Abstract
The investigation of community structures in networks is an important issue in many domains and disciplines. This problem is relevant for social tasks (objective analysis of relationships on the web), biological inquiries (functional studies in metabolic and protein networks), or technological problems (optimization of large infrastructures). Several types of algorithms exist for revealing the community structure in networks, but a general and quantitative definition of community is not implemented in the algorithms, leading to an intrinsic difficulty in the interpretation of the results without any additional nontopological information. In this article we deal with this problem by showing how quantitative definitions of community are implemented in practice in the existing algorithms. In this way the algorithms for the identification of the community structure become fully self-contained. Furthermore, we propose a local algorithm to detect communities which outperforms the existing algorithms with respect to computational cost, keeping the same level of reliability. The algorithm is tested on artificial and real-world graphs. In particular, we show how the algorithm applies to a network of scientific collaborations, which, for its size, cannot be attacked with the usual methods. This type of local algorithm could open the way to applications to large-scale technological and biological systems.
Evidence has rapidly grown in the past few years that very diverse systems in many different fields can be described as complex networks, i.e., assemblies of nodes and edges with nontrivial topological properties (1, 2). Examples range from technological systems [the Internet and the web (3, 4)] to biological [epidemiology (5, 6), metabolic networks (7–9), and ecological webs (10–13)] and social systems [scientific collaborations and structure of large organizations (14, 15)].
In this article we deal with a topological property of networks, the community structure, that has attracted a great deal of interest recently. The concept of community is common, and it is linked to the classification of objects in categories for the sake of memorization or retrieval of information. From this point of view the notion of community is general and, depending on the context, can be synonymous with module, class, group, cluster, etc. Among the many contexts where this notion is relevant it is worth mentioning the problem of modularity in metabolic or cellular networks (9, 16) or the problem of the identification of communities in the web (17). This last issue is relevant for the implementation of search engines of a new generation, content filtering, automatic classification, or the automatic realization of ontologies.
Given the relevance of the problem, it is crucial to construct efficient procedures and algorithms for the identification of the community structure in a generic network. This task, however, is highly nontrivial.
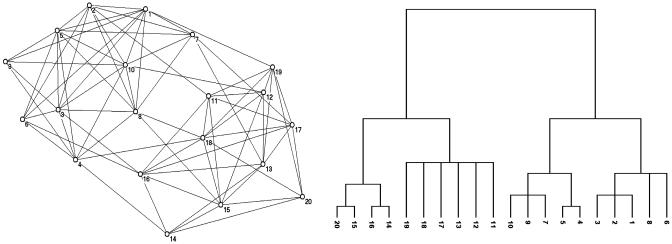
Qualitatively, a community is defined as a subset of nodes within the graph such that connections between the nodes are denser than connections with the rest of the network. The detection of the community structure in a network is generally intended as a procedure for mapping the network into a tree (Fig. 1). In this tree (called a dendrogram in the social sciences), the leaves are the nodes whereas the branches join nodes or (at higher level) groups of nodes, thus identifying a hierarchical structure of communities nested within each other.
Fig. 1.
A simple network (Left) and the corresponding dendrogram (Right).
Several algorithms to perform this mapping are known in the literature. The traditional method is the so-called hierarchical clustering (18). For every pair i,j of nodes in the network, one calculates a weight Wi,j, which measures how closely connected the vertices are. Starting from the set of all nodes and no edges, links are iteratively added between pairs of nodes in order of decreasing weight. In this way nodes are grouped into larger and larger communities, and the tree is built up to the root, which represents the whole network. Algorithms of this kind are called agglomerative.
For the other class of algorithms, called divisive, the order of construction of the tree is reversed: one starts with the whole graph and iteratively cuts the edges, thus dividing the network progressively into smaller and smaller disconnected subnetworks identified as the communities. The crucial point in a divisive algorithm is the selection of the edges to be cut, which have to be those connecting communities and not those within them. Very recently, Girvan and Newman (GN) have introduced a divisive algorithm where the selection of the edges to be cut is based on the value of their “edge betweenness” (19), a generalization of the centrality betweenness introduced by Anthonisse (20) and Freeman (21). Consider the shortest paths between all pairs of nodes in a network. The betweenness of an edge is the number of these paths running through it. It is clear that, when a graph is made of tightly bound clusters, loosely interconnected, all shortest paths between nodes in different clusters have to go through the few interclusters connections, which therefore have a large betweenness value. The single step of the GN detection algorithm consists in the computation of the edge betweenness for all edges in the graph and in the removal of those with the highest score. The iteration of this procedure leads to the splitting of the network into disconnected subgraphs that in their turn undergo the same procedure, until the whole graph is divided in a set of isolated nodes. In this way the dendrogram is built from the root to the leaves.
The GN algorithm represents a major step forward for the detection of communities in networks, since it avoids many of the shortcomings of traditional methods (19, 22). This fact explains why it has been quickly adopted in the past year as a sort of standard for the analysis of community structure in networks (23–27).
This article follows a different track by proposing an alternative strategy for the identification of the community structure. This complementary approach follows from the need to address the two following issues.
In general, algorithms define communities operationally as what the they find. A dendrogram, i.e., a community structure, is always produced by the algorithms down to the level of single nodes, independently from the type of graph analyzed. This feature is due to the lack of explicit prescriptionsto discriminate between networks that are actually endowed with a community structure and those that are not. As a consequence, in practical applications, one needs additional, nontopological information on the nature of the network to understand which of the branches of the tree have a real significance. Without such information it is not clear at all whether the identification of a community is reliable. Two noticeable proposals have been made to solve this problem. In particular, it is worth mentioning the approach proposed by Wilkinson and Huberman (24), which is limited to the lowest level of the community structure and specific to algorithms based on betweenness. More recently (22), Newman and Girvan introduced an a posteriori measure of the strength of the community structure, which they called modularity. More precisely, the modularity estimates the fraction of inward links in a community minus the expectation value of the same quantity in a network with the same community divisions but random connections between the nodes. This quantity definitely gives an indication of the strength of the community structure, even though the lack of the implementation of a quantitative definition of community does not allow the objective discrimination of meaningful communities.
The “edge betweenness algorithm” is computationally costly, as already remarked by Girvan and Newman (19, 22). Evaluating the score for all edges requires a time of the order of MN, where M is the number of edges and N the number of nodes. The iteration of the procedure for all M edges leads in the worst case to a total scaling of the computational time as M2N, which makes the analysis practically unfeasible already for moderately large networks (of the order of N = 10,000; ref. 22).
In this article we propose solutions to both these problems. First, we introduce a general criterion for deciding which of the subgraphs singled out by the detection algorithms are actual communities. We discuss in detail the case of two quantitative definitions of community. In this way, we transform the GN algorithm in a self-contained tool. Second, we present an alternative algorithm, based on the computation of local quantities, which gives, in controlled cases, results of accuracy similar to the GN method, but largely outperforming it from the point of view of computational speed. [It is worth mentioning that, after the completion of this work, Newman proposed a new agglomerative algorithm to address the issue of the computational efficiency (28).]
Quantitative Definitions of Community
The idea to solve the first of the problems discussed above is very simple: the algorithm that builds the tree just selects subgraphs that are candidates to be considered communities. One has then to check whether they are actually such by using a precise definition. If the subgraph does not meet the criterion, the subgraph isolated from the network is not a community, and the corresponding branch in the dendrogram should not be drawn.
As mentioned above, a community is generally thought of as a part of a network where internal connections are denser than external ones. To sharpen the use of detection algorithms a more precise definition is needed. Many possible definitions of communities exist in the literature (18). Here we consider explicitly the implementation in the algorithms of two plausible definitions of community which translate the sentence above into formulas.
The basic quantity to consider is ki, the degree of a generic node i, which in terms of the adjacency matrix Ai,j of the network G is ki = ∑jAi,j. (The adjacency matrix fully specifies the topology of the network. In the simplest case of an unweighted, undirected network, it is equal to 1 if i and j are directly connected; it is equal to zero otherwise.) If we consider a subgraph V ⊂ G, to which node i belongs, we can split the total degree in two contributions: 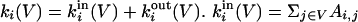 is the number of edges connecting node i to other nodes belonging to V.
is the number of edges connecting node i to other nodes belonging to V. 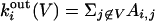 is clearly the number of connections toward nodes in the rest of the network.
is clearly the number of connections toward nodes in the rest of the network.
Definition of Community in a Strong Sense. The subgraph V is a community in a strong sense if
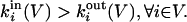 |
[1] |
In a strong community each node has more connections within the community than with the rest of the graph. This definition coincides with the one proposed in ref. 17 in the framework of the identification of web communities.
Definition of Community in a Weak Sense. The subgraph V is a community in a weak sense if
 |
[2] |
In a weak community the sum of all degrees within V is larger than the sum of all degrees toward the rest of the network.
Clearly a community in a strong sense is also a community in a weak sense, whereas the converse is not true.
It is worth mentioning that our definitions of community, although very natural, do not represent the only possible choice. Several other possible definitions, possibly more appropriate in some cases, exist and are described in ref. 18. Among them, for instance, the definition of the so-called LS-set goes in the direction of our strong definition even though it is extremely more stringent. An LS-set is a set of nodes such that each of its proper subsets has more ties to its complement within the set than outside. On the other hand, the definition of k-core is roughly, although not exactly, equivalent to our weak definition. A k-core is defined as a subgraph in which each node is adjacent to at least a minimum number, k, of the other nodes in the subgraph.
Self-Contained Algorithms
From the definitions given above, it is apparent that, if a network is randomly split in two parts, one very large and the other with only few nodes, the very large part almost always fulfills the definition of community. To deal with this problem, let us consider the Erdös–Renyi random graph (29). If we cut at random the graph in two parts containing αN and (1 - α)N nodes, respectively, it is easy to evaluate analytically the probability P(α) that the subgraph containing αN nodes fulfills the weak or the strong definition. It turns out that, as soon as N is sufficiently large, the probability is very close to a step function around α = 0.5. Hence, it is extremely likely that, in a random graph randomly cut in two parts, the largest one is a community according to the previous definitions. However, it is extremely unlikely that both subgraphs fulfill the definitions simultaneously; therefore, if we accept divisions only if both groups fulfill the definition of community, we correctly find that a random graph has no community structure. We extend this criterion to the general case: if less than two subgraphs obtained from the cut satisfy the definitions, then the splitting is considered to be an artifact and is disregarded.
We can now summarize the improved self-contained version of the GN algorithm.
Choose a definition of community.
Compute the edge betweenness for all edges and remove those with the highest score.
If the removal does not split the (sub-)graph go to point 2.
If the removal splits the (sub-)graph, test if at least two of the resulting subgraphs fulfill the definition. If they do, draw the corresponding part of the dendrogram.
Iterate the procedure (going back to point 2) for all the subgraphs until no edges are left in the network.
It is important to remark that the quantities appearing in Eqs. 1 and 2 must always be evaluated with respect to the full adjacency matrix. The application of this procedure to a network produces a tree, where every branch-splitting represents a meaningful (with respect to the definition) separation in communities.
It is now possible to blindly test the effectiveness of the GN algorithm. We have considered the “artificial” graph already discussed by Girvan and Newman. It is a simple network with N nodes divided into four groups: connections between pairs within a group are present with probability pin, whereas pairs of nodes in different groups are connected with probability pout. As the probability pout grows from zero, the community structure in the network becomes less well defined.
For every realization of the artificial graph, the application of the detection algorithm generates a tree. We consider the algorithm to be successful if the four communities are detected, each node is classified in the right community, and the communities are not further subdivided.
Fig. 2 presents the comparison of the fraction of successes for the modified GN algorithm with the expected value computed analytically. We see that the GN algorithm captures very well the existence of communities in a strong sense, whereas it performs less well for the weak definition. However, one should not be misguided by the quantity presented in Fig. 2. By looking at a softer measure of success, the fraction f of nodes not correctly classified, one realizes that, when the algorithm with weak definition seems to fail, it correctly identifies the four communities and it misclassifies only a few nodes up to much higher values of pout. The deviations from the theoretical behavior observed for small values of pout are due to the possibility that one or more of the four communities are further split in smaller subcommunities. This event, not taken into account in the analytical calculation, becomes very unlikely as the size of the system increases.
Fig. 2.
Test of the efficiency of the different algorithms in the analysis of the artificial graph with four communities. The construction of the graph is described in the text. Here N = 128 and pin is changed with pout to keep the average degree equal to 16. (Left) Strong definition: fraction of successes for the different algorithms compared with the analytical probability that four communities are actually defined. (Right) Weak definition: in addition to the same quantities plotted in Left, here we report, for every algorithm, the fraction f of nodes not correctly classified.
A Fast Algorithm
The GN algorithm is computationally expensive because it requires the repeated evaluation, for each edge in the system, of a global quantity, the betweenness, whose value depends on the properties of the whole system. Despite smart methods to compute the edge betweenness simultaneously for all edges (30, 31), the evaluation of such a quantity is the time-consuming part of the procedure. As a consequence, the time to completely analyze a network turns out to grow fast with its size, making the analysis unfeasible for networks larger than ≈10,000 nodes (22).
To overcome this problem we introduce a divisive algorithm that requires the consideration of local quantities only and is therefore much faster than the GN algorithm. The fundamental ingredient of a divisive algorithm is a quantity that can single out edges connecting nodes belonging to different communities. We consider the edge-clustering coefficient, defined, in analogy with the usual node-clustering coefficient, as the number of triangles to which a given edge belongs, divided by the number of triangles that might potentially include it, given the degrees of the adjacent nodes. More formally, for the edge-connecting node i to node j, the edge-clustering coefficient is
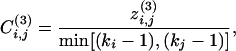 |
[3] |
where 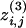 is the number of triangles built on that edge and min[(ki - 1),(kj - 1)] is the maximal possible number of them.
is the number of triangles built on that edge and min[(ki - 1),(kj - 1)] is the maximal possible number of them.
The idea behind the use of this quantity in a divisive algorithm is that edges connecting nodes in different communities are included in few or no triangles and tend to have small values of 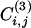 . On the other hand, many triangles exist within clusters. Hence, the coefficient
. On the other hand, many triangles exist within clusters. Hence, the coefficient 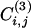 is a measure of how intercommunitarian a link is. A problem arises when the number of triangles is zero, because
is a measure of how intercommunitarian a link is. A problem arises when the number of triangles is zero, because 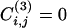 , irrespective of ki and kj {or even
, irrespective of ki and kj {or even 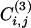 is indeterminate, when min[(ki - 1),(kj - 1)] = 0}. To remove this degeneracy we consider a slightly modified quantity by using, at the numerator, the number of triangles plus one:
is indeterminate, when min[(ki - 1),(kj - 1)] = 0}. To remove this degeneracy we consider a slightly modified quantity by using, at the numerator, the number of triangles plus one:
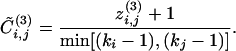 |
[4] |
By considering higher order cycles we can define, in much the same way, coefficients of order g as:
 |
[5] |
where 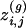 is the number of cyclic structures of order g the edge (i, j) belongs to, and
is the number of cyclic structures of order g the edge (i, j) belongs to, and 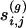 is the number of possible cyclic structures of order g that can be built given the degrees of the nodes.
is the number of possible cyclic structures of order g that can be built given the degrees of the nodes.
We can now define, for every g, a detection algorithm that works exactly as the GN method with the difference that, at every step, the removed edges are those with the smallest value of  . By considering increasing values of g, one can smoothly interpolate between a local and a nonlocal algorithm. Notice that the definition of
. By considering increasing values of g, one can smoothly interpolate between a local and a nonlocal algorithm. Notice that the definition of 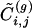 guarantees that nodes with only one connection are not considered as isolated communities by the algorithm, since for their unique edge
guarantees that nodes with only one connection are not considered as isolated communities by the algorithm, since for their unique edge 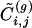 is infinite.
is infinite.
We have checked the accuracy of this algorithm by comparing its performance with the GN method. Fig. 2 reports the results for the artificial test graph with four communities. It turns out that, with respect to the strong definition, this algorithm is as accurate as the GN algorithm, both in the cycles of order g = 3 (triangles) and g = 4 (squares).
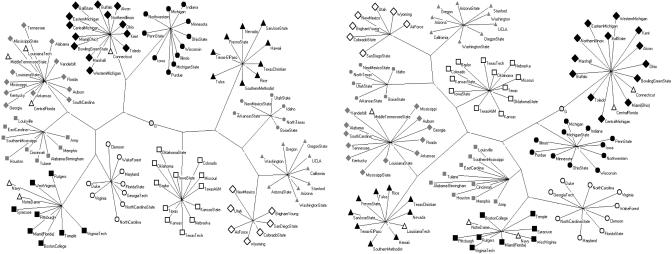
On the other hand, for the weak definition the best accuracy is achieved with this algorithm with g = 4. Another test is performed by considering the examples of social networks already studied by GN. Fig. 3 shows the trees resulting from the application of the GN algorithm and of the g = 4 algorithm to the network of college football teams. Again, the results are very similar, indicating that the local algorithm captures well the presence of communities in that network.
Fig. 3.
Plot of the dendrograms for the network of college football teams, obtained by using the GN algorithm (Left) and our algorithm with g = 4 (Right). Different symbols denote teams belonging to different conferences. In both cases, the observed communities perfectly correspond to the conferences, with the exception of the six members of the Independent conference, which are misclassified.
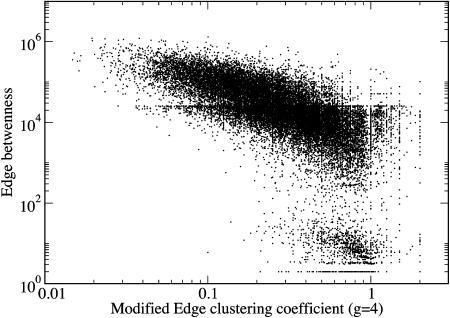
Additional insight into the relationship between the GN algorithm and this algorithm based on edge clustering is provided by Fig. 4, where the edge betweenness is plotted versus 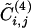 for each edge of the graph of scientific collaborations studied in ref. 15. It is clear that an anticorrelation exists between the two quantities; edges with low values of
for each edge of the graph of scientific collaborations studied in ref. 15. It is clear that an anticorrelation exists between the two quantities; edges with low values of 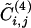 tend to have high values of betweenness. The anticorrelation is not perfect; the edge with minimum
tend to have high values of betweenness. The anticorrelation is not perfect; the edge with minimum 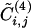 is not the one with maximal betweenness. Therefore, we expect the two algorithms to yield similar community structures although not perfectly coinciding.
is not the one with maximal betweenness. Therefore, we expect the two algorithms to yield similar community structures although not perfectly coinciding.
Fig. 4.
Edge betweenness vs. the modified edge-clustering coefficient 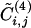 , for the network of scientific collaborations considered in Community Structure in a Network of Scientific Collaborations. Each dot represents an edge in the network. See Community Structure in a Network of Scientific Collaborations for details.
, for the network of scientific collaborations considered in Community Structure in a Network of Scientific Collaborations. Each dot represents an edge in the network. See Community Structure in a Network of Scientific Collaborations for details.
In this framework it is important to recall the parallel drawn by Burt (32) between betweenness centrality and the so-called redundancy. The definition of redundancy is very close to that of node clustering and, much in the spirit of our work, Burt first pointed out that nodes that belong to few loops are central in the betweenness sense.
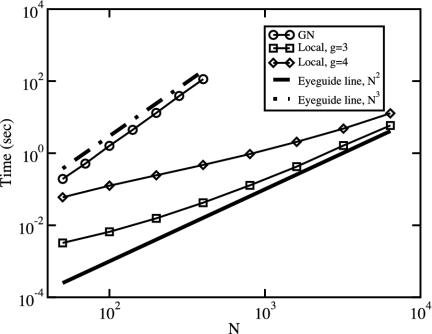
Let us now turn our attention to the question of the computational efficiency of our local algorithm. One can roughly estimate the scaling of the computational time as follows. When an edge is removed one has to check whether the whole system has been separated in disconnected components and update the value of 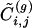 in a small neighborhood of the removed edge. The first operation requires a time of the order of M, the total number of edges present in the network, whereas the time required by the second operation does not scale with M. Because this operation has to be repeated for all edges, we can estimate the scaling of the total time as aM + bM2. We thus expect computational time to depend linearly on M for small systems and to crossover to an M2 regime for large sizes.
in a small neighborhood of the removed edge. The first operation requires a time of the order of M, the total number of edges present in the network, whereas the time required by the second operation does not scale with M. Because this operation has to be repeated for all edges, we can estimate the scaling of the total time as aM + bM2. We thus expect computational time to depend linearly on M for small systems and to crossover to an M2 regime for large sizes.
We have measured the velocity of this algorithm by computing the time needed to generate the whole tree for a random graph of increasing size N and fixed average degree (M ∼ N). Results, reported in Fig. 5, confirm that the algorithm based on the computation of the edge-clustering coefficient is much faster than the one based on edge betweenness, both for g = 3 and g = 4. The crossover between the initial linear dependence on N to the N2 growth for the local algorithm is evident for g = 3.
Fig. 5.
Plot of the average time (in seconds) needed to analyze a random graph of N nodes and fixed average degree 〈k〉 = 5. The time refers to the construction of the full tree down to single nodes (Fig. 1). No criterion to validate the communities was imposed. The runs are performed on a desktop computer with a 800-MHz CPU.
Community Structure in a Network of Scientific Collaborations
In this section we consider an application of the fast algorithm discussed in A Fast Algorithm to a network of scientific collaborations. In particular, we have considered the network of scientists who signed at least one paper submitted to the E-print Archive relative to Condensed Matter in the period 1995–1999 (http://xxx.lanl.gov/archive/cond-mat). Data have been kindly provided by Mark Newman (University of Michigan, Ann Arbor, MI).
The network includes 15,616 nodes (scientists) but it has a giant connected component that includes only n = 12,722 scientists. We have focused our attention on this giant component, and we have applied to it our algorithm for g = 3 and g = 4. The time needed for the generation of the dendrogram for g = 3 is ≈3 min on a desktop computer with a 800-MHz CPU. The algorithm detects, at the same time, the communities satisfying the weak and the strong criteria. At the end of the procedure one obtains, for every value of g, a list of communities identified in a weak sense and another list of communities identified in a strong sense. By definition, the second list is a subset of the first one. Fig. 6 reports the size distributions (for g = 3 and g = 4) of all the communities identified in a weak sense. These distributions feature a power-law behavior P(S) ≈ S-τ with τ ≃ 2, an indication of the self-similar community structure of this network (33). The exponent we observed coincides with the one obtained, using the GN algorithm, in the framework of a recently proposed model of social network formation (34).
Fig. 6.
Normalized size distribution of all the communities of scientists identified in a weak sense by the algorithm described in A Fast Algorithm for g = 3 (circles) and g = 4 (squares). In both cases, the behavior is well reproduced by a power law with exponent -2.
For what concerns a more detailed analysis of the communities found, validation of the results is far from trivial, because no quantitative criterion exists to assess their accuracy. One may directly inspect the dendrogram to answer questions like: Are the communities representative of real collaborations between the corresponding scientists? Do they identify specific research areas? Would a generic scientist agree about his or her belonging to a given community? Obviously, all these questions cannot be answered in a definitive and quantitative way. We have followed this path, in part, and we have checked several subsets of the network at different levels in the hierarchy. To the best of our knowledge, the results seem reasonable to us. Of course, this result does not represent a proof of the efficiency of the algorithm. We refer the reader to the detailed results of our analysis that we make available as additional supporting information.
Conclusions
The detection of the community structure in large complex networks is a promising field of research with many open challenges. The concept of community is qualitatively intuitive. However, to analyze a network it is necessary to specify quantitatively and unambiguously what a community is. Once a definition is given it is in principle possible to determine all subgraphs of a given network that fulfill the definition. However, in practice, this task is computationally out of reach even for small systems. Therefore, the search for the community structure has generally a more limited goal: selecting, among all possible communities, a subset of them organized hierarchically, a dendrogram. Divisive and agglomerative algorithms carry out this task. A comparison of the performances of such algorithms is nontrivial. In some simple cases, as the artificial graph with four subsets considered above, it is possible to assess quantitatively the validity of the results. In other cases, like the network of scientific collaborations, no quantitative measure exists to decide, given a precise definition of community, how good a dendrogram is. Typically, one may check whether the results appear sensible. However, this assessment is far from objective, being mediated by the observer's own perception and by his/her intuitive concept of community.
In this work we have proposed two improvements in the construction of the dendrogram. First, we have devised a way to implement, in a generic divisive algorithm, a quantitative definition of community. In this way algorithms become fully self-contained, i.e., they do not need nontopological input to generate the dendrogram. Then we have introduced a divisive algorithm, which is based on local quantities and therefore is extremely fast. Both these improvements have been tested successfully in controlled cases. The analysis of the large network of scientific collaborations gives results that appear reasonable. However, it is clear that, as discussed above, this statement is subjective and cannot be made more precise at present. Definitely, a quantitative measure for the evaluation of dendrograms would be a major step forward in this field.
At this point a remark is in order. So far, we have only discussed examples of the so-called social networks. It has been shown (35) that social networks substantially differ from other types of networks, namely technological or biological networks. Among other differences, they exhibit a positive correlation between the degree of adjacent vertices (assortativity), whereas most nonsocial networks are disassortative. Although these results are consistent with our and other's findings about community structures in social networks, they put into question the very existence of a community structure in nonsocial networks and the possibility of detecting it with the existing algorithms. From the perspective of our local algorithm, which relies on the existence of closed loops, disassortative networks could, in principle, be problematic because of the small number of short cycles. However, interesting insight comes from the study of the loops of arbitrary order (36). In particular, for four different types of networks (two social and assortative and two nonsocial and disassortative), measured values for the so-called average grid coefficient (the extension of the concept of clustering coefficient to cycles of order four) are two to four orders of magnitude larger than the corresponding coefficients of a random graph with the same average degree and size N. This argues in favor of the presence of some sort of hierarchical structure and well defined communities also in disassortative networks. It also hints that our algorithm could be fruitfully applied also to nonsocial (disassortative) networks, although future work is needed in this direction.
We believe that the elements presented in this article can be of great help in the analysis of networks. On the one hand, the implementation of a quantitative definition of community makes algorithms self-contained and allows analysis of the community structure based only on the network topology. On the other hand, the introduction of a class of local and fast algorithms could open the way for applications to large-scale systems.
Acknowledgments
We thank Mark Newman for providing data on the networks of college football teams and of scientific collaborations and Alain Barrat for useful suggestions and discussions.
Abbreviation: GN, Girvan–Newman.
References
- 1.Albert, R. & Barabási, A.-L. (2002) Rev. Mod. Phys. 74, 47-97. [Google Scholar]
- 2.Newman, M. E. J. (2003) SIAM Rev. 45, 167-256. [Google Scholar]
- 3.Albert, R., Jeong, H. & and Barabási, A.-L. (1999) Nature 401, 130-131. [Google Scholar]
- 4.Broder, A., Kumar, R., Maghoul, F., Raghavan, P., Rajagopalan, S., Stata, R., Tomkins, A. & Wiener, J. (2000) Comput. Networks 33, 309-320. [Google Scholar]
- 5.Moore, C. & Newman, M. E. J. (2000) Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Top. 61, 5678-5682. [DOI] [PubMed] [Google Scholar]
- 6.Pastor-Satorras, R. & Vespignani, A. (2001) Phys. Rev. Lett. 86, 3200-3203. [DOI] [PubMed] [Google Scholar]
- 7.Jeong, H., Tombor, B., Albert, R., Oltvai, Z. N. & Barabási, A.-L. (2000) Nature 407, 651-654. [DOI] [PubMed] [Google Scholar]
- 8.Wagner, A. & Fell, D. A. (2001) Proc. R. Soc. London Ser. B 268, 1803-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravasz, E., Somera, A., Mongru, D. A., Oltvai, Z. N. & Barabási A.-L. (2002) Science 297, 1551-1555. [DOI] [PubMed] [Google Scholar]
- 10.Montoya, J. M. & Solé, R. V. (2002) J. Theor. Biol. 214, 405-412. [DOI] [PubMed] [Google Scholar]
- 11.Dunne, J. A., Williams, R. J. & Martinez, N. D. (2002) Proc. Natl. Acad. Sci. USA 99, 12917-12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camacho, J., Guimerà, R. & Amaral, L. A. N. (2002) Phys. Rev. Lett. 88, 228102. [DOI] [PubMed] [Google Scholar]
- 13.Garlaschelli, D., Caldarelli, G. & Pietronero, L. (2003) Nature 423, 165-168. [DOI] [PubMed] [Google Scholar]
- 14.Redner, S. (1998) Eur. Phys. J. B 4, 131-134. [Google Scholar]
- 15.Newman, M. E. J. (2001) Proc. Natl. Acad. Sci. USA 98, 404-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rives, A. & Galitski, T. (2003) Proc. Natl. Acad. Sci. USA 100, 1128-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flake, G. W., Lawrence, S. R., Giles, C. L. & Coetzee, F. M. (2002) IEEE Computer 35, 66-71. [Google Scholar]
- 18.Wasserman, S. & Faust, K. (1994) Social Network Analysis (Cambridge Univ. Press, Cambridge, U.K.).
- 19.Girvan, M. & Newman, M. E. J. (2002) Proc. Natl. Acad. Sci. USA 99, 7821-7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anthonisse, J. M. (1971) Technical Report BN 9/71 (Stichting Mathematisch Centrum, Amsterdam).
- 21.Freeman, L. (1977) Sociometry 40, 35-41. [Google Scholar]
- 22.Newman, M. E. J. & Girvan, M. (2003) arXiv: cond-mat/0308217.
- 23.Holme, P., Huss, M. & Jeong, H. (2003) Bioinformatics 19, 532-538. [DOI] [PubMed] [Google Scholar]
- 24.Wilkinson, D. & Huberman, B. A. (2002) arXiv: cond-mat/0210147.
- 25.Gleiser, P. & Danon, L. (2003) arXiv: cond-mat/0307434.
- 26.Guimerà, R., Danon, L., Díaz-Guilera, A., Giralt, F. & Arenas, A. (2002) arXiv: cond-mat/0211498. [DOI] [PubMed]
- 27.Tyler, J. R., Wilkinson, D. M. & Huberman, B. A. (2003) arXiv: cond-mat/0303264.
- 28.Newman, M. E. J. (2003) arXiv: cond-mat/0309508.
- 29.Bollobas, B. (1985) Random Graphs (Academic, New York).
- 30.Newman, M. E. J. (2001) Phys. Rev. E 64, 016131. [DOI] [PubMed] [Google Scholar]
- 31.Brandes U. (2001) J. Math. Sociol. 25, 163-177. [Google Scholar]
- 32.Burt R. S. (1992) Structural Holes (Harward Univ. Press, Cambridge, MA).
- 33.Caldarelli G., Caretta Cartozo C., De Los Rios P. & Servedio V. D. P. (2003) arXiv: cond-mat/0311486. [DOI] [PubMed]
- 34.Boguña M., Pastor-Satorras R., Díaz-Guilera A. & Arenas A. (2003) arXiv: cond-mat/0309263.
- 35.Newman, M. E. J. & Park, J. (2003) Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Top. 68, 036122. [Google Scholar]
- 36.Caldarelli, G., Pastor-Satorras, R. & Vespignani A. (2002) arXiv: cond-mat/0212026.