SUMMARY
Clinical application of induced pluripotent stem (iPS) cells is limited by the low efficiency of iPS derivation and the fact that most protocols modify the genome to effect cellular reprogramming. Moreover, safe and effective means of directing the fate of patient-specific iPS cells towards clinically useful cell types are lacking. Here we describe a simple, non-integrating strategy for reprogramming cell fate based on administration of synthetic mRNA modified to overcome innate anti-viral responses. We show that this approach can reprogram multiple human cell types to pluripotency with efficiencies that greatly surpass established protocols. We further show that the same technology can be used to efficiently direct the differentiation of RNA-induced pluripotent stem (RiPS) cells into terminally differentiated myogenic cells. This technology represents a safe, efficient strategy for somatic cell reprogramming and directing cell fate that has broad applicability for basic research, disease modeling and regenerative medicine.
INTRODUCTION
The reprogramming of differentiated cells to pluripotency holds great promise as a tool for studying normal development, while offering hope that patient-specific induced pluripotent stem (iPS) cells could be used to model disease, or to generate clinically useful cell types for autologous therapies aimed at repairing deficits arising from injury, illness, and aging. Induction of pluripotency was originally achieved by Yamanaka and colleagues by enforced expression of four transcription factors, KLF4, c-MYC, OCT4, and SOX2 (KMOS) using retroviral vectors (Takahashi et al., 2007; Takahashi and Yamanaka, 2006). Viral integration into the genome initially presented a formidable obstacle to therapeutic use of iPS cells. The search for ways to induce pluripotency without incurring genetic change has thus become the focus of intense research effort. Towards this end, iPS cells have been derived using excisable lentiviral and transposon vectors, or through repeated application of transient plasmid, episomal, and adenovirus vectors (Chang et al., 2009; Kaji et al., 2009; Okita et al., 2008; Stadtfeld et al., 2008; Woltjen et al., 2009; Yu et al., 2009). iPS cells have also been derived using two DNA-free methods: serial protein transduction with recombinant proteins incorporating cell-penetrating peptide moieties (Kim et al., 2009; Zhou et al., 2009), and transgene delivery using the Sendai virus, which has a completely RNA-based reproductive cycle (Fusaki et al., 2009).
Despite such progress, considerable limitations accompany the non-integrative iPS derivation strategies devised thus far. For example, while DNA transfection-based methodologies are ostensibly safe, they nonetheless entail some risk of genomic recombination or insertional mutagenesis. In protein-based strategies, the recombinant proteins used are challenging to generate and purify in the quantities required (Zhou et al., 2009). Use of Sendai virus requires stringent steps to purge reprogrammed cells of replicating virus, and the sensitivity of the viral RNA replicase to transgene sequence content may limit the generality of this reprogramming vehicle (Fusaki et al., 2009). Importantly, methods that rely on repeat administration of transient vectors, whether DNA or protein-based, have so far shown very low iPS derivation efficiencies (Jia et al., 2010; Kim et al., 2009; Okita et al., 2008; Stadtfeld et al., 2008; Yu et al., 2009; Zhou et al., 2009), presumably due to weak or inconstant expression of reprogramming factors.
Here we demonstrate that repeated administration of synthetic messenger RNAs incorporating modifications designed to bypass innate anti-viral responses can reprogram differentiated human cells to pluripotency with conversion efficiencies and kinetics substantially superior to established viral protocols. Furthermore, this simple, non-mutagenic, and highly controllable technology is applicable to a range of tissue engineering tasks, exemplified here by RNA-mediated directed differentiation of RNA-iPS (RiPS) cells to terminally differentiated myogenic cells.
RESULTS
Development of modified-RNAs for directing cell fate
We manufactured mRNA using in vitro transcription (IVT) reactions templated by PCR amplicons (Figure S1). To promote efficient translation and boost RNA half-life in the cytoplasm, a 5′ guanine cap was incorporated by inclusion of a synthetic cap analog in the IVT reactions (Yisraeli et al., 1989). Within our IVT templates, the open reading frame (ORF) of the gene of interest is flanked by a 5′ untranslated region (UTR) containing a strong Kozak translational initiation signal, and an alpha-globin 3′ UTR terminating with an oligo(dT) sequence for templated addition of a polyA tail.
Cytosolic delivery of mRNA into mammalian cells can be achieved using electroporation or by complexing the RNA with a cationic vehicle to facilitate uptake by endocytosis (Audouy and Hoekstra, 2001; Elango et al., 2005; Holtkamp et al., 2006; Van den Bosch et al., 2006; Van Tendeloo et al., 2001). We focused on the latter approach, reasoning that this would allow for repeated transfection to sustain ectopic protein expression over the days to weeks required for cellular reprogramming. In preliminary experiments in which synthetic RNA encoding GFP was transfected into murine embryonic fibroblasts and human epidermal keratinocytes, a high, dose-dependent cytotoxicity was observed that was not attributable to the cationic vehicle, which was exacerbated on repeated transfection. These experiments revealed a serious impediment to achieving sustained protein expression by mRNA transfection, and highlighted a need to develop a technology that would permit sustained protein expression with mRNA with reduced cellular toxicity.
It is known that exogenous single-stranded RNA (ssRNA) activates antiviral defenses in mammalian cells through interferon and NF-κB dependent pathways (Diebold et al., 2004; Hornung et al., 2006; Kawai and Akira, 2007; Pichlmair et al., 2006; Uematsu and Akira, 2007). We sought approaches to reduce the immunogenic profile of synthetic RNA in order to increase the sustainability of RNA-mediated protein expression. Co-transcriptional capping yields a significant fraction of uncapped IVT products bearing 5′ triphosphates, which can be detected by the ssRNA sensor RIG-I (Hornung et al., 2006; Pichlmair et al., 2006), and have also been reported to activate PKR, a global repressor of protein translation (Nallagatla and Bevilacqua, 2008). We therefore treated synthesized RNA with a phosphatase, which resulted in modest reductions in cytotoxicity upon transfection (data not shown).
Eukaryotic mRNA is extensively modified in vivo, and the presence of modified nucleobases has been shown to reduce signaling by RIG-I and PKR, as well as by the less widely expressed but inducible endosomal ssRNA sensors TLR7 and TLR8 (Kariko et al., 2005; Kariko et al., 2008; Kariko and Weissman, 2007; Nallagatla and Bevilacqua, 2008; Nallagatla et al., 2008; Uzri and Gehrke, 2009). In an attempt to further reduce innate immune responses to transfected RNA, we synthesized mRNAs incorporating modified ribonucleoside bases. Complete substitution of either 5-methylcytidine (5mC) for cytidine, or pseudouridine (psi) for uridine in GFP-encoding transcripts markedly improved viability and increased ectopic protein expression, while the most significant improvement was seen when both modifications were used together (Figure 1A–C). These modifications dramatically attenuated interferon signaling as revealed by qRT-PCR for a panel of interferon response genes, although residual upregulation of some interferon targets was still detected (Figure 1D). It is known that cellular anti-viral defenses can self-prime through a positive-feedback loop involving autocrine and paracrine signaling by Type I interferons (Randall and Goodbourn, 2008). Media supplementation with a recombinant version of B18R protein, a Vaccinia virus decoy receptor for Type I interferons (Symons et al., 1995), further increased cell viability of RNA transfection in some cell types (data not shown). Synthesis of RNA with modified ribonucleotides and phosphatase treatment (henceforth, modified-RNAs), used in conjunction with media supplementation with the interferon inhibitor B18R allowed for high, dose-dependent levels of protein expression with high cell viability (Figure 1E).
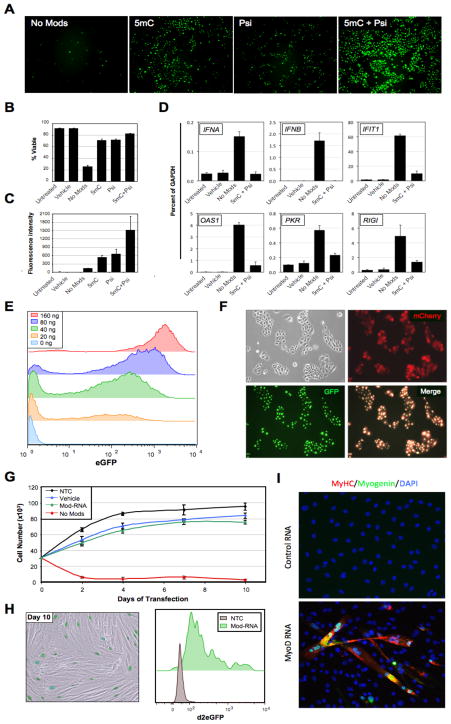
Figure 1. Modified-RNA overcomes anti-viral responses and can be used to direct cell fate.
(A) Microscopy images showing keratinocytes transfected 24 hours earlier with 400 ng/well of synthetic unmodified (No Mods), 5-methyl-cytosine modified (5mC), pseudouridine modified (Psi), or 5mC + Psi modified-RNA encoding GFP. (B) Percent viability and (C) mean fluorescence intensity of the cells shown in (A) as measured by flow cytometry. (D) Quantitative RT-PCR data showing expression of six interferon-regulated genes in BJ fibroblasts 24 hours after transfection with unmodified (No Mods), or modified (5mC + Psi) RNA encoding GFP (1200 ng/well), and vehicle and untransfected controls. (E) Flow cytometry histograms showing GFP expression in keratinocytes transfected with 0–160 ng of modified-RNA, 24 hours post transfection. (F) Microscopy images of keratinocytes co-transfected with modified-RNAs encoding GFP with a nuclear localization signal, and mCherry (cytosolic) proteins. (G) Growth kinetics of BJ fibroblasts transfected daily with unmodified, or modified-RNAs encoding a destabilized nuclear-localized GFP, and vehicle and untransfected controls for 10 days. (H) Sustained GFP expression of modified-RNA transfected cells described in (G) at day 10 of transfection shown by fluorescence imaging with bright field overlay (left panel), and flow cytometry (right panel). (I) Immunostaining for the muscle-specific proteins myogenin and myosin heavy chain (MyHC) in murine C3H/10T1/2 cell cultures 3 days after 3 consecutive daily transfections with a modified-RNA encoding MYOD. Error bars indicate s.d., n=3 for all panels. See also Figure S2.
Transfection of modified-RNA encoding GFP into six human cell types showed highly penetrant expression (50–90% positive cells), demonstrating the applicability of this technology to diverse cell types (Figure S2A). Simultaneous delivery of modified-RNAs encoding mCherry, and GFP containing a nuclear localization signal confirmed that generalized co-expression of multiple proteins could be achieved in mammalian cells, and that expressed proteins could be correctly localized to different cellular compartments (Figure 1F). Ectopic protein expression after RNA transfection is transient owing to RNA and protein degradation and the diluting effect of cell division. To establish the kinetics and persistence of protein expression, modified-RNA encoding GFP variants designed for high and low protein stability (Li et al., 1998) were synthesized and transfected into keratinocytes. Time course analysis by flow cytometry showed that protein expression persisted for several days for the high-stability variant, but peaked within 12 hours and decayed rapidly thereafter for the destabilized GFP (Figure S2B). These results indicated that a repetitive transfection regimen would be required to sustain high levels of ectopic expression for short-lived proteins over an extended time course. To address this and further examine the impact of repeated RNA transfection on cell growth and viability we transfected BJ fibroblasts for 10 consecutive days with either unmodified, or modified-RNAs encoding a destabilized nuclear GFP, and appropriate controls (Figure S3A). Daily transfection with modified-RNA permitted sustained protein expression without substantially compromising the viability of the culture beyond a modest reduction in growth kinetics that was largely attributable to the transfection reagent (Figure 1G–H, S2C). Microarray analysis of the cultures following the tenth and final transfection revealed that prolonged daily transfection with modified-RNA did not significantly alter the molecular profile of the transfected cells (Figure S2D), although upregulation of a number of interferon response genes was noted consistent with our previous observation that the RNA modifications did not completely abrogate interferon signaling (Figure 1D, Figure S2E). By contrast, repeated transfection with unmodified RNA severely compromised the growth and viability of the culture through elicitation of a massive interferon response (Figure 1D), indicating that the use of unmodified RNA was not a viable strategy for sustaining polypeptide expression in cells (Figure 1G).
To determine if modified-RNAs could be used to directly alter cell fate, we synthesized modified-RNA encoding the myogenic transcription factor MYOD (Davis et al., 1987) and transfected it into murine C3H10T1/2 cells over the course of 3 days, followed by continued culturing in a low serum media for an additional 3 days. The emergence of large, multi-nucleated myotubes that stained positive for the myogenic markers myogenin and myosin heavy chain (MyHC) provided proof of principle that transfection with modified-RNAs could be utilized to efficiently direct cell fate (Figure 1I).
Generation of induced pluripotent stem cells using modified-RNAs
We next sought to determine if induced pluripotent stem cells (iPS) could be derived using modified-RNAs. To this end, modified-RNAs encoding the four canonical Yamanaka factors, KLF4 (K), c-MYC (M), OCT4 (O), and SOX2 (S), were synthesized, and transfected into cells. Immuno-staining with antibodies directed against OCT4, KLF4 and SOX2 demonstrated that each of the factors was robustly expressed and correctly localized to the nucleus (Figure 2A). Expression kinetics was monitored by flow cytometry, which showed maximal protein expression 12 to 18 hours post-transfection, followed by rapid turnover of these transcription factors (Figure 2B). From this we concluded that daily transfections would be required to maintain high levels of expression of the Yamanaka factors during long-term, multi-factor reprogramming regimens.
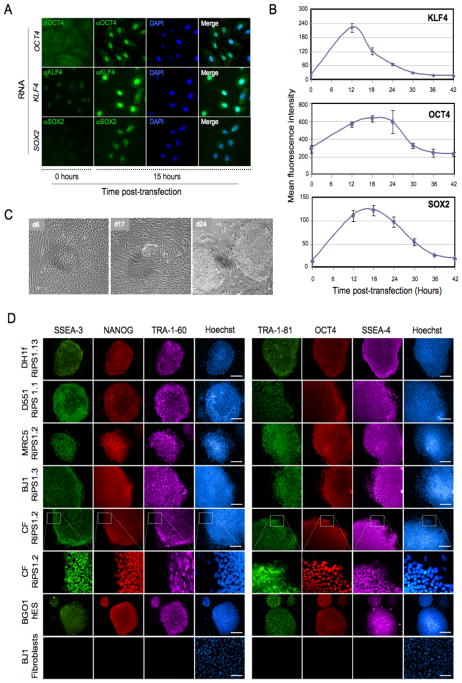
Figure 2. Generation of RNA-induced pluripotent stem cells (RiPS).
(A) Immunostaining for human KLF4, OCT4, and SOX2 proteins in keratinocytes 15 hours post-transfection with modified-RNA encoding KLF4, OCT4, or SOX2. (B) Time course showing kinetics and stability of KLF4, OCT4, and SOX2 proteins after modified-RNA transfection, assayed by flow cytometry following intracellular staining of reach protein. (C) Brightfield images taken during the derivation of RNA-iPS cells (RiPS) from dH1f fibroblasts showing early epitheliod morphology (day 6), small hES-like colonies (day 17), and appearance of mature iPS clones after mechanical picking and expansion (day 24). (D) Immunohistochemistry showing expression of a panel of pluripotency markers in expanded RiPS clones derived from dH1f fibroblasts, Detroit 551 (D551) and MRC-5 fetal fibroblasts, BJ post-natal fibroblasts, and cells derived from a skin biopsy taken from an adult cystic fibrosis patient (CF), shown also in high magnification. BG01 hES cells and BJ1 fibroblasts are included as positive and negative controls, respectively. See also Figure S3 and S4.
We next sought to establish a protocol to ensure sustained, high-level protein expression through daily transfection of modified-RNAs by exploring a matrix of conditions encompassing different transfection reagents, culture media, feeder cell types, and RNA doses (data not shown). Once optimized, we initiated long-term reprogramming experiments with human ES-derived dH1f fibroblasts, which display relatively efficient viral-mediated iPS cell conversion (Chan et al., 2009; Park et al., 2008). Low-oxygen (5% O2) culture conditions and a KMOS stoichiometry of 1:1:3:1 were employed, as these have been reported to promote efficient iPS conversion (Kawamura et al., 2009; Papapetrou et al., 2009; Utikal et al., 2009; Yoshida et al., 2009). Modified-RNA encoding a destabilized nuclear GFP was spiked into the KMOS RNA cocktail to allow visualization of continued protein expression throughout the experimental time course (Figure S3A). Widespread transformation of fibroblast morphology to a compact, epithelioid morphology was observed within the first week of modified-RNA transfection, followed by emergence of canonical hES-like colonies with tight morphology, well-defined borders, and prominent nucleoli towards the end of the second week of transfection (Figure 2C). RNA transfection was terminated on day 17, and colonies were mechanically picked three days later which were then expanded under standard ES culture conditions to establish 14 prospective iPS lines, designated dH1f-RiPS (RNA-derived iPS) 1-14.
We next attempted to reprogram somatically-derived cells to pluripotency using a similar reprogramming regimen. Anticipating that these cells might be more challenging to reprogram, we employed a five-factor cocktail including a modified-RNA encoding LIN28 (KMOSL), which has been shown to facilitate reprogramming (Yu et al., 2007; Hanna et al., 2009), and supplemented the media with valproic acid (VPA), a histone deacetylase inhibitor, which has been reported to increase reprogramming efficiency (Huangfu et al., 2008). Four human cell types were tested: Detroit 551 (D551) and MRC-5 fetal fibroblasts, BJ post-natal fibroblasts, and fibroblast-like cells cultured from a primary skin biopsy taken from an adult cystic fibrosis patient (CF cells). Daily transfection with the modified-RNA KMOSL cocktail gave rise to numerous hES-like colonies in the D551, BJ, CF and MRC5 cultures that were mechanically picked at day 18, 20, 21 and 25, respectively. More than 10 RiPS clones were expanded for each of the somatic lines, with notably very few clones failing to establish. Immunostaining confirmed the expression of OCT4, NANOG, TRA-1-60, TRA-1-81, SSEA3, and SSEA4 in all the RiPS lines examined (Figure 2D, Figure S3B). DNA fingerprinting confirmed parental origin of three RiPS clones from each somatic cell derivations, and all clones presented normal karyotypes (data not shown). Of note, additional experiments conducted in the presence or absence of VPA showed little difference in reprogramming efficiency (data not shown), and VPA was therefore not used in subsequent experiments.
Molecular characterization and functional potential of RiPS cells
A number of molecular and functional assays were performed to further assess whether the RiPS cells had been reprogrammed to pluripotency (Table S1). Multiple RiPS lines derived from each of the five starting cell types were evaluated by quantitative RT-PCR (qRT-PCR), and all demonstrated robust expression of the pluripotency-associated transcripts OCT4, SOX2, NANOG, and hTERT (Figure 3A). Bisulfite sequencing of the Oct4 locus revealed extensive demethylation relative to the parental fibroblasts, an epigenetic state equivalent to human ES cells (Figure 3B).
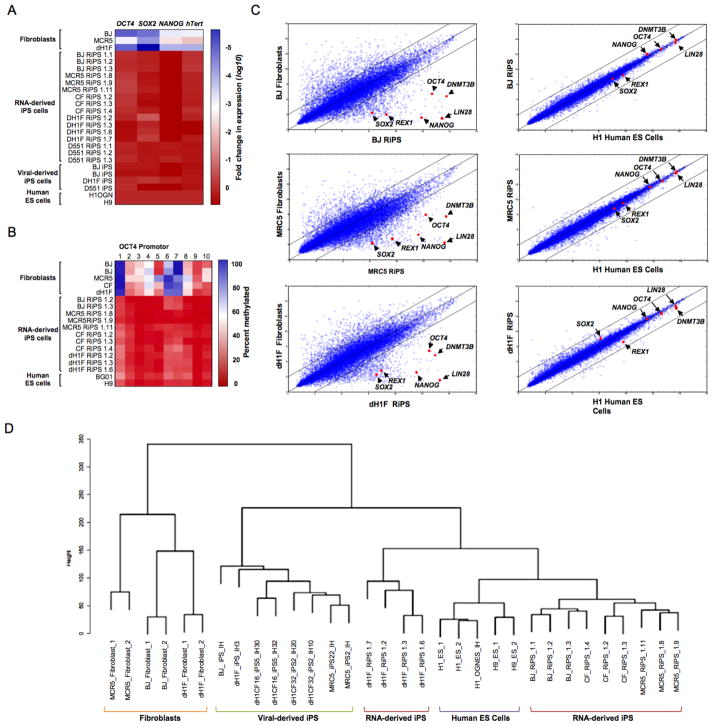
Figure 3. Molecular characterization of RiPS cells.
(A) Heatmap showing results of qRT-PCR analysis measuring the expression of pluripotency-associated genes in RiPS cell lines, parental fibroblasts and viral-derived iPS cells relative to hES cell controls. (B) Heatmap showing results of OCT4 promoter methylation analysis of RiPS cell lines, parental fibroblasts, and hES cell controls. (C) Global gene expression profiles of BJ-, MRC5- and dH1F-derived RiPS cells shown in scatter plots against parental fibroblasts and hES cells with pluripotency-associated transcripts indicated. (D) Dendrogram showing unsupervised hierarchical clustering of the global expression profiles for RiPS cells, parental fibroblasts, hES cells, and virus-derived iPS cells.
To gain more global insight into the molecular properties of RiPS cells, gene expression profiles of RiPS clones from multiple independent derivations were generated and compared to fibroblasts, human ES cells, and virally-derived iPS cell lines. These analyses revealed that all modified-RNA-derived iPS clones examined had a molecular signature that very closely recapitulated that of human ES cells while being highly divergent from the profile of the parental fibroblasts (Figure 3C). Importantly, pluripotency-associated transcripts including SOX2, REX1, NANOG, OCT4, LIN28 and DNMT3B were substantially upregulated in the RiPS cells compared to the parental fibroblast lines to levels comparable to hES cells (Figure 3C). Furthermore, when the transcriptional profiles were subjected to unsupervised hierarchical clustering analysis, all RiPS clones analyzed clustered more closely to hES than did virally-derived iPS cells suggesting that modified-RNA-derived iPS cells more fully recapitulated the molecular signature of human ES cells (Figure 3D).
To test the developmental potential of RiPS cells, embryoid bodies (EBs) were generated from multiple clones from five independent RiPS derivations, and beating cardiomyocytes were observed for the vast majority of the EBs (Table S1, Movie S1). Mesodermal potential was further evaluated in methylcellulose blood forming assays which showed that all lines tested were robustly able to differentiate into hematopoietic precursors capable of giving rise to colony numbers and a spectrum of blood colony types comparable to human ES cells (Figure 4A, Table S1). A subset of clones was further plated onto matrigel and differentiated into Tuj1-positive neurons (ectoderm), and alpha-fetoprotein-positive endodermal cells (Figure 4B, Table S1). Finally, tri-lineage differentiation potential was confirmed in vivo by the formation of teratomas from dH1F-, CF- and BJ-RiPS cells, that histologically revealed cell types of the three germ layers (Figure 4C, Figure S5, Table S1).
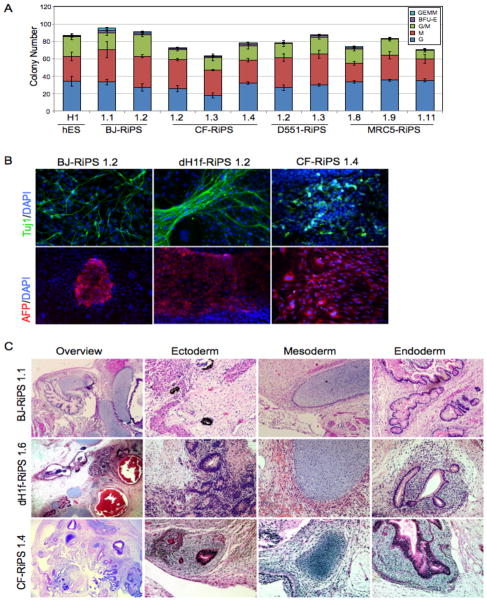
Figure 4. Trilineage differentiation of RiPS cells.
(A) Yield and typology of blood-lineage colonies produced by directed differentiation of embryoid bodies in methylcellulose assays with RiPS clones derived from BJ, CF, D551 and MCR5 fibroblasts, and a human ES (H1) control. (B) Immunostaining showing expression of the lineage markers Tuj1 (neuronal, ectodermal), and alpha-fetoprotein (epithelial, endodermal) in RiPS clones from 3 independent RiPS derivations subjected to directed differentiation. Hematoxylin and eosin staining of BJ-, CF- and dH1F-RiPS-derived teratomas showing histological overview, ectoderm (pigmented epithelia (BJ and CF), neural rosettes (dH1F)), mesoderm (cartilage, all), and endoderm (gut-like endothelium, all). For blood formation and methylcellulose assays, n=3 for each clone. See also Figure S5.
Taken together, these data demonstrate by the most stringent criteria available to human pluripotent cells (Chan et al., 2009; Smith et al., 2009), that modified-RNA-derived iPS clones from multiple independent derivations were reprogrammed to pluripotency, and closely recapitulated the functional and molecular properties of human ES cells.
Modified RNAs generate iPS cells at very high efficiency
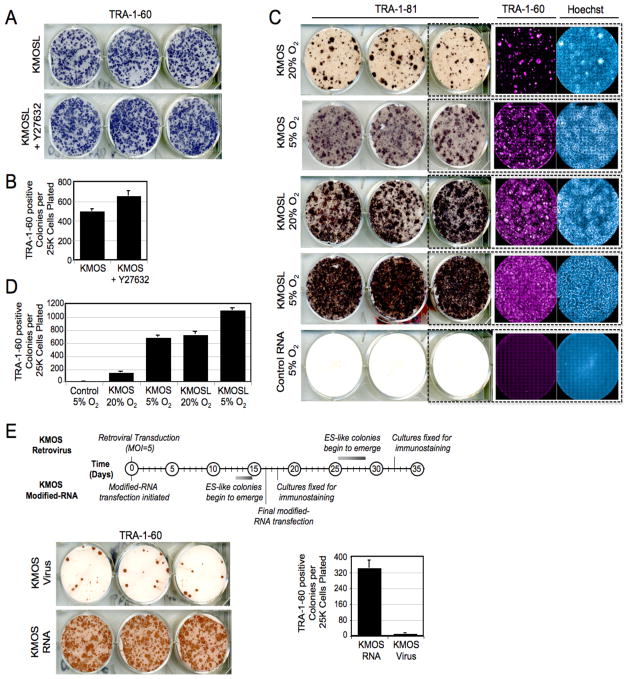
During the course of our experiments, we noted surprisingly high reprogramming efficiencies and rapid kinetics with which RiPS cells were generated. To quantify this more thoroughly, a number of reprogramming experiments were undertaken in which quantitative readout of efficiency was based on colony morphology and expression of the stringent pluripotency markers TRA-1-60 and TRA-1-81, (Chan et al., 2009; Lowry et al., 2008). In one set of experiments, BJ fibroblasts transfected with a five-factor modified-RNA cocktail (KMOSL), demonstrated an iPS conversion efficiency of over 2%, regardless of whether the cells were passaged in the presence or absence of Rho-associated kinase (ROCK), Y-27632 (Figure 5A–B, Table 1). This efficiency was two orders of magnitude higher than those typically reported for virus-based derivations. Moreover, in contrast to virus-mediated BJ-iPS derivations, in which iPS colonies typically take around 4 weeks to emerge, by day 17 of RNA transfection the plates had already become overgrown with ES-like colonies (Figure 5A).
Figure 5. Pluripotency induction by modified-RNAs is highly efficient.
(A) TRA-1-60 horseradish peroxidase (HRP) staining conducted at day 18 of a BJ-RiPS derivation with modified-RNAs encoding KMOSL and (B) frequency of TRA-1-60-positive colonies produced in the experiment relative to number of cells initially seeded. Error bars show s.d., n=6 for each condition. (C) TRA-181 HRP, TRA-160 immunofluorescence and Hoechst staining, and (D) colony frequencies for dH1f-RiPS experiments done using 4-factor (KMOS) and 5-factor (KMOSL) modified-RNA cocktails under 5% O2 or ambient oxygen culture conditions quantified at day 18. Control wells were transfected with equal doses of modified-RNA encoding GFP. (E) Kinetics and efficiency of retroviral and modified-RNA reprogramming. Timeline of colony formation (top panel), TRA-1-60 HRP immuno-staining (lower left panel), and TRA-1-60 positive colony counts (lower right panel) of dH1f cells reprogrammed using KMOS retroviruses (MOI=5 of each) or modified-RNA KMOS cocktails (n=3 for each condition). See also Figure S4.
Table 1. Quantification of reprogramming efficiency.
For each experimental condition, efficiency was calculated by dividing the average count of TRA-1-60-positive colonies per well by the initial number of cells plated, scaled to the fraction of cells replated in each well. Cultures were passaged at day 6 or 7 as indicated. The BJ experiment was started in a 10-cm dish, dH1f trials in individual wells of a 6-well plate. Colony counts are shown ± s.d., n=6, except in the RNA vs. Virus trial, where n=9 for virus, n=18 for RNA.
| Experiment | Cells plated | Split | Condition | Well fraction | Colonies/well | Efficiency (%) |
|---|---|---|---|---|---|---|
| BJ (KMOSL) | 300,000 | d7 | Y27632− | 1/24 | 249 ± 21 | 2.0 |
| Y27632+ | 1/24 | 326 ± 49 | 2.6 | |||
| 4-Factor (KMOS) vs. 5-Factor (KMOSL) | 50,000 | d6 | 4F 20% O2 | 1/6 | 48 ± 18 | 0.6 |
| 4F 5% O2 | 1/6 | 228 ± 30 | 2.7 | |||
| 5F 20% O2 | 1/6 | 243 ± 42 | 2.9 | |||
| 5F 5% O2 | 1/6 | 367 ± 38 | 4.4 | |||
| RNA vs. Virus (KMOS) | 100,000 | d6 | Virus | 1/3 | 13 ± 3.5 | 0.04 |
| RNA | 1/6 | 229 ± 39 | 1.4 |
We next evaluated the contributions of low-oxygen culture and LIN28 to the efficiency of RiPS derivation. The yield of TRA-1-60/TRA-1-81-positive colonies in the ambient (20%) oxygen condition was four-fold lower than in the cultures maintained at 5% O2 when using KMOS RNA, but this deficit was negated when LIN28 was added to the cocktail (Figure 5C–D, Table 1). The highest conversion efficiency (4.4%) was observed when low-oxygen culture and the five-factor KMOSL cocktail were combined.
To directly compare the kinetics and efficiency of our RiPS derivation protocol against an established viral protocol, we conducted an experiment in which dH1f fibroblasts were transfected with either KMOS modified-RNAs, or transduced with KMOS retroviruses in parallel. As had been observed in previous experiments, ES-like colonies began to emerge towards the end of the second week on the RNA-transfected cultures, and the plates became overgrown with ES-like colonies by the 16th and final day of transfection. By contrast, no ES-like colonies had appeared in the retrovirally transduced cultures by this timepoint, and colonies only began to emerge on the 24th day post-transduction, a time point consistent with previous reports describing iPS derivation by retroviruses (Lowry et al., 2008; Takahashi et al., 2007). The retroviral cultures were fixed for analysis on day 32. Both arms of the experiment were then immunostained for TRA-1-60 and colonies were counted. iPS derivation efficiencies were 1.4% and 0.04% for modified-RNA and retrovirus, respectively, corresponding to 36-fold higher conversion efficiency with the modified-RNA protocol (Figure 5E–F, Table 1). These experiments also revealed that the kinetics of modified-RNA iPS derivation were almost twice as fast as retroviral iPS derivation. Thus by the combined criteria of colony numbers and kinetics of reprogramming, the efficiency of modified-RNA iPS derivation greatly exceeds that of conventional retroviral approaches.
It should be noted that in the experiments described above, transfected fibroblast cultures were passaged once at an early time point (day 6 or 7) in order to promote fibroblast proliferation, which has been shown to facilitate reprogramming (Hanna et al., 2009). However, in preliminary experiments, RiPS cells were also efficiently derived from BJ and Detroit 551 fibroblasts in the absence of cell passaging indicating that splitting the culture during the reprogramming process was not required for modified-RNA iPS-derivation (Figure S4, and data not shown).
Utilization of modified-RNA to direct differentiation of pluripotent RiPS cells to a terminally-differentiated cell fate
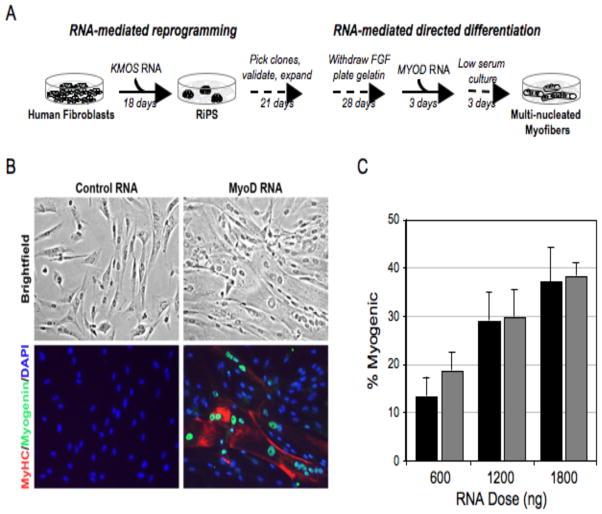
To realize the promise of iPS cell technology for regenerative medicine or disease modeling it is imperative that the multi-lineage differentiation potential of pluripotent cells be harnessed. Although progress has been made in directing the differentiation of pluripotent ES cells to various lineages by modulating the extracellular cytokine milieu, such protocols remain relatively inefficient. Given the high efficiency of iPS derivation by modified-RNAs, we reasoned that this technology might also be utilized to redirect pluripotent cells towards differentiated cell fates. To test this hypothesis, we subjected one of our validated RiPS lines to a simple in vitro differentiation protocol in which FGF was withdrawn, serum added, and the cells plated onto gelatin (Figure 6). Cells obtained under these conditions were then subjected to three consecutive days of transfection with a MYOD-encoding modified-RNA followed by an additional 3 days of culture in low serum conditions. The cultures were then fixed and immunostained for the myogenic markers myogenin and MyHC, which revealed a high percentage of large multi-nucleated myogenin and MyHC double positive myotubes (Figure 6).
Figure 6. Efficient directed differentiation of RiPS cells to terminally differentiated myogenic fate using modified-RNA.
(A) Schematic of experimental design. (B) Bright-field and immunostained images showing large, multi-nucleated, myosin heavy chain (MyHC) and myogenin positive myotubes in cells fixed three days after cessation of MYOD modified-RNA transfection. Modified-RNA encoding GFP was administered to the controls. (C) Penetrance of myogenic conversion relative to daily RNA dose. Black bars refer to an experiment in which cultures were plated at 104 cells/cm2, grey bars to cultures plated at 5x103 cells/cm2. Error bars show s.d. for triplicate wells.
Taken together, these experiments provide proof of principle that modified-RNAs can be used to both efficiently reprogram cells to a pluripotent state, and direct the fate of such cells to a terminally differentiated somatic cell type.
DISCUSSION
Using a combination of RNA modifications and a soluble interferon inhibitor to overcome innate anti-viral responses, we have developed a technology that enables highly efficient reprogramming of somatic cells to pluripotency, and can also be harnessed to direct the differentiation of pluripotent cells towards a desired lineage. Although it is relatively technically complex, the methodology described here offers several key advantages over established reprogramming techniques. By obviating the need to perform experiments under the stringent biological containment required for virus-based approaches, modified-RNA technology should make reprogramming accessible to a wider community of researchers. More fundamentally, since our technology is RNA-based, it completely eliminates the risk of genomic integration and insertional mutagenesis inherent to all DNA-based methodologies, including those that are ostensibly non-integrating. Moreover, our approach allows protein stoichiometry to be exquisitely regulated within cultures while avoiding the stochastic variation of expression typical of integrating vectors, as well as the uncontrollable effects of viral silencing. Given the stepwise character of the phenotypic changes observed during pluripotency induction (Chan et al., 2009; Smith et al., 2010), it seems likely that individual transcription factors play distinct, stage-specific roles during reprogramming. The unprecedented potential for temporal control over individual factor expression afforded by our technology should help researchers unravel these nuances, yielding insights that can be applied to further enhance the efficiency and kinetics of reprogramming.
The risk of mutagenesis is not the only safety concern holding back clinical exploitation of induced pluripotency, and it has become increasingly apparent that all iPS cells are not created equal with respect to epigenetic landscape and developmental plasticity (Hu et al., 2010; Miura et al., 2009). In this regard, we have applied the most stringent molecular and functional criteria for reprogramming human cells to pluripotency (Chan et al., 2009; Smith et al., 2009). Our results demonstrate that modified-RNA derived iPS clones from multiple independent derivations were fully reprogrammed to pluripotency, and that the resulting cells very closely recapitulated the functional and molecular properties of human ES cells. Our observation that modified-RNA derived iPS cells more faithfully recapitulated the global transcriptional signature of human ES cells than retrovirally-derived iPS cells is important as it suggests that RNA reprogramming may produce higher quality iPS cells, possibly owing to the fact that they are transgene-free.
The transient and non-mutagenic character of RNA-based protein expression could also deliver important clinical benefits outside the domain of lineage reprogramming. Indeed, the use of RNA transfection to express cancer or pathogen antigens for immunotherapy is already an active research area (Rabinovich et al., 2008; Rabinovich et al., 2006; Van den Bosch et al., 2006; Weissman et al., 2000), and such approaches may benefit from the non-immunogenic properties of modified-RNAs. One can readily envisage employing modified-RNA to transiently express surface proteins such as homing receptors to target cellular therapies toward specific organs, tissues, or diseased cells.
For tissue engineering to progress further into the clinic, there is a pressing need for safe and efficient means to redirect cell fate. This is doubly apparent when one considers that iPS cells are only a starting point for patient-specific therapies, and specification of clinically useful cell types is still required to produce autologous tissues for transplantation or for disease modeling. Importantly, we have demonstrated that our modified-RNA-based technology enables highly efficient reprogramming, and that it can equally be applied to efficiently redirect pluripotent cell fate to terminally differentiated fates without compromising genomic integrity. In light of these considerations, we believe that our approach has the potential to become a major enabling technology for cell-based therapies and regenerative medicine.
EXPERIMENTAL PROCEDURES
Construction of IVT templates
The pipeline for production of IVT template constructs and subsequent RNA synthesis is schematized in Figure S1. The oligonucleotide sequences used in the construction of IVT templates are shown in Table S2. All oligos were synthesized by Integrated DNA Technologies (Coralville, IA). ORF PCRs were templated from plasmids bearing human KLF4, c-MYC, OCT4, SOX2, human ES cDNA (LIN28), Clontech pIRES-eGFP (eGFP), pRVGP (d2eGFP) and CMV-MyoD from Addgene. The ORF of the low-stability nuclear GFP was constructed by combining the d2eGFP ORF with a 3′ nuclear localization sequence. PCR reactions were performed using HiFi Hotstart (KAPA Biosystems, Woburn, MA) per the manufacturer’s instructions. Splint-mediated ligations were carried out using Ampligase Thermostable DNA Ligase (Epicenter Biotechnologies, Madison, WI). UTR ligations were conducted in the presence of 200 nM UTR oligos and 100 nM splint oligos, using 5 cycles of the following annealing profile: 95°C for 10 seconds; 45°C for 1 minute; 50°C for 1 minute; 55°C for 1 minute; 60°C for 1 minute. A phosphorylated forward primer was employed in the ORF PCRs to facilitate ligation of the top strand to the 5′ UTR fragment. The 3′ UTR fragment was also 5′-phosphorylated using polynucleotide kinase (New England Biolabs, Ipswich, MA). All intermediate PCR and ligation products were purified using QIAquick spin columns (Qiagen, Valencia, CA) before further processing. Template PCR amplicons were sub-cloned using the pcDNA 3.3-TOPO TA cloning kit (Invitrogen, Carlsbad, CA). Plasmid inserts were excised by restriction digest and recovered with SizeSelect gels (Invitrogen) before being used to template tail PCRs.
Synthesis of modified-RNA
RNA was synthesized with the MEGAscript T7 kit (Ambion, Austin, TX), using 1.6 ug of purified tail PCR product to template each 40 uL reaction. A custom ribonucleoside blend was used comprising 3′-0-Me-m7G(5′)ppp(5′)G ARCA cap analog (New England Biolabs), adenosine triphosphate and guanosine triphosphate (USB, Cleveland, OH), 5-methylcytidine triphosphate and pseudouridine triphosphate (TriLink Biotechnologies, San Diego, CA). Final nucleotide reaction concentrations were 33.3 mM for the cap analog, 3.8 mM for guanosine triphosphate, and 18.8 mM for the other nucleotides. Reactions were incubated 3–6 hours at 37°C and DNAse-treated as directed by the manufacturer. RNA was purified using Ambion MEGAclear spin columns, then treated with Antarctic Phosphatase (New England Biolabs) for 30 minutes at 37°C to remove residual 5′-triphosphates. Treated RNA was re-purified, quantitated by Nanodrop (Thermo Scientific, Waltham, MA), and adjusted to 100 ng/uL working concentration by addition of Tris-EDTA (pH 7.0). RNA reprogramming cocktails were prepared by pooling individual 100 ng/uL RNA stocks to produce a 100 ng/uL (total) blend. The KMOS[L]+GFP cocktails were formulated to give equal molarity for each component except for OCT4, which was included at 3x molar concentration. Volumetric ratios used for pooling were as follows: 170:160:420:130:120[:90] (KLF4:c-MYC:OCT4:SOX2:GFP[:LIN28]).
Cells
Primary human neonatal epidermal keratinocytes, BJ human neonatal foreskin fibroblasts, MRC-5 human fetal lung fibroblasts, and Detroit 551 human fetal skin fibroblasts were obtained from ATCC (Manassas, VA). CF cells were obtained with informed consent from a skin biopsy taken from an adult cystic fibrosis patient. dH1f fibroblasts were sub-cloned from fibroblasts produced by directed differentiation of the H1-OGN human ES cell line as previously described (Park et al., 2008). BGO1 hES cells were obtained from BresaGen (Athens, GA). H1 and H9 hES cells were obtained from WiCell (Madison, Wi).
RNA transfection
RNA transfections were carried out using RNAiMAX (Invitrogen) or TransIT-mRNA (Mirus Bio, Madison, WI) cationic lipid delivery vehicles. RNAiMAX was used for RiPS derivations, the RiPS-to-myogenic conversion, and for the multiple cell-type transfection experiment documented in Figure S2. All other transfections were performed with TransIT-mRNA. For RNAiMAX transfections, RNA and reagent were first diluted in Opti-MEM basal media (Invitrogen). 100 ng/uL RNA was diluted 5x and 5 uL of RNAiMAX per microgram of RNA was diluted 10x, then these components were pooled and incubated 15 minutes at room temperature (RT) before being dispensed to culture media. For TransIT-mRNA transfections, 100 ng/uL RNA was diluted 10x in Opti-MEM and BOOST reagent was added (2 uL per microgram of RNA), then TransIT-mRNA was added (2 uL per microgram of RNA), and the RNA-lipid complexes were delivered to culture media after a 2-minute incubation at RT. RNA transfections were performed in Nutristem xeno-free hES media (Stemgent, Cambridge, MA) for RiPS derivations, Dermal Cell Basal Medium plus Keratinocyte Growth Kit (ATCC) for keratinocyte experiments, and Opti-MEM plus 2% FBS for all other experiments described. The B18R interferon inhibitor (eBioscience, San Diego, CA) was used as a media supplement at 200 ng/mL.
qRT-PCR
Transfected cells were lysed using 400 uL CellsDirect reagents (Invitrogen), and 20 uL of each lysate was taken forward to a 50 uL reverse transcription reaction using the VILO cDNA synthesis kit (Invitrogen). Reactions were purified on QIAquick columns (Qiagen). qRT-PCR reactions were performed using SYBR FAST qPCR supermix (KAPA Biosystems).
Reprogramming to pluripotency
Gamma-irradiated human neonatal fibroblast feeders (GlobalStem, Rockville, MD) were seeded at 33,000 cells/cm2. Nutristem media was replaced daily, four hours after transfection, and supplemented with 100 ng/mL bFGF and 200 ng/mL B18R (eBioscience, San Diego, CA). Where applied, VPA was added to media at 1 mM final concentration on days 8–15 of reprogramming. Low-oxygen experiments were carried out in a NAPCO 8000 WJ incubator (Thermo Scientific). Media were equilibrated at 5% O2 for approximately 4 hours before use. Cultures were passaged using TrypLE Select recombinant protease (Invitrogen). Y27632 ROCK inhibitor (Watanabe et al., 2007) was used at 10 uM in recipient plates until the next media change. The daily RNA dose applied in the RiPS derivations was 1200 ng per well (6-well plate format) or 8 ug to a 10-cm dish.
For RNA vs. retrovirus experiments, starting cultures were seeded with 100,000 cells in individual wells of a 6-well plate using fibroblast media (DMEM+10% FBS). The following day (day 1) KMOS RNA transfections were initiated in the RNA plate, and the viral plate was transduced with a KMOS retroviral cocktail (MOI=5 for each virus). All wells were passaged on day 6, using split ratios of 1:6 for the RNA wells and 1:3 for the virus wells. The conditions applied in the RNA arm of the trial were as in the initial RiPS derivation, including the use of Nutristem supplemented with 100 ng/mL bFGF, 5% O2 culture, and human fibroblast feeders. Ambient oxygen tension and other conventional iPS derivation conditions were used in the viral arm, the cells being grown in fibroblast media without feeders until the day 6 split, then being replated onto CF1 MEF feeders (GlobalStem) with a switch to hES media based on Knockout Serum Replacement (Invitrogen) supplemented with 10 ng/mL bFGF.
RiPS cell culturing
RiPS colonies were mechanically picked and transferred to MEF-coated 24-well plates with standard hES medium containing 5 uM Y27632 (BioMol, Plymouth Meeting, PA). The hES media comprised DMEM/F12 supplemented with 20% Knockout Serum Replacement (Invitrogen), 10 ng/mL of bFGF (Gembio, West Sacramento, CA), 1x non-essential amino acids (Invitrogen), 0.1mM β-ME (Sigma), 1mM L-glutamine (Invitrogen), plus antibiotics. Clones were mechanically passaged once more to MEF-coated 6-well plates, and then expanded using enzymatic passaging with collagenase IV (Invitrogen). For RNA and DNA preparation, cells were plated onto hES-qualified Matrigel (BD Biosciences) in mTeSR (Stem Cell Technologies, Vancouver, BC), and further expanded by enzymatic passaging using dispase (Stem Cell Technologies).
Immunostaining
Cells were fixed in 4% paraformaldehyde for 20 minutes. Washed cells were treated with 0.2% Triton X (Sigma) in PBS for 30 minutes. Cells were blocked with 3% BSA (Invitrogen) and 5% donkey serum (Sigma) for 2 hours at RT. Cells were stained in blocking buffer with primary antibodies at 4°C overnight. Cells were washed and stained with secondary antibodies and 1 ug/mL Hoechst 33342 (Invitrogen) in blocking buffer for 3 hours at 4°C or for 1 hour at RT, protected from light. Antibodies were used, at 1:100 dilution: TRA-1-60-Alexa Fluor 647, TRA-1-81-Alexa Fluor 488, SSEA-4-Alexa Fluor 647, and SSEA-3-Alexa 488 (BD Biosciences). Primary OCT4 and NANOG antibodies (Abcam, Cambridge, MA) were used at 0.5 ug/mL, and an anti-rabbit IgG Alexa Fluor 555 (Invitrogen) was used as the secondary. Images were acquired with a Pathway 435 bioimager (BD Biosciences). Live imaging was performed as described previously (Chan et al., 2009). For pluripotency factor time course experiments, transfected human keratinocytes were trypsinized, washed with PBS, and fixed in 4% paraformaldehyde for 10 minutes. Fixed cells were washed with 0.1M glycine, then blocked and permeabilized in PBS/0.5% saponin/1% goat serum (Rockland Immunochemicals, Gilbertsville, PA) for 20 minutes. Cells were incubated for 1 hour at RT with 1:100 diluted primary antibodies for KLF4, OCT4, SOX2 (Stemgent), washed, then for 45 minutes at RT with 1:200-diluted DyLight 488-labeled secondary antibodies (goat anti-mouse IgG+IgM and goat anti-rabbit IgG). Cells were suspended in PBS and analyzed by flow cytometry.
Gene expression analysis
RNA was isolated using the RNeasy kit (Qiagen) according to the manufacturer’s instructions. First-strand cDNA was primed with oligo(dT) primers and qPCR was performed with primer sets as described previously (Park et al., 2008), using Brilliant SYBR Green master mix (Stratagene, La Jolla, CA). For the microarray analysis, RNA probes were prepared and hybridized to Human Genome U133 Plus 2.0 oligonucleotide microarrays (Affymetrix, Santa Clara, CA) per the manufacturer’s instructions. Arrays were processed by the Coriell Institute Genotyping and Microarray Center (Camden, NJ). Microarray data will be uploaded to the GEO database at the time of publication. Gene expression levels were normalized with the Robust Multichip Average (RMA) algorithm. Hierarchical clustering was performed using the Euclidean distance with average linkage method. The similarity metric for comparison between different cell lines is indicated on the height of cluster dendrogram.
Bisulfite sequencing
DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer’s protocol. Bisulfite treatment of genomic DNA was carried out using EZ DNA Methylation™ Kit (Zymo Research, Orange, CA) according to the manufacturer’s protocol. For pyrosequencing analysis, the bisulfite treated DNA was first amplified by HotStar Taq Polymerase (Qiagen) for 45 cycles of (95°C 30 s; 53°C 30 s; 72°C 30 s). The analysis was performed by EpigenDx using the PSQ™96HS system according to standard procedures using primers that were developed by EpigenDx for the CpG sites at positions (−50) to (+96) from the start codon of the OCT4 gene.
Tri-lineage differentiation
Embryoid body (EB) hematopoietic differentiation was performed as previously described (Chadwick et al., 2003). Briefly, RiPS cells and hES cell controls were passaged with collagenase IV and transferred (3:1) in differentiation medium to 6-well low-attachment plates and placed on a shaker in a 37°C incubator overnight. Starting the next day, media was supplemented with the following hematopoietic cytokines: 10 ng/mL of interleukin-3 (R&D Systems, Minneapolis, MN) and interleukin-6 (R&D), 50 ng/mL of G-CSF (Amgen, Thousand Oaks, CA) and BMP-4 (R&D), and 300 ng/mL of SCF (Amgen) and Flt-3 (R&D). Media was changed every 3 days. On day 14 of differentiation, EBs were dissociated with collagenase B (Roche, Indianapolis, IN). 2x104 differentiated cells were plated into methylcellulose H4434 (Stem Cell Technologies) and transferred using a blunt needle onto 35mm dishes (Stem Cell Technologies) in triplicate and incubated at 37°C and 5°CO2 for 14 days. Colony Forming Units (CFUs) were scored based on morphological characteristics.
For neuronal differentiation, cells were differentiated at 70% confluency as a monolayer in neuronal differentiation medium (DMEM/F12, Glutamax 1%, B27-Supplement 1%, N2-Supplement 2%, P/S 1% and noggin 20ng/ml). After 7 days neuronal structures were visible. For endoderm differentiation (AFP stain), cells were differentiated as a monolayer in endoderm differentiation medium (DMEM, B27(-RA) and 100 ng/ml activin-a) for 7 days, then switched to growth medium (DMEM, 10% FBS, 1% P/S) and continued differentiation for 7 days. Antibodies used were as follows: Anti-β-Tubulin III (Tuj1) rabbit anti-human (Sigma, St. Louis, MO), 1:500; AFP (h-140) rabbit polyclonal IgG, (Santa Cruz Biotechnology, Santa Cruz, CA), 1:100 dilution. Secondary antibodies were conjugated to Alexa Fluor 488, or Alexa Fluor 594.
For cardiomyocyte differentiation, colonies were digested at 70% confluency using dispase and placed in suspension culture for embryoid body (EB) formation in differentiation medium (DMEM, 15% FBS, 100 uM ascorbic acid). After 11 days, EBs were plated to adherent conditions using gelatin and the same medium. Beating cardiomyocytes were observed 3 days after replating.
For teratomas, 2.5x106 cells were spun down, and all excess media was removed. In 20-week old female SCID mice, the capsule of the right kidney was gently elevated, and one droplet of concentrated cells was inserted under the capsule. Tumors harvested at 6–12 weeks were fixed in 4% PFA, run through an ethanol gradient, and stored in 70% ethanol. Specimens were sectioned and stained with H&E.
Myogenic differentiation of RiPS cells
Validated RiPS cells were plated into wells coated with 0.1% gelatin (Millipore, Billerica, MA), and cultured in DMEM+10% FBS for 4 weeks with passaging every 4–6 days using trypsin. The culture media was switched to Opti-MEM+2% FBS, and the cells were transfected with modified-RNA encoding either murine MYOD or GFP the following day, and for the following two days. Media was supplemented with B18R, and replaced 4 hours after each transfection. After the third and final transfection, the media was switched to DMEM+3% horse serum, and cultures were incubated for a further 3 days. Cells were then fixed in 4% PFA and immuno-stained as previously described (Shea et al., 2010). The percentage of myogenin-positive nuclei/total nuclei and nuclei/MyHC-positive myotubes was quantified, with a minimum of 500 nuclei counted per condition.
Technical Notes
Although reprogramming and directed differentiation using modified-RNAs are efficient processes, the protocols involved are nonetheless multi-stepped and complex. It is therefore advised that in efforts to apply this methodology, all steps of the protocols described herein are followed rigorously and quality controlled. Foremost amongst these: templates for RNA synthesis must be sequenced, and production of in vitro transcribed modified-RNAs must be quality controlled by gel electrophoresis and spectrophotometry. Critically, the expression of proteins with modified RNAs must be confirmed by immuno-staining. Modified RNAs must also be tested for immunogenicity at multiple points throughout the course of the experiment. Successful daily transfection must be monitored by inclusion of modified-RNA encoding a fluorescent reporter throughout the course of experiments. Any reagents involved in supporting pluripotency induction (media, feeder cells etc) should be tested for their ability to support the growth of pluripotent cells prior to the start of experiments. As is true of reprogramming by other methods, the quality of the starting cells (eg. passage number) impacts reprogramming using our technology.
Supplementary Material
Supplemental Figure S1 RNA production flowchart. To construct a template for RNA transcription reactions, the ORF of a gene of interest is first PCR amplified from a cDNA. Long oligonucleotides containing UTR sequences are then joined to the top strand of ORF amplicons by a thermostable DNA ligase, mediated by annealing to splint oligos which bring the desired single-stranded DNA (ssDNA) ends together. A T7 promoter is incorporated in the 5′ UTR fragment. The ssDNA product is amplified using generic primers and TA cloned. A polyA tail is added with a PCR reaction using a T120-heeled reverse primer, and the amplicons are used to template IVT reactions. Modified and unmodified nucleobases are used in the IVT reaction. An anti-reverse di-guanosine cap analog (ARCA) is included in the IVT reaction at four-fold higher concentration than guanosine triphosphate (GTP), as a result of which an estimated 80% of the product is capped. Spin-column purified IVT product is DNase-treated to eliminate the DNA template. Treatment with a phosphatase is used to remove immunonogenic 5′ triphosphate moieties from the uncapped RNA fraction. The completed modified-RNA is then re-purified for use in transfections. This figure supports data presented in main Figure 1.
Supplemental Figure S2 Penetrant and sustained protein expression mediated by modified-RNA transfection in diverse human cell types, and effects on cell viability and global gene expression. (A) Flow cytometry analysis showing penetrance of GFP expression 24-hour post-transfection of six human cell types transfected with 1000ng of modified-RNA encoding GFP. Cell types included: human epidermal keratinocytes (HEKs), adipose-derived stem cells (ADSCs), and four different human fibroblast types (BJ, Detroit 551, MRC-5 and dH1f). Error bars show s.d. for triplicate wells. (B) Expression time courses for cells transfected with modified-RNAs encoding high- and low-stability GFP variants (eGFP and d2eGFP, respectively), assayed by flow cytometry. (C) Annexin V staining at indicated days of BJ fibroblasts transfected daily over the course of 10 days. (D) Heatmap data from microarray analysis of BJ fibroblasts transfected for 10 consecutive days with modified-RNA encoding GFP, vehicle, or untransfected controls. A number of cell stress pathways are shown demonstrating that prolonged transfection with modified RNA does not significantly impact gene the molecular profile of transfected cells beyond upregulation of a number of interferon/NFκB highlighted in (E). (E) All genes upregulated greater than 2-fold in modified-RNA transfected cells versus untransfected cells (right) or vehicle transfected (left) showing induction of number of interferon/NFκB signaling genes consistent with the near but not absolute attenuation of interferon response shown in main Figure 1D. This figure supports data presented in main Figure 1.
Supplemental Figure S3 iPS-derivation from five human cell types. (A) Expression time course of low-stability nuclear GFP after a single transfection into keratinocytes, assessed by flow cytometry (right panel). Bright-field and GFP images (right panel) taken at four different time points during a reprogramming experiment. RNA-encoding the low-stability GFP analyzed in the left panel was spiked into the reprogramming cocktail (KMOSL) to visualize sustained protein expression from transfected modified-RNAs during iPS reprogramming (right panel). (B) Antibody stains of independent RiPS clones derived from cells taken from an adult cystic fibrosis patient (CF cells), BJ postnatal fibroblasts, MRC-5 and Detroit 551 fetal fibroblasts, and human ES-derived dH1f fibroblasts. Panels show cell-surface staining for SSEA-3, SSEA-4, TRA-1-60 and TRA-1-81, and intracellular staining for OCT4 and NANOG. Control stains of BG01 hES cells, dH1f and BJ fibroblasts are shown. Additional control stains show the specificity of the secondary antibody used for the OCT4 and NANOG intracellular stains. This figure supports data presented in main Figure 2.
Supplemental Figure S4 Efficient RiPS derivation from BJ fibroblasts without passaging. (A) Immunohistochemistry showing expression of pluripotency markers SSEA-4 and TRA-1-60 in a BJ fibroblast reprogramming experiment transfected for 16 days with 600ng per day of a KMOSL modified RNA cocktail containing a destabilized GFP spike-in. Cultures were fixed for staining at day 18. 50,000 BJ cells were originally seeded onto feeder cells and went unpassaged throughout the course of the experiment. (B) Quantification of TRA-1-60 colony count relative to the number of cells seeded. This figure supports data shown in main Figure 5.
Supplemental Figure S5 Teratoma formation and trilineage differentiation of modified-RNA derived iPS clones in vivo.
(A) Histology of hematoxylin and eosin stained teratomas from the indicated BJ and dH1F RiPS derivations showing structures derived from the three germ layers. This figure supports data shown in main Figure 4.
Supplemental Table 1: Pluripotency validation assays performed in this study. The table shows the RiPS clones that were validate in each assay. # Validated for immuno-staining for all of TRA-1-60, TRA-1-80, SSEA3, SSEA4, OCT4, NANOG. Ω Demethylation of the OCT4 promoter. In vitro differentiation including ^embryoid body formation, øtrilineage by directed differentiation, † beating cardiomyocytes, and * blood formation by CFC assays in methylcellulose. This table supports data presented in main Figure 4.
Supplemental Table S2 Oligonucleotides for IVT template construction. 5′ and 3′ UTR oligos are ligated to the top strand of gene-specific ORF amplicons to produce a basic template construct for cloning (Supplemental Figure S1). Underlined bases in the 5′ UTR oligo sequence indicate the upstream T7 promoter, and in the 3′ UTR oligo sequence show downstream restriction sites, introduced to facilitate linearization of template plasmids. Template PCR primers are used to amplify ligation products for sub-cloning. Tail PCR primers are used to append an oligo(dT) sequence immediately after the 3′ UTR to drive templated addition of a poly(A) tail during IVT reactions. Gene-specific ORF primers are used to capture the coding region (minus the start codon) from cDNA templates. Splint oligos mediate ligation of UTR oligos to the top strand of ORF amplicons. This table supports data in main Figure 2.
Supplemental Table S3 Primers for qRT-PCR analysis of interferon-regulated genes. This table supports data in main Figure 1.
Supplemental Movie 1 Embryoid bodies from independent RiPS derivations showing beating cardiomyocytes. Top row (left to right) BJ-RiPS1.1, BJ-RiPS1.2 and BJ-RiPS1.3 Middle row (left to right) CF-RiPS1.3, D551-RiPS1.2, MRC5-RiPS1.8, Bottom row (left to right) CF-RiPS1.4, D551-RiPS1.3, MRC5-RiPS1.9. This movie supports main figure 4.
Acknowledgments
The authors wish to thank Sun Hur, Victor Li, Laurence Daheron, Odelya Hartung, Alys Peisley, Sutheera Ratanasirintrawoot, Brad Hamilton, Chenmei Luo, Jonathan Kagan, Julie Sahalie, Alejandro De Los Angeles and Lior Zangi, for help, insight and suggestions. D.J.R., A.M., G.Q.D, A.S.B, C.C., and T.S.M. were supported by grants from the Harvard Stem Cell Institute. Y-H.L. is supported by the A*Star Institute of Medical Biology and Singapore stem cell consortium. T.A. was supported by the Roberto and Allison Mignone Fund for Stem Cell Research. J.J.C. is supported by Howard Hughes Medical Institute, SysCODE (Systems-based Consortium for Organ Design & Engineering) and NIH grant # RL1DE019021.
References
- Audouy S, Hoekstra D. Cationic lipid-mediated transfection in vitro and in vivo (review) Molecular membrane biology. 2001;18:129–143. [PubMed] [Google Scholar]
- Chadwick K, Wang L, Li L, Menendez P, Murdoch B, Rouleau A, Bhatia M. Cytokines and BMP-4 promote hematopoietic differentiation of human embryonic stem cells. Blood. 2003;102:906–915. doi: 10.1182/blood-2003-03-0832. [DOI] [PubMed] [Google Scholar]
- Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H, Miller JD, Hartung O, Rho J, Ince TA, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- Chang CW, Lai YS, Pawlik KM, Liu K, Sun CW, Li C, Schoeb TR, Townes TM. Polycistronic Lentiviral Vector for “Hit and Run” Reprogramming of Adult Skin Fibroblasts to Induced Pluripotent Stem Cells. Stem Cells. 2009;27:1042–1049. doi: 10.1002/stem.39. [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate Antiviral Responses by Means of TLR7-Mediated Recognition of Single-Stranded RNA. Science (New York, NY. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Elango N, Elango S, Shivshankar P, Katz MS. Optimized transfection of mRNA transcribed from a d(A/T)100 tail-containing vector. Biochemical and Biophysical Research Communications. 2005;330:958–966. doi: 10.1016/j.bbrc.2005.03.067. [DOI] [PubMed] [Google Scholar]
- Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proceedings of the Japan Academy. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtkamp S, Kreiter S, Selmi A, Simon P, Koslowski M, Huber C, Tureci O, Sahin U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108:4009–4017. doi: 10.1182/blood-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA Is the Ligand for RIG-I. Science (New York, NY. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Hu BY, Weick JP, Yu J, Ma LX, Zhang XQ, Thomson JA, Zhang SC. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nature biotechnology. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, Kay MA, et al. A nonviral minicircle vector for deriving human iPS cells. Nature methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Kariko K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariko K, Weissman D. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Current opinion in drug discovery & development. 2007;10:523–532. [PubMed] [Google Scholar]
- Kawai T, Akira S. Antiviral signaling through pattern recognition receptors. Journal of biochemistry. 2007;141:137–145. doi: 10.1093/jb/mvm032. [DOI] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, et al. Generation of Human Induced Pluripotent Stem Cells by Direct Delivery of Reprogramming Proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, Kain SR. Generation of Destabilized Green Fluorescent Protein as a Transcription Reporter. J Biol Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proceedings of the National Academy of Sciences. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Okada Y, Aoi T, Okada A, Takahashi K, Okita K, Nakagawa M, Koyanagi M, Tanabe K, Ohnuki M, et al. Variation in the safety of induced pluripotent stem cell lines. Nature biotechnology. 2009;27:743–745. doi: 10.1038/nbt.1554. [DOI] [PubMed] [Google Scholar]
- Nallagatla SR, Bevilacqua PC. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA (New York, NY. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallagatla SR, Toroney R, Bevilacqua PC. A brilliant disguise for self RNA: 5′-end and internal modifications of primary transcripts suppress elements of innate immunity. RNA biology. 2008;5 doi: 10.4161/rna.5.3.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science (New York, NY. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- Papapetrou EP, Tomishima MJ, Chambers SM, Mica Y, Reed E, Menon J, Tabar V, Mo Q, Studer L, Sadelain M. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proceedings of the National Academy of Sciences. 2009;106:12759–12764. doi: 10.1073/pnas.0904825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-Mediated Antiviral Responses to Single-Stranded RNA Bearing 5′-Phosphates. Science (New York, NY. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Rabinovich PM, Komarovskaya ME, Wrzesinski SH, Alderman JL, Budak-Alpdogan T, Karpikov A, Guo H, Flavell RA, Cheung NK, Weissman SM, et al. Chimeric Receptor mRNA Transfection as a Tool to Generate Antineoplastic Lymphocytes. Human gene therapy. 2008 doi: 10.1089/hum.2008.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich PM, Komarovskaya ME, Ye ZJ, Imai C, Campana D, Bahceci E, Weissman SM. Synthetic messenger RNA as a tool for gene therapy. Human gene therapy. 2006;17:1027–1035. doi: 10.1089/hum.2006.17.1027. [DOI] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Shea KL, Xiang W, LaPorta VS, Licht JD, Keller C, Basson MA, Brack AS. Sprouty1 regulates reversible quiescence of a self-renewing adult muscle stem cell pool during regeneration. Cell Stem Cell. 2010;6:117–129. doi: 10.1016/j.stem.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KP, Luong MX, Stein GS. Pluripotency: toward a gold standard for human ES and iPS cells. J Cell Physiol. 2009;220:21–29. doi: 10.1002/jcp.21681. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotech. 2010 doi: 10.1038/nbt.1632. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science (New York, NY. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons JA, Alcami A, Smith GL. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell. 1995;81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Uematsu S, Akira S. Toll-like Receptors and Type I Interferons. J Biol Chem. 2007;282:15319–15323. doi: 10.1074/jbc.R700009200. [DOI] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol, JVI. 2009:02449–02408. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bosch GA, Van Gulck E, Ponsaerts P, Nijs G, Lenjou M, Apers L, Kint I, Heyndrickx L, Vanham G, Van Bockstaele DR, et al. Simultaneous Activation of Viral Antigen-specific Memory CD4+ and CD8+ T-cells Using mRNA-electroporated CD40-activated Autologous B-cells. Journal of Immunotherapy. 2006;29:512–523. doi: 10.1097/01.cji.0000210385.48327.1e. 510.1097/1001.cji.0000210385.0000248327.0000210381e. [DOI] [PubMed] [Google Scholar]
- Van Tendeloo VF, Ponsaerts P, Lardon F, Nijs G, Lenjou M, Van Broeckhoven C, Van Bockstaele DR, Berneman ZN. Highly efficient gene delivery by mRNA electroporation in human hematopoietic cells: superiority to lipofection and passive pulsing of mRNA and to electroporation of plasmid cDNA for tumor antigen loading of dendritic cells. Blood. 2001;98:49–56. doi: 10.1182/blood.v98.1.49. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Nishikawa S-i, Muguruma K, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotech. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Weissman D, Ni H, Scales D, Dude A, Capodici J, McGibney K, Abdool A, Isaacs SN, Cannon G, Kariko K. HIV gag mRNA transfection of dendritic cells (DC) delivers encoded antigen to MHC class I and II molecules, causes DC maturation, and induces a potent human in vitro primary immune response. J Immunol. 2000;165:4710–4717. doi: 10.4049/jimmunol.165.8.4710. [DOI] [PubMed] [Google Scholar]
- Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009 doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yisraeli JK, Melton DA, James EDaJNA. Methods in Enzymology. Academic Press; 1989. Synthesis of long, capped transcripts in Vitro by SP6 and T7 RNA polymerases; pp. 42–50. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia Enhances the Generation of Induced Pluripotent Stem Cells. 2009. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin, Thomson JA. Human Induced Pluripotent Stem Cells Free of Vector and Transgene Sequences. Science (New York, NY. 2009:1172482. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science (New York, NY. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1 RNA production flowchart. To construct a template for RNA transcription reactions, the ORF of a gene of interest is first PCR amplified from a cDNA. Long oligonucleotides containing UTR sequences are then joined to the top strand of ORF amplicons by a thermostable DNA ligase, mediated by annealing to splint oligos which bring the desired single-stranded DNA (ssDNA) ends together. A T7 promoter is incorporated in the 5′ UTR fragment. The ssDNA product is amplified using generic primers and TA cloned. A polyA tail is added with a PCR reaction using a T120-heeled reverse primer, and the amplicons are used to template IVT reactions. Modified and unmodified nucleobases are used in the IVT reaction. An anti-reverse di-guanosine cap analog (ARCA) is included in the IVT reaction at four-fold higher concentration than guanosine triphosphate (GTP), as a result of which an estimated 80% of the product is capped. Spin-column purified IVT product is DNase-treated to eliminate the DNA template. Treatment with a phosphatase is used to remove immunonogenic 5′ triphosphate moieties from the uncapped RNA fraction. The completed modified-RNA is then re-purified for use in transfections. This figure supports data presented in main Figure 1.
Supplemental Figure S2 Penetrant and sustained protein expression mediated by modified-RNA transfection in diverse human cell types, and effects on cell viability and global gene expression. (A) Flow cytometry analysis showing penetrance of GFP expression 24-hour post-transfection of six human cell types transfected with 1000ng of modified-RNA encoding GFP. Cell types included: human epidermal keratinocytes (HEKs), adipose-derived stem cells (ADSCs), and four different human fibroblast types (BJ, Detroit 551, MRC-5 and dH1f). Error bars show s.d. for triplicate wells. (B) Expression time courses for cells transfected with modified-RNAs encoding high- and low-stability GFP variants (eGFP and d2eGFP, respectively), assayed by flow cytometry. (C) Annexin V staining at indicated days of BJ fibroblasts transfected daily over the course of 10 days. (D) Heatmap data from microarray analysis of BJ fibroblasts transfected for 10 consecutive days with modified-RNA encoding GFP, vehicle, or untransfected controls. A number of cell stress pathways are shown demonstrating that prolonged transfection with modified RNA does not significantly impact gene the molecular profile of transfected cells beyond upregulation of a number of interferon/NFκB highlighted in (E). (E) All genes upregulated greater than 2-fold in modified-RNA transfected cells versus untransfected cells (right) or vehicle transfected (left) showing induction of number of interferon/NFκB signaling genes consistent with the near but not absolute attenuation of interferon response shown in main Figure 1D. This figure supports data presented in main Figure 1.
Supplemental Figure S3 iPS-derivation from five human cell types. (A) Expression time course of low-stability nuclear GFP after a single transfection into keratinocytes, assessed by flow cytometry (right panel). Bright-field and GFP images (right panel) taken at four different time points during a reprogramming experiment. RNA-encoding the low-stability GFP analyzed in the left panel was spiked into the reprogramming cocktail (KMOSL) to visualize sustained protein expression from transfected modified-RNAs during iPS reprogramming (right panel). (B) Antibody stains of independent RiPS clones derived from cells taken from an adult cystic fibrosis patient (CF cells), BJ postnatal fibroblasts, MRC-5 and Detroit 551 fetal fibroblasts, and human ES-derived dH1f fibroblasts. Panels show cell-surface staining for SSEA-3, SSEA-4, TRA-1-60 and TRA-1-81, and intracellular staining for OCT4 and NANOG. Control stains of BG01 hES cells, dH1f and BJ fibroblasts are shown. Additional control stains show the specificity of the secondary antibody used for the OCT4 and NANOG intracellular stains. This figure supports data presented in main Figure 2.
Supplemental Figure S4 Efficient RiPS derivation from BJ fibroblasts without passaging. (A) Immunohistochemistry showing expression of pluripotency markers SSEA-4 and TRA-1-60 in a BJ fibroblast reprogramming experiment transfected for 16 days with 600ng per day of a KMOSL modified RNA cocktail containing a destabilized GFP spike-in. Cultures were fixed for staining at day 18. 50,000 BJ cells were originally seeded onto feeder cells and went unpassaged throughout the course of the experiment. (B) Quantification of TRA-1-60 colony count relative to the number of cells seeded. This figure supports data shown in main Figure 5.
Supplemental Figure S5 Teratoma formation and trilineage differentiation of modified-RNA derived iPS clones in vivo.
(A) Histology of hematoxylin and eosin stained teratomas from the indicated BJ and dH1F RiPS derivations showing structures derived from the three germ layers. This figure supports data shown in main Figure 4.
Supplemental Table 1: Pluripotency validation assays performed in this study. The table shows the RiPS clones that were validate in each assay. # Validated for immuno-staining for all of TRA-1-60, TRA-1-80, SSEA3, SSEA4, OCT4, NANOG. Ω Demethylation of the OCT4 promoter. In vitro differentiation including ^embryoid body formation, øtrilineage by directed differentiation, † beating cardiomyocytes, and * blood formation by CFC assays in methylcellulose. This table supports data presented in main Figure 4.
Supplemental Table S2 Oligonucleotides for IVT template construction. 5′ and 3′ UTR oligos are ligated to the top strand of gene-specific ORF amplicons to produce a basic template construct for cloning (Supplemental Figure S1). Underlined bases in the 5′ UTR oligo sequence indicate the upstream T7 promoter, and in the 3′ UTR oligo sequence show downstream restriction sites, introduced to facilitate linearization of template plasmids. Template PCR primers are used to amplify ligation products for sub-cloning. Tail PCR primers are used to append an oligo(dT) sequence immediately after the 3′ UTR to drive templated addition of a poly(A) tail during IVT reactions. Gene-specific ORF primers are used to capture the coding region (minus the start codon) from cDNA templates. Splint oligos mediate ligation of UTR oligos to the top strand of ORF amplicons. This table supports data in main Figure 2.
Supplemental Table S3 Primers for qRT-PCR analysis of interferon-regulated genes. This table supports data in main Figure 1.
Supplemental Movie 1 Embryoid bodies from independent RiPS derivations showing beating cardiomyocytes. Top row (left to right) BJ-RiPS1.1, BJ-RiPS1.2 and BJ-RiPS1.3 Middle row (left to right) CF-RiPS1.3, D551-RiPS1.2, MRC5-RiPS1.8, Bottom row (left to right) CF-RiPS1.4, D551-RiPS1.3, MRC5-RiPS1.9. This movie supports main figure 4.