Abstract
The Borrelia burgdorferi genome encodes five orthologues of the substrate binding protein oligopeptide permease A (OppA). It was previously shown that these genes are under the control of separate promoters and are differentially expressed under various environmental conditions. We were interested in determining whether there are also differences in substrate specificities among the proteins. The substrate specificities of recombinant proteins were determined by screening for high-affinity peptides by use of a combinatorial phage display heptapeptide library. Different heptapeptides with high affinities for OppA-1, OppA-2, and OppA-3 were identified. No heptapeptide binding OppA-4 or OppA-5 could be identified. Competitive binding assays were performed under various conditions to determine the substrate preferences of the OppA proteins. OppA-1 retained maximal activity over a broad range of pHs (5.5 to 7.5), whereas OppA-2 and OppA-3 showed peak activities at pHs below 5.5. OppA-1 and OppA-2 showed preferences for tripeptides over dipeptides and longer-chain peptides. Although a wide variety of amino acyl side chains were tolerated by all three OppA proteins, OppA-1 showed the broadest substrate specificity and was able to accommodate peptides composed of bulky hydrophobic residues; OppA-2 and OppA-3 showed preferences for peptides composed of small nonpolar amino acids. All three OppA proteins showed preferences for peptides composed of l- rather than d-amino acids. OppA-3 showed the greatest tolerance for changes in stereochemistry. Substantial differences in the substrate specificities of the OppA proteins of B. burgdorferi suggest that they may have distinct functions in the organism.
Peptide transport systems in bacteria have evolved to play an important role in diverse functions. Because of the importance of peptide acquisition, bacteria often have multiple peptide transport systems with overlapping functions. Many bacteria utilize exogenous peptides as a source of amino acids. However, beyond their role in nutrient acquisition, components of peptide transport systems help bacteria respond to changes in their environment by mediating functions such as quorum sensing, sporulation, pheromone transport, and chemotaxis (1, 16, 17, 19).
Borrelia burgdorferi is the causative organism of Lyme disease (26). Analysis of its genome has shown that it has machinery for the synthesis of only one amino acid, only two amino acid transport homologues, and only a single peptide transport system (8). This peptide transport system belongs to the superfamily of ABC transporters and bears a high degree of similarity to the oligopeptide permease (Opp) and dipeptide permease (Dpp) systems of bacteria such as Escherichia coli, Salmonella enterica serovar Typhimurium, and Lactococcus lactis (6, 11, 12, 20). All of these systems consist of a peptide binding protein (OppA or DppA), two transmembrane proteins (OppB and OppC or DppB and DppC), and two ATP binding proteins (OppD and OppF or DppD and DppF). The B. burgdorferi opp operon, however, differs in that it encodes two additional OppA-like proteins (OppA-2 and OppA-3). Two other OppA orthologues (OppA-4 and OppA-5) have been identified from borrelial plasmids (cp26 and lp-54, respectively). Lin et al. confirmed the ability of the B. burgdorferi OppA orthologues to transport tripeptides by using a complementation system in E. coli (18).
Wang et al. found that each of the five B. burgdorferi OppA orthologues is under the control of a separate promoter—which would allow for individual regulation of the expression of the proteins (29). B. burgdorferi undergoes a complex natural life cycle in which it encounters vast changes in environmental conditions, such as nutrient availability, temperature, and pH, as it moves from tick to mammalian hosts. The individual promoters have been shown to respond differentially to various environmental conditions (29). For example, the expression of OppA-4 and OppA-5 is highly up-regulated by the bacteria in a mouse host, while the expression of OppA-1 is relatively constitutive regardless of whether the bacteria are grown in vitro or are present in tick or mouse hosts.
The substrate specificities of the peptide permeases of other bacteria generally are thought to be dictated by the specificities of the substrate binding proteins (10), although recent studies with L. lactis also have suggested a role for the intrinsic membrane proteins (5). OppA proteins of other bacteria have been found to have very broad substrate specificities, with the ability to bind to ligands of diverse structures and lengths (7, 9, 28). Other peptide permeases, such as Dpp, have been shown to have much narrower substrate specificities. The maintenance in B. burgdorferi of five OppA orthologues, which are differentially expressed at different points in its life cycle, suggests that they may have distinct functions in the organism. We were interested in studying whether these proteins exhibit differences in biochemical properties that would provide the organism with an adaptive advantage under various environmental conditions. In this report, we describe our analysis of the peptide binding properties of three B. burgdorferi opp operon-encoded OppA proteins by use of a combinatorial phage display peptide library.
MATERIALS AND METHODS
Cloning of oppA genes.
B. burgdorferi B31 was grown to mid-logarithmic phase in BSK H medium (Sigma) at 37°C and harvested by centrifugation at 8,000 × g for 15 min. Total genomic DNA was isolated as previously described (14).
To clone oppA genes from B. burgdorferi genomic DNA, PCR was performed with the specific oligonucleotide primers listed in Table 1. Cycling parameters were as follows: denaturation at 95°C for 5 min followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min. PCR products were cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, Calif.) and electroporated into E. coli Top10 cells. Plasmid DNA was recovered with DNA miniprep columns (Qiagen, Valencia, Calif.). The inserted sequences were checked for PCR errors by automated DNA sequencing (ABI 3100; Applied Biosystems, Foster City, Calif.). Plasmids were digested with the appropriate restriction enzymes, and fragments were recovered from 1% low-melting-temperature agarose gels with Qiaquick (Qiagen) in accordance with the manufacturer's instructions. The recovered fragments were ligated into appropriately restricted pET 28c or pET 21b and electroporated into BL21 CodonPlus (DE3)-RIL (Stratagene, La Jolla, Calif.), which is a strain of bacteria containing additional copies of the argU, ileY, and leuW tRNA genes to correct for codon usage in B. burgdorferi gene sequences. A His6- C-terminal tag was incorporated into each of the recombinant proteins to aid in purification.
TABLE 1.
Oligonucleotide primers used for cloning borrelial oppA genesa
| Name | Sequence | PCR product (kb) |
|---|---|---|
| A1-F | 5′-ATACCATGGGCATGAAATATATAAAAA-3′ | oppAI (1.57) |
| A1-R | 5′-GCTCGAGTTTTTTAGTTTTAATA-3′ | |
| A2-F | 5′-ACATATGAAATTACAAAGGTC-3′ | oppAII (1.58) |
| A2-R | 5′-GCTCGAGTTTATTTTTTAATTTTA-3′ | |
| A3-F | 5′-ACCATATGAGCTTTAATAAAACTA-3′ | oppAIII (1.62) |
| A3-R | 5′-AGCTCGAGATTATGTTTTGCATT-3′ | |
| A4-F | 5′-ACATATGAAAATATTGATAAA-3′ | oppAIV (1.59) |
| A4-R | 5′-GCTCGAGTTTAATTGGTTTTAT-3′ | |
| A5-F | 5′-ACATATGATAATAAAAAAAAG-3′ | oppAV (1.59) |
| A5-R | 5′-GCTCGAGTTCTTCTATAGGTTT-3′ |
Underlining indicates restriction site NdeI (except for NcoI in A1-F) in the forward (F) primers and restriction site XhoI in the reverse (R) primers.
Preparation of recombinant OppA proteins.
Single colonies of bacterial strains transformed with plasmids containing genes for the individual OppA proteins were selected from plates and grown overnight in Luria-Bertani (LB) medium at 37°C. The overnight cultures then were seeded into larger flasks of LB medium at a dilution of 1:1,000 and grown to an optical density at 600 nm of 0.5. The cells were induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h at 25°C and finally harvested by centrifugation at 2,000 × g for 20 min at 4°C.
Cell pellets were suspended in buffer A (50 mM sodium phosphate buffer [pH 7.4], 100 mM NaCl, 0.5% dodecyl-β-d-maltoside, 10% glycerol). Lysozyme (final concentration, 100 μg/ml) was added to the suspension. The cell solution was rotated for 1 h at 30°C and then sonicated. The cell lysate was centrifuged at 2,000 × g, and the supernatant was collected. The supernatant was further clarified by a second centrifugation at 12,000 × g for 15 min. The supernatant was mixed with TALON resin (BD Biosciences Clontech, Palo Alto, Calif.) equilibrated in buffer A. The mixture was rotated at 4°C for 2 h. The resin was collected by centrifugation at 2,000 × g for 10 min at 4°C and then washed three times with a 10-fold volume of buffer B (50 mM sodium phosphate buffer [pH 7.4], 100 mM NaCl, 10% glycerol, 0.05% Triton X-100). The column was eluted with a gradient of increasing imidazole concentrations (5 to 250 mM) in buffer B. Fractions containing target proteins were determined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE).
Fractions containing recombinant proteins were pooled and dialyzed repeatedly against a 1,000-fold volume of buffer C (50 mM sodium phosphate buffer [pH 7.4], 100 mM NaCl, 10% glycerol) at 4°C. The dialyzed solution was mixed with TALON resin, and the initial purification steps were repeated. As a final purification step, the purified proteins were applied to a Sephacryl S-200 column; buffer C was used for equilibration and elution. Collected fractions were concentrated with Centricon YM-30 (Millipore, Billerica, Mass.) and stored in buffer C at 4°C.
Endogenous peptide ligands that may have been bound to recombinant OppA proteins were removed by using a modification of a previously described procedure (25). Purified proteins were dissolved in acetonitrile (50% [vol/vol]) containing trifluoroacetic acid (0.1% [wt/vol]). Proteins were dialyzed against solutions with progressively decreasing concentrations of acetonitrile and trifluoroacetic acid and then finally dialyzed against buffer C. Protein purity was checked by SDS-PAGE. Protein concentrations were estimated by spectrophotometry at a wavelength of 280 nm, and absorption coefficients were calculated for individual OppA proteins on the basis of amino acid sequences in ProtParam(ExPASy). The following absorption coefficients were obtained: 1.35 mg−1 cm−1 for OppA-1, 1.38 mg−1 cm−1 for OppA-2, 1.25 mg−1 cm−1 for OppA-3, 1.28 mg−1 cm−1 for OppA-4, and 1.25 mg−1 cm−1 for OppA-5.
IEF.
OppA protein samples were loaded onto IEF Phast Gels with a pH range of 3 to 9, and isoelectric focusing (IEF) was performed with a Phast separation control system in accordance with the manufacturer's instructions (Amersham Biosciences, Piscataway, N.J.). Gels were subsequently fixed with 20% trichloroacetic acid for 20 min at room temperature, followed by washing with a solution of 10% acetic acid and 30% methanol for 2 h at room temperature. Gels were stained with staining solution (0.02% Phast Gel Blue R, 0.1% copper sulfate, 10% acetic acid, 30% methanol) and destained with a solution of 10% acetic acid and 30% methanol in accordance with the manufacturer's instructions (Amersham Biosciences).
Western blotting.
Western blotting was performed as described previously (18) to confirm the identities of the purified proteins. Antibodies to the OppA proteins were kind gifts from James Bono and Patricia Rosa (Rocky Mountain Laboratories, Hamilton, Mont.).
Screening of the phage display peptide library.
A Ph.D.-7 phage display peptide library kit (New England Biolabs, Beverly, Mass.) was used to select for peptides that bind to the B. burgdorferi OppA proteins. Screening of the phage display peptide library was performed in accordance with the manufacturer's recommendations with the following modifications. Each recombinant OppA protein was diluted with 0.2% PBSA (50 mM sodium phosphate [pH 7.4], 50 mM NaCl, 0.2% bovine serum albumin [BSA]) to a final concentration of 10 μg/ml. Proteins were bound to wells of a Ni-nitrilotriacetic acid HisSorb plate (Qiagen) by adding 150 μl of protein solution to each well and incubating the samples for 2 h at 4°C. The plate was incubated with gentle shaking for 30 min at room temperature. Each well was washed six times with 0.1% Tween 20-Tris-buffered saline (TBST) prior to the addition of 100 μl of phage solution (2 × 1011 PFU). After 1 h of incubation, each well was washed 15 times with 0.1% TBST to remove unbound phage. Bound phage were eluted by the addition of 100 μl of elution solution (0.2 M glycine-HCl [pH 2.2], 1 mg of BSA/ml) and mixed promptly with 15 μl of 1 M Tris-HCl (pH 9.1). The eluted phage virions then were amplified in E. coli ER2738. Purified phage were collected from the supernatants of bacterial cultures. This entire procedure was repeated for three or four rounds to select for phage expressing peptides with the highest affinities for the recombinant proteins. The stringency of the washes in the subsequent rounds was increased by increasing the concentration of Tween 20 in the TBST washes to 0.2% and then 0.5%.
After the final round of screening, the eluted phage were allowed to infect E. coli and then were plated on LB agar supplemented with 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal)/ml and 83 μg of IPTG/ml. Positive plaques were selected and amplified individually. Single-stranded phage DNA was extracted in accordance with the manufacturer's instructions. DNA sequences encoding an exogenous heptapeptide were determined by direct DNA sequencing with a specific −96 gIII primer (New England Biolabs).
Competitive binding assay.
The ability of each phage to bind to OppA proteins was established by an enzyme immunoassay (EIA). OppA proteins were used to coat a 96-well Ni-nitrilotriacetic acid (HisSorb; Qiagen) plate as described above. After the plate was washed six times with 0.1% TBST, the amount of a recombinant protein bound to the wells was quantitated by the EIA with an antibody specific for the specific protein. A total of 109 PFU of a phage from a single clone then was added to each well. The plate was incubated with gentle rocking for 1 h at room temperature and then washed 15 times with 0.2% TBST. Horseradish peroxidase (HRP)-conjugated anti-M13 antibody (Amersham Biosciences) diluted in 0.1% TBST (1:5,000) then was added to each well. After incubation with gentle shaking for 1 h at 25°C, the plate was washed eight times with 0.1% TBST. HRP substrate solution (Bio-Rad, Hercules, Calif.) was added, and the OD405 was determined. The binding of phage selected by rounds of screening against a specific OppA protein was compared to the binding of unselected phage. The binding of phage to wells coated with BSA and treated in an identical manner was used as a control for nonspecific binding. All assays were performed in duplicate. For competition experiments, the same procedure was used, except that 20 to 50 μM peptide was added with the phage during the initial binding step. Peptides were purchased from Bachem or synthesized at the Tufts University Molecular Biology Core Facility. The competition efficiency was estimated with the following equation: percent inhibition = {1 − [(R405 − Cb405)/(Cp405 − Cb405)]} × 100. In this equation, R405 is the average value of each duplicate reaction at 405 nm, Cb405 is the average value for the blank control, and Cp405 is the average value for the positive control.
RESULTS
Expression, purification, and characterization of B. burgdorferi OppA proteins.
Recombinant OppA-1, OppA-2, OppA-3, OppA-4, and OppA-5 proteins were expressed with a C-terminal His6 tag. Each OppA protein was purified to homogeneity by metal ion affinity chromatography and gel filtration chromatography. The purities of the recombinant proteins were measured by scanning densitometry.
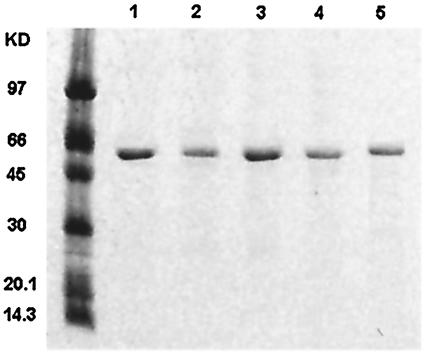
Several approaches were used to confirm the identity of each recombinant OppA. During purification, each OppA protein was examined by SDS-PAGE, and its mass was compared to the molecular mass predicted from the amino acid sequence. Then, Western blotting was used to detect the specific interaction between each OppA protein and antibody raised against a truncated OppA polypeptide. SDS-PAGE (Fig. 1) and Western blotting (data not shown) showed that the single, sharp protein band with the molecular mass (59.9 kDa for OppA-1, 60.7 kDa for OppA-2, 62.4 kDa for OppA-3, 60.8 kDa for OppA-4, and 61 kDa for OppA-5) predicted by sequence analysis interacted strongly with the OppA-specific antibody in each case. The pI for each pure OppA protein was established by isoelectric focusing. The pI for each purified protein (data not shown) was consistent with the value calculated on the basis of the amino acid sequence for each OppA protein (5.64 for OppA-1, 5.64 for OppA-2, 9.16 for OppA-3, 6.15 for OppA-4, and 5.5 for OppA-5).
FIG. 1.
Purification of B. burgdorferi OppA proteins. Histadine-tagged recombinant B. burgdorferi OppA proteins were expressed in E. coli. Recombinant proteins were purified by using a cobalt affinity column and a Sephacryl-200 gel filtration column. The purified products were analyzed by SDS-PAGE on a 12% acylamide gel and stained with Coomassie blue stain. Lanes: 1, OppA-1; 2, OppA-2; 3, OppA-3; 4, OppA-4; 5, OppA-5. The leftmost lane shows molecular mass markers.
Binding of phage expressing heptapeptides to B. burgdorferi OppA proteins.
Because the peptide binding characteristics of the OppA proteins were unknown, we used a combinatorial phage display peptide library to identify peptides with a high affinity for each OppA. In the phage library, all possible random combinations of seven amino acids are represented and expressed as a linear heptapeptide on the surface of the phage as part of a pIII fusion protein. The library contains approximately 70 copies of each of the 1.28 × 109 possible seven-residue sequences. Each virion displays an average of five copies of the encoded peptide on its surface. Phage expressing peptides with an affinity for a specific OppA are enriched in the pool of phage by mixing the library with the recombinant protein, washing away unbound phage, and eluting bound phage particles. Bound phage are amplified, pooled, and reselected against the recombinant protein three times. This strategy is designed to enrich for phage expressing peptides with the highest affinity for a specific protein and is not designed to evaluate the totality of peptides that may show lesser binding to that protein. Our goal in using the phage display library was to identify individual peptides that had a high affinity for a specific OppA and that could be used in subsequent assays to determine specificity.
After three rounds of screening, phage expressing three unique peptides, FGTRWFN, SGTKFFN, and FTTNQQD, were found to bind strongly to OppA-1. These three peptides were the only binding sequences found after the sequencing of more than 20 different phage. These sequences comprised >80% of sequenced phage after three rounds of panning; the remaining ∼20% of sequenced phage did not have a high affinity for OppA-1 on subsequent testing and likely represented nonspecifically bound phage. A similar screening of the phage library against OppA-2 and OppA-3 resulted in the identification of a single peptide with an affinity for OppA-2, HSMPTVL, and two other sequences, AVPQSAL and LTPPTSL, with a high binding affinity for OppA-3. In contrast, three rounds of screening of the phage library for binding to OppA-4 or OppA-5 did not result in the identification of phage expressing peptides with a high affinity for either of these proteins. Despite changes in the stringency of the wash conditions, pH, concentration of the phage, and other binding conditions, we were unable to identify any peptides that were expressed in the phage library and that bound to either OppA-4 or OppA-5.
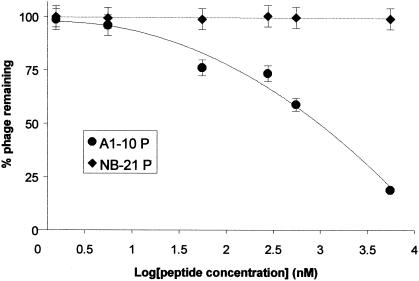
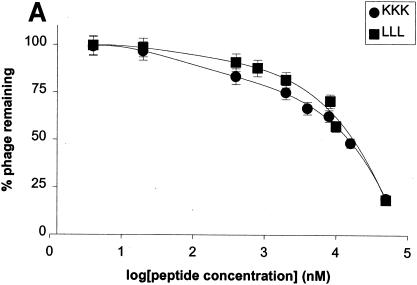
In order to confirm that binding to the OppA proteins was due to expression of the specific peptide and not to nonspecific binding of the phage, we first went back to the unscreened library and selected individual phage at random. None of over 20 randomly selected phage virions, which differed only in the sequence of the peptide expressed on their surface, bound to any OppA protein—suggesting that binding is not a result of the presence of the phage itself. Next, to further confirm binding specificity, we synthesized a 7-amino-acid peptide with a sequence identical to that expressed on phage A1-10. This peptide, A1-10P, was used in competitive inhibition experiments to determine whether the presence of the peptide could block the binding of the phage expressing the identical peptide. A peptide synthesized from the sequence of a nonbinding phage, NB-21P, was used as a control. A 6 μM concentration of peptide A1-10P inhibited the binding of phage A1-10 to OppA-1 by 91%; the same concentration of the negative control peptide NB-21P did not inhibit the binding of phage A1-10 to OppA-1. Inhibition by A1-10P was concentration dependent: increasing inhibition occurred with increasing peptide concentration (Fig. 2). The 50% inhibitory concentration for A1-10P is about 976 nM. A1-10P was also able to compete for binding of the other OppA-1-selected phage, A1-9 and A1-64, although inhibition was weaker than for A1-10 (30 and 37% versus 91% at a concentration of 6 μM). Again, the negative control peptide NB-21P did not inhibit OppA-1 binding to either A1-9 or A1-64. These results clearly demonstrate that binding in our assays was due to interactions of the OppA proteins with the expressed heptapeptides and not with other elements of the phage particle.
FIG. 2.
Dose-response plot of peptide competition assays. The ability of peptides to inhibit the binding of phage A1-10 to OppA-1 as a function of concentration was tested. A1-10 at 109 PFU was bound to OppA-1 in the presence of various concentrations of peptides. Quantitation of bound phage after washing was performed by an EIA. Results are shown for peptides A1-10P, which contains the same seven amino acids as those expressed by phage A1-10, and NB-21P, which contains amino acids identical to those of a heptamer expressed by a nonbinding phage. Error bars represent standard errors of the means.
Specificity of phage selected against an OppA protein for other OppA proteins.
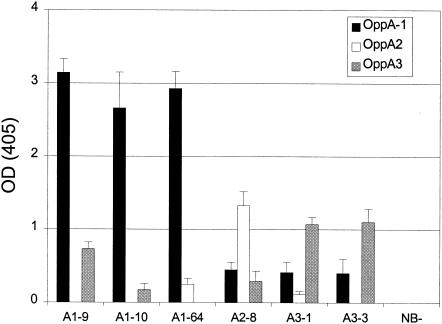
The specificity of the identified peptides for each OppA protein was tested by crossover binding experiments where phage selected against one protein were checked for binding to the other recombinant proteins. Amounts of bound phage were compared by an EIA (Fig. 3). OppA-1 showed only weak affinity for phage selected for binding to OppA-2 (A2-8) and OppA-3 (A3-1 and A3-3) compared with its affinity for phage selected for binding to OppA-1. OppA-2 showed slight binding to A1-64 and A3-1 but no binding to A1-9, A1-10, and A3-3. OppA-3 showed binding to A1-9 and A2-8 equivalent to the binding of phage selected against OppA-3 (A3-1 and A3-3). OppA-3 bound A1-10 very weakly and A1-64 not at all. Again, none of the OppA proteins showed any binding to NB-21. These results indicate that the borrelial OppA proteins have overlapping but nonidentical substrate specificities.
FIG. 3.
Binding of selected phage to heterologous OppA proteins. The ability of phage selected for binding to a specific OppA protein to bind to other OppA proteins was tested. Proteins were used to coat wells of a 96-well plate and were incubated in the presence of phage expressing a specific heptamer. Phage titers of 1010 PFU for A1-9, A1-10, and A1-64 and of 1011 PFU for A2-8, A3-1, A3-3, and NB-21 (NB-) were used. After the wells were washed, bound phage were quantitated with an HRP-conjugated anti-M13 antibody. The OD405 was determined after incubation with an HRP-labeled substrate. Error bars represent standard errors of the means from three experiments performed in duplicate.
Effects of pH on OppA ligand binding.
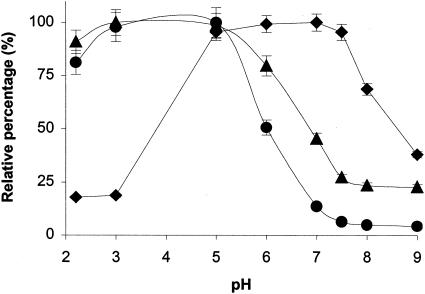
To investigate whether changes in pH affect the affinity of OppA for its peptide substrates, phage A1-9, which binds to both OppA-1 and OppA-3, was diluted to the final concentration of 109 PFU in different pH buffers (0.2 M glycine-HCl for pHs 2.2 and 3; 100 mM Tris-HCl for pHs 5, 6, 7. 7.5, 8, and 9). Binding assays were performed as before, and phage binding was measured by an EIA. The results are shown in Fig. 4. The binding of phage A1-9 to OppA-1 remained relatively constant over the range of pHs 5 to 7.5 but decreased at pHs below 5 and above 7.5. In contrast, OppA-3 showed the highest affinity for A1-9 at pHs 2.2 to 5, followed by a gradual decrease in binding at pHs higher than 5. At pH 8, binding of the phage was only 20% its maximum. The binding of A2-8 to OppA-2 at various pHs paralleled the results seen with OppA-3, with the highest binding in the pH range of 2.2 to 5 and decreased binding above pH 5. At pH 7, there was only approximately 10% maximal binding, and above pH 8, there was essentially no binding of phage to OppA-2.
FIG. 4.
Effect of pH on phage binding. The binding of A1-9 to OppA-1 (black diamonds) and OppA-3 (black triangles) and A2-8 to OppA-2 (black circles) was tested over a range of pHs. Phage were allowed to bind to OppA proteins in buffers with defined pHs prior to washing with binding buffer at the same pHs. The amount of bound phage was quantitated by an EIA with HRP-conjugated anti-M13 antibody. The relative percentage at each pH was calculated by dividing the value at each pH by the highest binding value. Each point represents the mean of a minimum of three experiments performed in duplicate; error bars represent standard errors of the means.
Determination of OppA substrate specificities by competitive inhibition.
Having shown that heptapeptides expressing the same sequence as the phage could successfully inhibit phage binding to OppA proteins, we next sought to define more systematically the substrate specificities of the OppA proteins by using a panel of small peptides with different biochemical characteristics. To confirm the validity of this approach, we first tested two tripeptides, His-Pro-Leu and Orn-Orn-Orn, that had been previously studied. Using a complementation model with E. coli, we found that B. burgdorferi OppA proteins were all capable of transporting His-Pro-Leu, while none of these proteins facilitated the transport of Orn-Orn-Orn (18). To establish the optimal conditions for performing the competitive assays and to minimize nonspecific binding, we first examined the binding of phage at various concentrations. Phage titers of 109 PFU for OppA-1 and 1010 PFU for OppA-2 and OppA-3, which are located in the linear portion of the binding curve, were selected for the competitive assays. The binding of A1-10, A2-8, and A3-3 to OppA-1, OppA-2, and OppA-3, respectively, was compared in the presence or absence of 20 or 50 μM His-Pro-Leu or Orn-Orn-Orn. His-Pro-Leu inhibited phage binding to OppA-1, OppA-2, and OppA-3 by 18, 12, and 17% at 20 μM and by 21, 20, and 42% at 50 mM, respectively. Orn-Orn-Orn did not inhibit phage binding to any of the proteins.
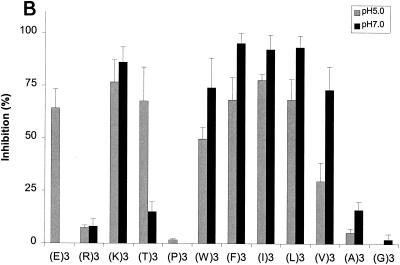
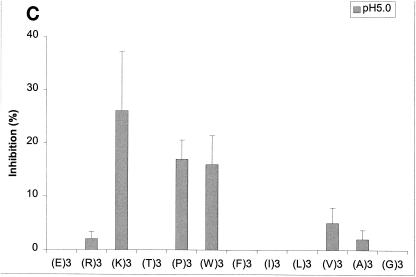
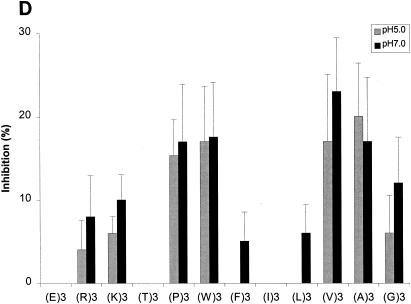
With confirmation that the results from our competitive inhibition assays were consistent with the results from previous functional assays, we next tested a series of peptides selected to assess the impact of various biochemical properties on the binding of these peptides to various OppA proteins. The tripeptides EEE, RRR, KKK, TTT, PPP,WWW, FFF, III, LLL, VVV, AAA, and GGG were each tested for the ability to inhibit the binding of phage A1-10, A2-8, and A3-3 to OppA-1, OppA-2, and OppA-3, respectively. Competitive binding assays for OppA-1 and OppA-3 were carried out at both pH 5.0 and pH 7.0, while assays for OppA-2 were performed only at pH 5.0 due to lack of binding of the phage at pH 7.0. A representative plot showing the direct correlation of inhibition with peptide concentration is shown in Fig. 5A. Data for all of the peptides at 20 and 50 μM concentrations are shown in Fig. 5B to D. Because different phage with different affinities for various OppA proteins were used, direct comparisons based on percent inhibition cannot be made. However, the ability of a specific peptide to compete for binding with a specific phage-OppA combination can be compared with the abilities of other peptides to compete for binding with the same phage-OppA combination, thus allowing relative comparisons of affinity. Inhibition of phage binding to OppA-1 was strongest with tripeptides with nonpolar, bulky hydrophobic residues (e.g., VVV, LLL, III, FFF, and WWW), although KKK also showed strong inhibition. The binding of EEE and TTT appeared to be highly pH dependent, with much higher inhibition at pH 5.0 than at pH 7.0. One possible explanation for this finding for EEE is that protonation of the carboxyl group of the side chain may occur at a low pH. In contrast to OppA-1, OppA-2 did not exhibit substantial binding to FFF, III, and LLL but did bind to PPP, which was a poor inhibitor for OppA-1. OppA-2 also showed an affinity for two peptides that also bound to OppA-1, KKK and WWW. Compared with OppA-1, OppA-3 showed a higher relative affinity for amino acids with small nonpolar side chains (GGG, AAA, and VVV). As with both OppA-1 and OppA-2, KKK and WWW also showed strong inhibitory properties. Like OppA-2, OppA-3 appeared to have a strong affinity for PPP.
FIG. 5.
Effects of amino acyl side chains on peptide binding. (A) Representative dose-response graph of peptide inhibition of binding of phage A1-10 to OppA-1. Data points represent the mean of a minimum of three experiments performed in duplicate; error bars represent standard errors of the means. (B to D) Abilities of a panel of 12 peptides [e.g., (E)3, which represents EEE] with various characteristics in their amino acyl side chains to compete for binding to OppA proteins. Assays were performed with 20 μM concentrations of peptides. Binding at both pH 5.0 and pH 7.0 was studied for OppA-1 (B) and OppA-3 (D). Only pH 5.0 was used for assays with OppA-2 (C) due to the low activity of the protein at pH 7.0. Phage A1-10, A2-8, and A3-3 were used as the competitive ligands for OppA-1, OppA-2, and OppA-3, respectively. Experiments were performed a minimum of three times in duplicate; error bars represent standard errors of the means.
Impact of peptide length on OppA binding.
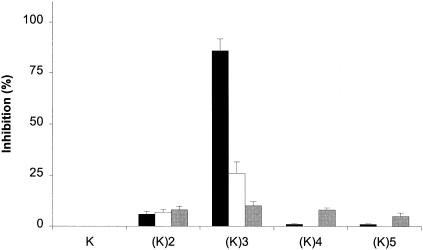
Using the competitive assays described above, we also tested the abilities of amino acids and peptides of various lengths to inhibit the binding of heptapeptide-expressing phage to OppA proteins. None of the amino acids tested, including l-lysine and l-tryptophan, whose tripeptides bound to all three OppA proteins, showed any inhibition of binding. Because the tripeptide KKK showed an affinity for all three OppA proteins, we tested lysine peptide chains of various lengths (Fig. 6). The lysine dipeptide showed some inhibition of specific phage binding to each of the OppA proteins; however, this inhibition was in the range of 5 to 10%, which is far less than that seen with the tripeptide KKK. Similarly, the tetrapeptide KKKK and the pentapeptide KKKKK also showed substantially less ability to compete for phage binding to any of the OppA proteins than the tripeptide.
FIG. 6.
Effects of peptide chain length on binding to OppA proteins. Competitive binding assays were performed with lysine-containing peptides of various lengths [e.g., (K)2, which represents KK]. The abilities to compete for binding were compared for A1-10 and OppA-1 (black bars), A2-8 and OppA-2 (white bars), and A3-3 and OppA-3 (gray bars). Assays were performed at pH 7.0 for OppA-1 and OppA-3 and at pH 5.0 for OppA-2. In each assay, a 20 μM concentration of amino acid or peptide was used as a competitor. Experiments were performed a minimum of three times in duplicate; error bars represent standard errors of the means.
Stereochemical specificity.
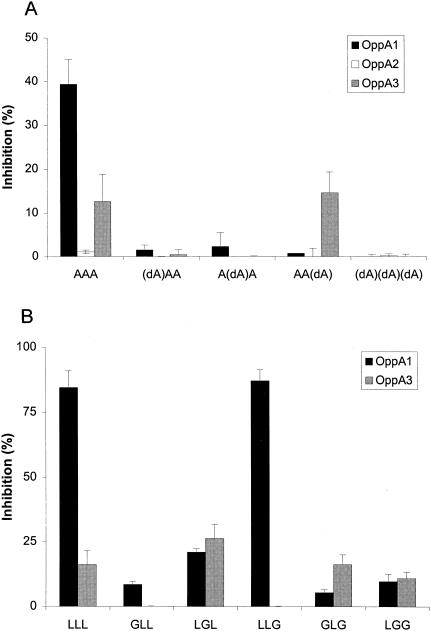
To investigate whether d-amino acids in a peptide sequence undermine the competitive ability of the peptide, four alanyl tripeptides with d-alanine at different positions were used as competitors. In agreement with the results obtained for other well-studied OppA proteins (4, 5, 8), the borrelial OppA-1, OppA-2, and OppA-3 proteins also showed strong stereochemical preference for l-residues in peptide binding (Fig. 7). The presence of d-alanine at either the first or the second position was sufficient to abolish the competitive ability of a tripeptide. The introduction of a d-amino acid at the third position did not affect the binding of phage A3-3 to OppA-3, although binding to OppA-1 was still significantly inhibited. These results may indicate some increased flexibility in the C-terminal residues accepted by OppA-3.
FIG. 7.
Effects of stereochemistry on peptide binding. A d-amino acid (A) or glycine (B) was used as a substitution at various positions of peptide chains containing alanine or leucine, respectively. The abilities of the different peptides to compete with A1-10, A2-8, and A3-3 for binding to OppA-1, OppA-2, and OppA-3, respectively, were tested. Assays for OppA-1 and OppA-3 were performed at pH 7.0; assays for OppA-2 were performed at pH 5.0. Peptides were studied at a concentration of 20 μM. Experiments were performed a minimum of three times in duplicate; error bars represent standard errors of the means. dA, d-alanine; A, l-alanine.
In support of a role for the stereochemistry of a peptide in binding to OppA proteins, we obtained similar results in studies of glycine replacement in leucyl tripeptides. Glycine lacks a chiral center and thus allows rotation of the peptide. We studied the effect of serial replacements of glycine into a leucine-containing peptide on competition with the binding of A1-10 to OppA-1 and OppA-3. OppA-2 was not tested due to low binding of the tripeptide LLL. The introduction of a glycine at the first or second position resulted in significantly reduced competition, whereas the introduction of a glycine at the third position did not appear to affect binding to OppA-1. In contrast, substitution of glycine did not appear to affect binding to OppA-3 in a regimented manner, as GLG and LGG competed for binding just as well as did LLL, but GLL and LLG showed greatly reduced competition.
DISCUSSION
To date, all of the crystallized substrate binding proteins in the OppA/DppA family of proteins have been found to share similar tertiary folding patterns (23, 24, 27, 28). The binding of peptide ligands has been described as occurring through a “Venus flytrap” mechanism, where the two domains of OppA engulf the peptide (21). The interaction between the peptide ligand and the binding protein occurs through the formation of parallel and antiparallel β sheets; specific acidic and basic residues on the binding protein interact with the N and C termini of the peptide to form salt bridges, stabilizing the interaction. The side chains of peptides are accommodated in large hydrated pockets that allow for great flexibility in binding. This flexibility is reflected in the broad substrate specificities exhibited by the OppA proteins of E. coli and S. enterica serovar Typhimurium (13, 28). However, small changes in the structures of the active binding site and hydrated pockets can confer a higher specificity for a ligand of a certain length or conformation, such as is seen with E. coli DppA (25).
In this study, we have shown that the OppA proteins of B. burgdorferi exhibit substantial differences in biochemical properties and in substrate specificities from both the OppA/DppA proteins of other bacteria and from each other. Like E. coli OppA, B. burgdorferi OppA-1, OppA-2, and OppA-3 did not bind to amino acids and showed only weak affinities for dipeptides (9). However, unlike E. coli OppA, which shows the highest affinity for oligopeptides two to five amino acids in length (28), the three opp operon-encoded B. burgdorferi OppA proteins showed significant affinities for heptamers attached to a bulky phage virion. In this aspect, they may be more similar to OppA of L. lactis. L. lactis OppA has been shown to have a predilection for longer peptides and is thought to enclose the first six amino acids of peptide ligands (7, 15). The remainders of longer peptides stick out and may interact with the surface of a binding protein. Interestingly, despite the strong binding of the longer heptamers to the B. burgdorferi OppA proteins, the tetrapeptide KKKK and the pentapeptide KKKKK did not compete as effectively for OppA-1 or OppA-2 binding as the tripeptide KKK. One possible explanation for this finding is a high selectivity in the amino acid composition at positions 4, 5, and 6 of peptide ligands that has been described for L. lactis OppA. In this scenario, proline at positions 4, 5, and 6 may not stabilize peptide binding as well as the amino acids found at these positions on the binding heptamers. One of the differences seen for the B. burgdorferi OppA proteins was that the binding of KK, KKKK, and KKKKK to OppA-3 was not substantially different from the binding of KKK.
Because we were unable to find a heptamer-expressing phage that bound to either OppA-4 or OppA-5, we cannot comment on the size predilection of these proteins. Previous studies showed that both OppA-4 and OppA-5 are capable of transporting tripeptides in a complementation model. Thus, it is tempting to speculate that OppA-4 and OppA-5 may be more similar to E. coli OppA in preferring tripeptides to longer peptides. However, other possible explanations for our inability to identify heptamers binding to OppA-4 and OppA-5 include improper folding of the recombinant protein, instability of the protein, or activity limited to binding conditions that were not used in testing. Although we were able to show that borrelial OppA-4 and OppA-5, which complemented auxotrophic OppA− E. coli grown on minimal medium in which the required amino acids were supplied only in the form of a tripeptide, were capable of replacing native OppA function to support growth, the amount of affinity and transport required to support growth may be much lower than that required to differentiate binding phage in the system used here. Both oppA-4 and oppA-5 are isolated genes on separate plasmids rather than part of the opp operon, and it is certainly possible that their main function is not oligopeptide binding.
Two of the few tripeptides that do not bind well to E. coli OppA, GGG and PPP, also did not bind well to B. burgdorferi OppA-1. It has been postulated that the rigid pyrrolidine rings of PPP may prevent adoption of the conformation necessary for binding (9). Conversely, the fairly unrestricted range of rotational angles for GGG may reduce the effective concentration of a particular conformation required for binding to OppA. In contrast to OppA-1, both OppA-2 and OppA-3 showed relatively high affinities for PPP, and OppA-3 also showed good affinity for GGG. OppA-3 also showed greater tolerance for the substitution of glycine at various positions of a competitor peptide than did OppA-1. These results suggest that binding to OppA-3 may be more tolerant of changes in stereochemistry than binding to OppA-1. Overall, there was a trend toward OppA-1 showing a preference for peptides composed of bulky hydrophobic residues (more similar to L. lactis OppA), whereas OppA-2 and OppA-3 favored the binding of peptides composed of small nonpolar amino acids (more similar to E. coli and S. enterica serovar Typhimurium OppA). However, these preferences are not absolute (e.g., OppA-3 binds to WWW, which has a large side chain, very well), and it is clear that multiple factors are at play in the interaction between a specific peptide and an OppA protein. Analysis of the key interacting residues in the OppA binding pocket supports the dissimilarity of substrate specificities between B. burgdorferi OppA-1 and S. enterica serovar typhimurium OppA and the similarity of substrate specificities between B. burgdorferi OppA-2 and OppA-3 and S. enterica serovar Typhimurium OppA.
OppA-1, OppA-2, and OppA-3 also differ in their activities at different pHs. Binding activity for OppA-1 peaked at pHs 5.5 to 7.5, whereas peak binding activity for OppA-2 and OppA-3 occurred at pHs 2.0 to 5.0. OppA-2, in particular, was essentially inactive at a physiologic pH of 7.4. B. burgdorferi is likely to encounter significant variations in environmental pH as it moves between its tick and mouse hosts. The pH in the tick midgut where B. burgdorferi resides ranges from 6.9 to 7.4, depending on the stage of feeding; the pH in the tick salivary gland through which B. burgdorferi passes on its way to entering its mammalian hosts is approximately 9.5 (3, 22). The pH in mammalian tissues is 7.4; however, tissue pH may be significantly lower in areas of inflammation due to metabolic acidosis and will also be lower in phagolysosomes of inflammatory cells, where B. burgdorferi may reside in transit (4).
Our data raise the possibility that different OppA binding proteins may be active during different portions of the B. burgdorferi life cycle. Previous studies showed that OppA-1 is the most highly expressed of the five B. burgdorferi OppA proteins and that its expression is constitutively maintained over a wide range of environmental conditions and in both tick and mouse hosts. In contrast, OppA-2 expression is over 100-fold lower than that of OppA-1 in vitro and is undetectable by reverse transcriptase PCR in B. burgdorferi isolated from fed and unfed ticks. The expression of OppA-3 is intermediate between that of OppA-1 and that of OppA-2; however, the expression of OppA-3 is higher in unfed than in fed ticks and is much higher (∼15-fold) in B. burgdorferi isolated from mouse tissue.
The broad substrate specificity of OppA-1, combined with its high constitutive expression and activity at the most physiologically relevant pH, suggests that it is likely to play a role in essential metabolic functions of the organism, such as nutrient acquisition. The more limited substrate specificities of OppA-2 and OppA-3 (and of OppA-4 and OppA-5, as judged by their inability to bind heptapeptides), combined with the alteration of their expression by environmental conditions, suggest that these substrate binding proteins may play more specialized roles in the organism. Differences in substrate specificity and activity may give the organism an advantage in adapting to specific environmental conditions. Oligopeptide and dipeptide permeases of other bacteria have been shown to be involved in cellular functions including but not limited to chemotaxis, quorum sensing, and conjugation (1, 16, 17, 19). For Listeria monocytogenes, OppA was shown to be critical to environmental adaptation, allowing the bacteria to grow at a low temperature and survive intracellularly (2). Many potential roles for the five B. burgdorferi OppA proteins are possible. For example, could the increased expression in mouse tissue and the activity at a low pH of OppA-2 be used for signaling the presence of inflammatory immune cells and activating cellular functions, such as negative chemotaxis, or allow the acquisition of specific nutrients not otherwise available to the organism? Further investigations into the functions of the B. burgdorferi OppA orthologues will be necessary to understand the full spectrum of their roles in microbial pathogenesis.
Acknowledgments
We thank James Bono and Patricia Rosa for the kind gift of anti-OppA antibodies, Bo Lin for technical advice, and Jenifer Coburn for help with review of the manuscript.
This research was funded by grants R01 AI44240 (to L.T.H.), R01 AI50043 (to L.T.H.), R01 AI05336 (to M.S.K.), and R21 AI37241 (to M.S.K.) from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Abouhamad, W. N., M. Manson, M. M. Gibson, and C. F. Higgins. 1991. Peptide transport and chemotaxis in Escherichia coli and Salmonella typhimurium: characterization of the dipeptide permease (Dpp) and the dipeptide-binding protein. Mol. Microbiol. 5:1035-1047. [DOI] [PubMed] [Google Scholar]
- 2.Borezee, E., E. Pellegrini, and P. Berche. 2000. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect. Immun. 68:7069-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowman, A. S., J. R. Sauer, K. Zhu, and J. W. Dillwith. 1995. Biosynthesis of salivary prostaglandins in the lone star tick, Amblyomma americanum. Insect Biochem. Mol. Biol. 25:735-741. [DOI] [PubMed] [Google Scholar]
- 4.Carroll, J. A., C. F. Garon, and T. G. Schwan. 1999. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect. Immun. 67:3181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charbonnel, P., M. Lamarque, J. Piard, C. Gilbert, VJuillard, and D. Atlan. 2003. Diversity of oligopeptide transport specificity in Lactococcus lactis species. J. Biol. Chem. 278:14832-14840. [DOI] [PubMed] [Google Scholar]
- 6.Detmers, F. J., E. R. Kunji, F. C. Lanfermeijer, B. Poolman, and W. N. Konings. 1998. Kinetics and specificity of peptide uptake by the oligopeptide transport system of Lactococcus lactis. Biochemistry 37:16671-16679. [DOI] [PubMed] [Google Scholar]
- 7.Detmers, F. J., F. C. Lanfermeijer, R. Abele, R. W. Jack, R. Tampe, W. N. Konings, and B. Poolman. 2000. Combinatorial peptide libraries reveal the ligand-binding mechanism of the oligopeptide receptor OppA of Lactococcus lactis. Proc. Natl. Acad. Sci. USA 97:12487-12492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 9.Guyer, C. A., D. G. Morgan, and J. V. Staros. 1986. Binding specificity of the periplasmic oligopeptide-binding protein from Escherichia coli. J. Bacteriol. 168:775-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higgins, C. F., and M. M. Gibson. 1986. Peptide transport in bacteria. Methods Enzymol. 125:365-377. [DOI] [PubMed] [Google Scholar]
- 11.Higgins, C. F., and M. M. Hardie. 1983. Periplasmic protein associated with the oligopeptide permeases of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 155:1434-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgins, C. F., M. M. Hardie, D. Jamieson, and L. M. Powell. 1983. Genetic map of the opp (oligopeptide permease) locus of Salmonella typhimurium. J. Bacteriol. 153:830-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiles, I. D., L. M. Powell, and C. F. Higgins. 1987. Peptide transport in Salmonella typhimurium: molecular cloning and characterization of the oligopeptide permease genes. Mol. Gen. Genet. 206:101-109. [DOI] [PubMed] [Google Scholar]
- 14.Hu, L. T., S. D. Pratt, G. Perides, L. Katz, R. A. Rogers, and M. S. Klempner. 1997. Isolation, cloning, and expression of a 70-kilodalton plasminogen binding protein of Borrelia burgdorferi. Infect. Immun. 65:4989-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanfermeijer, F. C., F. J. Detmers, W. N. Konings, and B. Poolman. 2000. On the binding mechanism of the peptide receptor of the oligopeptide transport system of Lactococcus lactis. EMBO J. 19:3649-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazazzera, B. A., J. M. Solomon, and A. D. Grossman. 1997. An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis. Cell 89:917-925. [DOI] [PubMed] [Google Scholar]
- 17.Leonard, B. A. B., A. Podbielski, P. J. Hedberg, and G. M. Dunny. 1996. Enterococcus faecalis pheromone binding protein, PrgA, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc. Natl. Acad. Sci. USA 93:260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin, B., S. A. Short, M. Eskildsen, M. S. Klempner, and L. T. Hu. 2001. Functional testing of putative oligopeptide permease (Opp) proteins of Borrelia burgdorferi: a complementation model in opp(−) Escherichia coli. Biochim. Biophys. Acta 1499:222-231. [DOI] [PubMed] [Google Scholar]
- 19.Manson, M. D., V. Blank, G. Brade, and C. F. Higgins. 1986. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature 321:253-256. [DOI] [PubMed] [Google Scholar]
- 20.Payne, J. W., and M. W. Smith. 1994. Peptide transport by micro-organisms. Adv. Microb. Physiol. 36:1-80. [DOI] [PubMed] [Google Scholar]
- 21.Quiocho, F. A., and P. S. Ledvina. 1996. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol. Microbiol. 20:17-25. [DOI] [PubMed] [Google Scholar]
- 22.Ribeiro, J. M. 1988. The midgut hemolysin of Ixodes dammini (Acari:Ixodidae). J. Parasitol. 74:532-537. [PubMed] [Google Scholar]
- 23.Sleigh, S. H., P. R. Seavers, A. J. Wilkinson, J. E. Ladbury, and J. R. Tame. 1999. Crystallographic and calorimetric analysis of peptide binding to OppA protein. J. Mol. Biol. 291:393-415. [DOI] [PubMed] [Google Scholar]
- 24.Sleigh, S. H., J. R. Tame, E. J. Dodson, and A. J. Wilkinson. 1997. Peptide binding in OppA, the crystal structures of the periplasmic oligopeptide binding protein in the unliganded form and in complex with lysyllysine. Biochemistry 36:9747-9758. [DOI] [PubMed] [Google Scholar]
- 25.Smith, M. W., D. R. Tyreman, G. M. Payne, N. J. Marshall, and J. W. Payne. 1999. Substrate specificity of the periplasmic dipeptide-binding protein from Escherichia coli: experimental basis for the design of peptide prodrugs. Microbiology 145:2891-2901. [DOI] [PubMed] [Google Scholar]
- 26.Steere, A. C. 2001. Lyme disease. N. Engl. J. Med. 345:115-125. [DOI] [PubMed] [Google Scholar]
- 27.Tame, J. R., E. J. Dodson, G. Murshudov, C. F. Higgins, and A. J. Wilkinson. 1995. The crystal structures of the oligopeptide-binding protein OppA complexed with tripeptide and tetrapeptide ligands. Structure 3:1395-1406. [DOI] [PubMed] [Google Scholar]
- 28.Tame, J. R., G. N. Murshudov, E. J. Dodson, T. K. Neil, G. G. Dodson, C. F. Higgins, and A. J. Wilkinson. 1994. The structural basis of sequence-independent peptide binding by OppA protein. Science 264:1578-1581. [DOI] [PubMed] [Google Scholar]
- 29.Wang, X. G., B. Lin, J. M. Kidder, S. Telford, and L. T. Hu. 2002. Effects of environmental changes on expression of the oligopeptide permease (opp) genes of Borrelia burgdorferi. J. Bacteriol. 184:6198-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]