Abstract
Copper is known to play a role in iron recycling from macrophages. To examine whether cellular copper status affects expression of the iron exporter ferroportin-1 (FPN1), J774 macrophage cells were exposed to 10–100 μM CuSO4 for up to 20 h. Copper treatment significantly increased FPN1 mRNA in a dose- and time-dependent manner. After 20 h, 100 μM CuSO4 up-regulated FPN1 transcript levels ≈13-fold compared to untreated controls. Induction was detected 8 h after copper treatment was initiated and markedly increased thereafter. A corresponding increase in FPN1 protein levels was observed upon copper treatment. Induction of J774 cell FPN1 expression by copper was also associated with a dose-dependent increase in 59Fe release after erythrophagocytosis of labeled red blood cells. Thus, a previously uncharacterized role for copper in the regulation of macrophage iron recycling is suggested by the induction of FPN1 gene expression and iron efflux by this metal.
Copper and iron metabolism are closely interrelated (1). For example, some of the pathophysiological consequences of copper deficiency, which manifest as microcytic and hypochromic anemia, support a critical role for this metal in the release of iron into circulation (2–7). It is widely accepted that the requirement for copper as a cofactor for ceruloplasmin is necessary for this plasma protein's ferroxidase activity and that oxidation of Fe2+ to Fe3+ by ceruloplasmin enables iron-binding by transferrin, thereby facilitating iron release (8, 9). Consistent with this model, ceruloplasmin knockout mice store excess iron in liver and the reticuloendothelial system (10). However, the absence of ceruloplasmin activity does not fully explain the function of copper in iron release because these animals display only very mild iron deficiency anemia (11). Moreover, acerulo-plasminemic patients who inherit loss-of-function mutations in the ceruloplasmin gene are also only slightly anemic (12).
Recently, a mammalian iron export protein ferroportin-1 (FPN1) was identified (13). FPN1 [also called MTP1 (14) or Ireg1 (15)] is abundantly expressed in the macrophages of liver, spleen, and bone marrow. The function of FPN1 as an iron exporter has been demonstrated in Xenopus oocytes wherein exogenous expression induces significant efflux of iron (13, 15). Overexpression of FPN1 in tissue culture cells also results in the depletion of cytosolic iron as shown by decreased levels of the iron storage protein ferritin (14). Interestingly, several different mutations of the human FPN1 gene have been unequivocally associated with excess iron deposits in macrophages (reviewed by Pietrangelo; ref. 16), implicating an essential role for FPN1 in iron recycling from erythrophagocytosed red cells. It has been found that iron loading increases FPN1 expression in macrophages of liver and lung (14, 17), as well as J774 macrophage cells (18). Studies in human intestinal CaCo-2 cells suggest that FPN1 expression may also be regulated by other metals (19, 20). However, a possible role for copper in the regulation of this iron exporter in macrophages has not been fully examined. The results presented here demonstrate that, upon copper-loading, murine J774 cells display a dose- and time-dependent increase in FPN1 mRNA levels. Furthermore, these effects correlate with increased FPN1 protein levels and are associated with increased iron efflux by J774 cells after erythrophagocytosis. The implication of these findings is that copper status can modulate macrophage iron release by means of the regulation of FPN1 expression.
Experimental Procedures
Northern Analysis of J774 Cell mRNA. J774 mouse macrophage cells were cultured in α-minimum essential medium (α-MEM) supplemented with 10% FBS and antibiotics as described (18). Briefly, to investigate the effects of various metals, cells were grown to ≈80% confluency in six-well plates and incubated in serum-containing α-MEM with CuSO4 or CuCl2, Fe-nitrilotriacetic acid (NTA) in a molar ratio of 1:4, or ZnCl2 added as detailed in Figs. 1, 2, 3, 4, 5. To verify the specificity of FPN1 mRNA induction by copper, cells were also incubated with or without CuSO4 in the presence or absence of triethylenetetramine (TRIEN, Sigma) for 20 h. After these treatments, total RNA was isolated by using RNA-Bee (Tel-Test, Freindswood, TX) according to the manufacturer's instructions. Twenty-five-microgram aliquots were fractionated on 0.9% denaturing form-aldehyde–agarose gels, transferred to Nytran-N membranes by using the Turboblotter transfer system (Schleicher & Schuell), and cross-linked to the membrane by UV irradiation. The blots were prehybridized at 42°C in a buffer (750 mM NaCl/150 mM Tris/114 mM Na2HPO4/45 mM NaH2PO4/4 mM Na4P2O7, pH 7.4), containing 50% deionized formamide, 10× Denhardt's solution, 10 mM EDTA, 0.1% SDS, and 100 μg/ml heat-denatured salmon sperm DNA for 4 h. Hybridization was performed for 16 h in the same buffer containing 10% dextran sulfate and the appropriate probe labeled with [α-32P]dCTP by random priming (1 × 106 cpm/ml). cDNA probes were prepared by using the following plasmids as templates: pCMV-SPORT6-mouse FPN1 (GenBank accession no. BE554084, American Type Culture Collection, Manassas, VA), pBluescriptSK(-)-human β-actin (GenBank accession no. AA173248, American Type Culture Collection), pBluescriptSK(+)-mouse metallothionein-I (GeneBank accession no. BG099916, American Type Culture Collection), and pT7T3D-Pac-rat transferrin receptor (TfR) (GenBank accession no. AI454017, Resgen, Huntsville, AL). After hybridization, membranes were washed four times at room temperature for 15 min each in 0.1× standard sodium chloride-sodium citrate (SSC, 1× SSC is 0.15 M NaCl, 0.015 M sodium citrate) and 0.15% SDS. Radioactivity was detected by PhosphorImaging (Personal Molecular Imager FX, Bio-Rad) and quantified by using quantity one software (version 4.1.0, Bio-Rad).
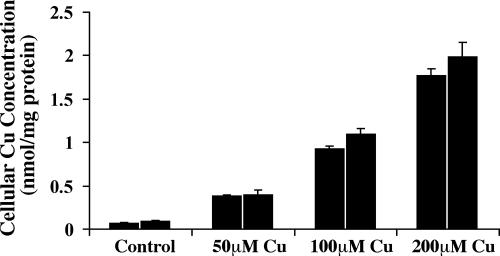
Fig. 1.
Dose-dependent copper loading of J774 cells. J774 macrophages were incubated with the indicated concentrations of CuSO4 for 20 h. Cells were extensively washed and digested with concentrated nitric acid overnight before copper analysis by inductively coupled plasma mass spectrometry (ICP-MS). Values were normalized to the cell protein content of the samples and shown are the means ± SD (n = 5) obtained from two individual experiments.
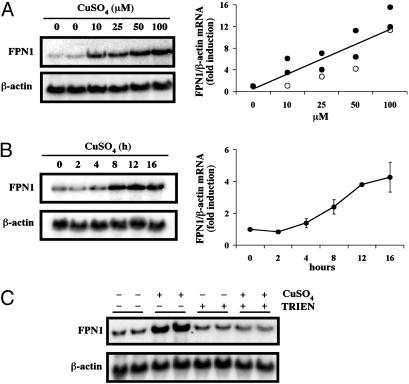
Fig. 2.
(A) Copper loading increases FPN1 mRNA levels in J774 cells in a dose-dependent manner. J774 cells were incubated with the indicated concentrations of CuSO4 for 20 h. Total RNA was isolated, electrophoresed, transferred to a Nytran N membrane, and hybridized with a 32P-labeled mouse FPN1 cDNA probe as described in Experimental Procedures. To control for sample loading, the membrane was stripped and reprobed for β-actin. Relative FPN1 mRNA levels after copper treatment were quantified by using quantity one software (Bio-Rad) and normalized to values for β-actin mRNA levels obtained in the same blot to determine fold induction compared to control (untreated cells). (Right) The relative FPN1 mRNA levels as a function of copper concentration (μM) after treatment with CuSO4 in two independent experiments (filled circles) and after treatment with CuCl2 in a separate experiment (open circles). (B) Copper loading increases in J774 cell FPN1 mRNA in a time-dependent manner. After incubation with 100 μM CuSO4 for the indicated times, total RNA was isolated and Northern analysis was performed. The fold induction of FPN1 mRNA was quantified as described for A, and Right shows mean values (±SE) determined as a function of time (h) in three separate experiments. (C) TRIEN blocks copper-mediated induction of FPN1 mRNA. J774 cells were treated with 20 μM CuSO4, 80 μM TRIEN, or both together for 20 h. After RNA isolation, Northern analysis was performed to detect FPN1 and β-actin mRNA. These results are representative of three independent experiments.
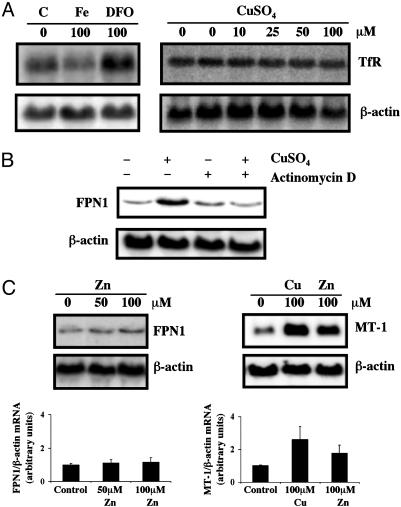
Fig. 3.
(A) Copper loading does not alter TfR expression in J774 macrophages. Cells were incubated with 100 μM Fe-NTA, 100 μM desferrioxamine, or the indicated concentrations of CuSO4 for 20 h. Total RNA was isolated, electrophoresed, transferred to a Nytran N membrane, and hybridized with a 32P-labeled mouse TfR cDNA probe. The membrane was stripped and reprobed with a mouse β-actin cDNA probe to control for loading and transfer. Data shown are representative of three independent experiments. (B) Actinomycin D blocks induction of FPN1 mRNA by copper. J774 cells were treated with 200 μM CuSO4, 0.5 μg/ml actinomycin D, or both agents together for 4 h. Total RNA was isolated for Northern analysis as described for A. Representative blot from one of four independent experiments is shown. (C) Zinc does not alter FPN1 mRNA levels. J774 cells were incubated with 50 and 100 μM ZnCl2 or 100 μM CuSO4 for 20 h. Northern analysis was performed to determine FPN1, metallothionein (MT-1), and β-actin mRNA levels as described for A. Levels of FPN1 and MT-1 mRNA were normalized to β-actin mRNA levels, and Lower shows the means (±SE) determined in three individual experiments.
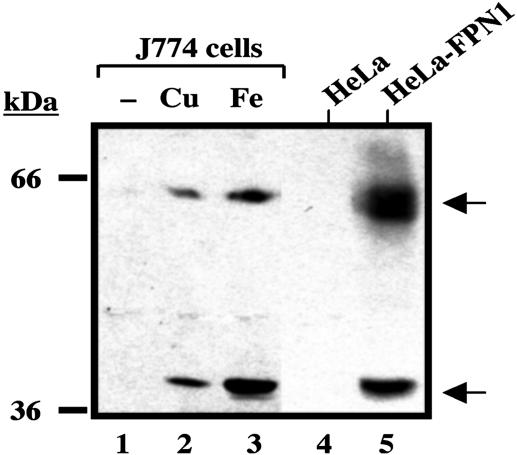
Fig. 4.
Copper loading increases FPN1 protein levels in J774 macrophages. J774 cells were incubated with CuSO4 or Fe-NTA for 48 h. HeLa cells were transiently transfected with pSPORT2 CMV vector containing full-length mouse FPN1 cDNA. Total cell lysates were collected, and samples of 100 μg (J774) or 1 μg (HeLa) of protein were resolved by electrophoresis on a 10% SDS/polyacrylamide gel, then transferred to nitrocellulose. Western blot analysis was then carried out as described in Experimental Procedures. Lane 1, control (untreated) J774 cells; lane 2, J774 cells treated with 200 μM CuSO4; lane 3, J774 cells treated with 200 μM Fe-NTA; lane 4, control (untreated) HeLa cells; and lane 5, HeLa cells transfected with full-length mouse FPN1 cDNA (HeLa-FPN1). A representative blot is shown with similar data obtained in at least three separate experiments.
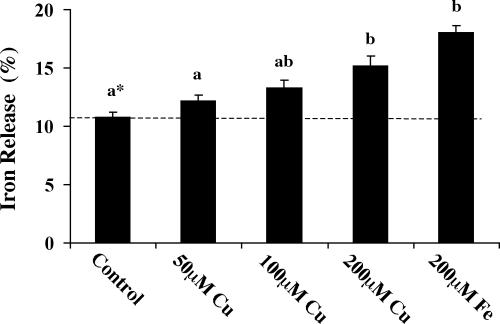
Fig. 5.
Copper loading increases iron release from J774 macrophages after erythrophagocytosis. J774 cells were treated with 50, 100, or 200 μM CuSO4 or 200 μM Fe-NTA for 48 h before release assays. Cells were washed free of the metals, and 59Fe-labeled, opsonized rat erythrocytes were added for 1.5 h at 37°C. After lysis of nonphagocytosed red blood cells, J774 cells were washed, resuspended in fresh culture media, and allowed to release iron into the media for 3 h. At the end of experiments, 59Fe activities in the collected media and cell lysates were measured by γ-counting. Iron release was calculated as percentage of 59Fe in the media versus total 59Fe measured (media plus cells). Values shown are the mean percentage iron release ± SEM (n = 9). *, Treatments not sharing the same superscript are significantly different from each other (ANOVA followed by Tukey's highly significant difference test, P < 0.05).
Western Blot Analysis. To determine the effect of copper on FPN1 protein levels, cells were grown to ≈25% confluency and incubated with 200 μM CuSO4 for 48 h. Cells were then washed with ice-cold PBS and scraped with lysis buffer [170 mM Tris·HCl, pH 6.8/2% SDS/5% glycerol/10% 2-mercaptoethanol, and protease inhibitors (Complete Mini protease inhibitor mixture, Roche Diagnostics, Mannheim, Germany)] before shearing DNA by sonication for 10 s. The addition of protease inhibitors was necessary to detect FPN1 immunoreactivity. Care was also taken not to heat samples before electrophoresis. As a control, HeLa cells were transiently transfected with a pSPORT2 CMV vector containing full-length mouse FPN1 cDNA (14) with Lipofectamine reagent (Invitrogen), and cell extracts were processed in the same fashion. Protein content of cellular extracts was determined by using the RC DC Protein Assay (Bio-Rad), and samples of cell extract protein (100 μg) were separated by electrophoresis on a 10% SDS/polyacrylamide gel, then transferred to an Optitran nitrocellulose membrane (Schleicher & Schuell). Equal loading and the integrity of transfer were confirmed by Ponceau-S staining. The membrane was blocked with 5% nonfat dry milk in Tris-buffered saline solution containing 0.01% Tween-20, pH 7.4 (TBS-T) for at least 2 h, and incubated with affinity-purified anti-FPN1 antibody (diluted to 1:2,500 in 5% nonfat dry milk/TBS-T) overnight at 4°C. Blots were then washed for 30 min with TBS-T, incubated with horseradish peroxidase-linked donkey anti-rabbit IgG antibody (1:2,000 dilution, Amersham Pharmacia) in 5% milk/TBS-T for 1 h, washed with TBS-T for 30 min, and visualized by using enhanced chemiluminescence (SuperSignal WestPico, Pierce).
Iron Release Assay. 59Fe-labeled red blood cells were prepared in vivo by injecting a male Sprague–Dawley rat (≈350 g) with 500 μCi 59Fe (Perkin–Elmer; 1 Ci = 37 GBq). To stimulate erythropoiesis, the rat was first subjected to repeated phlebotomy (≈3 ml) every other day for 5 weeks before injection of radioactive metal (hematocrit of the animal was ≈35%). One week after injection, blood was collected in heparin, and 59Fe-labeled red blood cells were separated and washed three times in Alsever's solution (Sigma). Animal protocols were in accordance with the National Institute of Health guidelines and approved by the Harvard Medical Area Standing Committee on Animals.
The erythrocytes were opsonized with rabbit anti-rat red blood cell IgG fraction (Research Diagnostics) at a dilution of 1:20 at 37°C for 15 min. The opsonized cells (2 × 107) were then added to control, copper-treated, or iron-treated J774 cells for 1.5 h at 37° with a final ratio of ≈10 red blood cells per J774 cell. Care was taken to limit erythrophagocytosis such that between 1 and 3 red blood cells were ingested per macrophage (21, 22). After uptake, J774 cells were briefly washed with distilled water (10 s) to lyse any nonphagocytosed 59Fe-labeled red blood cells. J774 cells were then quickly washed two times with Dulbecco's PBS (Sigma), resuspended in fresh α minimal essential medium supplemented with 10% FBS, and allowed to export iron into the media for 3 h. Cell viability at the end of the experiment was confirmed by the fact that >99% of the J774 macrophages excluded trypan blue, indicating that release of iron was not a consequence of cell death or damage. Culture media and cell lysates were collected, and 59Fe content of both fractions was measured by γ-counting. Iron release was calculated as percentage of 59Fe in the media versus total 59Fe measured (media plus cells). To confirm that increased efflux was caused by export of nonheme iron, in some experiments the amount of heme-bound iron released was determined by extraction using cyclohexanone (23). Briefly, samples were acidified by addition of 1 M HCl (final concentration = 0.13 M), and cyclohexanone was added at a 1.35:1 ratio (vol/vol). After 2 min of vigorous vortexing, samples were centrifuged at 1,000 × g for 10 min. Five milliliters from the organic layer were removed, and the amount of 59Fe extracted was measured by γ-counting.
Results
Copper Loading Increases FPN1 mRNA Levels in J774 Macrophages. Overnight treatment of J774 cells with CuSO4 resulted in a dose-dependent increase in cellular copper levels (Fig. 1). There was no evidence of cytotoxicity with treatment as high as 200 μM CuSO4 for up to 48 h as determined by exclusion of trypan blue staining, lack of morphological effects, and continued adherence of J774 cells to tissue culture plates. Untreated cells contained 0.1 nmol Cu/mg protein, whereas exposure to 200 μM CuSO4 increased the cellular copper content ≈20-fold (1.9 nmol Cu per mg of protein). Tissue concentrations of copper in normal humans and mice range from 0.02 to 0.19 nmol per mg of tissue (24, 25). However, patients with Wilson's disease have much higher copper levels in liver (>4 nmol Cu per mg of tissue) (26). Similarly, hepatic copper in Long–Evans Cinnamon (LEC) rats, an animal model of Wilson's disease, is ≈5 nmol Cu per mg of tissue (27).
Northern blot analyses demonstrated that, under our experimental conditions, copper-loading increased steady-state levels of J774 cell FPN1 mRNA in a dose-dependent manner (Fig. 2A). The induction of FPN1 mRNA was clearly evident at 10 μM CuSO4 and increased linearly upon incubation with concentrations up to 100 μM (Fig. 2 A). Similar induction was observed by using CuCl2, indicating that the effect was independent of the anion. Quantitation of FPN1 mRNA levels normalized to β-actin mRNA levels revealed a ≈13-fold increase by 100 μM copper compared to untreated controls. Induction of FPN1 mRNA levels was readily detected 8 h after copper exposure, with significantly increased expression observed by 12–16 h (Fig. 2B). To further verify that up-regulation of FPN1 mRNA levels was caused by copper loading, J774 cells were cotreated with the high-affinity copper chelator TRIEN (Fig. 2C). Consistent with previous experiments (Fig. 2 A and B), 20 μM CuSO4 alone markedly enhanced FPN1 mRNA levels compared to controls (P < 0.01, n = 6). This induction was completely blocked by the presence of 80 μM TRIEN (Fig. 2C), indicating that increased intracellular copper results in induction of FPN1 mRNA. Treatment with 80 μM TRIEN alone failed to decrease the level of FPN1 transcripts.
Induction of FPN1 mRNA by Copper in J774 Cells Is Not Mediated by Changes in Cellular Iron Status and Is Actinomycin D-Sensitive. Previous studies have shown that iron availability can affect FPN1 gene expression (14, 15, 17, 18). To determine whether FPN1 induction by copper was mediated through changes in intracellular iron concentration, TfR mRNA levels were measured. TfR expression is inversely correlated with cellular iron status (28). As shown in Fig. 3A, iron depletion by desferrioxamine upregulates TfR mRNA levels in J774 cells, whereas iron-loading by Fe-NTA down-regulates the transcript pool. However, treatment of J774 cells with up to 100 μM CuSO4 for 20 h did not alter TfR mRNA levels, indicating that the copper-mediated effect does not involve changes in cellular free iron. Furthermore, nonheme iron levels did not increase in these cells (data not shown). Collectively, these results show that copper increases FPN1 mRNA levels independent of changes in cellular iron status.
To further delineate the molecular basis for increased expression of FPN1 in response to copper, cells were treated for 4 h with 200 μM CuSO4 in the presence or absence of 0.5 μg/ml actinomycin D, an inhibitor of RNA synthesis. Actinomycin D blocked increased FPN1 expression in copper-treated cells (Fig. 3B), suggesting that this metal up-regulates FPN1 expression in a transcription-dependent manner. Copper is known to regulate transcription of other mammalian genes, e.g., metallothionein, via metal response elements (MREs) (29, 30), which provide binding sites for the MRE-binding transcription factor-1 (MTF-1) (31). Because sequence analysis indicated that the promoter region of FPN1 harbors several potential MREs (gene inspector software), the influence of zinc, a potent MTF-1 activator, was also examined by Northern analysis. However, although both copper and zinc up-regulated J774 cell metallothionein mRNA, treatment with up to 100 μM ZnCl2 did not affect FPN1 gene expression (Fig. 3C).
FPN1 Protein Levels Increase After Copper Treatment. Western blot analyses were performed to determine whether the enhanced FPN1 mRNA levels observed after copper treatment correlated with increased FPN1 protein expression (Fig. 4). J774 cell FPN1 is detected as two immunoreactive species with molecular masses of ≈65 kDa and ≈40 kDa (18). As reported (18), both 65- and 40-kDa bands were induced by high iron treatment of J774 cells (see Fig. 4 lane 3). Copper treatment increased both FPN1 species in similar fashion, albeit to a lesser extent (see Fig. 4 lane 2). The relationship between the two FPN1 bands is unknown, but addition of either copper or iron to control samples (HeLa cells transfected to express FPN1) did not increase the ratio of the 40-kDa species relative to the 65-kDa band (data not shown). The predicted mass of FPN1 (62 kDa) is close to the size of the larger species, therefore we speculate that the smaller 40-kDa band is a proteolytic fragment that is prominent in J774 cells because macrophages contain abundant protease activity. It is difficult to assess the fold increase in FPN1 protein upon copper treatment not only because the relationship between these two immunoreactive bands is unclear, but also because basal levels of FPN1 are not readily detected in J774 cells (see Fig. 4 lane 1 and ref. 21). Furthermore, FPN1 protein synthesis is known to be regulated by iron-responsive proteins because of the presence of a 5′ iron-responsive element present in its transcript (15, 32). Thus, copper-induced increases in FPN1 mRNA will not necessarily reflect the same magnitude of protein induction. Nonetheless, the results of Western blot analysis do confirm that FPN1 protein levels are enhanced in copper-treated cells.
Copper Loading Increases Iron Release from J774 Macrophages After Erythrophagocytosis. To investigate whether up-regulation of FPN1 expression by copper alters the release of iron from macrophages, efflux of 59Fe after erythrophagocytosis of 59Felabeled red blood cells by J774 cells was measured. Because ingestion of excess red blood cells can damage macrophages (21, 22), assay conditions were adjusted such that between one and three erythrocytes were phagocytosed per J774 cell. Cell viability was not affected under these conditions, and was confirmed after erythrophagocytosis as well as at the end of iron release experiments by trypan blue exclusion. In these experiments, the total 59Fe cpm measured in each well was within the range of control values, indicating that copper-loading did affect phagocytic uptake of the labeled red blood cells. Previous studies in our laboratory have shown that erythrophagocytosis up-regulates FPN1 due to erythrocyte-derived iron (18); therefore, export studies were limited to the first 3 h after phagocytosis to avoid any iron-induced effects (increased FPN1 protein levels are detectable 4 h after erythrophagocytosis). Iron efflux was measured in culture medium containing 10% FBS. Preliminary experiments showed that the amount of iron released was not influenced by the addition of up to 500 μg/ml apotransferrin in the presence or absence of 125 μg/ml ceruloplasmin, indicating that sufficient levels of serum factors necessary to support iron efflux were present in the assay. Basal efflux of radioisotope under our assay conditions was 10.8% in close agreement with previous assays of iron release (33). As shown in Fig. 5, copper treatment enhanced release of iron from J774 macrophages in a dose-dependent manner that correlated with the metal's induction of FPN1 mRNA and protein. For comparison, 15.2% of erythrocyte-derived iron was released in cells pretreated with 200 μM CuSO4. Under these limiting assay conditions (3 h after phagocytosis), this result represents a 41% increase in the rate of 59Fe export. Longer times of incubation (up to 24 h after phagocytosis) result in maximal release of only ≈21% red cell iron under basal conditions (33). Because both heme and nonheme iron is released after erythrophagocytosis (21, 22), the influence of copper treatment on the distribution of 59Fe between these fractions was assessed by determining the amount of released heme iron by cyclohexanone extraction (23). From the efflux media, an average of 5.7 ± 0.6% (n = 4) of total iron was extracted into the organic phase, but the amount of released iron partitioning into the cyclohexanone layer was not influenced by copper treatment. We cannot rule out the possibility that copper loading enhances the pool of nonheme 59Fe available for export (e.g., stimulates degradation of hemoglobin from phagocytosed red cells), but treatment of J774 cells with 200 μM Fe-NTA consistently increased iron efflux to a greater extent (≈18% red cell iron), in concordance with the greater induction of FPN1 protein by iron (Fig. 4). Combined, these data implicate that up-regulation of FPN1 by both metals facilitates recycling of iron from macrophages.
Discussion
Macrophages of the reticuloendothelial system are responsible for the recycling of iron through erythrophagocytosis of senescent red cells (34). Because very little iron is absorbed from the diet (≈1 mg/day), the demand for iron required for erythropoiesis and other physiological processes is mainly fulfilled by iron recycling (20–30 mg/day). Consequently, abnormal regulation of iron release from macrophages contributes to the development of common disorders of iron metabolism such as autosomal recessive hereditary hemochromatosis and the anemia of chronic disease, with unusually low or high cellular iron concentrations in macrophages, respectively. A central role for the recently discovered iron export protein FPN1 in iron recycling is confirmed by the excess macrophage iron stores observed in patients with mutations in the FPN1 gene (35–38). Regulation of FPN1 expression therefore appears to be critically important for the maintenance of iron homeostasis.
A role for copper in the release of iron stores is well established (39), and has been thought to primarily involve the need for this metal cofactor to support ceruloplasmin's ferroxidase activity thus facilitating loading of iron onto serum transferrin (8, 9). The results of this investigation indicate that, in addition to this function, copper can also regulate the release of iron from macrophages by modifying the expression of the export protein FPN1. Treatment of J774 cells with copper induces FPN1 mRNA levels in a time- and dose-dependent manner, with effects observed at concentrations within the normal range of serum concentrations (≈20 μM) (24). Importantly, the induction of FPN1 mRNA by copper is associated not only with increased FPN1 protein levels, but also a dose-dependent increase in the export of iron. All of these observations are consistent with the model that copper status can modulate iron release due to the metal's regulation of FPN1 gene expression. This model is also consistent with the observations that copper-deficient rodents, which have ironloaded but copper-depleted livers, display profound microcytic anemia (5, 6), whereas Cp-/- mice and ceruloplasmin-deficient Long–Evans Cinnamon rats, which have hepatic iron- and copper-loading, suffer only mild hematological disturbances (10, 11, 40).
From these positive effects of copper loading, one might infer that deficiency states would disrupt the normal pattern of FPN1 expression to suppress the release of iron, but it should be noted that treatment of J774 cells with TRIEN alone did not affect FPN1 levels. However, under these conditions (20 h incubation with 80 μM chelator), the activity of copper, zinc-superoxide dismutase, a marker of copper deficiency (41), was unaffected, and thus it is probable that treatment with TRIEN did not reduce cellular copper levels sufficiently enough to alter FPN1 expression. Future studies are required to more fully examine how copper deficiency impacts FPN1 levels in macrophages. From the results of this study, we conclude that copper sufficiency is necessary to maintain appropriate levels of FPN1 expression and regulation of macrophage iron export activity.
The copper-induced regulation of FPN1 expression in macrophages is sensitive to actinomycin D, and therefore could reflect a transcription-dependent process. Transcriptional regulation of the FPN1 gene in monocytes/macrophages has been reported in response to iron, IFN-γ, and lipopolysaccharide (18, 42). Although the exact mechanism responsible for upregulation of FPN1 mRNA levels by copper remains to be determined, it does not appear to depend on changes in intracellular iron. The presence of an iron-responsive element-like structure in the FPN1 mRNA predicts posttranscriptional regulation, an effect that is borne out in reporter gene studies of the FPN1 5′ region (32). The observation that copper does not alter macrophage TfR mRNA levels, which are known to be highly regulated via iron response element interactions, argues against the possibility that copper may indirectly control FPN1 expression by affecting cellular iron status. It is also unlikely that copper directly modulates FPN1 via transcriptional control due to MRE-binding transcription factor 1 activation, particularly because zinc, which strongly activates the binding of this MRE-binding factor to MREs (43), does not affect FPN1 expression. In contrast to our results, Sharp and colleagues have reported that both copper (20) and zinc (19) induce FPN1 in intestinal CaCo-2 cells. However, in our hands, zinc does not affect FPN1 mRNA levels in macrophage J774 cells (Fig. 3), and it is therefore possible that the copper effects that we report here reflect the activity of macrophage-specific regulatory factors present in J774 cells.
Finally, the potent positive effects of copper on FPN1 levels prompt us to consider whether this metal may also be a transport substrate for export by FPN1. Promiscuous substrate specificity of metal transporters has been documented before: for example, divalent metal transporter-1 appears to transport a broad range of divalent metals such that, in Xenopus oocytes expressing the factor, metals like Zn2+, Cd2+, Mn2+, Cu2+, Fe2+, Co2+, Ni2+, and Pb2+ generate positive inward currents accompanying the cotransport of H+ (44). The observation that copper, like iron, up-regulates FPN1 suggests that further studies are warranted to examine whether FPN1 acts as an export protein in macrophages for other metals including copper.
Acknowledgments
We thank Dr. Mitchell D. Knutson for his experimental advice and helpful discussions in preparing this manuscript. This work was supported by National Institutes of Health Grants DK56160 (to M.W.-R.) and DK59429 (to D.J.H.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FPN1, ferroportin-1; TRIEN, triethylenetetramine; TfR, transferrin receptor; NTA, nitrilotriacetic acid; MRE, metal response elements.
References
- 1.Winzerling, J. J. & Law, J. H. (1997) Annu. Rev. Nutr. 17, 501-526. [DOI] [PubMed] [Google Scholar]
- 2.Hart, E. B., Steenbock, H., Waddle, J. & Elvehjem, C. A. (1928) J. Biol. Chem. 77, 797-812. [PubMed] [Google Scholar]
- 3.Elvehjem, C. A. & Sherman, W. C. (1932) J. Biol. Chem. 98, 309-319. [Google Scholar]
- 4.Lee, G. R., Nacht, S., Lukens, J. N. & Cartwright, G. E. (1968) J. Clin. Invest. 47, 2058-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Owen, C. A., Jr. (1973) Am. J. Physiol. 224, 514-518. [DOI] [PubMed] [Google Scholar]
- 6.Williams, D. M., Kennedy, F. S. & Green, B. G. (1983) Br. J. Nutr. 50, 653-660. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, N. L., Keen, C. L., Lonnerdal, B. & Hurley, L. S. (1983) Biochem. Biophys. Res. Commun. 113, 127-134. [DOI] [PubMed] [Google Scholar]
- 8.Osaki, S., Johnson, D. A. & Frieden, E. (1966) J. Biol. Chem. 241, 2746-2751. [PubMed] [Google Scholar]
- 9.Osaki, S., Johnson, D. A. & Frieden, E. (1971) J. Biol. Chem. 246, 3018-3023. [PubMed] [Google Scholar]
- 10.Harris, Z. L., Durley, A. P., Man, T. K. & Gitlin, J. D. (1999) Proc. Natl. Acad. Sci. USA 96, 10812-10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto, K., Yoshida, K., Miyagoe, Y., Ishikawa, A., Hanaoka, K., Nomoto, S., Kaneko, K., Ikeda, S. & Takeda, S. (2002) Biochim. Biophys. Acta 1588, 195-202. [DOI] [PubMed] [Google Scholar]
- 12.Hellman, N. E., Kono, S., Mancini, G. M., Hoogeboom, A. J., De Jong, G. J. & Gitlin, J. D. (2002) J. Biol. Chem. 277, 46632-46638. [DOI] [PubMed] [Google Scholar]
- 13.Donovan, A., Brownlie, A., Zhou, Y., Shepard, J., Pratt, S. J., Moynihan, J., Paw, B. H., Drejer, A., Barut, B., Zapata, A., et al. (2000) Nature 403, 776-781. [DOI] [PubMed] [Google Scholar]
- 14.Abboud, S. & Haile, D. J. (2000) J. Biol. Chem. 275, 19906-19912. [DOI] [PubMed] [Google Scholar]
- 15.McKie, A. T., Marciani, P., Rolfs, A., Brennan, K., Wehr, K., Barrow, D., Miret, S., Bomford, A., Peters, T. J., Farzaneh, F., et al. (2000) Mol. Cell 5, 299-309. [DOI] [PubMed] [Google Scholar]
- 16.Pietrangelo, A. (2004) Blood Cells Mol. Dis. 32, 131-138. [DOI] [PubMed] [Google Scholar]
- 17.Yang, F., Wang, X., Haile, D. J., Piantadosi, C. A. & Ghio, A. J. (2002) Am. J. Physiol. 283, L932-L939. [DOI] [PubMed] [Google Scholar]
- 18.Knutson, M. D., Vafa, M. R., Haile, D. J. & Wessling-Resnick, M. (2003) Blood 102, 4191-4197. [DOI] [PubMed] [Google Scholar]
- 19.Yamaji, S., Tennant, J., Tandy, S., Williams, M., Singh Srai, S. K. & Sharp, P. (2001) FEBS Lett. 507, 137-141. [DOI] [PubMed] [Google Scholar]
- 20.Tennant, J., Stansfield, M., Yamaji, S., Srai, S. K. & Sharp, P. (2002) FEBS Lett. 527, 239-244. [DOI] [PubMed] [Google Scholar]
- 21.Kondo, H., Saito, K., Grasso, J. P. & Aisen, P. (1988) Hepatology 8, 32-38. [DOI] [PubMed] [Google Scholar]
- 22.Moura, E., Noordermeer, M. A., Verhoeven, N., Verheul, A. F. & Marx, J. J. (1998) Blood 92, 2511-2519. [PubMed] [Google Scholar]
- 23.Krantz, S. B. & Fried, W. (1968) J. Lab. Clin. Med. 72, 157-164. [PubMed] [Google Scholar]
- 24.Linder, M. C. (1991) Biochemistry of Copper (Plenum, New York).
- 25.Prohaska, J. R., Bailey, W. R., Gross, A. M. & Korte, J. J. (1990) J. Nutr. Biochem. 1, 149-154. [DOI] [PubMed] [Google Scholar]
- 26.Cuthbert, J. A. (1995) J. Invest. Med. 43, 323-336. [PubMed] [Google Scholar]
- 27.Hayashi, M., Kuge, T., Endoh, D., Nakayama, K., Arikawa, J., Takazawa, A. & Okui, T. (2000) Biochem. Biophys. Res. Commun. 276, 174-178. [DOI] [PubMed] [Google Scholar]
- 28.Eisenstein, R. S. (2000) Annu. Rev. Nutr. 20, 627-662. [DOI] [PubMed] [Google Scholar]
- 29.Durnam, D. M. & Palmiter, R. D. (1981) J. Biol. Chem. 256, 5712-5716. [PubMed] [Google Scholar]
- 30.Yagle, M. K. & Palmiter, R. D. (1985) Mol. Cell. Biol. 5, 291-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrews, G. K. (2000) Biochem. Pharmacol. 59, 95-104. [DOI] [PubMed] [Google Scholar]
- 32.Liu, X. B., Hill, P. & Haile, D. J. (2002) Blood Cells Mol. Dis. 29, 315-326. [DOI] [PubMed] [Google Scholar]
- 33.Rama, R., Sanchez, J. & Octave, J. N. (1988) Biochim. Biophys. Acta 968, 51-58. [DOI] [PubMed] [Google Scholar]
- 34.Knutson, M. & Wessling-Resnick, M. (2003) Crit. Rev. Biochem. Mol. Biol. 38, 61-88. [DOI] [PubMed] [Google Scholar]
- 35.Njajou, O. T., Vaessen, N., Joosse, M., Berghuis, B., van Dongen, J. W., Breuning, M. H., Snijders, P. J., Rutten, W. P., Sandkuijl, L. A., Oostra, B. A., et al. (2001) Nat. Genet. 28, 213-214. [DOI] [PubMed] [Google Scholar]
- 36.Montosi, G., Donovan, A., Totaro, A., Garuti, C., Pignatti, E., Cassanelli, S., Trenor, C. C., Gasparini, P., Andrews, N. C. & Pietrangelo, A. (2001) J. Clin. Invest. 108, 619-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Devalia, V., Carter, K., Walker, A. P., Perkins, S. J., Worwood, M., May, A. & Dooley, J. S. (2002) Blood 100, 695-697. [DOI] [PubMed] [Google Scholar]
- 38.Roetto, A., Merryweather-Clarke, A. T., Daraio, F., Livesey, K., Pointon, J. J., Barbabietola, G., Piga, A., Mackie, P. H., Robson, K. J. & Camaschella, C. (2002) Blood 100, 733-734. [DOI] [PubMed] [Google Scholar]
- 39.Crichton, R. R. & Pierre, J. L. (2001) Biometals 14, 99-112. [DOI] [PubMed] [Google Scholar]
- 40.Sugawara, N., Ohta, T., Lai, Y. R., Sugawara, C., Yuasa, M., Nakamura, M. & Tamura, M. (1999) Life Sci. 65, 1423-1431. [DOI] [PubMed] [Google Scholar]
- 41.Prohaska, J. R. (1991) J. Nutr. 121, 355-363. [DOI] [PubMed] [Google Scholar]
- 42.Ludwiczek, S., Aigner, E., Theurl, I. & Weiss, G. (2003) Blood 101, 4148-4154. [DOI] [PubMed] [Google Scholar]
- 43.Davis, S. R. & Cousins, R. J. (2000) J. Nutr. 130, 1085-1088. [DOI] [PubMed] [Google Scholar]
- 44.Gunshin, H., Mackenzie, B., Berger, U. V., Gunshin, Y., Romero, M. F., Boron, W. F., Nussberger, S., Gollan, J. L. & Hediger, M. A. (1997) Nature 388, 482-488. [DOI] [PubMed] [Google Scholar]