Abstract
Elg1 and Srs2 are two proteins involved in maintaining genome stability in yeast. After DNA damage, the homotrimeric clamp PCNA, which provides stability and processivity to DNA polymerases and serves as a docking platform for DNA repair enzymes, undergoes modification by the ubiquitin-like molecule SUMO. PCNA SUMOylation helps recruit Srs2 and Elg1 to the replication fork. In the absence of Elg1, both SUMOylated PCNA and Srs2 accumulate at the chromatin fraction, indicating that Elg1 is required for removing SUMOylated PCNA and Srs2 from DNA. Despite this interaction, which suggests that the two proteins work together, double mutants elg1Δ srs2Δ have severely impaired growth as haploids and exhibit synergistic sensitivity to DNA damage and a synergistic increase in gene conversion. In addition, diploid elg1Δ srs2Δ double mutants are dead, which implies that an essential function in the cell requires at least one of the two gene products for survival. To gain information about this essential function, we have carried out a high copy number suppressor screen to search for genes that, when overexpressed, suppress the synthetic lethality between elg1Δ and srs2Δ. We report the identification of 36 such genes, which are enriched for functions related to DNA- and chromatin-binding, chromatin packaging and modification, and mRNA export from the nucleus.
Genome stability is of primary importance for the survival and proper functioning of all organisms. During DNA replication, the activity of DNA polymerases may be compromised by lesions or by the presence of secondary structures in the DNA. This may cause replication stalling and even fork collapse. Cells must react promptly to repair or bypass the DNA damage and to reactivate DNA replication. A number of mechanisms have been lately linked to this cellular response (Lee and Myung 2008).
PCNA is a homotrimeric ring that encircles the double-stranded DNA (Krishna et al. 1994) and plays a central role in DNA replication by ensuring the processivity of the replicative DNA polymerases (Chilkova et al. 2007; Krishna et al. 1994). Many proteins interact with PCNA and use it as a platform that enables stable interactions with DNA. These proteins include factors involved in DNA replication, DNA repair, chromatin remodeling, and other DNA-related activities that are important for cell viability, cell division, and genomic stability (Warbrick 2000).
In response to DNA damage, PCNA can be mono-ubiquitinated at lysine 164 by the E2/E3 pair Rad6 and Rad18 (Hoege et al. 2002). Mono-ubiquitination of PCNA allows the binding of the translesion synthesis polymerases to PCNA, resulting in an error-prone repair mechanism (Acharya et al. 2008). Alternatively, PCNA can be further poly-ubiquitinated on the same lysine residue by a mechanism that also requires Ubc13-Mms2 (E2 heterodimer) and Rad5 (E3) (Ulrich and Jentsch 2000). This poly-ubiquitination coordinates an error-free repair mechanism, whose details are still unclear (Branzei et al. 2008). Interestingly, the same residue of PCNA (lysine 164) can be modified by the ubiquitin-like molecule SUMO. This modification takes place during S-phase or after high doses of DNA damage. An additional residue, lysine 127, can also be SUMOylated but not ubiquitinated. In contrast to mutations in lysine 164, those in lysine 127 do not lead to DNA damage sensitive phenotypes (Hoege et al. 2002).
Two proteins have been shown to preferentially interact with SUMOylated PCNA in yeast: Srs2 and Elg1 (Armstrong et al. 2012; Kolesar et al. 2012; Parnas et al. 2010; Pfander et al. 2005). Srs2 is a DNA helicase; depending on the assay used, Srs2 has been shown to promote (Aylon et al. 2003) or prevent (Schiestl et al. 1990) homologous recombination. Srs2 was found to be able to displace the Rad51 strand-exchange protein (homologous to bacterial RecA) from ssDNA (Krejci et al. 2003; Veaute et al. 2003). SUMOylated PCNA recruits Srs2 to replication forks, where the helicase appears to prevent unscheduled recombination events (Papouli et al. 2005; Pfander et al. 2005).
The Elg1 protein resembles the large subunit of Replication Factor C, a protein complex in charge of loading/unloading PCNA from DNA during replication. Mutations in ELG1 lead to a variety of genomic instability phenotypes, including, among others, hyper-recombination, chromosome loss, elongated telomeres, and increased telomeric silencing (Banerjee and Myung 2004; Banerjee et al. 2007; Bellaoui et al. 2003; Ben-Aroya et al. 2003; Kanellis et al. 2003). Similarly to Srs2, mutations in ELG1 suppress the sensitivity to DNA damaging agents of mutants of the post-replication repair pathway (Parnas et al. 2010). Both proteins are recruited to SUMOylated PCNA by a similar mechanism involving PCNA- (PIP) and SUMO-interacting motifs (Armstrong et al. 2012; Kolesar et al. 2012; Parnas et al. 2010; Pfander et al. 2005). Moreover, deletion of ELG1 leads to accumulation of Srs2 at the chromatin fraction, suggesting that Elg1 may play a role in Srs2 unloading, perhaps together with SUMOylated PCNA (Parnas et al. 2010). Remarkably, however, double deletion of the ELG1 and SRS2 genes results in a synthetic fitness reduction in haploid cells (Parnas et al. 2010) and in complete lethality in diploids (this study). These results imply that at least one of them must be active for viability. Because both proteins are active during DNA replication, it is likely that they provide alternative mechanisms to deal with specific DNA lesions or intermediates. However, alternative possibilities exist. Here we try to gain information about the function of these proteins by carrying out a genetic screen for genes that, when overexpressed, are able to rescue the synthetic lethality (SL) between elg1 and srs2 mutations.
Materials and Methods
Yeast strains
Strain OP738 (MATa/MATalpha elg1::KanMX/elg1:: KanMX srs2::KanMX srs2:: KanMX ade2/ade2 ade3/ade3 leu2/leu2 ura3/ura3 his3/his3 [p1313: CEN-LEU2-ELG1-ADE3]) was used for the genetic screen. Strain MK166 (lys2:: Ty1Sup ade2-1(o) can1-100(o) ura3-52 leu2-3, 112 his3del200 trp1del901 HIS3:: lys2:: ura3 his4:: TRP1:: his4) (Liefshitz et al. 1995) and its derivatives were used for testing sensitivity to DNA-damaging agents and to measure recombination levels.
Genetic screen
A high copy number library in YEp24 (a 2-μm URA3 plasmid) was used in the screen, carried out as described (Parnas et al. 2009). Each positive plasmid was extracted and retransformed into OP738. Only those that gave sectors were analyzed. Individual genes were subcloned into high copy number plasmids and retested.
GO (Gene Ontology) enrichment was calculated using an in-house score algorithm. The algorithm is a simple hypergeometric calculation; categories annotated with two few (≤3) or too many (≥500) genes were excluded from the enrichment calculation. Scores were corrected for multiple hypotheses testing an FDR threshold of 0.001.
Results and Discussion
SL between elg1Δ and srs2Δ
Elg1 and Srs2 physically interact with SUMOylated PCNA in a similar manner through their PIP and SUMO-interacting domains (Armstrong et al. 2012; Kolesar et al. 2012; Parnas et al. 2010; Pfander et al. 2005). In addition, Elg1 seems to be required for the unloading of Srs2 from chromatin (Parnas et al. 2010). We would thus expect mutations in ELG1 and SRS2 to show epistasis, such that the double mutant would not show a phenotype more severe than the single mutants. However, elg1Δ srs2Δ double mutants show, in comparison with each of the single mutants, increased sensitivity to DNA-damaging agents, such as the alkylating agent methylmethane sulfonate, the inhibitor of deoxynucleotides synthesis hydroxyurea, and the topoisiomerase inhibitor camptothecin (Figure 1).
Figure 1.
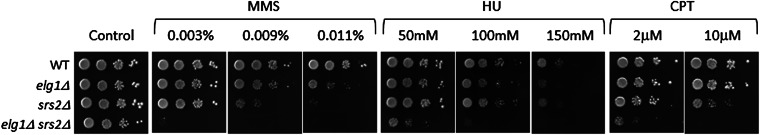
The elg1Δ and srs2Δ mutants display synergistic sensitivity to DNA-damaging agents. Ten-fold serial dilutions of logarithmic cultures were plated on SD-complete plates containing either methylmethane su\lfonate (MMS), hydroxyurea (HU), or camptothecin (CPT) at the concentrations shown.
Mutations in each of the two genes cause an increase in homologous recombination (Ben-Aroya et al. 2003; Liefshitz et al. 1998). To characterize the type of recombination events that occur in the single and double mutants, we used strain MK166 (Liefshitz et al. 1995). This strain allows the detection of both gene conversion events between repeated sequences (Ty elements) or ectopic single sequences (lys2 alleles) and direct-repeat recombination (DRR) at Ty elements and at the HIS4 locus (Liefshitz et al. 1995). Whereas gene conversion transfers information between alleles, DRR results in the deletion of information located between the two repeats (Agmon et al. 2009). Table 1 shows that despite the fact that each mutant is hyperrecombinant, compared with the wt, they show a very different pattern of homologous recombination. Whereas srs2Δ shows increased levels of gene conversion events, the elg1Δ mutant shows both high gene conversion and high DRR levels. The elg1Δ srs2Δ double mutant shows the same level of DRR as elg1Δ; however, it exhibits a synergistic effect for gene conversion: lys2 gene conversion is elevated eightfold, and the level of Ty ectopic conversion is increased 80-fold above that of the wt strain (Table 1). We interpret these results as follows: The activity of Srs2 prevents gene conversion events, probably by evicting the Rad51 recombination protein from the DNA (Krejci et al. 2003; Veaute et al. 2003). In the absence of Srs2, we see an increase in gene conversion events but not in DRR events, which are probably carried out by single strand annealing and do not require Rad51 (Krejci et al. 2003). In contrast, the absence of Elg1 promotes all types of recombination events, either because Elg1 does not allow the first steps of recombination to take place, or, as has been suggested (Davidson et al. 2012), because in its absence spontaneous DNA damage accumulates. The results of the double mutant, however, suggest that Elg1 activity restricts the level of gene conversion events of srs2 mutants, and thus both proteins must play some role during the actual recombination process.
Table 1. Recombination frequency.
| Strain | His+ (DRR) | Ty DRR | Ty GC | Lys+ (GC) |
|---|---|---|---|---|
| wt | 1 (10 × 10−6) | 1 (1 × 10−6) | 1 (0.5 × 10−6) | 1 (0.25 × 10−6) |
| srs2Δ | 1.1 | 1.1 | 3.5 | 2 |
| elg1Δ | 4.5 | 12 | 5 | 1.5 |
| elg1Δ srs2Δ | 4.4 | 11 | 80 | 8 |
Direct repeat recombination (DRR) is measured between tandem repeats at the HIS4 locus or between Ty elements. Gene conversion (GC) measures the transfer of information between Ty elements or two copies of the LYS2 gene carrying different mutations.
elg1Δ srs2Δ double mutants are extremely sick as haploids (Parnas et al. 2010), indicating that cells need at least one of the proteins to carry out a function important for fitness, probably some type of repair that is so common that it occurs often during DNA replication, such as fork stalling response. Previous work showed that many of the phenotypes of srs2Δ are stronger in diploids, and that several mutations affecting DNA repair produce a synthetic sick phenotype when combined with srs2Δ as haploids, but are completely lethal as diploids (Klein 2001). We thus crossed an elg1Δ srs2Δ haploid with either an elg1Δ haploid, an srs2Δ haploid or a double mutant elg1Δ srs2Δ of the opposite sex. Mating took place at normal frequencies. Zygotes were micromanipulated under the microscope to fixed positions in the plate and were then incubated for 48 hr. Thirty-two of 32 zygotes and 31 of 32 zygotes were able to form colonies in the first two crosses (with single elg1Δ and srs2Δ mutants, respectively), whereas only 2 of the 80 elg1/elg1srs2/srs2 zygotes formed viable colonies after micromanipulation. Microscopic observation of the zygotes showed that the majority produced between 0 and 2 cell divisions before dying. Thus, diploid elg1/elg1srs2/srs2 strains are inviable.
A high copy number screen for suppressors of the SL between elg1Δ and srs2Δ
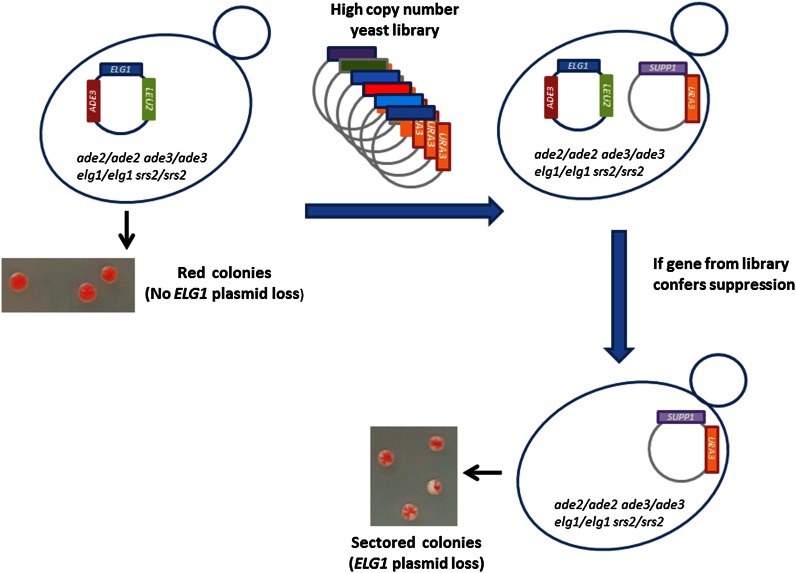
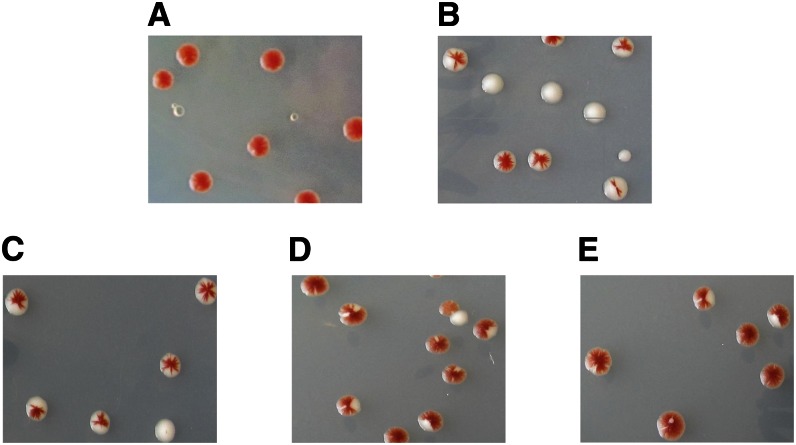
Our high copy number screen was based on a red/white colony phenotype (Koren et al. 2003; Koshland et al. 1985; Parnas et al. 2009). ade2 mutants form red colonies due to the accumulation of a red pigment. Mutations in the upstream-acting gene ADE3 prevent the accumulation of the red pigment and render the cells white. An ade2ade3elg1Δ srs2Δ diploid strain (OP738) kept alive by the presence of a LEU2-marked plasmid carrying the ELG1 gene as well as the ADE3 gene forms uniformly red colonies (as the ade3 mutation is complemented, and any cell that loses the plasmid dies; Figure 2). These cells were then transformed with a yeast genomic library cloned in a high copy number plasmid (containing the URA3 marker). Cells that received a plasmid with a gene that, when overexpressed, can suppress the SL phenotype of elg1Δ srs2Δ, are now able to lose the ELG1-ADE3-LEU2 plasmid, and therefore show white/red sectored colonies (Figures 2 and 3). The sectoring pattern (number and size of sectors) reflects the rate of viable plasmid loss, and thus provides a measure of the efficiency of suppression. Examples of such patterns are seen in Figure 3.
Figure 2.
Schematic representation of the screen. A diploid double mutant elg1Δ srs2Δ strain is kept alive by the presence of a plasmid carrying the ELG1 gene. The ADE3 marker on the plasmid confers a red pigment to the cells carrying it (as the strain is ade2 ade3). Since any cell that loses the plasmid during colony formation dies, all colonies are uniformly red. This strain was transformed with a high copy number library carrying random fragments of the yeast genome. Cells that received a plasmid that suppresses the SL phenotype (SUPP1) can now lose the ELG1-containing plasmid, becoming white. These cells create white or red/white sectored colonies.
Figure 3.
Examples of sectoring phenotypes observed in the presence of various high copy number plasmids. (A) Empty vector. (B) ELG1. (C) MOB1 (strong suppressor). (D) RTT109 (medium). (E) PAB1 (weak suppressor).
After screening ~50,000 yeast transformants, 92 candidates showed some level of colony sectoring. Plasmids (each containing a ~8 kb insert on average) were obtained from these colonies and retransformed into OP738. Only 54 of these candidates clearly exhibited sectors upon retransformation. These plasmids were sequenced to determine the genomic region responsible for the suppression. Many plasmids carried overlapping genomic fragments, and in total were found to represent 34 genomic regions. Subcloning experiments were then carried out to identify the genes responsible for the suppressive phenotype. With a few exceptions (RTT103 and NSE3; CHD1 and PAB1), only one gene per plasmid caused increased white sectoring. Table 2 shows the names and functions of the 36 genes identified.
Table 2. Genes in the screen.
| Gene Name | Short Description | Degree of Suppression |
|---|---|---|
| BAP2 | High-affinity branched aminoacid permease | Strong |
| ECO1 | Acetyltransferase required for Smc3p acetylation during the establishment of sister chromatid cohesion | Strong |
| HTB1 | Histone H2B | Strong |
| CDC34 | Ubiquitin-conjugating enzyme (E2) and catalytic subunit of SCF ubiquitin-protein ligase complex | Strong |
| CHD1 | Chromatin remodeller | Strong |
| CRM1 | Major karyopherin, involved in export of proteins, RNAs, and ribosomal subunits from the nucleus | Strong |
| MOB1 | Component of the mitotic exit network | Strong |
| DDC1 | Member of the 9-1-1 clamp essential for the DNA damage response | Strong |
| CST6 | Basic leucine zipper (bZIP) transcription factor | Strong |
| MPH1 | Helicase that functions in an error-free DNA damage bypass pathway | Strong |
| YKL044w | Gene of unknown function | Strong |
| SIS2 | Negative regulatory subunit of the protein phosphatase 1 Ppz1 | Strong |
| UBC11 | Ubiquitin-conjugating enzyme | Strong |
| NHP6A | High-mobility group non-histone chromatin protein | Strong |
| TPT1 | tRNA 2’-phosphotransferase, catalyzes the final step in yeast tRNA splicing | Strong |
| TAF3 | TFIID subunit involved in transcription initiation | Strong |
| RTT103 | Involved in transcription termination | Medium |
| SRL3 | Cytoplasmic protein that, when overexpressed, suppresses the lethality of a rad53 null mutation | Medium |
| RTT109 | Histone H3 acetylase | Medium |
| GID8 | Member of a ubiquitin ligase that carries out poly-ubiquitination. | Medium |
| GIS2 | Protein with seven cysteine-rich CCHC zinc-finger motifs | Medium |
| RTT106 | Histone chaperone | Medium |
| YOL114c | Gene of unknown function | Medium |
| IRC13 | Null mutant displays increased levels of spontaneous Rad52 foci | Medium |
| NUP1 | Nuclear pore complex (NPC) subunit, involved in protein import/export and in export of RNAs | Weak |
| PPS1 | Protein phosphatase with specificity for serine, threonine, and tyrosine | Weak |
| UBP14 | Ubiquitin-specific protease | Weak |
| NSE3 | Essential subunit of the Mms21-Smc5-Smc6 complex | Weak |
| PAB1 | Poly(A) binding protein, part of the 3′-end RNA-processing complex | Weak |
| DAM1 | Essential subunit of the Dam1 complex, involved in kinetochore movement | Weak |
| CPD1 | Cyclic nucleotide phosphodiesterase | Weak |
| UBP7 | Ubiquitin-specific protease that cleaves ubiquitin-protein fusions | Weak |
| RPB4 | Subunit of RNA polymerase involved in 3′-end RNA processing and export | Weak |
| RFX1 | Major transcriptional repressor of DNA-damage-regulated genes | Weak |
| HAS1 | ATP-dependent RNA helicase, localizes to nuclear pore | Weak |
| CNM67 | Component of the spindle pole body outer plaque | Weak |
The genes identified span a large number of functions, pathways and cellular location. Analysis of GO (gene onthology) term enrichment (Table 3) shows that this gene set is enriched for genes of chromosomal location, with functions related to DNA- and chromatin-binding and involved in biological processes related to DNA packaging and modification. Additionally, there is an enrichment for genes involved in regulatory mechanisms as well as genes that participate in the response to DNA damage. Below, we present the genes, divided into functional categories obtained by reading the literature regarding each of the genes. Naturally, this division is artificial, as many of the categories overlap, and the precise mechanism of suppression is still unknown.
Table 3. GO annotation enrichment.
| Description | GOID | Ontology | Intersection | Category Size | P Value |
|---|---|---|---|---|---|
| Chromosome | GO:0005694 | Cellular compartment | 11 | 393 | 2.33E-06 |
| Chromosomal part | GO:0044427 | Cellular compartment | 9 | 352 | 4.78E-05 |
| Structure-specific DNA binding | GO:0043566 | Molecular function | 5 | 101 | 1.47E-04 |
| Chromatin binding | GO:0003682 | Molecular function | 4 | 84 | 8.27E-04 |
| Regulation of mitotic cell cycle | GO:0007346 | Biological process | 6 | 105 | 1.33E-05 |
| Negative regulation of cellular process | GO:0048523 | Biological process | 10 | 422 | 3.17E-05 |
| Negative regulation of biological process | GO:0048519 | Biological process | 10 | 432 | 3.86E-05 |
| DNA repair | GO:0006281 | Biological process | 8 | 263 | 4.05E-05 |
| DNA packaging | GO:0006323 | Biological process | 4 | 61 | 2.49E-04 |
| Chromatin assembly or disassembly | GO:0006333 | Biological process | 4 | 66 | 3.36E-04 |
| Nucleosome organization | GO:0034728 | Biological process | 4 | 69 | 3.97E-04 |
| DNA conformation change | GO:0071103 | Biological process | 4 | 80 | 6.91E-04 |
| Negative regulation of transposition | GO:0010526 | Biological process | 2 | 8 | 7.22E-04 |
| Response to DNA damage stimulus | GO:0006974 | Biological process | 7 | 307 | 7.27E-04 |
| Negative regulation of cellular metabolic process | GO:0031324 | Biological process | 7 | 307 | 7.27E-04 |
| DNA replication-independent nucleosome organization | GO:0034724 | Biological process | 2 | 9 | 9.23E-04 |
An FDR correction of 0.001 was used
Genes affecting genome stability:
This category includes the following genes:
ECO1/CTF7:
Eco1 is a protein essential for sister chromatid cohesion (Skibbens et al. 1999; Toth et al. 1999). It has an acetyltransferase activity that acetylates the cohesin subunit Smc3. This acetylation has been shown to be critical for the establishment of cohesion during S phase (Unal et al. 2008). Eco1 interacts with PCNA through a PIP motif (Moldovan et al. 2006), thus effectively coordinating between cohesion establishment and DNA replication. It was found that loss of PCNA SUMOylation by deletion of the SUMO ligase SIZ1 rescued the temperature sensitivity of eco1 mutants, indicating a role for SUMO in this process (Moldovan et al. 2006). Elg1 has been shown to affect sister chromatid cohesion (Parnas et al. 2009) and genetic antagonistic interactions have been observed between Elg1 and Eco1: deletion of ELG1 partially rescues the phenotypes of a temperature sensitive eco1 mutant, whereas its overexpression enhances its phenotypes (Maradeo and Skibbens 2009; Maradeo and Skibbens 2010). Despite their common interaction with PCNA, no physical or genetic interactions have been detected between Eco1 and Srs2.
NSE3:
Nse3 is part of the Smc5-6 complex, which is structurally similar to cohesin, but whose precise function still remains enigmatic (Bustard et al. 2012). This complex was found to be required for a variety of DNA repair activities, including sister chromatid recombination, and mutants defective for its components exhibit an unusual amount of recombination intermediates (Bustard et al. 2012), a phenotype shared with both elg1Δ and srs2Δ mutants. Interestingly, the complex contains a SUMO-ligase and potentially a ubiquitin-ligase. A recent publication links cohesin SUMOylation by the Smc5-6 complex to cohesion establishment, in an Eco1-independent fashion (Almedawar et al. 2012).
MPH1:
Mph1 is the yeast homolog of FANCM, a human gene that, when mutated, leads to Fanconi anemia. Mph1 is a DNA helicase involved in error-free bypass of DNA lesions. It has been proposed that Mph1 may promote sister chromatid recombination as a way of bypassing lesions that stall fork progression (Ede et al. 2011). Interestingly, deletion of MPH1 rescues the DNA damage sensitivity of mutants of the Smc5-6 complex (Chavez et al. 2011; Chen et al. 2009), whereas Mph1 overexpression exacerbates the mutants’ phenotypes (Chen et al. 2009). Physical and negative genetic interactions have been observed between Mph1 and Srs2 (Chiolo et al. 2005; St Onge et al. 2007). mph1 mutants share with both elg1 and srs2 genome instability phenotypes (Daee et al. 2012), suggesting that Mph1 may be sharing repair functions with both genes.
DDC1:
Ddc1 forms part of a PCNA-like molecule (called the 9-1-1 clamp). Loading of this heterotrimeric ring onto DNA is essential for the activation of the DNA damage response (Melo et al. 2001). This clamp has been recently been proposed to play a pivotal role in an error-free lesion bypassing mechanism that operates in parallel to Srs2 (Karras et al. 2013).
These four genes seem to be exerting their suppressive effect by affecting processes that take place during DNA replication, at the fork. Their overexpression may prevent stalling or may be able to act in DNA repair, either replacing a common function of Elg1 and Srs2, or providing alternative repair mechanisms. Ddc1 overexpression may lead to a stronger or extended DNA checkpoint response, which may help cope with fork stalling. Alternatively, the lethality between elg1 and srs2 may be due to lesions left behind the fork in the absence of the two repair activities, and overexpression of proteins that participate in alternative repair mechanisms may be able to repair this lethal damage, rescuing the cells.
Genes affecting histones and chromatin:
This category includes the following genes:
HTB1:
DNA is packaged in eukaryotic cells around nucleosomes composed of two units each of the H2A-H2B and H3-H4 heterodimers. HTB1 encodes a copy of histone H2B. Ubiquitination of this histone on residue K123 is a prerequisite for histone H3 modification and is essential for a variety of cellular processes, including gene expression and DNA repair (Dover et al. 2002; Robzyk et al. 2000).
RTT106:
During DNA replication, nucleosomes ahead of the replication fork are disassembled, and the recently replicated DNA is reassembled into nucleosomes using both the old, parental histones, as well as newly synthesized histones. Deposition of newly synthesized H3–H4 requires histone chaperones, including Rtt106 (Li et al. 2012). The dimeric histones delivered by Rtt106 are marked by acetylation at lysine 56 of histone H3 (Su et al. 2012).
RTT109:
Rtt109 is a histone acetyltransferase that is required for proper acetylation of histone H3 at lysine 56 (H3K56), which occurs during S phase and DNA damage repair. The formation of H3K56ac by Rtt109 and its deposition by Rtt106 stabilize the advancing replication forks and allow the repair of lesions that occur during replication (Clemente-Ruiz et al. 2011).
CHD1:
Chd1 is a chromatin remodeler that belongs to the chromodomain-containing subfamily of Snf2-like proteins; these proteins are believed to be recruited through methylated histone lysine residues and to promote nucleosome remodeling. Interestingly, it was recently shown that Chd1 is required for histone H2B ubiquitination, which is required for histone restoration after RNA polymerase passage (Lee et al. 2012).
NHP6A:
Nhp6A is an abundant small protein that binds DNA nonspecifically and bends it sharply. By binding the minor groove of DNA with a single HMGB domain and wrapping around the DNA to contact the major groove, it helps remodeling the nucleosomes (Stillman 2010). A less-abundant, very similar protein, Nhp6B, exists. Both were found to be components, together with Spt16p and Pob3p, of the FACT complex, which plays important roles in transcription and DNA replication (Ruone et al. 2003).
Thus, overexpression of histone H2B, or of proteins that promote nucleosome re-establishment after DNA or RNA polymerase transit, alleviate the SL phenotype of elg1Δ and srs2Δ. We can think of three possible models for this: (1) mutations in these genes cause replication fork instability, which can be suppressed by accelerating replication in the presence of excess histone dimers ready to be assembled into the newly replicated DNA. (2) Alternatively, Srs2 and Elg1 may play a direct role in the regulation of histone levels in the cell, and their SL is alleviated when histone levels are increased by other means. (3) A third model, involving an effect of these chromatin regulators in transcription regulation (which is also affected by histone levels and deposition), seems to us less likely, although recent evidence supports a role for histone levels in the regulation of promoter fidelity (Silva et al. 2012).
It is interesting to note that elg1 mutants are synthetic sick with mutations in HTB1 (data not shown), RTT106 (Imbeault et al. 2008), RTT109 (Bellaoui et al. 2003), although not with chd1Δ, nhp6aΔ or the double nhp6aΔ nhp6bΔ (data not shown). In contrast, srs2Δ shows a synthetic fitness phenotype when combined with chd1Δ (Pan et al. 2006), rtt109Δ (Costanzo et al. 2010), but not with htb1Δ, rtt106Δ, nhp6aΔ or the double nhp6aΔ nhp6bΔ (data not shown). Thus, the spectrum of synthetic phenotypes between elg1Δ and srs2Δ only partially overlap, consistent with nonoverlapping functions for the two genes.
Genes affecting RNA end-processing and/or nuclear transport:
This category includes the following genes:
RTT103:
Transcription by RNA polymerase II (PolII) and transcript processing are coordinated by the phosphorylation status of the PolII C-terminal domain (CTD). Transcription termination requires, surprisingly, the activity of RNA nucleases such as Rat1, which is recruited by Rtt103 (Kim et al. 2004). This requires an interaction between Rtt103 and the Ser2-phosphorylated form of PolII CTD. It was recently found that phosphorylation of the Tyr-1 residue in the CTD prevents Rtt103 binding, thus enhancing transcription processivity. At the 3′ end of the transcript Tyr-1 levels drop, allowing Rtt103 binding and transcription termination followed by poly-adenylation (Mayer et al. 2012).
PAB1:
Pab1 associates with RNA poly(A) tails, mediating the interactions between the 5′ cap structure and the 3′ mRNA poly(A) tail. It plays a role in mRNA stability and stimulates the initiation of translation (Amrani et al. 2008; Amrani et al. 1997). In addition, Pab1 is essential in the coupling of 3′ RNA processing and nuclear export of the mRNA (Dunn et al. 2005).
RPB4:
Rpb4 is an RNA polymerase II subunit that forms a complex with Rpb7. Contrary to other subunits of RNA polymerase II, Rpb4 is not essential under normal conditions; however, it becomes essential under stress. Lately it was found that this complex plays an important role in coordinating transcription in the nucleus with mRNA degradation and translation in the cytoplasm (Harel-Sharvit et al. 2010).
GIS2:
Gis2 binds to a subset of mRNA molecules and promotes their translation by a still poorly understood mechanism (Sammons et al. 2011; Tsvetanova et al. 2010). Both Gis2 and its human ortholog, CNBP, mutations that cause muscular dystrophy, are part of stress granules (Rojas et al. 2012).
HAS1:
The HAS1 gene encodes an ATP-dependent RNA helicase that plays an important role in ribosomal RNA processing and nuclear export. Accordingly, it is highly enriched in nuclear pore complex fractions (Rout et al. 2000).
NUP1:
The Nup1 protein is a component of the central core of the nuclear pore complex. It plays a role in RNA and protein transport through the pore, by serving as a release factor for karyopherin (Aitchison and Rout 2012; Izaurralde and Adam 1998).
CRM1:
Crm1 is the main karyopherin protein, involved in export of proteins, RNAs, and ribosomal subunits from the nucleus. It plays a central role in cargo export (exportin function) (Aitchison and Rout 2012).
The common mechanism suggested by these genes is that the SL phenotype observed can be suppressed by increasing the levels of certain mRNA(s) as well as their transport from the nucleus (through the nuclear pore), perhaps causing an increase in the translation level of (a) target gene(s).
Spindle pole body and spindle checkpoints:
CNM67:
Cnm67 is a component of the spindle pole body outer plaque. It plays a central role in spindle orientation and in mitotic nuclear migration (Brachat et al. 1998). Cnm67 interacts physically and genetically with Gis2 (Scherrer et al. 2011; Wilmes et al. 2008). It also interacts with several kinetochore components, and mutations in CNM67 exhibit a synthetic fitness defect when combined with srs2 mutations (Pan et al. 2006).
DAM1:
Dam1 is a member of the DASH complex, which facilitates microtubule-kinetochore interactions, thereby playing an essential role in chromosome segregation. The DASH complex forms a ring around microtubules that stabilizes the microtubule and promotes its growth (Miranda et al. 2005). It also allows the kinetochore to preferentially bind to the microtubule plus end (Miranda et al. 2005). Dam1 is phosphorylated by the Ipl1 Aurora kinase; although this phosphorylartion does not seem to affect the binding activities of the complex, it is essential for the establishment of bipolar attachments. Notably, Dam1 is also methylated by the Set1 methyltransferase, the same enzyme in charge of methylating histone H3 at lysine4 (H3K4). Remarkably, both H3K4 and Dam1 methylation depend on prior ubiquitination of histone H2B (Latham et al. 2011). Mutations in DAM1 show synthetic fitness defects with mutations in RTT103.
MOB1:
Mob1 is a member of the mitotic exit network. It is located at the spindle pole body, where it binds and regulates the Dbf2 protein kinase. Activation by Cdc15p-dependent phosphorylation leads to a change in its location, inducing cytokinesis and cell separation (Mah et al. 2001). Interestingly, Mob1 is important for the proper localization of the Ipl1 Aurora kinase (Stoepel et al. 2005), which phosphorylates Dam1. Thus, mitotic exit network activity is important for proper kinetochore activity, and controls the spindle checkpoint (Tan et al. 2005).
The mechanism by which these genes may suppress the SL of elg1Δ srs2Δ cells could be related to the spindle checkpoint, which gets activated as a secondary response to DNA damage and prevents adaptation to DNA damage in srs2Δ mutants (Dotiwala et al. 2010).
Protein modification:
A number of genes encoding protein modifiers were identified; these include ubiquitin-related proteins, as well as a protein phosphatase and a regulator of another phosphatase. The targets of these proteins that allow suppression of the SL phenotype is still unclear:
CDC34:
Cdc34 encodes a ubiquitin-conjugating enzyme (E2) that is the catalytic subunit of the SCF ubiquitin-protein ligase complex. This complex is in charge of targeting proteins for degradation, particularly those involved in cell cycle progression. Cdc34 levels are increased under conditions of DNA replication stress, suggesting that it plays an important role in coping with this type of genotoxic situation (Tkach et al. 2012).
UBC11:
Ubc11 is a ubiquitin-conjugating enzyme (E2). Its E3 partners, targets or functions are currently unknown.
GID8:
Gid8 is part of another multiprotein complex that functions as an E3 ubiquitin ligase to ubiquitinate proteins to send them to degradation by the proteasome (Menssen et al. 2012).
UBP14:
Ubp14 is a deubiquitinating protein that disassembles free poly-ubiquitin chains, thus controlling the rate of degradation by the proteasome (Amerik et al. 1997). Interestingly, Ubp14 collaborates with the Gid complex (Regelmann et al. 2003) and mutations in this gene are synthetic lethal with mutations in Cdc34 (Cocklin et al. 2011).
UBP7:
Ubp7 is another deubiquitination enzyme of unknown function.
PPS1:
Pps1 is a phosphatase that plays a role during S-phase (Ernsting and Dixon 1997). Its targets remain unknown.
SIS2:
Sis2/Hal3 binds to the C-terminal catalytic domain of the serine/threonine phosphatase Ppz1 and strongly inhibits its activity (de Nadal et al. 1998). Decreased activity of Ppz1 leads to salt tolerance, compromised cell integrity and accelerated G1/S transition (Munoz et al. 2003; Posas et al. 1995). In addition, SIS2/HAL3 has a role in the biosynthetic pathway of coenzyme A, as part of a different complex, the PPCDC (phosphopantothenoylcysteine decarboxylase complex), which also includes the Cab3 and Vhs3 subunits (Ruiz et al. 2009). However, transformation of the elg1Δ srs2Δ strain with plasmids overexpressing these two subunits conferred no suppression. We thus conclude that it is likely that its interaction with Ppz1 phosphatase is the one responsible for the suppression (although we cannot rule out the possibility that Sis2, and only Sis2, is the limiting factor in the activity of the CoA synthesis complex). Overexpression of a phosphatase repressor may increase the level of a phosphorylated protein, which may be limiting in the elg1Δ srs2Δ strains.
tRNA splicing:
Two genes were found, encoding successive steps in tRNA splicing.
TPT1:
TPT1 is an essential gene that encodes tRNA 2′-phosphotransferase, which catalyzes the final step of tRNA splicing (Culver et al. 1997). tRNA splicing is carried out by a multisubunit endonuclease, followed by ligation. Tpt1 catalyzes the transfer of the splice junction 2’-phosphate from the ligated tRNA to NAD to produce ADP-ribose 1′′-2′′ cyclic phosphate (Appr > p) (Culver et al. 1997). In a conditional lethal tpt1 mutant, 2’-phosphorylated tRNAs accumulate, and the tRNAs show altered posttranscriptional modification (Spinelli et al. 1997).
CPD1:
Each tRNA splicing reaction produces an equimolar amount of ADP-ribose 1′′,2′′-cyclic phosphate (Appr > p). This molecule is converted into ADP-ribose-1′′-phosphate (Appr1p) by Cpd1 (Shull et al. 2005). It is estimated that ∼500 000 tRNA splicing events take place per Saccharomyces cerevisiae generation (Shull et al. 2005).
The significance of having found these two related enzymes in our screen is not clear. One possibility is that Appr1p or its derivatives may serve as signaling molecules in the cell, and an increase in its level may somehow be beneficial. We note that Tpt1 was identified as a histone H3-interacting protein (Gilmore et al. 2012).
Transcription factors:
The genes in this category are as follows:
CST6:
CST6 was identified in a search for genes that are important for chromosome stability (CST genes) (Ouspenski et al. 1999). Interestingly, this screen also identified, among others, PAB1, as well as ubiquitin and histone genes (Ouspenski et al. 1999). Cst6 is a transcription factor activator, a member of the S. cerevisiae ATF/CREB family (Garcia-Gimeno and Struhl 2000). Its targets are still unknown.
RFX1:
RFX1 encodes the major transcriptional repressor of some DNA-damage-regulated genes. It helps recruit the repressors Tup1 and Cyc8 to the promoters of the relevant genes, such as the subunits of the ribonucleotide reductase enzyme. Upon DNA damage Rfx1 becomes phosphorylated and is released from the genes, allowing the recruitment of SAGA or TFIID subunits required for their expression (Ghosh and Pugh 2011).
TAF3:
Taf3 is a subunit of the TFIID complex, required for gene expression. It has lately been found that TFIID is preferentially recruited to promoters that lack TATA boxes, whereas SAGA binds preferentially TATA-containing regulatory sequences (Rhee and Pugh 2012).
The detection of these genes, involved in transcriptional regulation, as suppressors of the SL phenotype of elg1Δ and srs2Δ, implies that the suppression is possible by transcriptional activation or repression of target genes. However, the identity of the regulated genes, albeit, is still to be found.
Additional genes:
BAP2:
Bap2 encodes a high-affinity leucine permease that can also function as a branched-chain amino acid permease involved in the uptake of isoleucine and valine (Grauslund et al. 1995).
SRL3:
The SRL3 gene was identified in a screen for high copy number suppressors of the essential function of the DNA damage response genes MEC1 (ATR in humans) and RAD53 (CHK2) (Desany et al. 1998). Its exact function remains enigmatic.
IRC9, IRC13:
These two genes were identified in a screen for mutants exhibiting increased levels of Rad52 foci. Both are considered “dubious ORFs” by the Saccharomyces Gene Database (http://www.yeastgenome.org).
YOL114c:
YOL114c is an uncharacterized ORF.
YKL044w:
This small ORF is present within the UTR of the PHD1 gene. Evidence for its being translated comes from ribosomal profiling experiments (Brar et al. 2012).
In summary, we have carried out a screen for genes that, when overexpressed, can suppress the SL of an elg1Δ srs2Δ strain. We did not isolate plasmids carrying the ELG1 or SRS2 genes; however, this is an expected result: overexpression of these genes is slightly toxic to the cells, and these genes tend to be underrepresented when screening high copy number libraries (Leon Ortiz et al. 2011; Parnas et al. 2009). The coverage used (>x20) allowed us to identify a relatively large number of candidate genes, which were confirmed one by one by re-cloning and re-testing. Although most genes were identified only once, several of the genomic regions were independently isolated several times, and some (e.g., PAB1) were identified six times. Thus, we are confident that our results represent a significant fraction of the genes that affect the genetic interactions between SRS2 and ELG1. It appears that there is a large number of ways in which the SL phenotype can be bypassed. Our results suggest that chromatin modification and sister chromatid cohesion are processes related to the function defective in elg1Δ srs2Δ double mutants. In addition, increased transcription/translation and nuclear export seem to be able to bypass the synthetic phenotype, possibly by affecting an effector protein(s). We believe that the results obtained from this overexpression screen will provide valuable information for further research needed to better understand the functions carried out by these genes and their interactions.
Acknowledgments
We thank Daniel Kornitzer, steve Elledge, Dan Spatt, Patrick Hartzog, Dan Patrick, Charles Cole, Alain Verreault, Tim Formosa, Ed Hurt, Joaquin Arino, Fred Winston, Steve Jackson, and Robert Skibbens, for plasmids. We also thank all the Kupiec lab members for ideas, encouragement and support. This work was supported by grants from the Israel Science Foundation, the Thyssen Foundation, and the Ministry of Science and Technology to M.K.
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Acharya N., Yoon J. H., Gali H., Unk I., Haracska L., et al. , 2008. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase eta in translesion DNA synthesis. Proc. Natl. Acad. Sci. USA 105: 17724–17729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agmon N., Pur S., Liefshitz B., Kupiec M., 2009. Analysis of repair mechanism choice during homologous recombination. Nucleic Acids Res. 37: 5081–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison J. D., Rout M. P., 2012. The yeast nuclear pore complex and transport through it. Genetics 190: 855–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almedawar S., Colomina N., Bermudez-Lopez M., Pocino-Merino I., Torres-Rosell J., 2012. A SUMO-dependent step during establishment of sister chromatid cohesion. Curr. Biol. 22: 1576–1581 [DOI] [PubMed] [Google Scholar]
- Amerik A., Swaminathan S., Krantz B. A., Wilkinson K. D., Hochstrasser M., 1997. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J. 16: 4826–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N., Minet M., Le Gouar M., Lacroute F., Wyers F., 1997. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol. 17: 3694–3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N., Ghosh S., Mangus D. A., Jacobson A., 2008. Translation factors promote the formation of two states of the closed-loop mRNP. Nature 453: 1276–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong A. A., Mohideen F., Lima C. D., 2012. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature 483: 59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y., Liefshitz B., Bitan-Banin G., Kupiec M., 2003. Molecular dissection of mitotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 23: 1403–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Myung K., 2004. Increased genome instability and telomere length in the elg1-deficient Saccharomyces cerevisiae mutant are regulated by S-phase checkpoints. Eukaryot. Cell 3: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Sikdar N., Myung K., 2007. Suppression of gross chromosomal rearrangements by a new alternative replication factor C complex. Biochem. Biophys. Res. Commun. 362: 546–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellaoui M., Chang M., Ou J., Xu H., Boone C., et al. , 2003. Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. EMBO J. 22: 4304–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aroya S., Koren A., Liefshitz B., Steinlauf R., Kupiec M., 2003. ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc. Natl. Acad. Sci. USA 100: 9906–9911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachat A., Kilmartin J. V., Wach A., Philippsen P., 1998. Saccharomyces cerevisiae cells with defective spindle pole body outer plaques accomplish nuclear migration via half-bridge-organized microtubules. Mol. Biol. Cell 9: 977–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Vanoli F., Foiani M., 2008. SUMOylation regulates Rad18-mediated template switch. Nature 456: 915–920 [DOI] [PubMed] [Google Scholar]
- Brar G. A., Yassour M., Friedman N., Regev A., Ingolia N. T., et al. , 2012. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335: 552–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustard D. E., Menolfi D., Jeppsson K., Ball L. G., Dewey S. C., et al. , 2012. During replication stress, non-SMC element 5 (NSE5) is required for Smc5/6 protein complex functionality at stalled forks. J. Biol. Chem. 287: 11374–11383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A., Agrawal V., Johnson F. B., 2011. Homologous recombination-dependent rescue of deficiency in the structural maintenance of chromosomes (Smc) 5/6 complex. J. Biol. Chem. 286: 5119–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. H., Choi K., Szakal B., Arenz J., Duan X., et al. , 2009. Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc. Natl. Acad. Sci. USA 106: 21252–21257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilkova O., Stenlund P., Isoz I., Stith C. M., Grabowski P., et al. , 2007. The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA. Nucleic Acids Res. 35: 6588–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I., Carotenuto W., Maffioletti G., Petrini J. H., Foiani M., et al. , 2005. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol. Cell. Biol. 25: 5738–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Ruiz M., Gonzalez-Prieto R., Prado F., 2011. Histone H3K56 acetylation, CAF1, and Rtt106 coordinate nucleosome assembly and stability of advancing replication forks. PLoS Genet. 7: e1002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocklin R., Heyen J., Larry T., Tyers M., Goebl M., 2011. New insight into the role of the Cdc34 ubiquitin-conjugating enzyme in cell cycle regulation via Ace2 and Sic1. Genetics 187: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., et al. , 2010. The genetic landscape of a cell. Science 327: 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver G. M., McCraith S. M., Consaul S. A., Stanford D. R., Phizicky E. M., 1997. A 2’-phosphotransferase implicated in tRNA splicing is essential in Saccharomyces cerevisiae. J. Biol. Chem. 272: 13203–13210 [DOI] [PubMed] [Google Scholar]
- Daee D. L., Ferrari E., Longerich S., Zheng X. F., Xue X., et al. , 2012. Rad5-dependent DNA repair functions of the Saccharomyces cerevisiae FANCM protein homolog Mph1. J. Biol. Chem. 287: 26563–26575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson M. B., Katou Y., Keszthelyi A., Sing T. L., Xia T., et al. , 2012. Endogenous DNA replication stress results in expansion of dNTP pools and a mutator phenotype. EMBO J. 31: 895–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E., Clotet J., Posas F., Serrano R., Gomez N., et al. , 1998. The yeast halotolerance determinant Hal3p is an inhibitory subunit of the Ppz1p Ser/Thr protein phosphatase. Proc. Natl. Acad. Sci. USA 95: 7357–7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desany B. A., Alcasabas A. A., Bachant J. B., Elledge S. J., 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12: 2956–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotiwala F., Harrison J. C., Jain S., Sugawara N., Haber J. E., 2010. Mad2 prolongs DNA damage checkpoint arrest caused by a double-strand break via a centromere-dependent mechanism. Curr. Biol. 20: 328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover J., Schneider J., Tawiah-Boateng M. A., Wood A., Dean K., et al. , 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277: 28368–28371 [DOI] [PubMed] [Google Scholar]
- Dunn E. F., Hammell C. M., Hodge C. A., Cole C. N., 2005. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes Dev. 19: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ede C., Rudolph C. J., Lehmann S., Schurer K. A., Kramer W., 2011. Budding yeast Mph1 promotes sister chromatid interactions by a mechanism involving strand invasion. DNA Repair (Amst.) 10: 45–55 [DOI] [PubMed] [Google Scholar]
- Ernsting B. R., Dixon J. E., 1997. The PPS1 gene of Saccharomyces cerevisiae codes for a dual specificity protein phosphatase with a role in the DNA synthesis phase of the cell cycle. J. Biol. Chem. 272: 9332–9343 [DOI] [PubMed] [Google Scholar]
- Garcia-Gimeno M. A., Struhl K., 2000. Aca1 and Aca2, ATF/CREB activators in Saccharomyces cerevisiae, are important for carbon source utilization but not the response to stress. Mol. Cell. Biol. 20: 4340–4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Pugh B. F., 2011. Sequential recruitment of SAGA and TFIID in a genomic response to DNA damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 31: 190–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore J. M., Sardiu M. E., Venkatesh S., Stutzman B., Peak A., et al. , 2012. Characterization of a highly conserved histone related protein, Ydl156w, and its functional associations using quantitative proteomic analyses. Mol. Cell. Proteomics. 11: M111:011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grauslund M., Didion T., Kielland-Brandt M. C., Andersen H. A., 1995. BAP2, a gene encoding a permease for branched-chain amino acids in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1269: 275–280 [DOI] [PubMed] [Google Scholar]
- Harel-Sharvit L., Eldad N., Haimovich G., Barkai O., Duek L., et al. , 2010. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell 143: 552–563 [DOI] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S., 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Imbeault D., Gamar L., Rufiange A., Paquet E., Nourani A., 2008. The Rtt106 histone chaperone is functionally linked to transcription elongation and is involved in the regulation of spurious transcription from cryptic promoters in yeast. J. Biol. Chem. 283: 27350–27354 [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Adam S., 1998. Transport of macromolecules between the nucleus and the cytoplasm. RNA 4: 351–364 [PMC free article] [PubMed] [Google Scholar]
- Kanellis P., Agyei R., Durocher D., 2003. Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr. Biol. 13: 1583–1595 [DOI] [PubMed] [Google Scholar]
- Karras G. I., Fumasoni M., Sienski G., Vanoli F., Branzei D., et al. , 2013. Noncanonical role of the 9-1-1 clamp in the error-free DNA damage tolerance pathway. Mol. Cell 49: 536–546 [DOI] [PubMed] [Google Scholar]
- Kim M., Krogan N. J., Vasiljeva L., Rando O. J., Nedea E., et al. , 2004. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432: 517–522 [DOI] [PubMed] [Google Scholar]
- Klein H. L., 2001. Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Delta with other DNA repair genes in Saccharomyces cerevisiae. Genetics 157: 557–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesar P., Sarangi P., Altmannova V., Zhao X., Krejci L., 2012. Dual roles of the SUMO-interacting motif in the regulation of Srs2 sumoylation. Nucleic Acids Res. 40: 7831–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A., Ben-Aroya S., Steinlauf R., Kupiec M., 2003. Pitfalls of the synthetic lethality screen in Saccharomyces cerevisiae: an improved design. Curr. Genet. 43: 62–69 [DOI] [PubMed] [Google Scholar]
- Koshland D., Kent J. C., Hartwell L. H., 1985. Genetic analysis of the mitotic transmission of minichromosomes. Cell 40: 393–403 [DOI] [PubMed] [Google Scholar]
- Krejci L., Van Komen S., Li Y., Villemain J., Reddy M. S., et al. , 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309 [DOI] [PubMed] [Google Scholar]
- Krishna T. S., Kong X. P., Gary S., Burgers P. M., Kuriyan J., 1994. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79: 1233–1243 [DOI] [PubMed] [Google Scholar]
- Latham J. A., Chosed R. J., Wang S., Dent S. Y., 2011. Chromatin signaling to kinetochores: transregulation of Dam1 methylation by histone H2B ubiquitination. Cell 146: 709–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Garrett A. S., Yen K., Takahashi Y. H., Hu D., et al. , 2012. Codependency of H2B monoubiquitination and nucleosome reassembly on Chd1. Genes Dev. 26: 914–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. Y., Myung K., 2008. PCNA modifications for regulation of post-replication repair pathways. Mol. Cells 26: 5–11 [PMC free article] [PubMed] [Google Scholar]
- Leon Ortiz A. M., Reid R. J., Dittmar J. C., Rothstein R., Nicolas A., 2011. Srs2 overexpression reveals a helicase-independent role at replication forks that requires diverse cell functions. DNA Repair (Amst.) 10: 506–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Burgess R., Zhang Z., 2012. All roads lead to chromatin: Multiple pathways for histone deposition. Biochim. Biophys. Acta 1819: 238–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefshitz B., Parket A., Maya R., Kupiec M., 1995. The role of DNA repair genes in recombination between repeated sequences in yeast. Genetics 140: 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefshitz B., Steinlauf R., Friedl A., Eckardt-Schupp F., Kupiec M., 1998. Genetic interactions between mutants of the ’error-prone’ repair group of Saccharomyces cerevisiae and their effect on recombination and mutagenesis. Mutat. Res. 407: 135–145 [DOI] [PubMed] [Google Scholar]
- Mah A. S., Jang J., Deshaies R. J., 2001. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA 98: 7325–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maradeo M. E., Skibbens R. V., 2009. The Elg1-RFC clamp-loading complex performs a role in sister chromatid cohesion. PLoS ONE 4: e4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maradeo M. E., Skibbens R. V., 2010. Replication factor C complexes play unique pro- and anti-establishment roles in sister chromatid cohesion. PLoS ONE 5: e15381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A., Heidemann M., Lidschreiber M., Schreieck A., Sun M., et al. , 2012. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 336: 1723–1725 [DOI] [PubMed] [Google Scholar]
- Melo J. A., Cohen J., Toczyski D. P., 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15: 2809–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen R., Schweiggert J., Schreiner J., Kusevic D., Reuther J., et al. , 2012. Exploring the topology of the Gid complex, the E3 ubiquitin ligase involved in catabolite-induced degradation of gluconeogenic enzymes. J. Biol. Chem. 287: 25602–25614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J. J., De Wulf P., Sorger P. K., Harrison S. C., 2005. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 12: 138–143 [DOI] [PubMed] [Google Scholar]
- Moldovan G. L., Pfander B., Jentsch S., 2006. PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell 23: 723–732 [DOI] [PubMed] [Google Scholar]
- Munoz I., Simon E., Casals N., Clotet J., Arino J., 2003. Identification of multicopy suppressors of cell cycle arrest at the G1-S transition in Saccharomyces cerevisiae. Yeast 20: 157–169 [DOI] [PubMed] [Google Scholar]
- Ouspenski I. I., Elledge S. J., Brinkley B. R., 1999. New yeast genes important for chromosome integrity and segregation identified by dosage effects on genome stability. Nucleic Acids Res. 27: 3001–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Ye P., Yuan D. S., Wang X., Bader J. S., et al. , 2006. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124: 1069–1081 [DOI] [PubMed] [Google Scholar]
- Papouli E., Chen S., Davies A. A., Huttner D., Krejci L., et al. , 2005. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 19: 123–133 [DOI] [PubMed] [Google Scholar]
- Parnas O., Zipin-Roitman A., Mazor Y., Liefshitz B., Ben-Aroya S., et al. , 2009. The ELG1 clamp loader plays a role in sister chromatid cohesion. PLoS ONE 4: e5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnas O., Zipin-Roitman A., Pfander B., Liefshitz B., Mazor Y., et al. , 2010. Elg1, an alternative subunit of the RFC clamp loader, preferentially interacts with SUMOylated PCNA. EMBO J. 29: 2611–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B., Moldovan G. L., Sacher M., Hoege C., Jentsch S., 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433 [DOI] [PubMed] [Google Scholar]
- Posas F., Camps M., Arino J., 1995. The PPZ protein phosphatases are important determinants of salt tolerance in yeast cells. J. Biol. Chem. 270: 13036–13041 [DOI] [PubMed] [Google Scholar]
- Regelmann J., Schule T., Josupeit F. S., Horak J., Rose M., et al. , 2003. Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae: a genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways. Mol. Biol. Cell 14: 1652–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee H. S., Pugh B. F., 2012. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483: 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robzyk K., Recht J., Osley M. A., 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287: 501–504 [DOI] [PubMed] [Google Scholar]
- Rojas M., Farr G. W., Fernandez C. F., Lauden L., McCormack J. C., et al. , 2012. Yeast Gis2 and Its human ortholog CNBP are novel components of stress-induced RNP granules. PLoS ONE 7: e52824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., et al. , 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148: 635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A., Gonzalez A., Munoz I., Serrano R., Abrie J. A., et al. , 2009. Moonlighting proteins Hal3 and Vhs3 form a heteromeric PPCDC with Ykl088w in yeast CoA biosynthesis. Nat. Chem. Biol. 5: 920–928 [DOI] [PubMed] [Google Scholar]
- Ruone S., Rhoades A. R., Formosa T., 2003. Multiple Nhp6 molecules are required to recruit Spt16-Pob3 to form yFACT complexes and to reorganize nucleosomes. J. Biol. Chem. 278: 45288–45295 [DOI] [PubMed] [Google Scholar]
- Sammons M. A., Samir P., Link A. J., 2011. Saccharomyces cerevisiae Gis2 interacts with the translation machinery and is orthogonal to myotonic dystrophy type 2 protein ZNF9. Biochem. Biophys. Res. Commun. 406: 13–19 [DOI] [PubMed] [Google Scholar]
- Scherrer T., Femmer C., Schiess R., Aebersold R., Gerber A. P., 2011. Defining potentially conserved RNA regulons of homologous zinc-finger RNA-binding proteins. Genome Biol. 12: R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R. H., Prakash S., Prakash L., 1990. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics 124: 817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull N. P., Spinelli S. L., Phizicky E. M., 2005. A highly specific phosphatase that acts on ADP-ribose 1’’-phosphate, a metabolite of tRNA splicing in Saccharomyces cerevisiae. Nucleic Acids Res. 33: 650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. C., Xu X., Kim H. S., Fillingham J., Kislinger T., et al. , 2012. The replication-independent histone H3–H4 chaperones HIR, ASF1, and RTT106 co-operate to maintain promoter fidelity. J. Biol. Chem. 287: 1709–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R. V., Corson L. B., Koshland D., Hieter P., 1999. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 13: 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli S. L., Consaul S. A., Phizicky E. M., 1997. A conditional lethal yeast phosphotransferase (tpt1) mutant accumulates tRNAs with a 2’-phosphate and an undermodified base at the splice junction. RNA 3: 1388–1400 [PMC free article] [PubMed] [Google Scholar]
- St Onge R. P., Mani R., Oh J., Proctor M., Fung E., et al. , 2007. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat. Genet. 39: 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman D. J., 2010. Nhp6: a small but powerful effector of chromatin structure in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1799: 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoepel J., Ottey M. A., Kurischko C., Hieter P., Luca F. C., 2005. The mitotic exit network Mob1p-Dbf2p kinase complex localizes to the nucleus and regulates passenger protein localization. Mol. Biol. Cell 16: 5465–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D., Hu Q., Li Q., Thompson J. R., Cui G., et al. , 2012. Structural basis for recognition of H3K56-acetylated histone H3–H4 by the chaperone Rtt106. Nature 483: 104–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A. L., Rida P. C., Surana U., 2005. Essential tension and constructive destruction: the spindle checkpoint and its regulatory links with mitotic exit. Biochem. J. 386: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach J. M., Yimit A., Lee A. Y., Riffle M., Costanzo M., et al. , 2012. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 14: 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A., Ciosk R., Uhlmann F., Galova M., Schleiffer A., et al. , 1999. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13: 320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetanova N. G., Klass D. M., Salzman J., Brown P. O., 2010. Proteome-wide search reveals unexpected RNA-binding proteins in Saccharomyces cerevisiae. PLoS ONE 5: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich H. D., Jentsch S., 2000. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 19: 3388–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal E., Heidinger-Pauli J. M., Kim W., Guacci V., Onn I., et al. , 2008. A molecular determinant for the establishment of sister chromatid cohesion. Science 321: 566–569 [DOI] [PubMed] [Google Scholar]
- Veaute X., Jeusset J., Soustelle C., Kowalczykowski S. C., Le Cam E., et al. , 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312 [DOI] [PubMed] [Google Scholar]
- Warbrick E., 2000. The puzzle of PCNA’s many partners. Bioessays 22: 997–1006 [DOI] [PubMed] [Google Scholar]
- Wilmes G. M., Bergkessel M., Bandyopadhyay S., Shales M., Braberg H., et al. , 2008. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol. Cell 32: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]