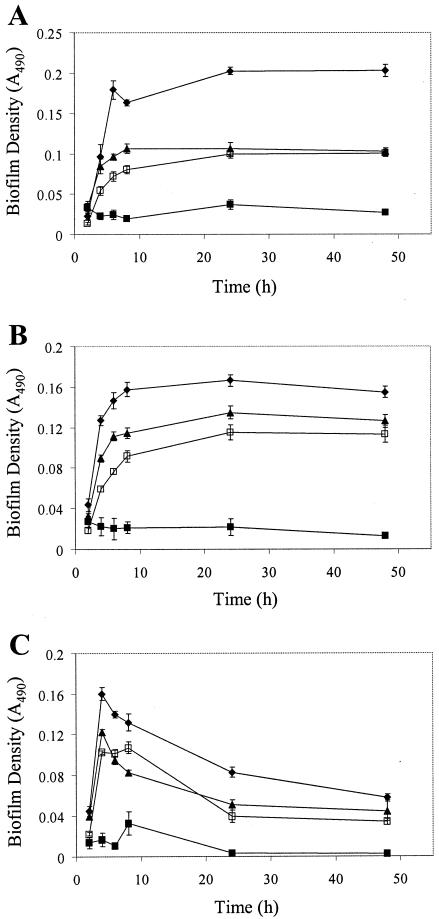
Microtiter plate analyses of biofilm formation by these strains as a function of time revealed that V583 and MMH594 exhibited biofilm formation phenotypes which were very similar to each other and which closely mimicked those of OG1RF, although with a slightly reduced magnitude (Fig. 4). Thus, these independent clinical isolates not only exhibited the ability to form biofilms but also retained the medium-dependent biofilm formation and maintenance phenotypes observed with OG1RF, indicating that the behavior of biofilm formation and the mechanisms in place for its control likely are common to most isolates of E. faecalis. JH2 was poor at forming biofilms in most growth media in which it was evaluated.
FIG. 4.
Biofilm development by various strains of E. faecalis in the wells of microtiter plates. The experiment was performed as described in Materials and Methods. All data are presented as the mean absorbance values for seven replicate wells ± standard deviations from a representative experiment. Diamonds, OG1RF; solid squares, JH2; triangles, V583; open squares, MMH594. (A) Biofilm accumulation in TSB. (B) Biofilm accumulation in M17. (C) Biofilm accumulation in THYE.
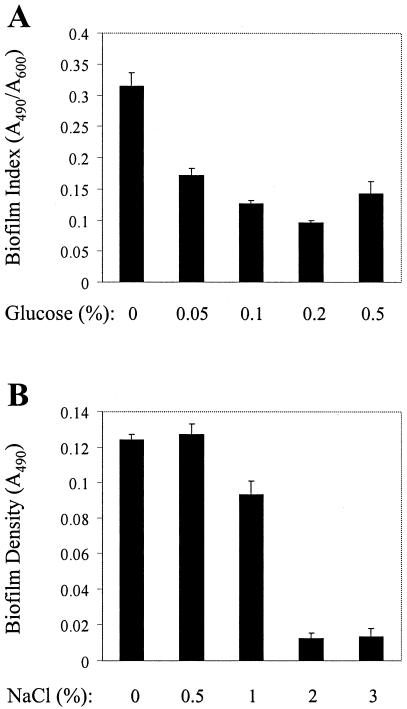
Previous work has indicated that the availability of nutrients is one of several factors modulating biofilm formation by various bacterial species (22, 25, 29, 32, 46). Furthermore, our results with laboratory media of different compositions suggest that nutrient availability or composition may be involved in regulating biofilm formation, maintenance, or detachment by E. faecalis (Fig. 3). To test whether nutrient concentration affects biofilm formation by E. faecalis, we used the microtiter plate assay to grow biofilms of OG1RF in M9YE medium supplemented with increasing concentrations of glucose. Because the total yield of cells after 24 h of growth varied depending on the glucose concentration in the medium, direct comparison of the biofilm accumulation under these disparate conditions does not reflect only the biofilm-forming ability of the bacteria subjected to a given growth condition; rather, the biofilm accumulation reflects a composite measure of both the ability of the bacteria to form a biofilm and the ability of the bacteria to generate additional biomass (through cell doublings) from a given amount of nutrients. In other words, when comparing biofilm accumulation under growth conditions in which biomass yield is drastically different, the proportion of total biomass that is found in the biofilm is a better measure for comparing biofilm formation than the raw biofilm accumulation. Therefore, for these experiments we expressed biofilm accumulation as a “biofilm index,” for which the biofilm density (A490) was normalized to the total cell yield (A600) in a given growth medium, to produce a relative measure of the tendency of bacteria to engage in biofilm formation. A similar normalization procedure has been used previously to characterize biofilm formation by Staphylococcus epidermidis (9). The results in Fig. 5A show that as the glucose concentration of the growth medium decreases, the biofilm indices for OG1RF increase, indicating that the relative tendency of OG1RF to form biofilms in nutrient-rich environments is reduced compared with its tendency in nutrient-poor environments. A similar trend was observed during biofilm formation by strains V583 and MMH594 (not shown).
FIG. 5.
Effect of environmental conditions on biofilm formation of OG1RF. (A) Effect of nutrient concentration on biofilm formation. Biofilms were grown in microtiter plates for 24 h in M9YE supplemented with the indicated concentration of glucose, as described in Materials and Methods. All data are presented as the mean values of seven replicates ± standard deviations from a representative experiment. (B) Effect of medium osmolarity on biofilm formation. Biofilms were grown in microtiter plates for 24 h in M9YE supplemented with the indicated concentration of NaCl, as described in Materials and Methods. All data are presented as the mean absorbance values for seven replicate wells ± standard deviations from a representative experiment.
To determine if a similar effect of nutrient concentration modulated biofilm formation in other disparate growth media, we used microtiter plate assays to evaluate the effect of nutrient dilution in complex laboratory media. Biofilm accumulation was assessed for OG1RF grown in complex media prepared according to standard recipes or in the same media diluted to 1/4 strength with sterile water. The results showed that nutrient dilution leads to increases in the biofilm index for most media tested (not shown), consistent with the results described above. A similar trend was observed during biofilm formation of strain V583 (not shown), suggesting that the molecular mechanism mediating the enhancement of biofilm accumulation in nutrient-poor media is conserved in independent strains.
In addition to nutrient availability, other environmental factors, including the osmolarity of the growth medium, are known to affect biofilm formation by diverse bacterial species (25, 29, 35). Using microtiter plate assays, we evaluated the effect of increasing NaCl concentrations in various media on biofilm formation by OG1RF. The results shown in Fig. 5B demonstrate that biofilm formation was abolished by growth in a medium with high osmolarity, despite the fact that the total yield of cells was not affected by growth in the presence of increased amounts of NaCl, suggesting that E. faecalis monitors the environmental conditions and modulates biofilm formation in response. Biofilm formation was also abolished when cells were grown in various complex media supplemented with 3% NaCl (not shown), indicating that this is not a medium-dependent phenomenon. A similar phenotype was observed during biofilm formation of strains V583 and MMH594 (not shown).
Diffusible, self-produced extracellular signaling molecules are known to control the development of mature biofilm architecture in diverse bacterial species (8, 24). In an attempt to identify a soluble biofilm-promoting factor from E. faecalis, we took advantage of the observation that strain JH2 was poor at forming biofilms in M17 (Fig. 4). We assayed cell-free, conditioned-medium supernatants from biofilm-forming cultures of OG1RF for the ability to enhance biofilm formation by JH2. Sterile, conditioned supernatants from OG1RF or JH2 were added to fresh M17 at 20% (vol/vol), and the resulting media were used as substrates for biofilm formation by JH2 in the microtiter plate assay. Although JH2-conditioned supernatant had no effect on JH2 biofilm formation, the OG1RF-conditioned culture supernatant was able to enhance biofilm formation by JH2 4.5-fold (Fig. 6), indicating that the OG1RF-conditioned supernatant contained biofilm-promoting activity. The OG1RF-conditioned supernatant did not, under any conditions tested, fully restore biofilm formation by JH2 to a level similar to that observed with OG1RF.
FIG. 6.
Characterization of biofilm-promoting activity found in spent culture supernatants of OG1RF. Biofilms were grown in microtiter plates for 24 h as described in Materials and Methods. All data are presented as the mean absorbance values for seven replicate wells ± standard deviations from a representative experiment. Column 1, biofilm of OG1RF in 100% fresh M17; column 2, biofilm of JH2 in 100% fresh M17; column 3, biofilm of JH2 in 80% fresh M17 and 20% OG1RF-conditioned M17; column 4, biofilm of JH2 in 80% fresh M17 and 20% JH2-conditioned M17; column 5, biofilm of JH2 in 80% fresh M17 and 20% heat-treated OG1RF-conditioned M17 (55°C for 10 min); column 6, same as column 5, but with heating at 80°C for 10 min; column 7, biofilm of JH2 in 80% fresh M17 and 20% OG1RF-conditioned M17 (YM10 flowthrough fraction); column 8, biofilm of JH2 in 80% fresh M17 and 20% OG1RF-conditioned M17 (YM10 retentate fraction); column 9, biofilm of JH2 in 80% fresh M17 and 20% TX5264-conditioned M17 (gelE sprE+); column 10, biofilm of JH2 in 80% fresh M17 and 20% TX5243-conditioned M17 (gelE+ sprE).
In order to determine the identity of the biofilm-promoting activity, we performed a preliminary characterization of its properties. Heating of the OG1RF-conditioned supernatant for 10 min at 80°C resulted in a substantial loss of biofilm-promoting activity from the conditioned supernatant (Fig. 6), suggesting that the activity is proteinaceous in nature. To estimate the approximate molecular mass of the biofilm-promoting protein, we used Microcon centrifugal filtration devices to fractionate the OG1RF-conditioned supernatant. The majority of the biofilm-promoting activity was retained by a YM-10 membrane (Fig. 6), suggesting that the apparent molecular mass of the activity is greater than the exclusion limit of the filter (10 kDa). Such a large apparent molecular mass is consistent with the possibility that the biofilm-promoting activity is a protein.
A recent report demonstrated that a secreted metalloprotease of E. faecalis, GelE, performs a variety of extracellular functions, such as degradation of misfolded surface proteins and determination of supernatant pheromone levels (45). These authors also found that GelE contributes to the determination of cellular chain length of strain OG1RF. To explain this observation, they suggested that GelE proteolytically activates one or more cell wall hydrolases responsible for peptidoglycan cleavage required for cell separation. The apparent involvement of GelE in a wide variety of extracellular biological processes, in combination with our previous observations that the biofilm-promoting activity found in OG1RF-conditioned supernatants exhibited properties consistent with those of a protein, led us to hypothesize that GelE itself was the biofilm-promoting protein found in OG1RF-conditioned supernatants. This hypothesis was consistent with the observation that strain JH2, which is known to be Gel−, was poor at forming biofilms (Fig. 4). To test this hypothesis, we prepared a conditioned supernatant from strain TX5264, an isogenic mutant of OG1RF carrying an in-frame deletion of most of the gelE gene, and tested this supernatant for the presence of biofilm-promoting activity. As a control, we also tested a conditioned supernatant from strain TX5243, an isogenic derivative of OG1RF carrying an insertion mutation in sprE, a secreted serine protease encoded immediately downstream from gelE. The results shown in Fig. 6 demonstrate that the conditioned supernatant from the sprE mutant possesses essentially 100% of the biofilm-promoting activity found in the conditioned supernatant from OG1RF, whereas the conditioned supernatant from the gelE mutant lacks the majority of the biofilm-promoting activity. Thus, our results indicate that the GelE protease is responsible for the biofilm-promoting activity found in OG1RF-conditioned supernatants.
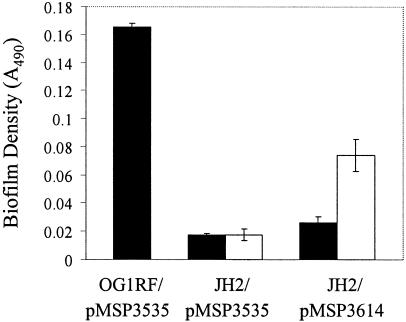
If GelE enhances biofilm formation by E. faecalis, then heterologous expression of GelE in the Gel− strain JH2 might be expected to restore biofilm formation to this biofilm-deficient isolate. To test this hypothesis, we used plasmid pMSP3614, carrying a cloned version of OG1RF gelE under the control of a nisin-inducible promoter (45), to express gelE in JH2. An agar plate-based assay confirmed that GelE was efficiently secreted from this strain in the presence of inducer (not shown). Using the microtiter plate assay to evaluate biofilm formation by this strain, we found that biofilm formation was partially restored in an inducer-dependent manner (Fig. 7), consistent with our previous results implicating GelE in enhancement of JH2 biofilm formation.
FIG. 7.
Expression of gelE in JH2 partially rescues its biofilm formation defect. Biofilms were grown in microtiter plates for 24 h as described in Materials and Methods. All data are presented as the mean absorbance values for seven replicate wells ± standard deviations from a representative experiment. OG1RF or JH2 containing either a plasmid harboring a cloned, nisin-inducible copy of gelE (pMSP3614) or the parental plasmid lacking gelE (pMSP3535) was grown in the absence (black bars) or presence (white bars) of nisin to induce gelE expression.
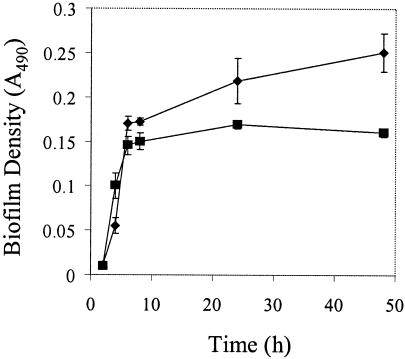
To determine if GelE also enhances biofilm formation of OG1RF, we used the microtiter plate assay to monitor biofilm accumulation by the isogenic gelE mutant of OG1RF (TX5264). A slight defect in biofilm accumulation, relative to OG1RF, was observed when cells were cultivated in TSB (Fig. 8), consistent with a role for GelE in promoting biofilm formation. Biofilms of the gelE mutant (TX5264) accumulated to a significantly higher level than those of JH2, suggesting that although GelE contributes to biofilm formation, OG1RF possesses at least one GelE-independent pathway that leads to development of biofilms. Because JH2 is unable to form biofilms comparable to those of TX5264 under identical conditions, it appears that JH2 possesses at least one deficiency, in addition to its Gel− phenotype, that contributes to its biofilm-defective phenotype.
FIG. 8.
An isogenic gelE mutant of OG1RF exhibits a defect in biofilm formation. Biofilms were grown in TSB in microtiter plates as described in Materials and Methods. All data are presented as the mean absorbance values for seven replicate wells ± standard deviations from a representative experiment. Biofilm accumulation is shown for the following strains: diamonds, OG1RF; squares, TX5264 (gelE negative).
DISCUSSION
The results presented here demonstrate that E. faecalis is fully capable of biofilm formation in the absence of the esp gene and, indeed, in the absence of the entire E. faecalis pathogenicity island previously identified for MMH594. In fact, although several strains examined in this report are not isogenic, our results suggest that the presence of this pathogenicity island (with or without the gene encoding Esp) does not confer any dramatically enhanced ability to form biofilms, given that multiple, independent isolates exhibited similar biofilm-forming phenotypes under the conditions we tested (Fig. 4). Furthermore, two independent methods of analysis, employing significantly different conditions for biofilm growth (the chemostat-based biofilm fermentor and the microtiter plate biofilm assay), both indicated that OG1RF, lacking the entire pathogenicity island, possesses the machinery necessary for biofilm formation. Thus, it appears unlikely that the essential genetic determinants conferring the biofilm formation phenotype are located within this E. faecalis pathogenicity island. These biofilm determinants, potentially including any genes required for production of extracellular polymeric matrix and genes whose products participate in sensory transduction to regulate the biofilm response, will likely be common to most strains of E. faecalis. The possibility remains, however, that Esp or other gene products encoded by the characterized pathogenicity island can influence biofilm formation under certain environmental conditions that were not tested here.
The dense OG1RF biofilm formed on glass surfaces in the biofilm fermentor contains 100-fold more CFU/cm2 than an S. mutans biofilm cultured under identical conditions. In contrast, the OG1RF planktonic cell density in the fermentor was only 15-fold higher than that of S. mutans (not shown). Because S. mutans is an oral bacterium that depends on biofilm growth for survival and persistence in its natural ecosystem of dental plaque, the relatively high biofilm cell density achieved by OG1RF suggests that E. faecalis is adept at growth in the biofilm state. Furthermore, the formation of biofilms by E. faecalis OG1RF appears to progress through characteristic stages of development that were previously observed during biofilm development of distantly related bacterial species: initial bacterial adherence to the substratum, microcolony formation, and maturation into complex three-dimensional structures apparently containing water channels (Fig. 2). Thus, control mechanisms similar to those described for other bacteria may operate to control biofilm development in E. faecalis.
Modulation of biofilm development by environmental conditions has been observed for diverse bacterial species. For example, the nutrient content of the growth medium influences biofilm formation by Escherichia coli (32), P. aeruginosa (22), Pseudomonas fluorescens (29), Streptococcus gordonii (25), and S. mutans (46). Similarly, the osmolarity of the growth medium has also been observed to modulate biofilm development for P. fluorescens (29), S. gordonii (25), and S. aureus (35), although the effect on biofilm development is species specific. The results of our microtiter plate analyses, demonstrating the medium dependence of biofilm formation (Fig. 3), suggest that environmental control of biofilm development also occurs for E. faecalis. Because growth in certain media promoted the formation and extended maintenance of biofilms, whereas growth in other media promoted only the initial biofilm formation but not extended maintenance of surface-adherent biofilms, we hypothesize that E. faecalis cells monitor their physiological state and/or the nascent environmental conditions and use this information to regulate biofilm development. Thus, we speculate that after roughly 4 h of biofilm accumulation in BHI or THYE medium, an unknown signal, specific to growth in these media, initiates the dispersion of preformed biofilm. An alternative possibility is that a medium-specific signal which occurs during growth in M17, TSB, and M9YE media but not in BHI and THYE media promotes the maintenance of preformed biofilms. Because both V583 and MMH594 exhibit the same medium-dependent biofilm accumulation phenotype as OG1RF (Fig. 4), it is likely that the sensory processes regulating this behavior will be common to all strains of E. faecalis.
Although the specific signals that are sensed to regulate the medium-specific biofilm phenotypes shown in Fig. 3 are unknown, other, defined environmental conditions were also found to modulate biofilm development by E. faecalis. Increasing the osmolarity of the growth medium inhibited biofilm formation without affecting the growth of the cells (Fig. 5), as has been previously observed with P. fluorescens and S. gordonii, suggesting that E. faecalis also monitors the osmotic strength of the environment. Nutrient concentration was found to influence biofilm development (Fig. 5), as growth in culture media in which the nutrients were more dilute tended to promote proportionally higher levels of biofilm formation relative to growth in nutrient-rich media. Taken together, our results argue that E. faecalis uses one or more sensory systems to monitor changes in its physiology or in the prevailing environmental conditions to regulate its ability to form biofilms. However, by using the microtiter plate biofilm assay system, we also identified environmental conditions that did not influence biofilm development of OG1RF or V583. Growth under anaerobic conditions yielded similar amounts of biofilm, as did growth under aerobic conditions, and growth at 30°C yielded similar amounts of biofilm as growth at 37°C (not shown). These results were independent of the composition of the growth medium used (we tested all growth media used for Fig. 3).
We also evaluated whether the presence of the pheromone-responsive conjugative plasmid pCF10 in OG1RF had any effect on biofilm development, using both the microtiter plate assay system and the biofilm fermentor system. pCF10 belongs to a class of conjugative plasmids that encodes a surface protein, Asc10, responsible for cell-cell adhesion and aggregation during conjugation (27). We found little difference in the amount of biofilm formed by OG1RF carrying pCF10 relative to that formed by plasmid-free OG1RF in either the biofilm fermentor or the microtiter plate system (not shown). However, it is worth noting that, at least in planktonic cultures, the expression of Asc10 is repressed in the absence of pheromone secreted by pCF10-free recipient cells (27). Our results indicate that pCF10-encoded functions do not alter biofilm development in the absence of pCF10-free recipient cells. Further work is required to determine if pCF10-encoded functions modulate biofilm formation in cultures that contain mixtures of pCF10-free and pCF10-containing cells.
Because soluble, self-produced, extracellular signals have been found to regulate biofilm development by several species of bacteria (8, 24), we searched for such a signal in conditioned culture supernatants of biofilm-forming E. faecalis. We identified a soluble protein, of relatively high apparent molecular mass, which was heat sensitive and was absent from the culture supernatant of a gelE mutant (Fig. 6). Taken together, these results indicated that GelE, a secreted metalloprotease, was responsible for the biofilm-promoting activity. This proposal is consistent with the observation that the Gel− E. faecalis strain JH2 is poor at forming biofilms in the microtiter plate system (Fig. 4). The inducible heterologous expression of gelE in JH2 led to enhanced biofilm formation (Fig. 7). Lastly, an otherwise isogenic gelE mutant exhibited less biofilm accumulation than OG1RF when grown in TSB (Fig. 8). Therefore, we conclude that GelE enhances biofilm development by E. faecalis. Furthermore, because the gelE mutant does form biofilms and these biofilms accumulate to a significantly higher level than those of JH2, we propose that multiple pathways function to regulate biofilm formation in E. faecalis.
The mechanism by which GelE enhances biofilm development is unknown. In one possible model, GelE might participate in the production of an extracellular signaling peptide by proteolytically processing an inactive secreted peptide precursor to a mature, active form. This processed peptide would then be capable of serving as a signal for enhanced biofilm formation. In an alternative model for GelE function, GelE might proteolytically activate another surface protein involved in some aspect of the regulation or execution of biofilm development, such as a protein that participates in the secretion of extracellular polymeric matrix material. If this putative secretion component is synthesized as an inactive precursor that requires proteolytic cleavage to become functional, processing by GelE would then be necessary for the secretion of matrix material for biofilm formation. Proteolytic processing of inactive exoprotein precursors has already been proposed as a function for GelE during activation of a cell wall hydrolase (autolysin) involved in cell separation of E. faecalis (45).
The fsr locus encodes a cell density-dependent sensory system in E. faecalis that is known to positively regulate the expression of gelE in E. faecalis (33, 34). A recent report (S. K. Pillai, G. Sakoulas, H. S. Gold, C. Wennersten, G. M. Eliopoulos, R. C. Moellering, Jr., and R. T. Inouye, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol., abstr. B-089, 2003) indicated that fsr mutants of OG1RF exhibited a 25 to 30% decrease in biofilm accumulation in a microtiter plate assay. Consequently, Pillai et al. suggested that the fsr system regulates the expression of unknown genes involved in biofilm formation. However, because gelE is not expressed in fsr mutants (34), and because we observed a reduction of approximately 25% in biofilm accumulation by the gelE mutant under conditions similar to those employed by Pillai et al. (Fig. 8), it is possible that the absence of gelE expression in the fsr mutants used by Pillai et al. is sufficient to account for the biofilm defect they observed. While it remains uncertain if the biofilm phenotypes of fsr mutants are substantially different from those of the gelE mutant, our results support the hypothesis that the absence of gelE expression in fsr mutants is primarily responsible for their biofilm defect. Thus, we suggest that the fsr cell density-dependent sensing system of E. faecalis participates in regulation of biofilm development via control of gelE expression.
In summary, we have used independent experimental approaches to characterize biofilm formation by E. faecalis. Our results demonstrate that E. faecalis forms robust biofilms and that biofilm development is modulated by the prevailing environmental conditions. Neither the esp gene product nor the remainder of the genes encoded by the known E. faecalis pathogenicity island is required for the development of biofilms with complex architecture. We have found that a secreted metalloprotease, GelE, enhances biofilm formation of E. faecalis but that, otherwise, the genetic determinants controlling biofilm development remain unknown. The results described in this study set the stage for future work focused on identifying and characterizing these determinants.
Acknowledgments
This work was supported by NIH grant HL51987 to G.M.D. C.J.K was supported by NIH training grant HD07381-12 and by NRSA grant F32-AI56684.
REFERENCES
- 1.Bae, T., B. Kozlowicz, and G. M. Dunny. 2002. Two targets in pCF10 DNA for PrgX binding: their role in production of Qa and prgX mRNA and in regulation of pheromone-inducible conjugation. J. Mol. Biol. 315:995-1007. [DOI] [PubMed] [Google Scholar]
- 2.Baldassarri, L., L. Bertuccini, M. G. Ammendolia, C. R. Arciola, and L. Montanaro. 2001. Effect of iron limitation on slime production by Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 20:343-345. [DOI] [PubMed] [Google Scholar]
- 3.Baldassarri, L., R. Cecchini, L. Bertuccini, M. G. Ammendolia, F. Iosi, C. R. Arciola, L. Montanaro, R. Di Rosa, G. Gherardi, G. Dicuonzo, G. Orefici, and R. Creti. 2001. Enterococcus sp. produces slime and survives in rat peritoneal macrophages. Med. Microbiol. Immunol. (Berlin) 190:113-120. [DOI] [PubMed] [Google Scholar]
- 4.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183-190. [DOI] [PubMed] [Google Scholar]
- 5.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 6.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 7.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 9.Deighton, M., and R. Borland. 1993. Regulation of slime production in Staphylococcus epidermidis by iron limitation. Infect. Immun. 61:4473-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donlan, R. M. 2001. Biofilm formation: a clinically relevant microbiological process. Clin. Infect. Dis. 33:1387-1392. [DOI] [PubMed] [Google Scholar]
- 11.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunny, G., C. Funk, and J. Adsit. 1981. Direct stimulation of the transfer of antibiotic resistance by sex pheromones in Streptococcus faecalis. Plasmid 6:270-278. [DOI] [PubMed] [Google Scholar]
- 13.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunny, G. M., and D. B. Clewell. 1975. Transmissible toxin (hemolysin) plasmid in Streptococcus faecalis and its mobilization of a noninfectious drug resistance plasmid. J. Bacteriol. 124:784-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunny, G. M., R. A. Craig, R. L. Carron, and D. B. Clewell. 1979. Plasmid transfer in Streptococcus faecalis: production of multiple sex pheromones by recipients. Plasmid 2:454-465. [DOI] [PubMed] [Google Scholar]
- 16.Foley, I., and P. Gilbert. 1997. In-vitro studies of the activity of glycopeptide combinations against Enterococcus faecalis biofilms. J. Antimicrob. Chemother. 40:667-672. [DOI] [PubMed] [Google Scholar]
- 17.Gold, O. G., H. V. Jordan, and J. van Houte. 1975. The prevalence of enterococci in the human mouth and their pathogenicity in animal models. Arch. Oral Biol. 20:473-477. [DOI] [PubMed] [Google Scholar]
- 18.Heilmann, C., C. Gerke, F. Perdreau-Remington, and F. Gotz. 1996. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect. Immun. 64:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huycke, M. M., C. A. Spiegel, and M. S. Gilmore. 1991. Bacteremia caused by hemolytic, high-level gentamicin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 35:1626-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacob, A. E., G. J. Douglas, and S. J. Hobbs. 1975. Self-transferable plasmids determining the hemolysin and bacteriocin of Streptococcus faecalis var. zymogenes. J. Bacteriol. 121:863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacob, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 23.Li, Y. H., P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Y. H., N. Tang, M. B. Aspiras, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marrie, T. J., and J. W. Costerton. 1984. Morphology of bacterial attachment to cardiac pacemaker leads and power packs. J. Clin. Microbiol. 19:911-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olmsted, S. B., S. M. Kao, L. J. van Putte, J. C. Gallo, and G. M. Dunny. 1991. Role of the pheromone-inducible surface protein Asc10 in mating aggregate formation and conjugal transfer of the Enterococcus faecalis plasmid pCF10. J. Bacteriol. 173:7665-7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 29.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 30.Paulsen, I. T., L. Banerjei, G. S. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 31.Pfaller, M., D. Davenport, M. Bale, M. Barrett, F. Koontz, and R. M. Massanari. 1988. Development of the quantitative micro-test for slime production by coagulase-negative staphylococci. Eur. J. Clin. Microbiol. Infect. Dis. 7:30-33. [DOI] [PubMed] [Google Scholar]
- 32.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 33.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2001. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 183:3372-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 68:2579-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rachid, S., K. Ohlsen, U. Wallner, J. Hacker, M. Hecker, and W. Ziebuhr. 2000. Alternative transcription factor sigma(B) is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182:6824-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richards, M. J., J. R. Edwards, D. H. Culver, and R. P. Gaynes. 2000. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control Hosp. Epidemiol. 21:510-515. [DOI] [PubMed] [Google Scholar]
- 37.Sahm, D. F., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandoe, J. A., I. R. Witherden, J. H. Cove, J. Heritage, and M. H. Wilcox. 2003. Correlation between enterococcal biofilm formation in vitro and medical-device-related infection potential in vivo. J. Med. Microbiol. 52:547-550. [DOI] [PubMed] [Google Scholar]
- 39.Shankar, N., A. S. Baghdayan, and M. S. Gilmore. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746-750. [DOI] [PubMed] [Google Scholar]
- 40.Shankar, V., A. S. Baghdayan, M. M. Huycke, G. Lindahl, and M. S. Gilmore. 1999. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect. Immun. 67:193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sifri, C. D., E. Mylonakis, K. V. Singh, X. Qin, D. A. Garsin, B. E. Murray, F. M. Ausubel, and S. B. Calderwood. 2002. Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect. Immun. 70:5647-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 43.Tannock, G. W., and G. Cook. 2002. Enterococci as members of the intestinal microflora of humans, p. 101-132. In M. S. Gilmore (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, D.C.
- 44.Toledo-Arana, A., J. Valle, C. Solano, M. J. Arrizubieta, C. Cucarella, M. Lamata, B. Amorena, J. Leiva, J. R. Penades, and I. Lasa. 2001. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 67:4538-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waters, C. M., M. H. Antiporta, B. E. Murray, and G. M. Dunny. 2003. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J. Bacteriol. 185:3613-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida, A., and H. K. Kuramitsu. 2002. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 68:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]