Abstract
The enteropathogen Campylobacter jejuni is a major worldwide health and economic burden, being one of the leading causes of bacterial gastroenteritis and commonly linked to postinfectious onset of autoimmune disease. Chickens are a major vector for human infection and even though variation in avian colonization level is heritable, no previous studies have identified regions of the genome associated with colonization resistance. We performed a genome-wide association study of resistance to C. jejuni colonization in the avian intestine by controlling for population structure, which revealed a risk locus with genome-wide significance spanning the T-cadherin (CDH13) gene. A second possible risk locus was also identified close to calmodulin (CALM1), a calcium-activated modulator of cadherin function. In addition, gene expression analysis of mRNA sequencing profiles revealed that the relative expression of the two genes is significantly associated with colonization resistance. Functional studies have previously demonstrated involvement of cadherins and calmodulin in C. jejuni intracellular invasion and colonization of human intestinal epithelial cells in vitro. Consistent with this finding, our analysis reveals that variation surrounding these genes is associated with avian colonization resistance in vivo and highlights their potential as possible targets for control of the bacterium in avian and human populations.
Keywords: GWAS, intestinal homeostasis, epithelial cell invasion, cadherin, calmodulin, immunity
Campylobacter jejuni is one of the main causes of bacterial gastrointestinal infection worldwide (Scallan et al. 2011; EFSA 2012) and represents a major public health burden. Campylobacteriosis is usually self-limiting but has been linked to postinfectious onset of the autoimmune diseases Guillain-Barré syndrome (McCarthy and Giesecke 2001; Tam et al. 2007), Miller Fisher syndrome (Koga et al. 2005), and reactive arthritis (Hill Gaston and Lillicrap 2003; Townes et al. 2008). Consumption of contaminated poultry is a common source of human infection. Although pathogenic in humans, C. jejuni is an intestinal commensal in most mammalian and avian hosts and frequently colonizes the chicken intestine at extremely high densities of 108–1010 CFU/g (Van Deun et al. 2008a; Lamb-Rosteski et al. 2008; Meade et al. 2009).
Chickens are born free of C. jejuni and only begin to acquire the bacterium at an average age of 2−3 wk (Newell and Fearnley 2003; van Gerwe et al. 2009). Once one individual in a flock becomes colonized, the bacterium spreads quite rapidly, with >95% of the flock colonized within several days (van Gerwe et al. 2009). However, C. jejuni colonization levels in the gastrointestinal tract can vary substantially between individual chickens, both within and between populations. Several previous studies have established that this variation in C. jejuni colonization level can be heritable, with significant differences in susceptibility observed between chicken lines (Stern et al. 1990; Boyd et al. 2005; Li et al. 2008). Although a quantitative trait loci mapping study has been referred to (Kaiser 2010), no attempts to identify the genes involved in this resistance by linkage mapping, genome-wide association or candidate-gene analysis have yet been published. The primary site of C. jejuni colonization in the chicken is within the cecum, where it populates the mucus layer overlying epithelial cells (Beery et al. 1988; Meade et al. 2009). Although epithelial cell attachment was not documented in vivo (Beery et al. 1988), attachment and invasion do occur in vitro and are thought to be necessary for successful colonization (Byrne et al. 2007; Hermans et al. 2011) and several adhesins involved have been identified (Hermans et al. 2011). The invasiveness of different strains of C. jejuni into chicken epithelial cells is correlated with systemic colonization (van Deun et al. 2008a), which commonly occurs in the spleen, liver, and bursa of Fabricius (Cox et al. 2005; van Deun et al. 2008a; Lamb-Rosteski et al. 2008; Meade et al. 2009) even though commensal bacteria do not normally disseminate systemically. Both transcellular and paracellular translocation have been described in human epithelial cells in vitro (Konkel et al. 1992; Grant et al. 1993; Monteville and Konkel 2002; van Deun et al. 2008b; Hu et al. 2008; Kalischuk et al. 2009; Boehm et al. 2012), and the translocation route in chicken epithelial cells is unknown. Strain invasiveness into human cells is correlated with chicken intestinal colonization potential (Hänel et al. 2004).
In several studies, researchers have analyzed gene expression in the chicken cecum after C. jejuni infection and have demonstrated a proinflammatory response (Borrmann et al. 2007; Smith et al. 2008; Larson et al. 2008), lymphocyte involvement (Li et al. 2010; Shaughnessy et al. 2011; Li et al. 2011), and activation of Toll-like receptors (de Zoete et al. 2010), the mitogen-activated protein kinase pathway, and small GTPase-mediated signal transduction (Li et al. 2011). We have previously described the separation of a single population of 255 4-wk-old Barred Rock chickens into those resistant and susceptible to C. jejuni colonization 48 hr postinfection and have investigated the caecal gene expression profiles of resistant and susceptible chickens by using high-throughput short-read sequencing of mRNA (RNAseq) (Connell et al. 2012). Resistance was associated with significantly increased expression of genes involved in the innate immune response, cytokine signaling, B-cell and T-cell activation, immunoglobulin production, and the renin-angiotensin system. In this complementary study, a genome-wide analysis of association with colonization status was performed with 194 chickens, selected from both ends of the colonization spectrum and including the 28 transcriptionally profiled birds. These were genotyped on a chicken 60K single-nucleotide polymorphism (SNP) chip (Groenen et al. 2011). Using a case-control model incorporating population stratification, we identified a significantly associated resistance locus comprising four SNPs in a region of chromosome 11, which suggests involvement of the CDH13 (T-cadherin) gene in resistance to colonization. A second putative locus was also identified on chromosome 5 close to the CALM1 (calmodulin) gene, a mediator of calcium signaling that modulates cadherin function. Investigation of gene expression in cases and controls revealed that the relative expression of these two genes is associated with colonization status. Together, this information strongly suggests a functional role for these genes in establishment of C. jejuni colonization within the avian host.
Materials and Methods
Population history
The population of chickens used in the study was obtained from a line maintained at the University of Saskatchewan, Saskatoon, Canada. The Barred Rock is a dual-purpose breed which was developed in the middle of the 19th century. The line was acquired in the 1920s, and was initially selected but was subsequently maintained unselected from 1965 to 2003. Mild selection for egg production was performed after 2003. The procedure for selection involved maintaining 80 hens per generation. A total of 50 hens and roosters from the same dams were then selected for reproduction based on egg production. Some genetic substructure within the population is therefore expected due to violations of several assumptions of Hardy-Weinberg equilibrium. This population substructure was characterized before association analysis, and tests used to detect significance of association were adjusted to incorporate genetic stratification identified within the population.
Sample selection, SNP genotyping, and quality control
Caecal colonization levels were estimated for all 255 chickens in the population 48 hr postinoculation and caecal samples collected as reported previously (C. jejuni strain NCTC11168v1) (Connell et al. 2012). Caecal samples from 196 individuals were selected for phenol-chloroform genomic DNA extraction (Sambrook and Russell 2001) and genotyping with the 60K chicken SNP chip (Groenen et al. 2011). Samples were genotyped commercially using the standard protocol from Illumina Infinium iSelect Beadchips implementing BeadStudio Genotyping v3.0.19.0. Forty birds with no or very minimal colonization (C. jejuni−resistant cases) and 128 birds with very high levels of colonization >107 CFU/g (C.jejuni-susceptible controls) were selected for case−control association analysis (Figure 1). Of the total number of 57,636 SNP positions queried on the chip, 25,385 SNPs were polymorphic in this population. Average identity by state (IBS) was calculated from all SNP genotypes in the R package GenABEL (Aulchenko et al. 2007). Strict quality control filtering of the data was carried out before association analysis. In association studies of complex diseases, where single alleles are expected to only have a small effect on the trait under study, genotyping errors are more likely to result in false-positive or false-negative findings of association; therefore, SNPs with low minor allele frequency (≤1.8%) were removed. All SNPs with call rate <95% were excluded. Two individuals with call rate <95% were excluded because they may represent poor-quality samples with unreliable genotypes. All SNPs exhibiting substantially higher or lower heterozygosity than expected (P value ≤ 10−7) under Hardy-Weinberg equilibrium were also excluded. Autosomal heterozygosity was also checked to ensure all individuals had moderate autosomal heterozygosity but no individuals had excessively high heterozygosity [false-discovery rate (FDR) <1%]. Genetic relatedness of all possible pairs of individuals was calculated to identify individuals too closely related to be used in the association study. A matrix of IBS values was created from all filtered SNPs. No pair of individuals were >95% IBS. Subsequently, SNPs were filtered so that SNPs in complete linkage disequilibrium (LD) on the same chromosome were represented by only one tag SNP. Each set of SNPs in complete LD was tested as a unit using these tag SNPs.
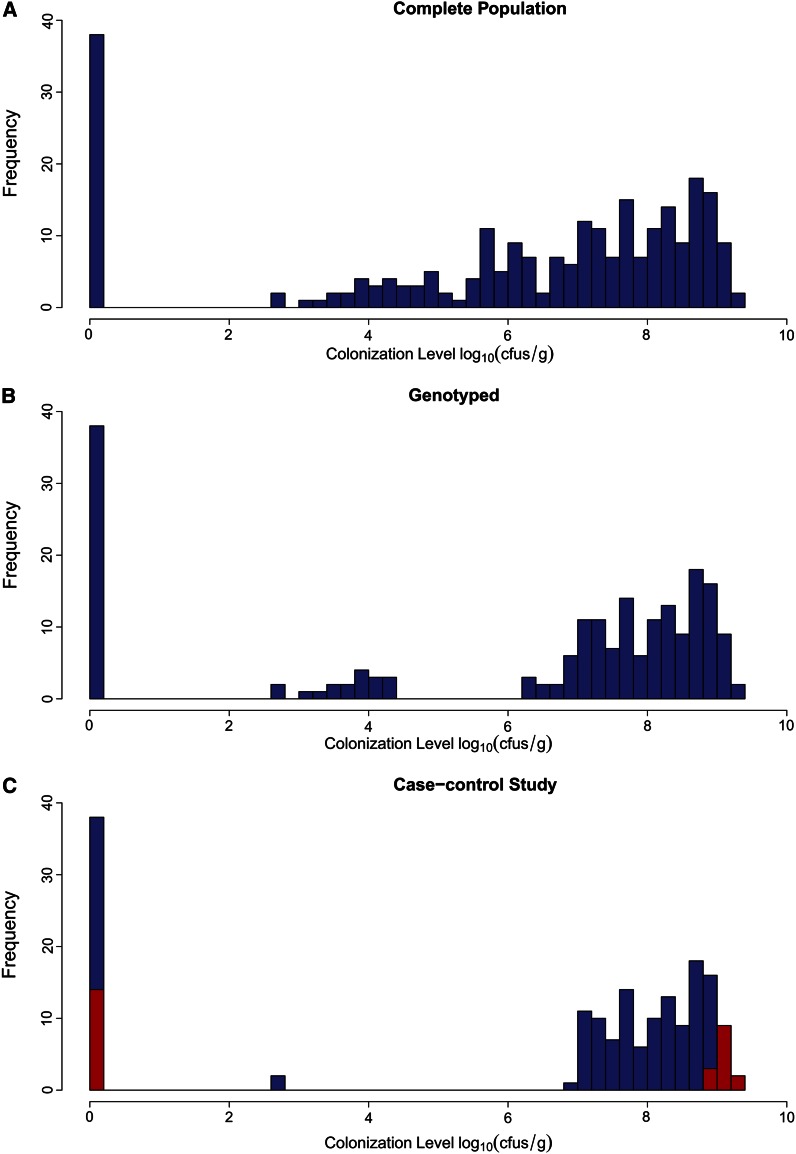
Figure 1.
Colonization status of chickens selected for the genome-wide association study. (A) The initial population of 255 chickens had highly variable colonization levels; 196 of these were selected for genotyping using the 60K chicken SNP chip. Their colonization levels are illustrated in (B). A total of 168 chickens from opposite ends of the colonization spectrum (i.e., all individuals with colonization level ≤400 CFU/g and all individuals with colonization level ≥107 CFU/g) were selected to conduct the case-control association study. Two individuals were filtered due to genotyping call-rate <95%. Colonization levels of all 166 individuals which passed quality control filtering for the case-control study are plotted in (C). Forty of these had no or very minimal colonization (resistant cases), and 126 had high colonization levels (susceptible controls). Birds for which RNAseq expression profiles are available are highlighted in red at the extremes of the distribution. The lower limit of detection = 400 CFU/g.
Detection of population stratification and case−control association analysis
To investigate any population stratification that may be present in the population, a matrix of genomic kinship was generated from all pairs of 194 individuals in the population that passed quality control filtering using genotypes of all filtered tag SNPs. This matrix of genomic kinship was converted to a distance matrix that was used to carry out classical multidimensional scaling analysis. All analysis of population stratification was conducted in GenABEL (Aulchenko et al. 2007). The qtscore function in GenABEL was implemented to carry out logistic regression of the trait onto SNP genotypes using the first three principal components of the genomic kinship matrix as covariates in the regression model, accounting for the genetic substructure within the population (Price et al. 2010). Overinflation of the test statistic was investigated by comparison of the distribution of the test statistic to that expected under the null hypothesis [inflation factor λ (Amin et al. 2007)]. Consistent deviation is indicative of inflation due to population substructure.
Gene expression analysis
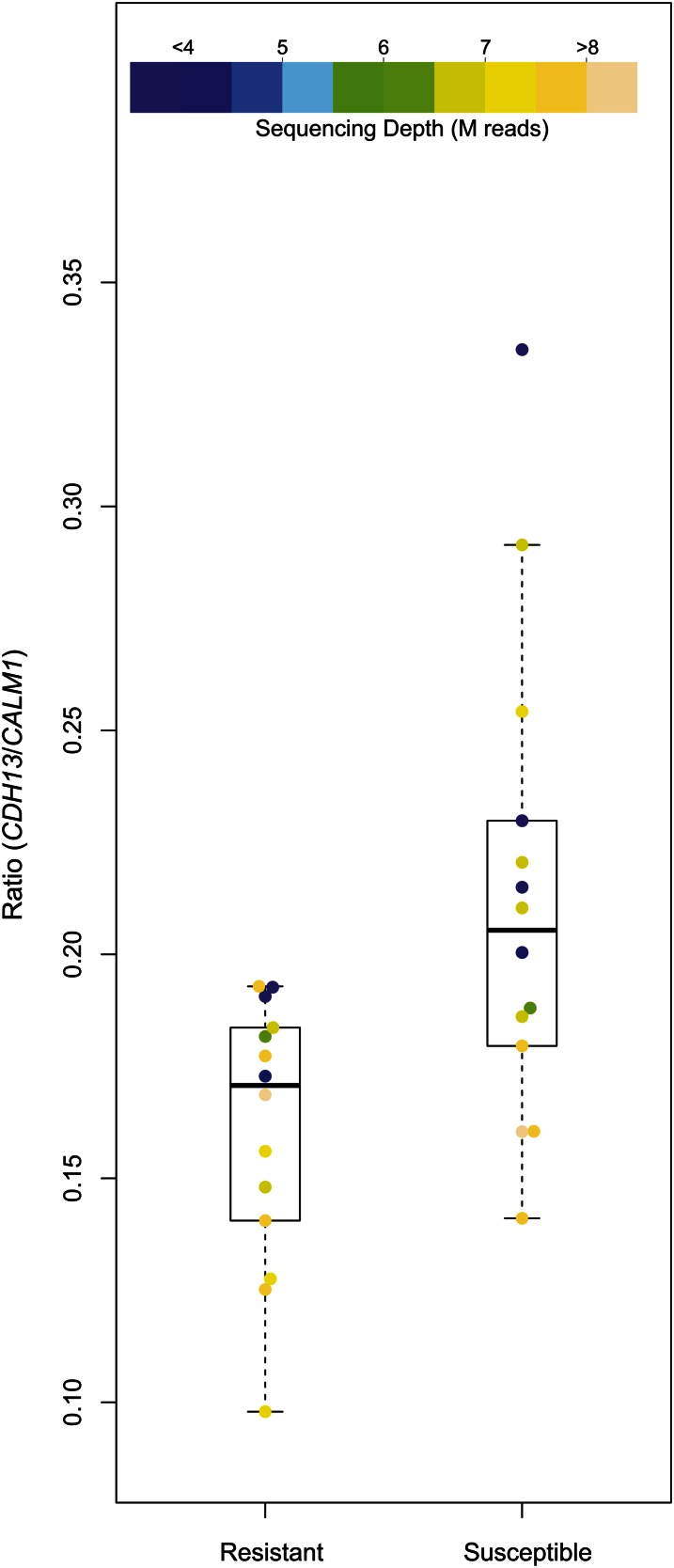
Read counts of gene expression were first corrected using upper quartile normalization and relative log expression in edgeR (Robinson et al. 2010). No correlation between SNP genotypes and relative expression levels was found. Similarly, no correlation between SNP genotypes in both genes was found. Significant differences in the ratios of CDH13/CALM1 expression (Figure 6) were observed between cases and controls (Mann-Whitney U-test, P = 0.004). All calculations were performed in R using the “lm” and “wilcox.test” functions.
Figure 6.
Relative expression levels of CDH13 and CALM1. Boxplots of resistant vs. susceptible birds reveal separation of the population based on CDH13/CALM1 expression (Mann-Whitney U-test, P = 0.004) and no bias in relative ratios due to sequencing depth. The ratio of CDH13/CALM1 expression for all 28 transcriptionally profiled birds is plotted, with shading reflecting depth of sequencing.
Results
Genotyping and quality control
Querying of the study population of 196 chickens using a 60K Illumina iSelect chip [37] revealed that of 57,636 SNP positions, just 25,385 were polymorphic in this population. All SNP genotypes and colonization status of individuals are detailed in Supporting Information, File S1 and File S2, respectively. Average IBS calculated using all SNP genotypes was 91.5% for all pairs of individuals, attesting to the high inbreeding present in the population due to its population history and maintenance as an inbred population since the 1920s. A total of 16,871 SNPs and 194 chickens passed quality control filtering (call-rate >95%, minor allele frequency < 1.8%, Hardy-Weinberg P-value < 10−7). After filtering out SNPs on the same chromosome in complete LD, 5609 nonredundant tag SNPs remained and were used in the analysis of population substructure and genome-wide association testing.
Multidimensional scaling reveals population stratification
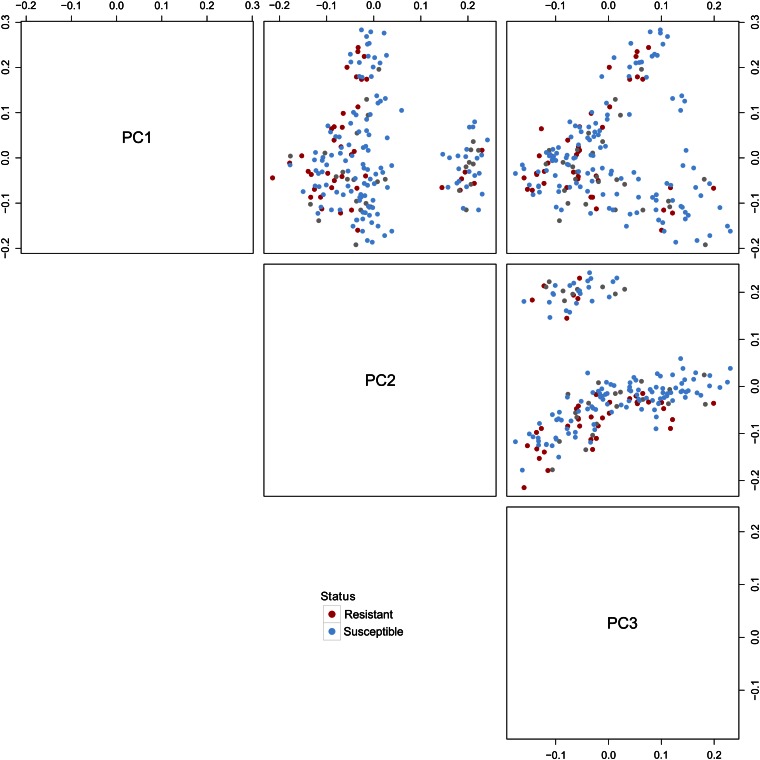
Population substructure can induce spurious associations in genetic association studies. It is therefore essential that population stratification is identified and adjusted for, especially in genome-wide association studies of complex traits in which SNPs may have only a small effect on the trait being analyzed. To characterize genetic substructure within the population, genomic kinship was investigated using the R package GenABEL (Aulchenko et al. 2007). First, a matrix of IBS values was calculated between all pairs of 194 individuals which passed quality control filtering using genotypes of the 5609 tag SNPs. Classical multidimensional scaling analysis was performed on the distance matrix derived from this matrix of kinship values. The major stratification within the population was resolved by the first three dimensions and these are plotted in Figure 2. This plot reveals some substructure within the population which is most likely due to breeding procedures, particularly those used to select for increased egg production (see Materials and Methods). Resistant cases and susceptible controls are distributed randomly throughout the plots indicating that they do not cluster together genetically.
Figure 2.
Principal components revealing population stratification. All 194 individuals that passed quality control filtering are represented. Colonization-susceptible individuals from the case-control study are shaded blue and colonization-resistant birds shaded red. Gray circles represent individuals from the center of the colonization spectrum that were not used in the association study. All autosomal markers were used to construct a kinship matrix of genetic relatedness or identity by descent (IBD). This matrix was converted to a distance matrix and classical multidimensional scaling performed to detect any population substructure. There is evidence for some stratification of the population which was resolved by the first three principal components (PC1, PC2, and PC3). The percentage variation which is represented by each of these principal components is 18.6% (PC1), 16.8% (PC2), and 11.1% (PC3). The first two principal components reveal separation of the population into two subpopulations—one large group with 157 individuals and one smaller group of 37 individuals.
Case−control study identifies loci associated with colonization resistance
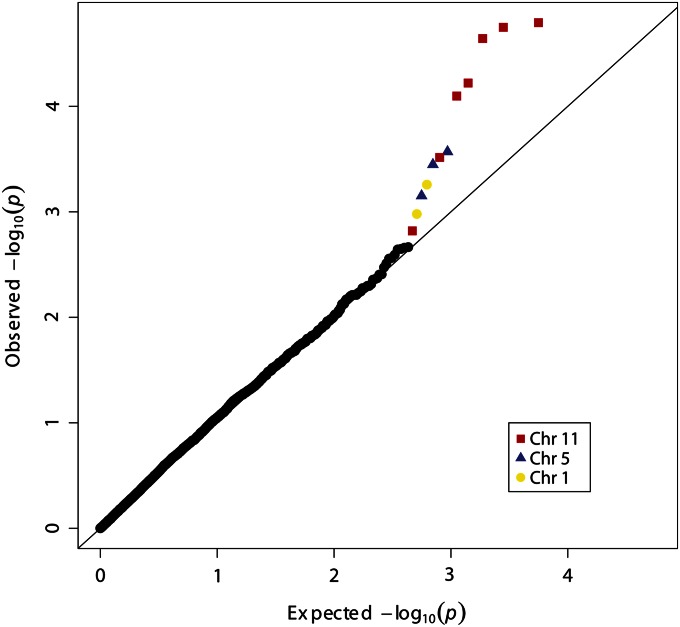
The qtscore function in GenABEL was applied to perform logistic regression of colonization status onto SNP genotypes. Population stratification was corrected for by using the first three principal components of the kinship matrix as covariates in the regression model. This yields a genomic inflation factor λ = 1.06 (compared with λ = 1.42 without adjustment for population stratification) indicating that the major substructure has been adjusted for. The quantile-quantile plot of observed P-values vs. the expected null distribution (Figure 3) did not reveal deviation from the null, apart from 12 SNP outliers at the tail of the distribution exhibiting the most significant P-values, which all lie within three regions of the genome—on chromosomes 11, 5, and 1. When these SNPs are not included the genomic inflation factor λ = 0.995. Combined, this finding suggests that these SNPs may represent truly associated regions of the genome and deviations are not due to over-inflation of the test statistic.
Figure 3.
Quantile-Quantile plot of observed vs. expected P-values. The q-q plot of observed P-values shows they follow the expected null distribution apart from 12 SNPs with the most significant P-values. These are located within three regions: chromosomes 11 (red squares), 5 (blue triangles), and 1 (yellow circles).
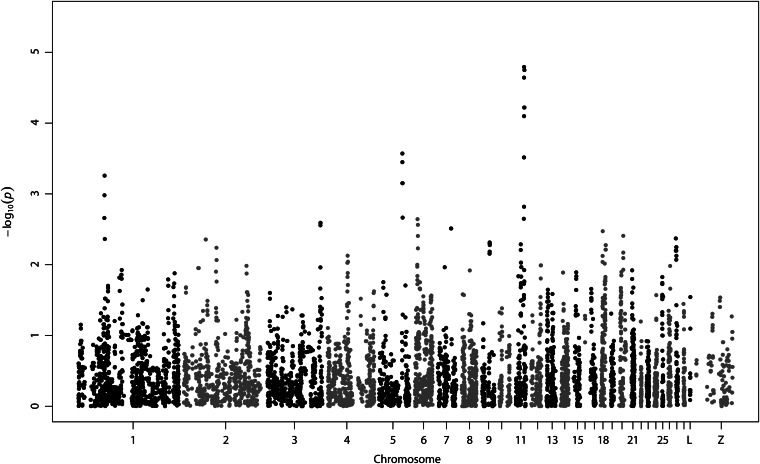
The genome-wide plot of SNP association significance is shown in Figure 4, where these three regions are again visible outliers in this plot, displaying high association P-values. The most significant associations identified involved SNPs on chromosome 11, where four SNPs reached genome-wide significance, after we controlled for multiple testing (FDR = 0.04), and odds ratios of 2.33−2.50 (Table 1). These SNPs are all in high LD, with two adjacent SNPs being in complete LD. They are located within a region spanning 80 kbp, are intergenic, and surround the CDH13 gene on chromosome 11 (Figure 5).
Figure 4.
Manhattan plot of genome-wide association. All 16,871 SNPs that passed quality control are shown. Genomic location is plotted against −log10(P). Four SNPs on chromosome 11 reached genome-wide significance (FDR = 0.04).
Table 1. Case-control study: top SNPs.
| SNP ID | Chr | Position | N | Allele Frequency | Odds Ratio (95% Confidence Interval) | P | FDR |
|---|---|---|---|---|---|---|---|
| Gga_rs15622247 | 11 | 17594419 | 166 | 0.40/0.21 | 2.50 (1.45−4.30) | 1.61E-05 | 0.0426 |
| GGaluGA079086 | 11 | 18376977 | 166 | 0.40/0.22 | 2.33 (1.35−3.99) | 1.78E-05 | 0.0426 |
| Gga_rs15622362 | 11 | 17655435 | 166 | 0.40/0.21 | 2.50 (1.41−4.19) | 2.28E-05 | 0.0426 |
| Gga_rs14027234 | 11 | 17657890 | 166 | 0.40/0.21 | 2.50 (1.41−4.19) | 2.28E-05 | 0.0426 |
SNPs exhibiting the most significant association with colonization status are listed, along with chromosome position. Four SNPs exhibited genome-wide significance after FDR control for multiple testing (FDR = 0.04). Minor allele frequencies in cases/controls and odds ratios with 95% confidence intervals are shown. Two SNPs (Gga_rs15622362 and Gga_rs14027234) are in complete LD. SNP, single-nucleotide polymorphism; FDR, false-discovery rate; LD, linkage disequilibrium.
Figure 5.
Regional plot of significant association on chromosome 11. Four SNPs with genome-wide significance FDR < 0.05 are highlighted red. These four SNPs are intergenic but surround the T-cadherin (CDH13) gene. Two of these SNPs are in complete LD, situated very close to each other, and are almost indistinguishable on the plot. The physical positions of all Refseq genes within the region are depicted below the plot.
Association with gene expression profiles
We have previously analyzed the gene expression profiles of 28 of these chickens (14 resistant cases with no C. jejuni colonization and 14 controls with the highest C. jejuni colonization levels) through mRNA sequencing (i.e., RNAseq) (Connell et al. 2012). All data from RNA sequencing gene expression analysis, including raw sequence files, can be downloaded from NCBI GEO accession no. GSE44341. As the second most significant region identified in this study approaches significance and surrounds another Ca2+-regulated gene, CALM1, associations between gene expression and SNP genotypes in both regions were investigated. No correlation with SNP genotypes was detected. Neither of these genes was differentially expressed between cases and controls. However relative CDH13/CALM1 gene expression is significantly associated with resistance to colonization (Mann-Whitney U-test, P = 0.004), with resistant birds displaying lower CDH13 and higher CALM1 relative expression levels (Figure 6).
Discussion
Several studies have demonstrated that resistance to C. jejuni colonization of the chicken gastrointestinal tract can be heritable, but this is the first to identify specific regions of the genome associated with colonization resistance. Association analysis treating colonization status as a binomial trait was performed and identified a resistance locus for C. jejuni colonization on chromosome 11 containing four significantly associated SNPs. This locus centers on the CDH13 (T-cadherin) gene, suggesting a functional role for this gene in the colonization process. In previous studies, authors have demonstrated a direct interaction between C. jejuni HtrA protease and another member of the cadherin superfamily, E-cadherin (cadherin 1, CDH1), in which cleavage of the protein facilitates transmigration across polarized epithelial cells in vivo (Hoy et al. 2012; Boehm et al. 2012). Because T-cadherin is expressed on the apical surface of chicken intestinal epithelial cells in vivo (Koller and Ranscht 1996) and has a highly similar ectodomain to E-cadherin, the interaction of T-cadherin with C. jejuni in the intestinal lumen is highly likely and variations altering the function or expression of this gene may plausibly affect caecal colonization status.
Cadherins are a superfamily of calcium-dependent proteins with prominent roles in homophilic cell−cell adhesion and maintenance of structural and functional tissue integrity (Halbleib and Nelson 2006). Classical cadherins are composed of an ectodomain containing 5 extracellular cadherin repeats, a transmembrane anchor, and an intracellular domain. The most extensively studied of the cadherins, E-cadherin (cadherin 1, CDH1), is a cell adhesion molecule and tumor suppressor that localizes to adherens junctions on the basolateral membrane of epithelial cells. Despite its typical location below tight junctions, it interacts with several bacteria, facilitating internalization and colonization. The protein can act as a receptor for Listeria monocytogenes and Candida albicans, allowing these pathogens to gain access into the cell (Mengaud et al. 1996; Phan et al. 2007). Cleavage of the protein is also a common feature of epithelial cell invasion. Candida albicans (Frank and Hostetter 2007; Villar et al. 2007), Helicobacter pylori (Hoy et al. 2010; Hoy et al. 2012), Shigella flexneri (Sansonetti et al. 1994; Hoy et al. 2012), Porphyromonas gingivalis (Katz et al. 2000; Katz et al. 2002), Bacteroides fragilis (Wu et al. 1998), and enteropathogenic Escherichia coli (Hoy et al. 2012) all cleave E-cadherin on the surface of human epithelial cells, leading to loss of the adherens junction and cell−cell adhesion, thus disrupting the epithelial barrier. Streptococcus pneumoniae (Anderton et al. 2007) and Clostridium botulinum (Sugawara et al. 2010) also produce proteins that bind and disrupt E-cadherin−mediated cell−cell adhesion without cleavage. A direct interaction with C. jejuni HtrA protease has recently been shown in which E-cadherin is cleaved by the bacterial protease, resulting in inhibition of intercellular epithelial adhesion (Hoy et al. 2012). Deletion of C. jejuni HtrA leads to loss of E-cadherin cleavage and highly defective invasion and paracellular transmigration (Hoy et al. 2012; Boehm et al. 2012).
Inactivation of E-cadherin in the mouse intestine has indicated that the protein plays a crucial role in maintenance of homeostasis not only through development of epithelial cell adherens junctions and desmosomes but also in maturation and localization of Paneth and goblet cells (Schneider et al. 2010). These cells are involved in the first line of host defense against pathogenic invasion and secrete a variety of protective antimicrobial molecules into the intestinal lumen in response to pathogenic challenge (Bevins and Salzman 2011; Gill et al. 2011; Forman et al. 2012). Inactivation of E-cadherin subsequently results in increased susceptibility to intestinal bacterial infection (Schneider et al. 2010).
T-cadherin is a unique member of the cadherin family which lacks the highly conserved cytoplasmic and transmembrane domains, and attaches to the cell membrane through a glycosylphosphatidylinositol anchor (Philippova et al. 2009). In vivo, T-cadherin is expressed on the apical surface of chicken intestinal epithelial cells (Koller and Ranscht 1996) (unlike classical cadherins, which are expressed basolaterally). The ectodomain (extracellular domain) shares high sequence homology with those of classical cadherins such as E-cadherin. It also lacks the cytoplasmic region, which is usually required for classical cadherin-mediated cell−cell adhesion but induces homophilic cell−cell adhesion through an alternative binding mechanism (Vestal and Ranscht 1992; Ciatto et al. 2010). The protein may play a major role in signal transduction due to its location within lipid rafts which regulate signal transduction (Simons and Toomre 2000). Due to its expression on the apical surface of polarized epithelial cells at the interface with the intestinal lumen and the high sequence homology of its ectodomain with that of E-cadherin, we hypothesize that it may interact with C. jejuni and consequentially affect resistance to colonization either by facilitating internalization or inciting a protective host response.
Although the locus exhibiting the second-most significant association with colonization resistance did not reach genome-wide significance after multiple testing adjustment (FDR = 0.24), it localized close to the calmodulin (CALM1) gene, which encodes a modulator of cadherin-mediated cell-cell adhesion (Li et al. 1999). Calmodulin is a mediator of calcium signaling that binds calcium Ca2+ ions and subsequently regulates the activity of calcium-dependent enzymes, including cadherins, involved in a wide variety of cellular processes. An increase in cytosolic Ca2+ concentration is associated with intracellular invasion and pathogenicity of a wide range of bacteria including C. jejuni (Pace et al. 1993; Hu et al. 2005; Gekara et al. 2007; Kim et al. 2008; Bandyopadhaya et al. 2009; Asmat et al. 2011; Konar and Ghosh 2012). Invasion of human intestinal epithelial cells by C. jejuni in vitro requires release of Ca2+ from intracellular stores and inhibition of calmodulin using a calmodulin agonist inhibits C. jejuni intracellular epithelial cell invasion in vitro (Hu et al. 2005). This finding suggests there may be a relationship with the association observed surrounding the T-cadherin gene, although no correlation between SNP genotypes from the two regions was found. This association study was carried out subsequent to a study on a subset of the birds analyzed here which characterized differences in gene expression between resistant and susceptible chickens. Correlation between the SNP genotypes observed here and gene expression levels was not found and neither of these genes displayed significantly differential expression. However, a significant correlation between colonization status and the ratio of expression levels of the two genes is seen. Interestingly, the gene displaying the most significant differential expression in this study, which demonstrated fourfold higher expression in resistant birds (padj = 5.6e−20), encodes an epithelial calcium-activated chloride channel (LOC424523)(Connell et al. 2012).
To our knowledge, this is the first study to identify a gene significantly associated with resistance to C. jejuni colonization of the chicken. The study was based on a population of Barred Rock chickens, a dual-purpose breed that originated in the United States in the 1800s and one of the founder breeds of the broiler industry. The breed has contributed significantly to the genetics of modern commercial broiler and brown egg layer breeds, most notably in extensive use of the White Plymouth Rock, which was developed from the Barred Rock, in maternal commercial broiler grandparent lines (Crawford 1990). Due to the high inbreeding present within the population, the likelihood of shared mechanisms of resistance to colonization and repetition of these findings with commercial populations is uncertain. The resistance locus identified centers on T-cadherin, and although of marginal significance and requiring further validation, suggests this gene plays a prominent role in the colonization process. This is supported by the importance of the related protein, E-cadherin in epithelial cell invasion of several bacteria, including C. jejuni and in the maintenance of intestinal homeostasis. A putative risk locus centering on the calmodulin gene, which can regulate cadherin function, and previous evidence for differential expression of a calcium activated chloride channel associated with colonization provide additional evidence for Ca2+-regulated cadherin involvement. C. jejuni is one of the most commonly reported causes of food-borne illness worldwide and chickens are a major vector for infection. We believe the results reported here highlight the significance of cadherins, and specifically T-cadherin, in control of the bacterium within the chicken cecum.
Supplementary Material
Acknowledgments
This research was funded by The Irish Department of Agriculture and Food’s Food Institutional Research Measure (http://www.agriculture.gov.ie/research/foodinstitutionalresearchmeasurefirm) - Grant No: 06_RDD_486.
Footnotes
Communicating editor: D. Schneider
Literature Cited
- Amin N., van Duijn C. M., Aulchenko Y. S., 2007. A genomic background based method for association analysis in related individuals. PLoS ONE 2: e1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderton J. M., Rajam G., Romero-Steiner S., Summer S., Kowalczyk A. P., et al. , 2007. E-cadherin is a receptor for the common protein pneumococcal surface adhesin A (PsaA) of Streptococcus pneumoniae. Microb. Pathog. 42: 225–236 [DOI] [PubMed] [Google Scholar]
- Asmat T. M., Agarwal V., Räth S., Hildebrandt J. P., Hammerschmidt S., 2011. Streptococcus pneumoniae infection of host epithelial cells via polymeric immunoglobulin receptor transiently induces calcium release from intracellular stores. J. Biol. Chem. 286: 17861–17869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko Y. S., Ripke S., Isaacs A., van Duijn C. M., 2007. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23: 1294–1296 [DOI] [PubMed] [Google Scholar]
- Bandyopadhaya A., Das D., Chaudhuri K., 2009. Involvement of intracellular signaling cascades in inflammatory responses in human intestinal epithelial cells following Vibrio cholerae infection. Mol. Immunol. 46: 1129–1139 [DOI] [PubMed] [Google Scholar]
- Beery J. T., Hugdahl M. B., Doyle M. P., 1988. Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl. Environ. Microbiol. 54: 2365–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevins C. L., Salzman N. H., 2011. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9: 356–368 [DOI] [PubMed] [Google Scholar]
- Boehm M., Hoy B., Rohde M., Tegtmeyer N., Bæk K. T., et al. , 2012. Rapid paracellular transmigration of Campylobacter jejuni across polarized epithelial cells without affecting TER: role of proteolytic-active HtrA cleaving E-cadherin but not fibronectin. Gut Pathog 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrmann E., Berndt A., Hänel I., Köhler H., 2007. Campylobacter-induced interleukin-8 responses in human intestinal epithelial cells and primary intestinal chick cells. Vet. Microbiol. 124: 115–124 [DOI] [PubMed] [Google Scholar]
- Boyd Y., Herbert E. G., Marston K. L., Jones M. A., Barrow P. A., 2005. Host genes affect intestinal colonisation of newly hatched chickens by Campylobacter jejuni. Immunogenetics 57: 248–253 [DOI] [PubMed] [Google Scholar]
- Byrne C. M., Clyne M., Bourke B., 2007. Campylobacter jejuni adhere to and invade chicken intestinal epithelial cells in vitro. Microbiology 153: 561–569 [DOI] [PubMed] [Google Scholar]
- Ciatto C., Bahna F., Zampieri N., VanSteenhouse H. C., Katsamba P. S., et al. , 2010. T-cadherin structures reveal a novel adhesive binding mechanism. Nat. Struct. Mol. Biol. 17: 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell S., Meade K. G., Allan B., Lloyd A. T., Kenny E., et al. , 2012. Avian resistance to Campylobacter jejuni colonization is associated with an intestinal immunogene expression signature identified by mRNA sequencing. PLoS ONE 7: e40409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox N. A., Hofacre C. L., Bailey J. S., Buhr R. J., Wilson J. L., et al. , 2005. Presence of Campylobacter jejuni in various organs one hour, one day, and one week following oral or intracloacal inoculations of broiler chicks. Avian Dis. 49: 155–158 [DOI] [PubMed] [Google Scholar]
- Crawford R. D., 1990. Poultry genetic resources: evolution, diversity and conservation, pp. 43–60 in Poultry Breeding and Genetics, edited by Crawford R. D., Elsevier, New York [Google Scholar]
- de Zoete M. R., Keestra A. M., Roszczenko P., van Putten J. P., 2010. Activation of human and chicken toll-like receptors by Campylobacter spp. Infect. Immun. 78: 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority. European Center for Disease Prevention and Control, 2012 The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2010. EFSA J. 10: 2597. Available at: www.efsa.europa.eu/efsajourna Accessed: April 11, 2013.
- Forman R. A., deSchoolmeester M. L., Hurst R. J., Wright S. H., Pemberton A. D., et al. , 2012. The goblet cell is the cellular source of the anti-microbial angiogenin 4 in the large intestine post Trichuris muris infection. PLoS ONE 7: e42248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank C. F., Hostetter M. K., 2007. Cleavage of E-cadherin: a mechanism for disruption of the intestinal epithelial barrier by Candida albicans. Transl. Res. 149: 211–222 [DOI] [PubMed] [Google Scholar]
- Gekara N. O., Westphal K., Ma B., Rohde M., Groebe L., et al. , 2007. The multiple mechanisms of Ca2+ signalling by listeriolysin O, the cholesterol-dependent cytolysin of Listeria monocytogenes. Cell. Microbiol. 9: 2008–2021 [DOI] [PubMed] [Google Scholar]
- Gill N., Wlodarska M., Finlay B. B., 2011. Roadblocks in the gut: barriers to enteric infection. Cell. Microbiol. 13: 660–669 [DOI] [PubMed] [Google Scholar]
- Grant C. C., Konkel M. E., Cieplak W., Tompkins L. S., 1993. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect. Immun. 61: 1764–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen M. A., Megens H. J., Zare Y., Warren W. C., Hillier L. W., et al. , 2011. The development and characterization of a 60K SNP chip for chicken. BMC Genomics 12: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbleib J. M., Nelson W. J., 2006. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 20: 3199–3214 [DOI] [PubMed] [Google Scholar]
- Hänel I., Müller J., Müller W., Schulze F., 2004. Correlation between invasion of Caco-2 eukaryotic cells and colonization ability in the chick gut in Campylobacter jejuni. Vet. Microbiol. 101: 75–82 [DOI] [PubMed] [Google Scholar]
- Hermans D., Van Deun K., Martel A., Van Immerseel F., Messens W., et al. , 2011. Colonization factors of Campylobacter jejuni in the chicken gut. Vet. Res. 42: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill Gaston J. S., Lillicrap M. S., 2003. Arthritis associated with enteric infection. Best Pract. Res. Clin. Rheumatol. 17: 219–239 [DOI] [PubMed] [Google Scholar]
- Hoy B., Löwer M., Weydig C., Carra G., Tegtmeyer N., et al. , 2010. Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion. EMBO Rep. 11: 798–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy B., Geppert T., Boehm M., Reisen F., Plattner P., et al. , 2012. Distinct roles of secreted HtrA proteases from gram-negative pathogens in cleaving the junctional protein and tumor suppressor E-cadherin. J. Biol. Chem. 287: 10115–10120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Raybourne R. B., Kopecko D. J., 2005. Ca2+ release from host intracellular stores and related signal transduction during Campylobacter jejuni 81–176 internalization into human intestinal cells. Microbiology 151: 3097–3105 [DOI] [PubMed] [Google Scholar]
- Hu L., Tall B. D., Curtis S. K., Kopecko D. J., 2008. Enhanced microscopic definition of Campylobacter jejuni 81–176 adherence to, invasion of, translocation across, and exocytosis from polarized human intestinal Caco-2 cells. Infect. Immun. 76: 5294–5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser P., 2010. Advances in avian immunology–prospects for disease control: a review. Avian Pathol. 39: 309–324 [DOI] [PubMed] [Google Scholar]
- Kalischuk L. D., Inglis G. D., Buret A. G., 2009. Campylobacter jejuni induces transcellular translocation of commensal bacteria via lipid rafts. Gut Pathog 1: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz J., Sambandam V., Wu J. H., Michalek S. M., Balkovetz D. F., 2000. Characterization of Porphyromonas gingivalis-induced degradation of epithelial cell junctional complexes. Infect. Immun. 68: 1441–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz J., Yang Q. B., Zhang P., Potempa J., Travis J., et al. , 2002. Hydrolysis of epithelial junctional proteins by Porphyromonas gingivalis gingipains. Infect. Immun. 70: 2512–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. V., Pearce D., Kim K. S., 2008. Ca(2+)/calmodulin-dependent invasion of microvascular endothelial cells of human brain by Escherichia coli K1. Cell Tissue Res. 332: 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga M., Gilbert M., Li J., Koike S., Takahashi M., et al. , 2005. Antecedent infections in Fisher syndrome: a common pathogenesis of molecular mimicry. Neurology 64: 1605–1611 [DOI] [PubMed] [Google Scholar]
- Koller E., Ranscht B., 1996. Differential targeting of T- and N-cadherin in polarized epithelial cells. J. Biol. Chem. 271: 30061–30067 [DOI] [PubMed] [Google Scholar]
- Konar M., Ghosh S., 2012. Enteroaggregative Escherichia coli induced increase in intracellular calcium concentration modulates cytoskeletal F-actin rearrangement and bacterial entry in INT-407 cells. Microb. Pathog. 52: 278–284 [DOI] [PubMed] [Google Scholar]
- Konkel M. E., Mead D. J., Hayes S. F., Cieplak W., 1992. Translocation of Campylobacter jejuni across human polarized epithelial cell monolayer cultures. J. Infect. Dis. 166: 308–315 [DOI] [PubMed] [Google Scholar]
- Lamb-Rosteski J. M., Kalischuk L. D., Inglis G. D., Buret A. G., 2008. Epidermal growth factor inhibits Campylobacter jejuni-induced claudin-4 disruption, loss of epithelial barrier function, and Escherichia coli translocation. Infect. Immun. 76: 3390–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson C. L., Shah D. H., Dhillon A. S., Call D. R., Ahn S., et al. , 2008. Campylobacter jejuni invade chicken LMH cells inefficiently and stimulate differential expression of the chicken CXCLi1 and CXCLi2 cytokines. Microbiology 154: 3835–3847 [DOI] [PubMed] [Google Scholar]
- Li X., Swaggerty C. L., Kogut M. H., Chiang H., Wang Y., et al. , 2008. The paternal effect of Campylobacter jejuni colonization in ceca in broilers. Poult. Sci. 87: 1742–1747 [DOI] [PubMed] [Google Scholar]
- Li X., Swaggerty C. L., Kogut M. H., Chiang H. I., Wang Y., et al. , 2010. Gene expression profiling of the local cecal response of genetic chicken lines that differ in their susceptibility to Campylobacter jejuni colonization. PLoS ONE 5: e11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Y., Swaggerty C. L., Kogut M. H., Chiang H. I., Wang Y., et al. , 2011. Caecal transcriptome analysis of colonized and non-colonized chickens within two genetic lines that differ in caecal colonization by Campylobacter jejuni. Anim. Genet. 42: 491–500 [DOI] [PubMed] [Google Scholar]
- Li Z., Kim S. H., Higgins J. M., Brenner M. B., Sacks D. B., 1999. IQGAP1 and calmodulin modulate E-cadherin function. J. Biol. Chem. 274: 37885–37892 [DOI] [PubMed] [Google Scholar]
- McCarthy N., Giesecke J., 2001. Incidence of Guillain-Barré syndrome following infection with Campylobacter jejuni. Am. J. Epidemiol. 153: 610–614 [DOI] [PubMed] [Google Scholar]
- Meade K. G., Narciandi F., Cahalane S., Reiman C., Allan B., et al. , 2009. Comparative in vivo infection models yield insights on early host immune response to Campylobacter in chickens. Immunogenetics 61: 101–110 [DOI] [PubMed] [Google Scholar]
- Mengaud J., Ohayon H., Gounon P., Mege R.-M., Cossart P., 1996. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84: 923–932 [DOI] [PubMed] [Google Scholar]
- Monteville M. R., Konkel M. E., 2002. Fibronectin-facilitated invasion of T84 eukaryotic cells by Campylobacter jejuni occurs preferentially at the basolateral cell surface. Infect. Immun. 70: 6665–6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell D. G., Fearnley C., 2003. Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 69: 4343–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace J., Hayman M. J., Galán J. E., 1993. Signal transduction and invasion of epithelial cells by S. typhimurium. Cell 72: 505–514 [DOI] [PubMed] [Google Scholar]
- Phan Q. T., Myers C. L., Fu Y., Sheppard D. C., Yeaman M. R., et al. , 2007. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 5: e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippova M., Joshi M. B., Kyriakakis E., Pfaff D., Erne P., et al. , 2009. A guide and guard: the many faces of T-cadherin. Cell. Signal. 21: 1035–1044 [DOI] [PubMed] [Google Scholar]
- Price A. L., Zaitlen N. A., Reich D., Patterson N., 2010. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 11: 459–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W., 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- Sansonetti P. J., Mounier J., Prévost M. C., Mège R. M., 1994. Cadherin expression is required for the spread of Shigella flexneri between epithelial cells. Cell 76: 829–839 [DOI] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R. M., Angulo F. J., Tauxe R. V., Widdowson M. A., et al. , 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17: 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M. R., Dahlhoff M., Horst D., Hirschi B., Trülzsch K., et al. , 2010. A key role for E-cadherin in intestinal homeostasis and Paneth cell maturation. PLoS ONE 5: e14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaughnessy R. G., Meade K. G., McGivney B. A., Allan B., O’Farrelly C., 2011. Global gene expression analysis of chicken caecal response to Campylobacter jejuni. Vet. Immunol. Immunopathol. 142: 64–71 [DOI] [PubMed] [Google Scholar]
- Simons K., Toomre D., 2000. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1: 31–39 [DOI] [PubMed] [Google Scholar]
- Smith C. K., Abuoun M., Cawthraw S. A., Humphrey T. J., Rothwell L., et al. , 2008. Campylobacter colonization of the chicken induces a proinflammatory response in mucosal tissues. FEMS Immunol. Med. Microbiol. 54: 114–121 [DOI] [PubMed] [Google Scholar]
- Stern N. J., Meinersmann R. J., Cox N. A., Bailey J. S., Blankenship L. C., 1990. Influence of host lineage on cecal colonization by Campylobacter jejuni in chickens. Avian Dis. 34: 602–606 [PubMed] [Google Scholar]
- Sugawara Y., Matsumura T., Takegahara Y., Jin Y., Tsukasaki Y., et al. , 2010. Botulinum hemagglutinin disrupts the intercellular epithelial barrier by directly binding E-cadherin. J. Cell Biol. 189: 691–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam C. C., O’Brien S. J., Petersen I., Islam A., Hayward A., et al. , 2007. Guillain-Barré syndrome and preceding infection with campylobacter, influenza and Epstein-Barr virus in the general practice research database. PLoS ONE 2: e344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townes J. M., Deodhar A. A., Laine E. S., Smith K., Krug H. E., et al. , 2008. Reactive arthritis following culture-confirmed infections with bacterial enteric pathogens in Minnesota and Oregon: a population-based study. Ann. Rheum. Dis. 67: 1689–1696 [DOI] [PubMed] [Google Scholar]
- Van Deun K., Pasmans F., Ducatelle R., Flahou B., Vissenberg K., et al. , 2008a Colonization strategy of Campylobacter jejuni results in persistent infection of the chicken gut. Vet. Microbiol. 130: 285–297 [DOI] [PubMed] [Google Scholar]
- Van Deun K., Pasmans F., Van Immerseel F., Ducatelle R., Haesebrouck F., 2008b Butyrate protects Caco-2 cells from Campylobacter jejuni invasion and translocation. Br. J. Nutr. 100: 480–484 [DOI] [PubMed] [Google Scholar]
- van Gerwe T., Miflin J. K., Templeton J. M., Bouma A., Wagenaar J. A., et al. , 2009. Quantifying transmission of Campylobacter jejuni in commercial broiler flocks. Appl. Environ. Microbiol. 75: 625–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestal D. J., Ranscht B., 1992. Glycosyl phosphatidylinositol–anchored T-cadherin mediates calcium-dependent, homophilic cell adhesion. J. Cell Biol. 119: 451–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar C. C., Kashleva H., Nobile C. J., Mitchell A. P., Dongari-Bagtzoglou A., 2007. Mucosal tissue invasion by Candida albicans is associated with E-cadherin degradation, mediated by transcription factor Rim101p and protease Sap5p. Infect. Immun. 75: 2126–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Lim K. C., Huang J., Saidi R. F., Sears C. L., 1998. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc. Natl. Acad. Sci. USA 95: 14979–14984 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.