Abstract
Imaging mass spectrometry is an emerging technique of great potential for investigating the chemical architecture in biological matrices. Although the potential for studying neurobiological systems is evident, the relevance of the technique for application in neuroscience is still in its infancy. In the present Review, a principal overview of the different approaches, including matrix assisted laser desorption ionization and secondary ion mass spectrometry, is provided with particular focus on their strengths and limitations for studying different neurochemical species in situ and in vitro. The potential of the various approaches is discussed based on both fundamental and biomedical neuroscience research. This Review aims to serve as a general guide to familiarize the neuroscience community and other biomedical researchers with the technique, highlighting its great potential and suitability for comprehensive and specific chemical imaging.
Keywords: Imaging mass spectrometry, SIMS, MALDI, neurotransmitters
The human brain is the most complex and heterogeneous organ. It is involved in a vast variety of body functions ranging from motor control, touch sensing, vision, hearing, smelling, hormone regulation, and many more. In no other organ are the signaling mechanisms between different cells so poorly understood. The nervous system maintains its functionality through a vast number of chemical substances orchestrating neurological processes. Complex biological and pathological processes in the nervous system involve the translocation of a wide range of chemical species. Probing the spatiotemporal dynamics of ongoing biochemical processes is therefore essential to deepen our understanding of complex biological mechanisms including signal transduction processes within distinct regions of the central nervous system (CNS) as well as the CNS with peripheral systems.
A major objective when studying molecular mechanisms and interactions at subcellular levels is the acquisition of molecular images (e.g., topographical and temporal information of molecular abundance distribution). The main challenge in molecular imaging is to achieve appropriate temporal and spatial resolution with high precision, specificity, and sensitivity. However, this is significantly hampered due to the lack of effective research tools available to study the chemistry of these complex systems. Hence, the development of analytical technologies that facilitate subcellular analysis with high molecular specificity and at high spatial resolution is of essential relevance in order to deepen our understanding of inter- and intracellular molecular processes.
During the last 20 years, mass spectrometry (MS) has gained significant relevance in biological and biomedical research, especially for large scale, comprehensive protein profiling, termed proteomics.1 The introduction of soft ionization techniques such as matrix assisted laser desorption ionization (MALDI)2 and electrospray ionization (ESI)3 afforded MS analysis of large biomolecules such as proteins, peptides and lipids. Indeed, the development of these soft ionization techniques for mass spectrometry that allow fast, sensitive, and specific analysis of larger biomolecules including proteins and peptides was the major catalyst for the immense growth in relevance of proteomics. While tissue proteomics facilitates protein identification and quantitation, spatial information within the respective tissue compartment is not obtained. Given the complexity of the human nervous system, the spatial information of protein distribution is of major interest in order to resolve ongoing molecular mechanisms.
Imaging mass spectrometry (IMS) is a powerful approach for comprehensive analysis of spatial intensity distribution profiles of molecular species in biological tissue and single cells.4 In contrast to common molecular biology techniques, including chemical staining, immuno-based imaging approaches (immunohistochemistry, IHC; confocal laser scanning microscopy, CLSM), and nucleotide detection (in situ PCR), IMS does not require any a priori knowledge of the potential target species. It is furthermore not dependent on antibody or primer availability and specificity. IMS features high molecular specificity and allows comprehensive, multiplexed detection, identification, and localization of hundreds of proteins, peptides, and lipids in biological tissues samples.4 Over the past decade, IMS has slowly evolved as a relevant, alternative approach in biomedical research for studying proteins, peptides, and lipids in disease pathology, pharmacotherapy, as well as fundamental biological processes.5,6 The technique allows matching of histological features of a biological tissue sample to molecular localization patterns and can therefore also be referred to as molecular histology.7,8 Although, theoretically, one can carry out untargeted studies with IMS, in general it is necessary to have target molecules in mind in choosing the matrix best suited to provide the required sensitivity.
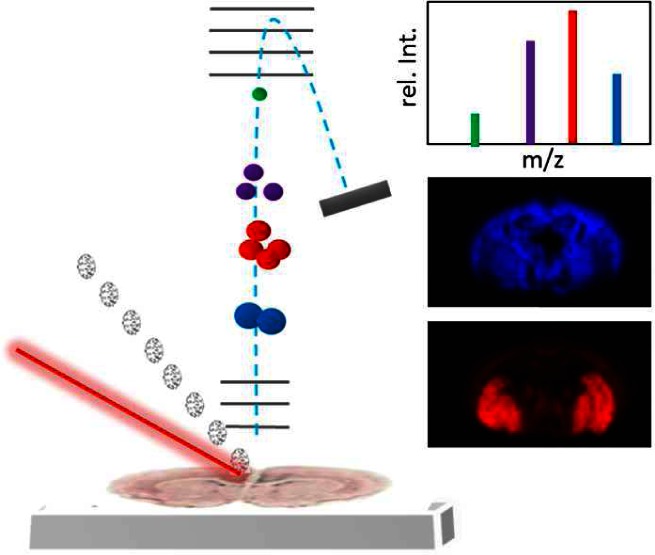
Imaging mass spectrometry can be performed with different probes to desorb and ionize molecular species directly from a biological sample (Figure 1a). The most prominent approaches include, laser desorption/ionization (LDI) based techniques including MALDI, and time-of-flight -secondary ion mass spectrometry (TOF-SIMS), in which an ion beam is employed to sputter molecular species into the gas phase.9 In addition, another, electrospray based desorption (DESI, nanoscale DESI),10 has become rather popular recently mostly for images with larger resolution pixels. Different imaging MS technologies have various strengths and limitations particularly with respect to spatial resolution and molecular information.9 Other aspects include sample throughput, sample preparation, economical and experimental efforts as well as data handling obstacles. A major consideration in IMS, is the effect of ion-suppression or matrix effects that can affect ionization yield and thereby the quantitative information obtained. Appropriate internal and external controls are necessary to account for these limitations, some of which are presented and discussed below. All of these different features have to be taken into account when designing an IMS based experimental study. The aim of the present Review is therefore to provide an overview of current IMS techniques and to discuss their suitability for biomolecular imaging with particular focus on neuroscience research. Furthermore, different methodological concepts and pitfalls in sample preparation, data acquisition, and data handling will be discussed. Finally an overview of some few examples of IMS based fundamental and translational neuroscience research is provided. The scope of this Review does not permit us to be comprehensive, and we apologize to those wonderful works we were forced to omit.
Figure 1.
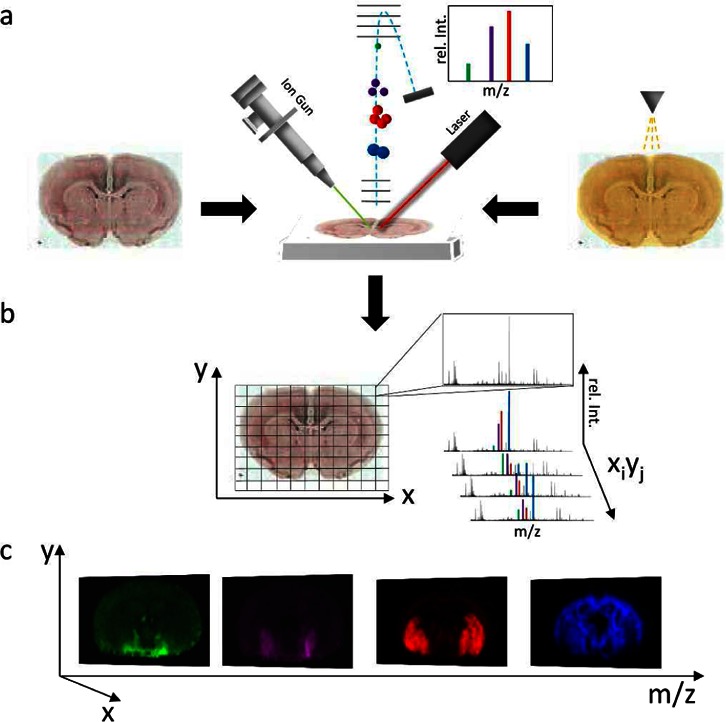
Principle of imaging mass spectrometry. (a) For SIMS IMS, tissue sections are probed with an ion beam (top left). In contrast, MALDI IMS requires precoating with matrix (indicated by a matrix sprayer, top right) before systematic scanning with a laser probe. For both techniques, molecular species are desorbed and ionized followed by mass analysis (top middle) using, e.g., a time-of-flight (TOF) analyzer. In TOF, a mass spectrum is generated where the molecular mass corresponds to flight time (m/z ∝ t2) and number of molecules that reach the detector correspond to signal intensity. (b) An acquisition raster is defined for the tissue section, predefining spatial resolution. Individual mass spectra are acquired for every xi,yj coordinate of the tissue section according to the predefined raster. (c) Single ion images are generated by mapping the intensity of an individual ion signal (m/z; rel.Int) over the whole tissue slide.
Principle of Imaging Mass Spectrometry
In IMS, the molecular intensity distribution in biological samples is obtained by sequential acquisition of mass spectra from predefined, individual spots (pixel array) over a biological tissue section or a single cell (Figure 1b). Molecular ion images are generated by plotting the intensity distribution of a distinct molecular species over the analyzed MS analysis array (Figure 1c). The point-to-point distance defines the limit for lateral resolution of the IMS analysis. Spatial resolution is mainly limited by the probe (ionbeam, SIMS or laser, MALDI); however, lateral diffusion during sample preparation has a major impact as well. In SIMS, lateral resolution is also compromised by the mass resolution and essentially the limit of detection. Similarly, high spatial resolution MALDI IMS experiments are limited by the sensitivity of the method to the respective compounds of interest.9,11
Acquired images can involve enormous data amounts, as there is a complete spectrum for each pixel; hence, these often comprise file sizes of multiple gigabytes. Therefore, spatial resolution and pixel number need to be in a practical balance. Analysis of imaging data can be performed using multiple multivariate statistical analysis (MVSA) tools. This is an excellent approach for unbiased interrogation of molecular intensity distributions and potential localization to anatomical features of interest. Multiple image analysis strategies have been reported for TOF SIMS.12−14 Similarly, at least one hierarchical clustering based MVSA-solution has been demonstrated for MALDI IMS data.15 MVSA allows segmentation of a biological sample in distinct regions of interest by detecting variances and correlations in the multivariate data that are encompessed in factors. From the corresponding scores and loadings, the variables contributing the most (i.e., mass peak values) to these variances can be deduced. However, generation of MVSA results does require mandatory verification by manual inspection of respective single ion images. All generated IMS results have to be further validated to rule out false positive findings as a result of e.g. suppression effects. Validation of peak identity and quantity can be achieved by targeted ESI- LCMS/MS experiments for intact lipids and neuropeptides as well as small intact proteins. Protein changes can be validated in situ with immune-based methods. This is, however, largely dependent on antibody availability and specificity and difficult to compare in the case of truncated protein isoforms. Protein peak identification protocols have been investigated thoroughly in previous studies.16−18 This includes, for example, direct on tissue digestion followed by imaging and fragmentation of tryptic peptides. Identified peptide species are then confirming protein identity by showing similar distribution pattern as the corresponding intact protein.16 Alternatively, a straightforward top down proteomic approach was demonstrated as a convenient strategy to identify unknown protein peaks. The workflow was based on protein prefractionation using reversed phase liquid chromatography (LC) followed by MALDI interrogation of collected protein fractions (LC-MALDI). Those protein fractions that contained a protein peak of interest were then subjected to subsequent trypsination and identified by LC-ESI-MS/MS based peptide separation and fragmentation.17,18 While imaging mass spectrometry is a very powerful approach, this highlights the need for complementary validation strategies in order to fully exploit the potential of the technique.
MALDI Imaging MS
Since its introduction,2 MALDI TOF-MS has significantly impacted the field of protein analysis in molecular biological and biomedical research. MALDI MS is a soft ionization technique permitting analysis of intact macromolecules including proteins and peptides. The technique is based on laser irradiation induced desorption and ionization of analyte molecules with the help of a crystalline UV absorbing matrix (Figure 1a). MALDI MS, in general, is characterized by its superior mass range, sensitivity, high resolution, and mass accuracy as well its robustness and insensitivity to sample impurities.
In 1997, Caprioli and co-workers introduced a MALDI TOF MS based approach for spatial profiling of large molecular species in mammalian tissue samples.19 The technique, hence referred to as MALDI imaging MS or MALDI IMS, is based on the application of matrix solution to a thaw mounted tissue section followed by MALDI MS analysis in a quadratic pattern with a spot to spot distance defining the spatial resolution (Figure 1b). The technique is particularly well suited for medium to large biomolecules including glycolipids, neuropeptides, and proteins. Detection of small molecules such as drugs, metabolites, and neurotransmitters is, however, hampered by interference of matrix cluster ions. This limitation can be overcome by matrix free laser desoption/ionization (LDI) approaches as well as other strategies including, for example, use of a stable isotope modified matrix or MS/MS methodologies just to name a few.20−22
Spatial resolution in MALDI IMS, is essentially dependent on three factors: the laser beam focus, the matrix crystal, and lateral diffusion of analyte molecules resulting during sample preparation. Typically, a practical spatial resolution of 20–50 μm can be achieved; however, different efforts to push these limits with respect to the probe have been reported.23,24 Here a spot to spot distance, smaller than the laser diameter, can be used, exposing the sample to only a fraction of the beam. This oversampling approach is, however, highly dependent on homogeneous and rigorous sample ablation to avoid interference with adjacent compounds.23 Another high resolution MALDI IMS approach is based on efforts to focus the laser.24 For both MALDI and oversampling MALDI, method sensitivity, data file size, and acquisition time have to be considered. In addition, sample preparation is also an important factor affecting spatial resolution.
Sample Preparation for MALDI IMS
For surface analysis in general and MALDI in particular, sample preparation is one of the most critical issues. Various target preparation parameters have an important impact on final data quality, particularly with respect to signal intensity, reproducibility, as well as lateral resolution. The sample preparation workflow in MALDI IMS comprises the whole chain of sample treatment starting with appropriate tissue retrieval and tissue storage over sectioning, sample wash, and until final matrix application. Each of these steps is critical and might impact the final data significantly. In particular, matrix application is a cornerstone in MALDI IMS experimental workflow schemes and has to be tailored according to many factors, including tissue origin and molecular target.
As for all tissue imaging experiments, tissue retrieval from animal or human sources is critical to data quality. This is of particular relevance, since commonly used perfusion and fixation strategies cannot be applied in mass spectrometry due to interference of the polymeric fixation agents such as paraffin. Solutions to overcome these obstacles, including in situ trypsination following paraffin removal or alternatively use of a reactive matrix 2,4-dinitrophenylhydrazine, have been presented.25
Fresh frozen tissue samples are, however, by far the most commonly used samples for IMS. In terms of sample dissection, such as, for intact brain, quick performance is essential, since post mortem delays by as short as 3 min result in severe degradation of neuropeptides.26 One solution for overcoming this shortcoming is heat stabilization, where quick sample heating to 95 °C leads to efficient protease inactivation.27 This approach, however, impacts the sample morphology, which in turn hampers its suitability in IMS. This is primarily due to difficulties in maintaining tissue integrity during sectioning, although an elegant workaround using collection on conductive carbon tape has been presented recently.28
Cryosections of fresh frozen tissue are thaw mounted onto conductive glass slides. Here, the collected sections have to be dried immediately before storage at −80 °C to prevent damage by water condensation during freezing (29)
A further critical step in sample handling is the choice of appropriate washing protocols. Lipid species typically do not require any advanced washing steps, whereas drugs, neuropeptides, and proteins require optimized washing protocols for signal enhancement. These involve pH sensitive cleanup as well as organic solvents for precipitation of peptides and proteins while washing off remaining lipids that potentially interfere with neuropeptide signals prior to matrix application.29,30
In terms of matrix application, currently there are three approaches available, each with its own strength and limitations with respect to simplicity, cost, resulting signal intensity, spatial resolution, and signal reproducibility. In general, there is a trade-off between signal quality and lateral resolution based on the correlation of vertical and lateral infusion with increased droplet size. The most straightforward approach involves manual application of matrix solution employing an airbrush sprayer. While this is a quite cost-effective solution, it is severely hampered by its lack in reproducibility as well its susceptibility to sample diffusion as well as limited extraction efficiency. A more controlled way to deposit matrix is to use a nebulizer-based sample application available in a commercial ImagePrep device (Bruker Daltonics, Bremen, Germany). Here, pneumatic aerosol formation and subsequent matrix application is monitored by a light scattering sensor below the sample glass to estimate matrix thickness. Although this is not as economical as the bare nebulizer, this technology has significantly advanced the aerosol-based matrix application in terms of reproducibility and signal quality. Still, one major disadvantage of this solution remains in its rather low extraction efficiency, as a consequence of reduced vertical diffusion. The most controlled approach for sample application involves deposition by a chemical inkjet (ChiP, Shimadzu, Kyoto, Japan) or an pneumatic vertical spotter (Portrait, Labcite, Sunnyvale, CA). This approach uses accurate deposition of distinct droplets in a geometrical pattern, thereby avoiding lateral diffusion of the analyte.31 The spotting approach is particularly beneficial for low abundant species, including neuropeptides.32 Although this approach is limited in terms of spatial resolution, the best data quality in terms of signal strength and reproducibility is achieved.
A plethora of different matrices has been reported for MALDI IMS of various molecular species. Different matrices are well suited for different compounds, and the choice depends largely on the targeted substance. Recent advances in matrix developments have been aimed at overcoming various limitations of the standard compounds, including crystal size, reproducibility, interfering matrix cluster, mass range, and signal quality. Here, ionic matrices were found to give highly homogeneous matrix layers, facilitating high spatial resolution imaging of lipids and neuropeptides.33 However, this approach is limited in terms of signal reproducibility as well as extraction efficiency and hence sensitivity.
Further approaches have been developed to overcome the limitations in various matrix application protocols and to control crystal size. These include various sublimation techniques as well as dry coating.34,35 Both of these techniques give very small crystals and allow high spatial resolution, but the compromise is poorer signal intensity and reproducibility. In contrast to all matrix application techniques, various matrix free laser desorption technologies have been demonstrated to be powerful alternatives. These include development and application of functional surfaces to facilitate desorption and ionization. These range from bare porous silicon or advanced nanoinitiator compounds as well as functionalized graphene and carbon nanotube surfaces.20,36,37 However, to date, no single biological study using these approaches beyond proof of principle has been reported.
Secondary Ion Mass Spectrometry Imaging
Secondary ion mass spectrometry (SIMS) is a very powerful imaging technique providing chemical information with high spatial resolution (<50 nm). Briefly, a focused ion beam impacts the sample surface resulting in the ejection of secondary species including positive- and negative ions as well as neutral molecules and elements. The desorbed and ionized compounds are then analyzed by means of mass spectrometry using different mass analyzers such as magnetic sector field and time-of-flight (TOF) analyzers.38 Chemical information about the surface is generated while spatial resolution is maintained, by either collecting spectra in a raster pattern or using a position sensitive detector. The result is an image with chemical information contained in each pixel.
An important characteristic of SIMS, which sets it apart from other biological imaging methods, is that the sample can be analyzed without chemical perturbation. That is to say no labels or matrixes are needed to acquire SIMS signal, however, some studies do require isotopic labels to obtain molecular information. The technique does, however, have limitations with respect to the mass range (<1500 Da) as a result of in source fragmentation. Furthermore, as it is a surface sensitive technique, extreme care must be taken during sample preparation to prevent any possible ambient contamination.
To overcome these limitations, the design of new primary ion sources is one of the main assignments in instrumental development. The ultimate goal is to achieve spatial resolution of a few nanometers while improving secondary ion yield efficiency and molecular integrity.
Today the available primary ion sources comprise atomic, polyatomic, macromolecular cluster, and gas cluster ions that have different advantages and limitations. The choice of projectile depends on the respective application mode that covers the whole range from inorganic surfaces to biological tissue and cells.
Choice of Projectiles and Modes of Operation
SIMS is an inherently destructive process. However, when irradiation with a distinct primary ion doses (maximum of 1013 ions/cm2) causes a small degree of surface and subsurface destruction, this is defined as the static limit. In contrast, in dynamic SIMS, a primary ion dose that exceeds the static limit is used. Typically, reactive primary ion beams such as O2+ and Cs+ are used in this mode, giving high secondary ion yield to enhance sensitivity; however, this also results in higher fragmentation of the ions. This limits the mass range of the molecular analytes to atomic species and very small fragments. However, higher sensitivity is achieved, making this technique well suited for studying inorganic/elemental surface characteristics, where molecular information is not required.38,39 To extract molecular information, isotopic labeling strategies have been developed. Multiple isotope imaging mass spectrometry (MIMS) allows imaging of several molecular species using this destructive technique.40 Furthermore, high ion doses erode the sample and can be used to obtain information in the third dimension for 3D imaging. Due to abundant signal in a very small spot, dynamic SIMS can provide very high lateral resolution for imaging.39
Static SIMS sidesteps this issue by using an extremely low primary ion dose below the static limit, typically 1013 ions/cm2,9 to disregard any damage done to the sample surface. Under the static condition, less than 1% of the surface receives an ion impact, ensuring that mass spectra are not collected from a damaged surface. For all intents and purposes, the measurement does not disturb the pristine condition of the sample. This is achieved by using polyatomic liquid metal ion guns (LMIG).41 These ultrabright probes provide extremely focused ion beams (<50 nm) but can be used at low doses, allowing formation and detection of molecular ions. These finely focused beams offer the potential to map ions in a mass range up to m/z 1500; atomic species, lipids, amino acid, and fatty acids can be mapped with high resolution over cell and tissue surfaces. The relatively low availability of biomolecular ions, due to in source fragmentation and poor ionization efficiency, can limit the useful size of the probe. Increases in sensitivity therefore require an increase in primary ion dose with the effect of sample degradation.42 This has been significantly improved with the development of macromolecular ion beams such as C60 and SF6. Although these beams do not compare in terms of probe size (∼1 um), energy is deposited at even shallower depths than with cluster polyatomic LMIGs, thereby decreasing surface damage and sample degradation.43,44 In addition, this facilitates high resolution depth profiling, which is achieved by stepwise etching and analysis of molecular surface layers.42,45 Current developments have provided large size Arn cluster ion sources (n < 1000–5000), attracting great interest. These beams provide higher ion yield efficiency and less subsurface damage. While these are obvious advantages with respect to molecular mass range and depth profiling resolution, there are still limitations concerning ion beam focus and lateral resolution.46
A general outcome of these developments is that the minimal damage accumulated when using macromolecular beams and gas cluster beams is blurring the line between dynamic (destructive) and static (nondestructive) SIMS as previously defined by the static limit. The advent of cluster ion beams has significantly impacted the area of molecular depth profiling and 3D imaging MS.42
Molecular 3D biological imaging is essentially the combination of static and dynamic modes. By taking advantage of the minimal damage accumulation observed for molecular and gas cluster beams, it is now possible to erode a sample and uncover relatively intact molecular layers and then chemically analyze them. The imaging can be done using a focused eroding beam, or a dual beam strategy can be employed, where the surface is etched with a defocused beam and subsequently analyzed with a focused beam. This mode of analysis opens up SIMS to numerous biological applications, as it allows simultaneous detection of biological molecules inside biosystems without any further chemical labeling.42,45,47−49
Image Generation in SIMS
Typically biological SIMS imaging is carried out in microprobe mode. This approach is characterized by rastering a focused primary ion beam across the surface of the sample and correlating the chemical information to the beam position. Spatial resolution is dependent on several factors; however, in general the most important factor is the size of the probe. While there are physical limitations to focusing a probe, the main issue is ionization efficiency. In essence, decreasing the probe size reduces the amount of material available to be ionized and eventually the probability of creating an ion vanishes. This principle is summed up with the concept of useful resolution. Simply put, useful resolution is related to the square area required detecting a given analyte. Many factors contribute to this limit including transmission, analyte concentration, and ionization efficiency, but ultimately there is little advantage to focusing a primary ion beam beyond this point. One consequence of this is that dynamic SIMS, which tracks smaller more abundant ions, enjoys higher lateral resolution than static SIMS.
Another approach to SIMS images is the microscope mode. The detector in this case is position sensitive, and therefore, probe size becomes unimportant. Useful lateral resolution still applies in this case. As a result, most of the biological work using this type of imaging has been done using dynamic SIMS.38
Sample Preparation
Appropriate sample preparation protocols play an essential role for a reliable measurement in the SIMS technique. The main challenge here is preservation of morphology and chemical information of a sample while exposing it to high vacuum condition.
The most common and convenient sample preparation method is freeze-drying. Briefly, freshly obtained biological tissue is plunge frozen to keep it intact. The frozen tissue is then cryosectioned, typically at −20 °C, into 10–20 μm slices. The tissue slices are then thawed onto conductive substrates and subsequently inserted into mbar vacuum where the water from the samples is sublimated slowly to dryness. Cellular samples can be prepared in a similar procedure in which the cells are sectioned by using an ultramicrotome, which reduces the thickness down to several micrometers. Alternatively, cells can also be freeze-dried directly after they are plunge frozen. Freeze-drying enables the analysis to be carried out at room temperature, which keeps sample handling simple and easy. The method is well suited for tissue imaging; however, when subcellular information is of interest, freeze-drying can cause redistribution of biological molecules on the subcellular size scale.27
Freeze fracture has been considered the ideal method for maintaining chemical and structural information for subcellular biological imaging.50,51 Tissue or cellular samples are plunge frozen in either a sandwich assembly50,52 or a spring-loaded device, which is opened in the instrument under vacuum.53,54 The sample surface is exposed to provide intact chemical information. The protocol protects the sample from possible environmental contamination and produces a thin layer of condensed water, which enhances molecular ion signals. In addition, the method can be used to fracture the inner compartments of the cells for subcellular analysis. Elaborate sample handling required for cryogenic conditions is the main drawback of the method. There is also an issue with obtaining a suitable fracture plane. Rough surfaces and buried samples often hinder analysis.
In order to overcome the limitations of the above methods, the main limitations being possible contamination from ambient environment in freeze-drying and topography issues in freeze fracture, etching by elevated temperature55 or by use of a C6056 cluster ion gun have been employed. These approaches remove the top layer of sample without damaging the sublayer and flatten the sample surface, respectively, eliminating the topography problem. In etching by temperature or so-called freeze etching, temperature is increased to a critical point where the top layer of condensed water slowly sublimates leaving an intact layer below. In etching using a C60 projectile beam, the top layer is removed by sputtering with C60. The methods are good problem solving strategies, however, there are some minor disadvantages, namely long analysis time using freeze etching and care in controlling etching using C60 gun to obtain flat surface.51
Fundamental IMS based Applications in Neuroscience
The contribution of IMS to basic neurobiological study has been somewhat sparse. The reasons for this include limited access to the technology, relatively low sample throughput, and difficult sample preparation. These have been reviewed elsewhere and represent the main focus of IMS research.57,58 This work has in fact produced many impressive brain tissue27,59−63 and single neuron64,65 chemical images. This area has generated a large body of spatio-chemical information, but in general a link to basic neurological function is difficult to make. However, there are several examples that have produced information of general interest to this field.
MALDI imaging affords the mapping of biological molecules over a large mass range spanning from intact lipids to proteins. Although great strides have been made to increase lateral resolution, single cell analysis is somewhat challenging, thus keeping most work in the realm of brain tissue analysis. The images produced have hundreds of compound peaks with unique localizations that give indication of functionality. The information dense data collected often contains several unknown peaks, which might represent neurologically important peptides and proteins especially considering specific localizations of these molecules. Universally, peak identification can be a sticking point when attempting to make convincing conclusions about these images considering the specificity of labeling strategies used in other methods. To aid in identification, Taban et al. have employed the high mass resolution of Fourier transform ion cyclotron resonance mass spectrometry. The MS/MS capability in combination with ultrahigh mass accuracy afforded by MALDI-Q-FTICR-MS (ApexQ, Bruker Daltonics, Bremen, Germany) was used allowing examination of fragment ions directly from the tissue sample to positively identify it as the neuropeptide vasopressin.25 The localization also verified the identity. This study highlighted the exploratory possibilities of IMS for fundamental studies and the need for powerful tools for validation.
It can be argued that the most important role SIMS imaging has played in biology thus far has been lipid analysis.66−71 Primarily, this strength is afforded by the relative abundance of these molecules in biological systems. In terms of neurobiology, this tends to lead toward membrane controlled processes, namely, exocytosis. It is theorized, most notably with the stalk model,72 that lipids might have specific roles during fusion, which require them to concentrate in specific areas based on molecular structure. This phenomenon is theoretically ideal for probing with SIMS imaging; however, the small size of the structure limits the availability of the molecules that can be analyzed and the three-dimensional configuration of the structure makes it challenging to investigate using a surface sensitive technique. Although these challenges could be overcome as groups continue to work on increasing ion yields and acquisition of 3D SIMS images.73
To study lipid domains during membrane fusion, the central process of exocytosis, a large-scale model was used in the single cell organism Tetrahymena thermopile. This species undergoes sexual reproduction by fusing to a second cell and transferring genetic material via hundreds of fusion pores that are formed along the membrane interfaces. This event creates a curvature gradient between the relatively flat cell membrane and the highly curved bilayer at the junction. The large size scale of the junction compensates for low ionization efficiencies, which make extracting chemical information from individual fusion events at this point prohibitive. SIMS imaging was used to image mated Tetrahymena cells, and a chemical gradient was observed between the cell and the junction.67 Specifically, the concentration of the lamellar phosphatidylcholine (PC) lipids was greatly reduced in the junction, indicating that the curvature gradient was linked to a demixing of the lipid components. To establish whether the curvature gradient was induced or if it was induced by the observed lipid reorganization, a time dependent study was also carried out. The appearance of the chemical gradient was shown to lag behind the formation of the curvature gradient. It was proposed that the local change in composition was driven by the change in curvature giving evidence for the so-called curvature-coupling hypothesis (Figure 2a).69
Figure 2.
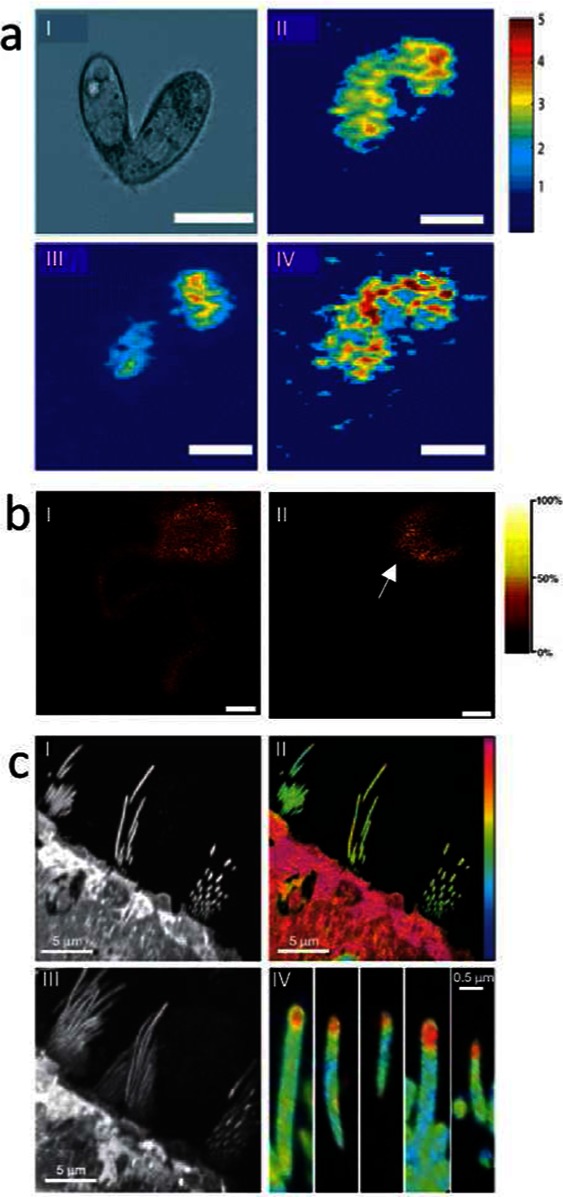
Fundamental neuroscience research imaging mass spectrometry. (a) SIMS imaging reveals distribution of lipid species that are coupled to membrane curvature in mating cells.69 (I) Differential interference contrast microscopy image of a mating cell pair. (II–IV) SIMS ion images of a triturated pair of mating T. thermophila for different lipid species, including C5H9 (I), PC (II), and 2-AEP (IV) (the intensity in the 2-AEP image has been multiplied by factor 3) (scale bar = 25 um). (b) Application of SIMS for imaging single neurons allows subcellular localization of vitamin E. (Reproduced with permission from ref (65). Copyright 2005 American Chemical Society.) (I) Neuronal lipid distribution illustrated by the choline fragment trimethylethenamine (m/z 86), derived from sphingosine and phosphatidylcholin (PC), showing homogeneous distribution throughout soma and neurite. (II) Interestingly, vitamin E was found to distribute to the soma–neurite junction, suggesting an important role in neuronal communication (II) (scale bar = 100 μm). (c) Multi isotope imaging MS was employed at 30 nm resolution to demonstrate protein turnover in hair cells.80 (I) SIMS ionimage of protein fragment CN (m/z 26) of utricle; day 56 and ration CN15/CN14 (m 27/m 26) showing low incorporation in stereocilia. (III) Projection of a three-dimensional stack of image (I). (IV) CN15/CN14 Ratio image from (III) reveals high protein turnover to occur at a high rate at the tip of hair cells.
Curvature-coupling might be an important factor that determines the kinetics of individual exocytotic events. This hypothesis puts forward the idea that a membrane component can sense curvature by virtue of its own intrinsic curvature. Simply put, molecules will tend to localize to areas that match their curvature. The upswing of this is that these molecules will lower the resistance to bending by increasing the local spontaneous curvature thus affecting geometric curvature of the structure. There is somewhat of a controversy as to whether or not lipids are actually large enough to sense membrane curvature; however, it was recently demonstrated that the addition of DOPE (a lipid with a strong negative intrinsic curvature) decreased the diameter of a lipid nanotube by presumably being preferentially sorted into the negatively curved inner leaflet of the tube.17 There is, however, a fair amount of evidence that curvature-coupling is involved in exocytosis or at the very least the intrinsic curvature has an effect. In these experiments, individual exocytotic events were monitored at secretory cells using amperometry with submillisecond temporal resolution. The outer leaflets of the cells were bathed with exogenous lipids and the resulting changes in release can largely be attributed to the intrinsic curvature of the bathing lipid.
By expanding upon relative quantitative methods developed in the SIMS community,40,74,75 a deuterium labeling scheme has been reported to estimate the amount of lipid that was incorporated into these augmented secretory cells.76 The method used the same incubation procedure and cell line used in several of the studies described here62,75,77 but substituted the exogenous lipids for lipids with deuterated acyl chains. The deuterium label supplied a unique peak that was used to track the relative amount of the added lipid. The yield for this ion was calculated from a pure sample and used to calibrate the signal from the treated cell. This was also done for the endogenous PC lipids and the ratio of these signals was used to calculate the relative amount of exogenous lipid. The values were then corrected by taking the known PC concentration of the cell into account. The results suggest that the observed changes in exocytosis were brought about by very small changes (0.5–1.3%) in lipid composition.76 The implication is that subtle lipid heterogeneity across the membrane of a cell could be a powerful regulatory mechanism for these events. Indeed, events measured at these cells are fairly heterogeneous and work has shown that locations on a single cell can be more active, so-called hot spots.59,77 This is a very exciting potential target for this method but remains difficult to image; although there are examples of subcellular imaging, the length scales here are a significant hurdle when considering the current ionization yields.78
The difficulty in extracting subcellular chemical information is most simply overcome by choosing a large cell for analysis. This approach is illustrated in the imaging of large neurons extracted from the mollusk Aplysia californica. In one notable study, Monroe et al. used TOF-SIMS imaging to chemically map the membrane of these cells and found that vitamin E is strongly associated with the soma–neurite junction (Figure 2b).65 It is known that vitamin E deficiencies are related to neurological disease, and historically this has been attributed to protection from oxidative stress. Clinical trials, however, do not overwhelmingly support this conclusion, suggesting that the role of vitamin E is more complex, possibly having a more fundamental role.79 The specific localization of vitamin E in the Aplysia neuron strongly indicates that there is some underling function possibly critical to healthy neuronal function. This experiment exhibits an important advantage to IMS in that little prior knowledge about the sample is needed. Certainly parameters need to be adjusted for the type of sample, but in general if the surface component can be ionized in the gas phase the result will be a chemical map of the surface.
The ultimate potential of SIMS for subcellular chemical imaging has been reported recently by the Lechene group.80 Using multi-isotope imaging MS (MIMS), the protein turnover on the hair cells of the auditory somatosensory system in the inner ear was studied at nanometer scale. Animals were fed 15N-labeled leucine, and the isotopic ratio of the percent and the location of the new protein was determined. Using the dynamic mode of SIMS, they were able to generate sufficient signal to see a distinct increase in metabolism at the tip of the hair, an area only a few 100 nm long. The combination of protein sensitivity and high lateral resolution that MIMS provides presents the real possibility of following chemical changes of the dense protein structures in neural cellular substructures including, for example, synaptic terminals and post synaptic density (Figure 2c).
Biomedical Applications of Imaging Mass Spectrometry in Neuroscience
Imaging MS has great potential in exploring biomolecular mechanisms underlying neurodegenerative disorders. However, so far only few studies have been reported to answer questions relevant to clinical and molecular neuroscience. As discussed above, this is probably related to the limited interface between neurochemical and analytical chemistry research not to mention the sparse overlap of research topics and interests in mass spectrometry in analytical chemistry and neurobiology in general.
Arguably there is certain skepticism among neuroscientists toward mass spectrometry based proteomics and imaging. One reason is that there has been a vast amount of untargeted profiling studies using proteomics for neurological samples. The lack of a hypothesis beyond the detection of potential differences or biomarker discovery is certainly alien to classical mechanistic investigations common in neuroscience. It is a central aim of this section of the review to overcome this skepticism by discussing studies that have demonstrated the value and power of IMS in molecular neurobiology.
Imaging of Neurotransmitters and Low Molecular Weight Metabolites
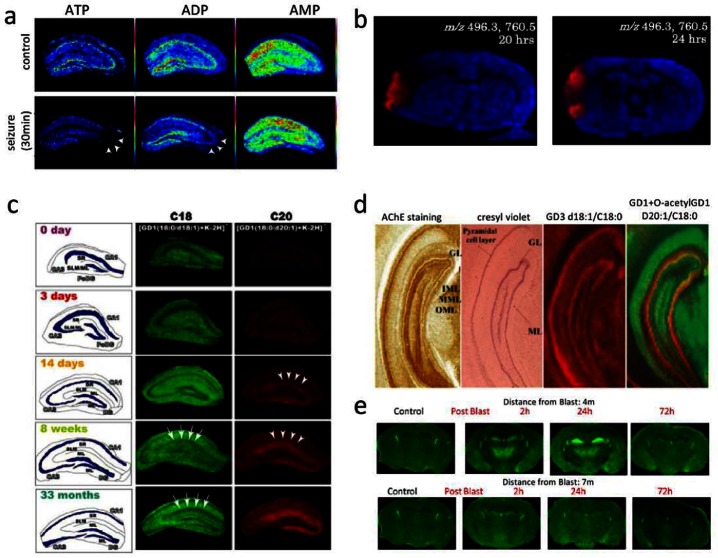
Analysis of low molecular weight compounds (<500 Da), including neurotransmitters, using MALDI IMS is hampered due to interference of clusters of ions in the sample matrix. There have been recent attempts to overcome this limitation using tandem mass spectrometry81 or a stable isotope modified MALDI matrix for detection of acetylcholine.21 Another elegant approach using titanium dioxide nanoparticles TiO2 based laser desorption ionization permitted detection of aminobutyric acid (GABA) in situ.82 The potential of MALDI IMS for comprehensive metabolite profiling was demonstrated in a study on limbic, kainite induced seizure.83 Here, ion intensity patterns of ATP, ADP, and AMP were analyzed for visualization of spatiotemporal energy dynamics of hippocampal neurons. The results show CA3 selective ATP and ADP depletion and diminished energy recovery following seizures, suggesting a CA3-nerve cell specific energy metabolism (Figure 3a).
Figure 3.
Imaging mass spectrometry in clinical and molecular neuroscience for low molecular weight compounds and lipids. (a) Investigation of energy metabolism in hippocampus formation reveals selective ATP/ADP turnover in CA3 following induced seizure (ATP/ADP, arrows).83 (b) IMS based evaluation of ischemia shows regional selective increase of LPC (m/z 496) following injury 20 and 24 h.85 (c) Application of MALDI imaging for mapping ganglioside species reveals aging dependent regional increase of C20 gangliosides in the molecular layer (ML) of the dentate gyrus (DG) (14 days and 18 weeks, C20 arrows).91 (d) Using a similar approach, IMS revealed characteristic distributions of gangliosides in the hippocampus based on their ceramide core. In addition, in situ characterization of unknown glycolipid modifications as demonstrated by the discovery of O-acetylation (right overlay image). (Reproduced with permission from ref (92). Copyright 2011 American Chemical Society.) (e) IMS based ganglioside imaging reveals selective C20-ganglioside increase in, e.g., the hippocampal formation following blast induced mild TBI (2 and 24 h past blast, at 4 m but not 7 m distance). (Reproduced with permission from ref (93). Copyright 2013 American Chemical Society.)
Lipids and Gangliosides
As discussed above, the detection of lipid species by means of IMS is aided by their relatively high abundance. There has been a debate about the relevance of elucidating lipid distributions in situ beyond elucidating structural and topological features. However, the relevance of lipid species in neurophysiological mechanisms such as biogenic lipid signaling84 or membrane curvature modulation in cellular communication has been well established. IMS has, for example, been used to monitor localized production of the biogenic lipid, lyso-phophatidylcholine (LPC, 16:0) during injury following cerebral ischemia (Figure 3b).85 Other studies employed IMS to investigate abnormal cerebral phospholipid regulation in schizophrenia,86 cerebral edema,87 as well as infantile neuroaxonal dystrophy.88 Another study based on SIMS imaging investigated Aβ-plaques in a transgenic mouse model of AD.89 Using this high resolution imaging technology alongside immunostaining approaches and electron microscopy showed that cholesterol was selectively localized to the inclusions. This is consistent with previous findings and hypotheses involving transmembrane cholesterol–amyloid precursor protein interaction and suggests this has a key role in amyloid beta formation and AD pathology.90
MALDI IMS has also been demonstrated as a powerful technique for detection of different glycolipids and particularly gangliosides, which are known to be involved in different CNS and PNS pathologies including AD, GM1- and GM2-gangliosidosis, Sandhoff AB, and Guillan Barré syndrome. Here, the technique has a particular advantage over immune-based approaches, since its molecular specificity allows comprehensive characterization and differentiation of the ceramide core as well as the oligosaccharide side chain. In an earlier study on ganglioside distributions in hippocampal formation, IMS was used to reveal distinct localizations of gangliosides based on the ceramide long-chain base (LCB) and sphingoid base. Here, C18-LCB gangliosides (C18-GM1 and C18-GD1) were found to distribute rather uniformly throughout the prefrontal brain, while C20-gangliosides were found to localize along the entorhinal-hippocampal projection. This includes the entorhinal cortex (EC) and the molecular layer (ML) of the dentate gyrus (DG), suggesting that EC neurons exclusively express C20-ceramide carrying gangliosides.91 Gangliosides are thought to play an essential role in memory formation, neurogenesis, and synaptic transmission. Altered ganglioside expression might indicate altered neuronal stem cell proliferation, neuronal function, and potential neurodegeneration. Indeed, in the same study, the authors showed age-related accumulation of C20-GD1 in the molecular layer of the DG as well as the sum lacunosum stratum (SLM) and the stratum radium (SR) (Figure 3c). This suggests that the sphingoid base specific accumulation of gangliosides (in this case C20) in EC and its output structures (ML/SLM) might be related to age dependent neurological disease, including Alzheimer’s disease, where selective degeneration of EC neurons is observed in early stages.
IMS is a not only well suited to elucidate sphingoid base specific ganglioside distributions but is also a powerful technique to identify saccharide side chain modifications such as O-acetylation in situ. This was demonstrated in a recent study, where IMS of the hippocampus revealed differential abundance of gangliosides in axonal and dendritic parts of pyramidal neurons in the ML of the DG (Figure 3d).92 Moreover, O-acetylation was observed only in the granular layer (GL) of the DG. De novo identification and in situ localization of lipid modifications such acetylation highlight the strength of IMS over classic antibody-based imaging techniques.
The relevance of IMS based elucidation of spatiotemporal ganglioside regulations in the brain was further demonstrated in a recent study of the same group. Here, MALDI imaging MS revealed upregulation of ganglioside GM2 in hippocampus, thalamus, and hypothalamus following blast-induced mild traumatic brain injury (Figure 3e).93 This is of particular interest since no neurological or structural signs of brain injury can be observed using standard techniques, significantly hampering diagnosis and prognosis of mild traumatic brain injury (TBI).
Neuropeptides and Proteins
In situ detection and quantification of neuropeptides is of central interest in neuroscience research though hampered by many methodological shortcomings. Typically, antibody-based techniques are currently the dominant method used for in situ neuropeptide detection. However, this approach is significantly limited by antibody specificity. This is particularly relevant for opioid peptides that differ in only a few C-terminal amino acids, which compromises the reliability of IHC results significantly. Another limitation in immuno-based approaches is the inherent limited throughput, allowing only detection of one or two species at a time. Furthermore, there are well-known quantification issues, permitting only semiquantitative information to be retrieved from signal intensity in immunohistochemistry based experiments. Mass spectrometry based techniques, including LC-MS based peptide quantification in tissue extracts, have proven to be a powerful approach to analysis of peptides and proteins; however, in situ analysis is advantageous in order to preserve spatial information. Imaging MS based neuropeptide detection has significant advantages over immuno-based tequniques with respect to molecular specificity. However, the technique is mainly impacted by sample preparation and resulting sensitivity and reproducibility issues and requires stringent protocols to ensure acquisition of quality data.
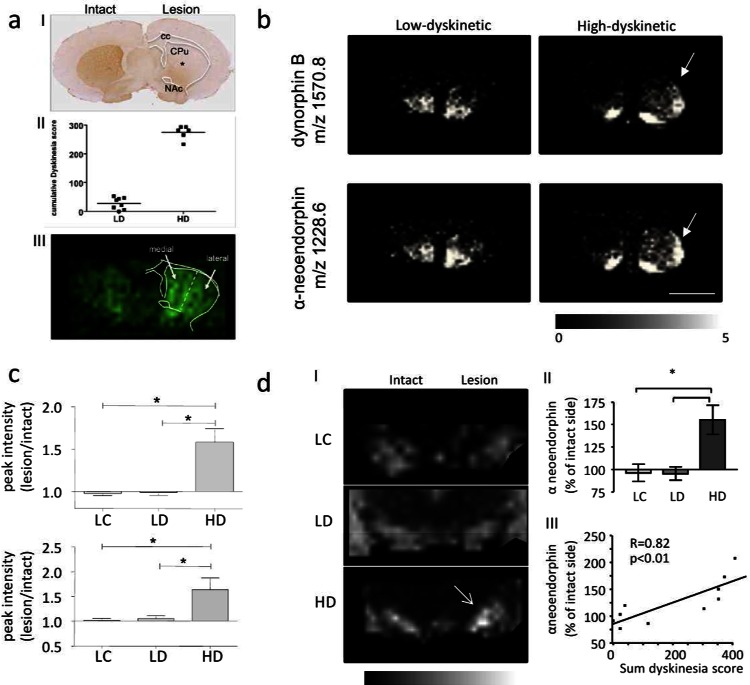
The inherent power of MALDI MS for multiplexed, untargeted in situ peptide identification and quantification has been exploited in two recent studies reported by the Andersson lab.94,95 Here, IMS was employed to characterize spatial regulations of prodynorphin peptides in L-DOPA induced dyskinesia in experimental Parkinsons’s disease. L-DOPA is the major pharmacotherapy for symptomatic Parkinsons’s disease treatment but is accompanied by severe side effects, including abnormal involuntary movements, that is, dyskinesia. Elevated prodynorphin levels have been previously associated with L-DOPA induced dyskinesia; however, the distinct prodynorphin processing products remained uncharacterized due to the lack of specific antibodies. IMS analysis showed that dynorphin B and alpha neoendorphin were significantly elevated in the dorsal lateral striatum of high but not low dyskinetic animals (Figure 4b,c). Moreover, both opioid peptides correlated positively with LID severity. Similarly, dynorphin B and alpha neoendorphin were found to be elevated in substantia nigra reticulata the major output structure of the direct GABA-ergic output pathway in rat (Figure 4d).95 Finally, a selective N-terminal processing of these dynorphin peptides in the striatum involving N-terminal cleavage of tyrosine was observed. Interestingly, these des-Tyr dynorphins correlate with LID severity suggesting an LID related mechanism of dynorphin inactivation. Moreover, des-Tyr dynorphins were earlier reported to show NMDA receptor affinity pointing toward alternative opioid signaling mechanisms in LID.94
Figure 4.
Imaging mass spectrometry of neuropeptides in L-DOPA induced dyskinesia. (a) (I) Unilateral 6-OHDA injection leads to dopamine depletion (as illustrated by TH immunostaining*); (II) L-DOPA therapy results in two distinct groups with low and high-dyskinesia; (III) MALDI IMS and assignment of ROI and spectra extraction. (b,c) Similarly, dynorphin peptides were found significantly increased in the dorso-lateral striatum of high- (HD) compared to low -dyskinetic (LD) animals and lesion controls (LC). (For panels (a)–(c), this research was originally published in Molecular Cellular Proteomics. Hanrieder et al. Mol. Cell Proteomics2011; M111.009308. Copyright the American Society for Biochemistry and Molecular Biology.32) (d) (I,II) MALDI IMS reveals significant increase of dynorphin peptide (here alpha neoendorphin) in the substantia nigra (SN). (III) Dynorphin peptide intensity correlated positively with dyskinesia severity.95
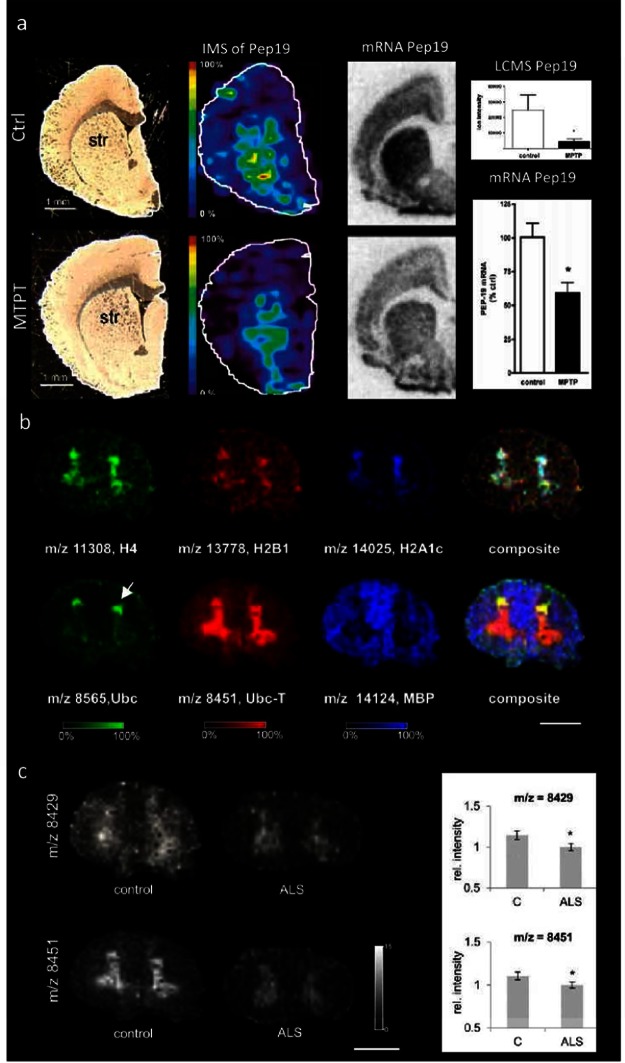
In the area of PD research, MALDI Imaging has been successfully employed to image proteins. Here, IMS was used to elucidate striatal protein distributions in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) injected rats96 (Figure 5a). The results showed a striatal decrease of Pep19, a neuronal calmodulin binding protein, which in turn suggested altered calcium homeostasis that might underlie cytotoxic events in neurodegeneration. Application of IMS to human material is particularly challenging, since it is difficult to account for technical issues that impact sample quality and increase variation. These parameters include, for example, standardized sample collection and storage as well as postmortem delay. In a recent study, postmortem spinal cord sections (thoracic) of patients that suffered from amyotrophic lateral sclerosis (ALS) were analyzed by means of MALDI IMS.18 Here, characteristic protein patterns consistent with major histological features were observed (Figure 5b). Moreover, significantly decreased intensities were observed for two previously unknown protein peaks in ALS compared to controls (Figure 5c). In order to characterize these unknown peak identities, an extensive bottom up proteomic approach was employed that was based on chromatographic separation and fractionation of intact protein tissue extracts followed by systematic MALDI MS inspection of collected protein fractions (LC-MALDI) and subsequent trypsination and LC-MS/MS based identification. With this technique, one of the peaks was identified as C-terminally truncated ubiquitin (ubc 1–76 and ubc-T 1–74), where two glycine residues were removed. This points toward an altered protein turnover in ALS. These findings highlight the potential of IMS for studies of peptides and proteins in neurodegeneration, since in situ differentiation of C-terminal truncations cannot be achieved with antibody based techniques due to limited specificity. Moreover, this study also points out one of the major obstacles in IMS of intact proteins, since mass accuracy and resolution do not permit unequal identification of intact protein peaks and require extensive identification and validation strategies. A further study employed MALDI IMS for monitoring hippocampal protein changes in a neurotoxicology experiment on cognitive and systematic effects of neonatal exposure beta-methylamino-l-alanine (BMAA) in rats.98 The results showed long-term changes including a decreased expression of proteins involved in energy metabolism and intracellular signaling in the adult hippocampus at a dose 150 mg/kg. Developmental exposure to a higher dose (460 mg/kg) also induced changes in the expression of S100β, histones, calcium- and calmodulin-binding proteins, and guanine nucleotide-binding proteins. Interestingly, at this dose, severe lesions in the adult hippocampus including neuronal degeneration, cell loss, calcium deposits, and astrogliosis were observed that were not observed for the lower dose. Finally, the technique has been demonstrated to be a valuable tool for biochemical characterization of anatomical structures in the brain. Here, the potential of MALDI IMS based protein mapping has been exploited for establishing a novel structural definition of the claustrum and the insula.97 G-protein gamma 2 subunit (Gng2) was detected as novel claustrum specific protein marker, which in turn revealed that the claustrum projects to cortical but not subcortical sites.
Figure 5.
Imaging mass spectrometry of proteins in neurodegenerative diseases. (a) In experimental Parkinson’s disease, MALDI IMS reveals decrease of striatal Pep19 levels in MPTP lesioned rats, as verified by in situ hybridization and LCMS in tissue extracts. (Reproduced with permission from ref (96). Copyright 2006 American Chemical Society.) (b,c) Analysis of proteins in post mortem human spinal cord using MALDI IMS. (b) Spinal cord proteins show characteristic distribution patterns that are consistent with anatomical regions, including gray (top panel) and white matter (lower panel). Here histones (H4, H2B1, H2A1c) as well as ubiquitin (Ubc) and truncated ubiquitin 1–74 (Ubc-T) were found to localize to the gray matter. In contrast, myelin basic protein shows localization to the white matter (scale bar = 1 cm). Interestingly, ubiquitin and its C-terminally truncated metabolite Ubc-T show different localizations, where Ubc is distributed to the dorsal horn (arrow) compared to Ubc-T beeing more homogeneously distributed throughout the gray matter. (c) Multivariate statistics reveal significant decreases for two protein peaks, including Ubc-T (m/z 8451) in the gray matter of ALS patients compared to controls18 (scale bar = 1 cm).
Challenges and Perspectives
A central aspect in using IMS for biological studies is its relatively low throughput. With an appropriate study design, including a sufficient number of biological and technical replicates, utilization of this technology generates a tremendous workload. Of course, IMS offers the possibility to monitor hundreds of molecular species in parallel; however, this leads to a high quantity of data. Given the large number of potential sources of variation, acquisition of reproducible, high quality data poses a major challenge. This in turn puts high demands on standardized sample collection, preservation procedures, and sample storage in order to minimize variation. In MALDI, the laser absorbing matrix is key to its success, but it is also a limiting factor. Hence, development of alternative more reproducible matrix application methods and even matrix free approaches is a central objective in IMS method development. While there have been exciting approaches for matrix free laser desorption ionization imaging, their suitability for large scale IMS experiments have so far not been demonstrated and require further investigation. Matrix free approaches based on, for example, functionalized surfaces or nanoparticles should have a major advantage over classical matrix approaches in that they facilitate detection of low molecular weight compounds including, for example, neurotransmitters.81
In SIMS, instrumental developments are also needed and a central focus has been on improving the projectiles for imaging. The development of cluster ion sources has had a significant impact, since it facilitates analysis of intact biomolecular species. Currently, a lot of effort has been placed on development and application of argon cluster ion probes that facilitate even softer molecular desorption. This should improve analysis of larger molecular species as well as depth profiling to an even higher degree.46
Analysis is improved with greater sensitivity for imaged species, and thus, a main concern in desorption ionization techniques, particularly in SIMS, is ionization efficiency. Since the ionization efficiency in SIMS is only about 10–4 (0.01%), the need for optimization in this area is evident. Sensitivity is largely impacted by the ionization efficiency, especially with respect to increasing spatial resolution and the static limit in TOF-SIMS.
In addition to spatial resolution, mass resolution and identification are key parameters. Targeted fragmentation (MS/MS) has been routinely used for structural identification in MALDI, mostly with a technique called post source decay. However, post source decay is significantly inferior to collision induced dissociation based MS/MS methods, and MS/MS has been used sparingly in SIMS.
Technical improvements of appropriate software tools are badly needed for both MALDI and SIMS for handling the vast amount of data generated in imaging MS. Today, data handling in IMS is still mostly at an experimental rather than commercial level. This area needs to be unified and made collectively useful, and to date the commercial aspects seem to be limiting real progress in this direction.
Imaging mass spectrometry is a powerful alternative and complementary approach to existing analytical tools commonly used in molecular biology and clinical research. Using IMS for studying the spatio-dynamic distribution pattern of molecular species that orchestrate neuronal function might give further insight into ongoing fundamental mechanisms of the nervous system including development, learning, and memory as well as cognition and behavior. Although the potential of the technique has been demonstrated successfully, a number of challenges remain to be addressed in order for the technology to be more widely used by facilitating its acceptance and awareness in molecular biology and biomedical neuroscience research in particular. Importantly, there is still a lack of understanding between the different points of view in analytical chemistry as well as ion physics on the one side and biomedical and neuroscience research on the other. One reason for this is the different approaches to the design of experiments. While mass spectrometry typically has a focus on method development and proof of principle, neuroscience research is hypothesis driven and relies on rather conservative techniques to answer the scientific questions. In conclusion, imaging mass spectrometry is a powerful technique with great potential for the investigation of spatial and temporal aspects of chemical information in neurobiological systems. Although still in development, this technique holds the promise for many new applications in neuroscience research.
The Swedish Research Council (J.H. and A.G.E.), the Royal Swedish Academy of Sciences KVA (J.H.), the Wenner-Gren Foundations (J.H.), and the Swedish Chemical Society (J.H.) are gratefully acknowledged for financial support. A.G.E. further acknowledges the NIH, the European Research Council (Advanced grant) and Wallenberg Foundation (Wallenberg Fellow).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Aebersold R.; Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207. [DOI] [PubMed] [Google Scholar]
- Karas M.; Hillenkamp F. (1988) Laser Desorption Ionization of Proteins with Molecular Masses Exceeding 10000 Da. Anal. Chem. 60, 2299–2301. [DOI] [PubMed] [Google Scholar]
- Fenn J. B.; Mann M.; Meng C. K.; Wong S. F.; Whitehouse C. M. (1989) Electrospray Ionization for Mass-Spectrometry of Large Biomolecules. Science 246, 64–71. [DOI] [PubMed] [Google Scholar]
- Cornett D. S.; Reyzer M. L.; Chaurand P.; Caprioli R. M. (2007) MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat. Methods 4, 828–833. [DOI] [PubMed] [Google Scholar]
- McDonnell L. A.; Heeren R. M. A. (2007) Imaging mass spectrometry. Mass Spectrom. Rev. 26, 606–643. [DOI] [PubMed] [Google Scholar]
- Schwamborn K.; Caprioli R. M. (2010) MALDI Imaging Mass Spectrometry - Painting Molecular Pictures. Mol. Oncol. 4, 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornett D. S.; Mobley J. A.; Dias E. C.; Andersson M.; Arteaga C. L.; Sanders M. E.; Caprioli R. M. (2006) A novel histology-directed strategy for MALDI-MS tissue profiling that improves throughput and cellular specificity in human breast cancer. Mol. Cell. Proteomics 5, 1975–1983. [DOI] [PubMed] [Google Scholar]
- Schwamborn K.; Caprioli R. M. (2010) INNOVATION Molecular imaging by mass spectrometry - looking beyond classical histology. Nat. Rev. Cancer 10, 639–646. [DOI] [PubMed] [Google Scholar]
- Vickerman J. C. (2011) Molecular imaging and depth profiling by mass spectrometry--SIMS, MALDI or DESI?. Analyst 136, 2199–2217. [DOI] [PubMed] [Google Scholar]
- Girod M.; Shi Y.; Cheng J. X.; Cooks R. G. (2010) Desorption electrospray ionization imaging mass spectrometry of lipids in rat spinal cord. J. Am. Soc. Mass Spectrom. 21, 1177–1189. [DOI] [PubMed] [Google Scholar]
- Benabdellah F.; Seyer A.; Quinton L.; Touboul D.; Brunelle A.; Laprevote O. (2010) Mass spectrometry imaging of rat brain sections: nanomolar sensitivity with MALDI versus nanometer resolution by TOF-SIMS. Anal. Bioanal. Chem. 396, 151–162. [DOI] [PubMed] [Google Scholar]
- Tyler B. J.; Rayal G.; Castner D. G. (2007) Multivariate analysis strategies for processing ToF-SIMS images of biomaterials. Biomaterials 28, 2412–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham D. J.; Castner D. G. (2012) Multivariate analysis of ToF-SIMS data from multicomponent systems: the why, when, and how. Biointerphases 7, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson A.; Fletcher J. S.; Vickerman J. C. (2009) A comparison of PCA and MAF for ToF-SIMS image interpretation. Surf. Interface Anal. 41, 666–674. [Google Scholar]
- Deininger S.-O.; Ebert M. P.; Fuetterer A.; Gerhard M.; Roecken C. (2008) MALDI Imaging Combined with Hierarchical Clustering as a New Tool for the Interpretation of Complex Human Cancers. J. Proteome Res. 7, 5230–5236. [DOI] [PubMed] [Google Scholar]
- Groseclose M. R.; Andersson M.; Hardesty W. M.; Caprioli R. M. (2007) Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. J. Mass Spectrom. 42, 254–262. [DOI] [PubMed] [Google Scholar]
- Bashkirov P. V.; Chekashkina K. V.; Akimov S. A.; Kuzmin P. I.; Frolov V. A. (2011) Variation of Lipid Membrane Composition Caused by Strong Bending. Biol. Membr. 28, 145–152. [Google Scholar]
- Hanrieder J.; Ekegren T.; Andersson M.; Bergquist J. (2013) MALDI Imaging mass spectrometry of human post mortem spinal cord in amyotrophic lateral sclerosis. J. Neurochem. 124, 695–707. [DOI] [PubMed] [Google Scholar]
- Caprioli R. M.; Farmer T. B.; Gile J. (1997) Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 69, 4751–4760. [DOI] [PubMed] [Google Scholar]
- Northen T. R.; Yanes O.; Northen M. T.; Marrinucci D.; Uritboonthai W.; Apon J.; Golledge S. L.; Nordstrom A.; Siuzdak G. (2007) Clathrate nanostructures for mass spectrometry. Nature 449, 1033–1036. [DOI] [PubMed] [Google Scholar]
- Shariatgorji M.; Nilsson A.; Goodwin R. J. A.; Svenningsson P.; Schintu N.; Banka Z.; Kladni L.; Hasko T.; Szabo A.; Andren P. E. (2012) Deuterated Matrix-Assisted Laser Desorption Ionization Matrix Uncovers Masked Mass Spectrometry Imaging Signals of Small Molecules. Anal. Chem. 84, 7152–7157. [DOI] [PubMed] [Google Scholar]
- Nilsson A.; Fehniger T. E.; Gustavsson L.; Andersson M.; Kenne K.; Marko-Varga G.; Andren P. E. (2010) Fine Mapping the Spatial Distribution and Concentration of Unlabeled Drugs within Tissue Micro-Compartments Using Imaging Mass Spectrometry. PLoS One 5, e1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurchen J. C.; Rubakhin S. S.; Sweedler J. V. (2005) MALDI-MS imaging of features smaller than the size of the laser beam. J. Am. Soc. Mass Spectrom. 16, 1654–1659. [DOI] [PubMed] [Google Scholar]
- Spengler B.; Hubert M. (2002) Scanning microprobe matrix-assisted laser desorption ionization (SMALDI) mass spectrometry: Instrumentation for sub-micrometer resolved LDI and MALDI surface analysis. J. Am. Soc. Mass Spectrom. 13, 735–748. [DOI] [PubMed] [Google Scholar]
- Taban I. M.; Altelaar A. F. M.; Van der Burgt Y. E. M.; McDonnell L. A.; Heeren R. M. A.; Fuchser J.; Baykut G. (2007) Imaging of peptides in the rat brain using MALDI-FTICR mass spectrometry. J. Am. Soc. Mass Spectrom. 18, 145–151. [DOI] [PubMed] [Google Scholar]
- Steinhauser M. L.; Bailey A. P.; Senyo S. E.; Guillermier C.; Perlstein T. S.; Gould A. P.; Lee R. T.; Lechene C. P. (2012) Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature 481, 516–U131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjovall P.; Lausmaa J.; Johansson B. (2004) Mass spectrometric imaging of lipids in brain tissue. Anal. Chem. 76, 4271–4278. [DOI] [PubMed] [Google Scholar]
- Goodwin R. J. A.; Nilsson A.; Borg D.; Langridge-Smith P. R. R.; Harrison D. J.; Mackay C. L.; Iverson S. L.; Andren P. E. (2012) Conductive carbon tape used for support and mounting of both whole animal and fragile heat-treated tissue sections for MALDI MS imaging and quantitation. J. Proteomics 75, 4912–4920. [DOI] [PubMed] [Google Scholar]
- Hanrieder J.; Ljungdahl A.; Andersson M. (2012) MALDI imaging mass spectrometry of neuropeptides in Parkinson’s disease. J. Visualized Exp. 10, e3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariatgorji M.; Kallback P.; Gustavsson L.; Schintu N.; Svenningsson P.; Goodwin R. J. A.; Andren P. E. (2012) Controlled-pH Tissue Cleanup Protocol for Signal Enhancement of Small Molecule Drugs Analyzed by MALDI-MS Imaging. Anal. Chem. 84, 4603–4607. [DOI] [PubMed] [Google Scholar]
- Aerni H. R.; Cornett D. S.; Caprioli R. M. (2006) Automated acoustic matrix deposition for MALDI sample preparation. Anal. Chem. 78, 827–834. [DOI] [PubMed] [Google Scholar]
- Hanrieder J.; Ljungdahl A.; Faelth M.; Mammo S. E.; Bergquist J.; Andersson M. (2011) L-DOPA-induced Dyskinesia is Associated with Regional Increase of Striatal Dynorphin Peptides as Elucidated by Imaging Mass Spectrometry. Mol. Cell. Proteomics 10, M111.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire R.; Tabet J. C.; Ducoroy P.; Hendra J. B.; Salzet M.; Fournier I. (2006) Solid ionic matrixes for direct tissue analysis and MALDI Imaging. Anal. Chem. 78, 809–819. [DOI] [PubMed] [Google Scholar]
- Yang J.; Caprioli R. M. (2011) Matrix Sublimation/Recrystallization for Imaging Proteins by Mass Spectrometry at High Spatial Resolution. Anal. Chem. 83, 5728–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puolitaival S. M.; Burnum K. E.; Cornett D. S.; Caprioli R. M. (2008) Solvent-free matrix dry-coating for MALDI Imaging of phospholipids. J. Am. Soc. Mass Spectrom. 19, 882–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J.; Buriak J. M.; Siuzdak G. (1999) Desorption-ionization mass spectrometry on porous silicon. Nature 399, 243–246. [DOI] [PubMed] [Google Scholar]
- Kim Y. K.; Na H. K.; Kwack S. J.; Ryoo S. R.; Lee Y.; Hong S.; Jeong Y.; Min D. H. (2011) Synergistic Effect of Graphene Oxide/MWCNT Films in Laser Desorption/Ionization Mass Spectrometry of Small Molecules and Tissue Imaging. ACS Nano 5, 4550–4561. [DOI] [PubMed] [Google Scholar]
- Pacholski M. L.; Winograd N. (1999) Imaging with mass spectrometry. Chem. Rev. 99, 2977–3006. [DOI] [PubMed] [Google Scholar]
- Guerquin-Kern J. L.; Wu T. D.; Quintana C.; Croisy A. (2005) Progress in analytical imaging of the cell by dynamic secondary ion mass spectrometry (SIMS microscopy). Biochim. Biophys. Acta 1724, 228–238. [DOI] [PubMed] [Google Scholar]
- Lechene C.; Hillion F.; McMahon G.; Benson D.; Kleinfeld A. M.; Kampf J. P.; Distel D.; Luyten Y.; Bonventre J.; Hentschel D.; Park K.; Ito S.; Schwartz M.; Benichou G.; Slodzian G. (2006) High-resolution quantitative imaging of mammalian and bacterial cells using stable isotope mass spectrometry. J. Biol. 5, 20–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touboul D.; Kollmer F.; Niehuis E.; Brunelle A.; Laprevote O. (2005) Improvement of biological time-of-flight-secondary ion mass spectrometry imaging with a bismuth cluster ion source. J. Am. Soc. Mass Spectrom. 16, 1608–1618. [DOI] [PubMed] [Google Scholar]
- Fletcher J. S.; Vickerman J. C. (2010) A new SIMS paradigm for 2D and 3D molecular imaging of bio-systems. Anal. Bioanal. Chem. 396, 85–104. [DOI] [PubMed] [Google Scholar]
- Garrison B. J.; Postawa Z.; Ryan K. E.; Vickerman J. C.; Webb R. P.; Winograd N. (2009) Internal energy of molecules ejected due to energetic C60 bombardment. Anal. Chem. 81, 2260–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J. S.; Conlan X. A.; Jones E. A.; Biddulph G.; Lockyer N. P.; Vickerman J. C. (2006) TOF-SIMS analysis using C60. Effect of impact energy on yield and damage. Anal. Chem. 78, 1827–1831. [DOI] [PubMed] [Google Scholar]
- Fletcher J. S. (2009) Cellular imaging with secondary ion mass spectrometry. Analyst 134, 2204–2215. [DOI] [PubMed] [Google Scholar]
- Rabbani S.; Barber A. M.; Fletcher J. S.; Lockyer N. P.; Vickerman J. C. (2011) TOF-SIMS with argon gas cluster ion beams: a comparison with C60+. Anal. Chem. 83, 3793–3800. [DOI] [PubMed] [Google Scholar]
- Fletcher J. S.; Rabbani S.; Henderson A.; Blenkinsopp P.; Thompson S. P.; Lockyer N. P.; Vickerman J. C. (2008) A new dynamic in mass spectral imaging of single biological cells. Anal. Chem. 80, 9058–9064. [DOI] [PubMed] [Google Scholar]
- Fletcher J. S.; Vickerman J. C.; Winograd N. (2011) Label free biochemical 2D and 3D imaging using secondary ion mass spectrometry. Curr. Opin. Chem. Biol. 15, 733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J. S.; Vickerman J. C. (2013) Secondary ion mass spectrometry: characterizing complex samples in two and three dimensions. Anal. Chem. 85, 610–639. [DOI] [PubMed] [Google Scholar]
- Cannon D. M.; Pacholski M. L.; Winograd N.; Ewing A. G. (2000) Molecule specific imaging of freeze-fractured, frozen-hydrated model membrane systems using mass spectrometry. J. Am. Chem. Soc. 122, 603–610. [Google Scholar]
- Kurczy M. E.; Piehowski P. D.; Parry S. A.; Jiang M.; Chen G.; Ewing A. G.; Winograd N. (2008) Which is more important in bioimaging SIMS experiments-The sample preparation or the nature of the projectile?. Appl. Surf. Sci. 255, 1298–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S.; Morrison G. H. (1992) Sample Preparation of Animal-Tissues and Cell-Cultures for Secondary Ion Mass-Spectrometry (SIMS) Microscopy. Biol. Cell 74, 31–42. [DOI] [PubMed] [Google Scholar]
- Lanekoff I.; Kurczy M. E.; Hill R.; Fletcher J. S.; Vickerman J. C.; Winograd N.; Sjovall P.; Ewing A. G. (2010) Time of Flight Mass Spectrometry Imaging of Samples Fractured In Situ with a Spring-Loaded Trap System. Anal. Chem. 82, 6652–6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J. S.; Lockyer N. P.; Vickerman J. C. (2011) Developments in molecular SIMS depth profiling and 3D imaging of biological systems using polyatomic primary ions. Mass Spectrom. Rev. 30, 142–174. [DOI] [PubMed] [Google Scholar]
- Piehowski P. D.; Kurczy M. E.; Willingham D.; Parry S.; Heien M. L.; Winograd N.; Ewing A. G. (2008) Freeze-etching and vapor matrix deposition for ToF-SIMS imaging of single cells. Langmuir 24, 7906–7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurczy M. E.; Piehowsky P. D.; Willingham D.; Molyneaux K. A.; Heien M. L.; Winograd N.; Ewing A. G. (2010) Nanotome Cluster Bombardment to Recover Spatial Chemistry After Preparation of Biological Samples for SIMS Imaging. J. Am. Soc. Mass Spectrom. 21, 833–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubakhin S. S.; Hatcher N. G.; Monroe E. B.; Heien M. L.; Sweedler J. V. (2007) Mass spectrometric imaging of the nervous system. Curr. Pharm. Des. 13, 3325–3334. [DOI] [PubMed] [Google Scholar]
- Heeren R. M. A.; McDonnell L. A.; Amstalden E.; Luxembourg S. L.; Altelaar A. F. M.; Piersma S. R. (2006) Why don’t biologists use SIMS? A critical evaluation of imaging MS. Appl. Surf. Sci. 252, 6827–6835. [Google Scholar]
- Altelaar A. F. M.; van Minnen J.; Jimenez C. R.; Heeren R. M. A.; Piersma S. R. (2005) Direct molecular Imaging of Lymnaea stagnalis nervous tissue at subcellular spatial resolution by mass spectrometry. Anal. Chem. 77, 735–741. [DOI] [PubMed] [Google Scholar]
- Crecelius A. C.; Cornett D. S.; Caprioli R. M.; Williams B.; Dawant B. M.; Bodenheimer B. (2005) Three-dimensional visualization of protein expression in mouse brain structures using imaging mass spectrometry. J. Am. Soc. Mass Spectrom. 16, 1093–1099. [DOI] [PubMed] [Google Scholar]
- McDonnell L. A.; Piersma S. R.; Altelaar A. F. M.; Mize T. H.; Luxembourg S. L.; Verhaert P.; van Minnen J.; Heeren R. M. A. (2005) Subcellular imaging mass spectrometry of brain tissue. J. Mass. Spectrom. 40, 160–168. [DOI] [PubMed] [Google Scholar]
- Todd P. J.; McMahon J. M.; McCandlish C. A. (2004) Secondary ion images of the developing rat brain. J. Am. Soc. Mass Spectrom. 15, 1116–1122. [DOI] [PubMed] [Google Scholar]
- Jackson S. N.; Ugarov M.; Egan T.; Post J. D.; Langlais D.; Schultz J. A.; Woods A. S. (2007) MALDI-ion mobility-TOFMS imaging of lipids in rat brain tissue. J. Mass. Spectrom. 42, 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis J. T.; Nie H. Y.; Taylor A. R.; Walzak M. J.; Chang W. H.; MacFabe D. F.; Lau W. M. (2008) ToF-SIMS cluster ion imaging of hippocampal CA1 pyramidal rat neurons. Appl. Surf. Sci. 255, 1126–1130. [Google Scholar]
- Monroe E. B.; Jurchen J. C.; Lee J.; Rubakhin S. S.; Sweedler J. V. (2005) Vitamin E imaging and localization in the neuronal membrane. J. Am. Chem. Soc. 127, 12152–12153. [DOI] [PubMed] [Google Scholar]
- Kraft M. L.; Weber P. K.; Longo M. L.; Hutcheon I. D.; Boxer S. G. (2006) Phase separation of lipid membranes analyzed with high-resolution secondary ion mass spectrometry. Science 313, 1948–1951. [DOI] [PubMed] [Google Scholar]
- Ostrowski S. G.; Van Bell C. T.; Winograd N.; Ewing A. G. (2004) Mass spectrometric imaging of highly curved membranes during Tetrahymena mating. Science 305, 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sostarecz A. G.; McQuaw C. M.; Ewing A. G.; Winograd N. (2004) Phosphatidylethanolamine-induced cholesterol domains chemically identified with mass spectrometric imaging. J. Am. Chem. Soc. 126, 13882–13883. [DOI] [PubMed] [Google Scholar]
- Kurczy M. E.; Piehowski P. D.; Van Bell C. T.; Heien M. L.; Winograd N.; Ewing A. G. (2010) Mass spectrometry imaging of mating Tetrahymena show that changes in cell morphology regulate lipid domain formation. Proc. Natl. Acad. Sci. U.S.A. 107, 2751–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarelli M. K.; Winograd N. (2011) Lipid imaging with time-of-flight secondary ion mass spectrometry (ToF-SIMS). Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 1811, 976–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauser M. L.; Bailey A. P.; Senyo S. E.; Guillermier C.; Perlstein T. S.; Gould A. P.; Lee R. T.; Lechene C. P. (2012) Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature 481, 516–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik L. (1996) Non-bilayer lipids and biological fusion intermediates. Chem. Phys. Lipids 81, 203–213. [DOI] [PubMed] [Google Scholar]
- Fletcher J. S.; Rabbani S.; Henderson A.; Blenkinsopp P.; Thompson S. P.; Lockyer N. P.; Vickerman J. C. (2008) A New Dynamic in Mass Spectral Imaging of Single Biological Cells. Anal. Chem. 80, 9058–9064. [DOI] [PubMed] [Google Scholar]
- Ostrowski S. G.; Kurczy M. E.; Roddy T. P.; Winograd N.; Ewing A. G. (2007) Secondary ion MS imaging to relatively quantify cholesterol in the membranes of individual cells from differentially treated populations. Anal. Chem. 79, 3554–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M. S. (2008) Towards quantitative chemical imaging with ToF-SIMS. Appl. Surf. Sci. 255, 992–996. [Google Scholar]
- Lanekoff I.; Sjovall P.; Ewing A. G. (2011) Relative Quantification of Phospholipid Accumulation in the PC12 Cell Plasma Membrane Following Phospholipid Incubation Using TOF-SIMS Imaging. Anal. Chem. 83, 5337–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.; Adams K. L.; Luber S. J.; Eves D. J.; Heien M. L.; Ewing A. G. (2008) Spatially and temporally resolved single-cell exocytosis utilizing individually addressable carbon microelectrode arrays. Anal. Chem. 80, 1394–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J. S.; Lockyer N. P.; Vickerman J. C. (2011) Molecular SIMS imaging; spatial resolution and molecular sensitivity: Have we reached the end of the road? Is there light at the end of the tunnel?. Surf. Interface Anal. 43, 253–256. [Google Scholar]
- Rimbach G.; Minihane A. M.; Majewicz J.; Fischer A.; Pallauf J.; Virgli F.; Weinberg P. D. (2002) Regulation of cell signalling by vitamin E. Proc. Nutr. Soc. 61, 415–425. [DOI] [PubMed] [Google Scholar]
- Zhang D. S.; Piazza V.; Perrin B. J.; Rzadzinska A. K.; Poczatek J. C.; Wang M.; Prosser H. M.; Ervasti J. M.; Corey D. P.; Lechene C. P. (2012) Multi-isotope imaging mass spectrometry reveals slow protein turnover in hair-cell stereocilia. Nature 481, 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura Y.; Zaima N.; Setou M.; Ito S.; Yao I. (2012) Visualization of acetylcholine distribution in central nervous system tissue sections by tandem imaging mass spectrometry. Anal. Bioanal. Chem. 403, 1851–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivas K.; Hayasaka T.; Sugiura Y.; Setou M. (2011) Method for Simultaneous Imaging of Endogenous Low Molecular Weight Metabolites in Mouse Brain Using TiO2 Nanoparticles in Nanoparticle-Assisted Laser Desorption/Ionization-Imaging Mass Spectrometry. Anal. Chem. 83, 7283–7289. [DOI] [PubMed] [Google Scholar]
- Sugiura Y.; Taguchi R.; Setou M. (2011) Visualization of Spatiotemporal Energy Dynamics of Hippocampal Neurons by Mass Spectrometry during a Kainate-Induced Seizure. PLoS One 6, e17952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M.; Rashid M. H.; Fujita R.; Contos J. J. A.; Chun J.; Ueda H. (2004) Initiation of neuropathic pain requires lysophosphatidic acid receptor signaling. Nat. Med. 10, 712–718. [DOI] [PubMed] [Google Scholar]
- Koizumi S.; Yamamoto S.; Hayasaka T.; Konishi Y.; Yamaguchi-Okada M.; Goto-Inoue N.; Sugiura Y.; Setou M.; Namba H. (2010) Imaging Mass Spectrometry Revealed the Production of Lyso-Phosphatidylcholine in the Injured Ischemic Rat Brain. Neuroscience 168, 219–225. [DOI] [PubMed] [Google Scholar]
- Matsumoto J.; Sugiura Y.; Yuki D.; Hayasaka T.; Goto-Inoue N.; Zaima N.; Kunii Y.; Wada A.; Yang Q.; Nishiura K.; Akatsu H.; Hori A.; Hashizume Y.; Yamamoto T.; Ikemoto K.; Setou M.; Niwa S.-i. (2011) Abnormal phospholipids distribution in the prefrontal cortex from a patient with schizophrenia revealed by matrix-assisted laser desorption/ionization imaging mass spectrometry. Anal. Bioanal. Chem. 400, 1933–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi S.; Hayasaka T.; Goto-Inoue N.; Doi K.; Setou M.; Namba H. (2012) Imaging Mass Spectrometry Evaluation of the Effects of Various Irrigation Fluids in a Rat Model of Postoperative Cerebral Edema. World Neurosurg. 77, 153–159. [DOI] [PubMed] [Google Scholar]
- Beck G.; Sugiura Y.; Shinzawa K.; Kato S.; Setou M.; Tsujimoto Y.; Sakoda S.; Sumi-Akamaru H. (2011) Neuroaxonal Dystrophy in Calcium-Independent Phospholipase A(2)beta Deficiency Results from Insufficient Remodeling and Degeneration of Mitochondrial and Presynaptic Membranes. J. Neurosci. 31, 11411–11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sole-Domenech S.; Sjovall P.; Vukojevic V.; Fernando R.; Codita A.; Salve S.; Bogdanovic N.; Mohammed A. H.; Hammarstrom P.; Nilsson K. P.; Laferla F. M.; Jacob S.; Berggren P. O.; Gimenez-Llort L.; Schalling M.; Terenius L.; Johansson B. (2012) Localization of cholesterol, amyloid and glia in Alzheimer’s disease transgenic mouse brain tissue using time-of-flight secondary ion mass spectrometry (ToF-SIMS) and immunofluorescence imaging. Acta Neuropathol. 125, 145–157. [DOI] [PubMed] [Google Scholar]
- Barrett P. J.; Song Y.; Van Horn W. D.; Hustedt E. J.; Schafer J. M.; Hadziselimovic A.; Beel A. J.; Sanders C. R. (2012) The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science 336, 1168–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura Y.; Shimma S.; Konishi Y.; Yamada M. K.; Setou M. (2008) Imaging Mass Spectrometry Technology and Application on Ganglioside Study; Visualization of Age-Dependent Accumulation of C20-Ganglioside Molecular Species in the Mouse Hippocampus. PLoS One 3, e3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colsch B.; Jackson S. N.; Dutta S.; Woods A. S. (2011) Molecular Microscopy of Brain Gangliosides: Illustrating their Distribution in Hippocampal Cell Layers. ACS Chem. Neurosci. 2, 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A. S.; Colsch B.; Jackson S. N.; Post J.; Baldwin K.; Roux A.; Hoffer B.; Cox B. M.; Hoffer M.; Rubovitch V.; Pick C. G.; Schultz J. A.; Balaban C. (2013) Gangliosides and Ceramides Change in a Mouse Model of Blast Induced Traumatic Brain Injury. ACS Chem. Neurosci. 10.1021/cn300216h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanrieder J.; Ljungdahl A.; Falth M.; Mammo S. E.; Bergquist J.; Andersson M. (2011) L-DOPA-induced dyskinesia is associated with regional increase of striatal dynorphin peptides as elucidated by imaging mass spectrometry. Mol. Cell. Proteomics 10, M111 009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungdahl A.; Hanrieder J.; Faelth M.; Bergquist J.; Andersson M. (2011) Imaging Mass Spectrometry Reveals Elevated Nigral Levels of Dynorphin Neuropeptides in L-DOPA-Induced Dyskinesia in Rat Model of Parkinson’s Disease. PLoS One 6, e25653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skold K.; Svensson M.; Nilsson A.; Zhang X. Q.; Nydahl K.; Caprioli R. M.; Svenningsson P.; Andren P. E. (2006) Decreased striatal levels of PEP-19 following MPTP lesion in the mouse. J. Proteome Res. 5, 262–269. [DOI] [PubMed] [Google Scholar]
- Karlsson O.; Berg A.-L.; Lindstrom A.-K.; Handrieder J.; Arnerup G.; Roman E.; Bergquist J.; Lindquist N. G.; Brittebo E. B.; Andersson M. (2012) Neonatal Exposure to the Cyanobacterial Toxin BMAA Induces Changes in Protein Expression and Neurodegeneration in Adult Hippocampus. Toxicol. Sci. 130, 391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur B. N.; Caprioli R. M.; Deutch A. Y. (2009) Proteomic Analysis Illuminates a Novel Structural Definition of the Claustrum and Insula. Cereb. Cortex 19, 2372–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]